Abstract
One-dimensional 1H NMR experiments were conducted for aqueous solutions of glycosaminoglycan oligosaccharides to measure the amide proton temperature coefficients and activation energy barriers for solvent exchange, and evaluate the effect of pH on the solvent exchange properties. A library of mono- and oligosaccharides was prepared by enzymatic depolymerization of amide-containing polysaccharides and by chemical modification of heparin and heparan sulfate saccharides including members that contain a 3-O-sulfated glucosamine residue. The systematic evaluation of this saccharide library facilitated assessment of the effects of structural characteristics, such as size, sulfation number and site, and glycosidic linkage, on amide proton solvent exchange rates. Charge repulsion by neighboring negatively charged sulfate and carboxylate groups was found to have a significant impact on the catalysis of amide proton solvent exchange by hydroxide. This observation leads to the conclusion that solvent exchange rates must be interpreted within the context of a given chemical environment. On their own, slow exchange rates do not conclusively establish the involvement of a labile proton in a hydrogen bond and additional supporting experimental evidence such as reduced temperature coefficients is required.
Graphical Abstract
INTRODUCTION
In this work we explore the effects of structural characteristics, such as length, sulfation number and site, and glycosidic linkage on the amide proton solvent exchange rates of saccharides derived from glycosaminoglycans (GAGs). The GAGs are a family of structurally related, unbranched anionic polysaccharides comprised of chondroitin sulfate, dermatan sulfate, heparan sulfate, heparin, hyaluronic acid and keratan sulfate.1 GAGs are classified based on their major disaccharide repeat subunits, in which a hexose residue is linked through the glycosidic bond to a hexosamine residue containing an amide, amino or sulfamate (N-sulfo) group. GAGs are important to many life-essential physiological processes such as cell development,2,3 inflammation,4–7 differentiation,8 and adhesion,9,10 including the pathological conditions of tumor growth, 10–13 and viral14–16 and microbial infections.17
Hydrogen bonds play an important role in many chemical and biological processes, therefore, the development of reliable methods for the identification of hydrogen bonds in GAGs is critically important to understanding their behavior. Reduced amide proton temperature coefficients (Δδ/ΔT) in the range of 2 – 4 ppb/K are well-established indicators of amide proton hydrogen bonds in peptides and proteins.18,19 Similarly, variable temperature experiments have been used in studies of GAG oligosaccharides to probe the involvement of labile protons in hydrogen bonds and to the investigate the kinetics and thermodynamics of solvent exchange in aqueous solution.20–23 For example, Langeslay et al. used line-shape analysis of the sulfamate group 1H NMR resonances to complement temperature coefficient results and identify a hydrogen bond between the sulfamate group and the adjacent 3-O-sulfo group of the internal GlcNS3S6S residue of the synthetic heparin pentasaccharide Arixtra (Figure 1q).20
Figure 1.
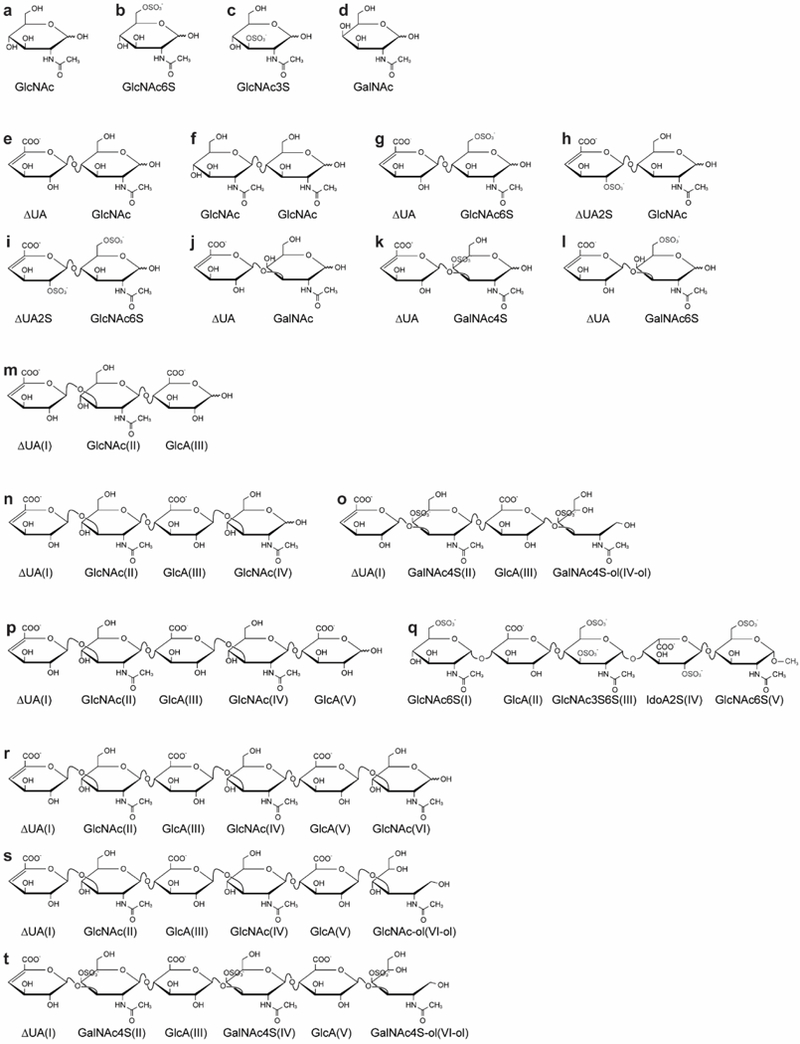
Structures of the library of amide-containing saccharides. For oligosaccharide descriptions refer to Table 1. The corresponding residue number starting from the non-reducing end is indicated in each oligosaccharide by Roman numerals.
For the slowly exchanging GAG sulfamate and amide protons, 1H NMR resonance line widths are related to the rate constant (kex) of the exchange reaction with the water solvent. Consequently, resonance line widths measured as a function of temperature can be fit with the Eyring-Polanyi equation (eq 1), followed by calculation of the energy barrier (ΔG‡ in kcal/mol) associated with solvent exchange:24–26
| (1) |
where kB is the Boltzmann constant, T is the temperature in Kelvin, h is Planck’s constant, and R is the ideal gas constant.
In prior work, labile saccharide protons with a comparatively higher energy barrier (by 2 – 3 kcal/mol or more) compared to other sites were proposed to be protected from exchange through involvement in a hydrogen bond. For example, Langeslay et al. used the ΔG‡ of the Arixtra GlcNS3S6S(III) sulfamate proton (ΔG‡ = 18.1 kcal/mol) as evidence for participation in a hydrogen bond as it is about 3 kcal/mol larger than the values determined for the other Arixtra sulfamate protons (ΔG‡ = 15.1 – 15.7 kcal/mol).20
The more rapidly exchanging GAG hydroxyl and amino protons have also been studied using the 2D 1H NMR exchange spectroscopy (EXSY) experiment.21,22,27,28 The EXSY cross-peak intensity depends on the exchange rates and spin relaxation properties of the detected nuclei. By integrating the cross-peak volumes, a build-up curve can be generated by plotting cross peak intensity as a function of the EXSY mixing time. Exchange rates are extracted by fitting the build-up curve with the Bloch equation modified to account for contributions from radiation damping to the longitudinal relaxation rate. 20,29,30
Using EXSY results, Beecher et al. reported three hydrogen bonds involving the Arixtra hydroxyl protons.21 Importantly, there was agreement between the reduced temperature coefficients and decreased exchange rates for only of the two hydroxyl protons. The protons of the hydroxyl group attached to the third carbon of the IdoA2S residue (OH3) behaved inconsistently, having a reduced exchange rate but large temperature coefficients. In other words, the exchange rate suggested a hydrogen bond involving the IdoA2S OH3, but the temperature coefficient did not support its presence.
Because temperature coefficients are a well-established indicator of hydrogen bonds in carbohydrates, proteins and peptides,20,21,31–34 we questioned the identification of a hydrogen bond by slower exchange rates alone without support from the temperature coefficient measurements. In this work, the solvent exchange properties of the labile amide protons in a library of GAG saccharides are examined and discussed as they relate to hydrogen bond participation in mono- and oligosaccharides with systematic variations in length, sulfo group number and position, uronic acid chirality (glucuronic or iduronic acid), type of glycosidic linkage, and other elements of structure. Additionally, the polyelectrolyte character of selected oligosaccharides is explored at physiological pH and the experimental findings reinforced by the results of molecular dynamics simulations.
EXPERIMENTAL AND THEORETICAL METHODS
Materials and Reagents.
Arixtra (Fondaparinux sodium), formulated as prefilled syringes for clinical use, was obtained from the University Pharmacy and Department of Pharmacy Administration of Semmelweis University (Budapest, Hungary). The cation exchange resin Dowex 50WX8 hydrogen form (50 – 100 mesh); 3-(trimethylsilyl)-1-propanesulfonic acid-d6 sodium salt (DSS-d6, 98% D); 99.5% acetic anhydride ((CH3CO)2O); chondroitin sulfate A sodium salt from bovine trachea (btCS-A); enzymes Chondroitinase ABC from Proteus vulgaris and Hyaluronidase from Streptomyces hyalurolyticus; monosaccharides N-acetyl-D-glucosamine (GlcNAc), N-acetyl- D-glucosamine-6-O-sulfate (GlcNAc6S), N-acetyl- D-galactosamine (GalNAc), D-glucosamine-3-O-sulfate (GlcN3S), and D-glucosamine-2-N-sulfate (GlcNS); ammonium bicarbonate (NH4HCO3); Tris-HCl; and 35 wt% deuterated hydrochloric acid (DCl, 99% D) were purchased from Sigma-Aldrich (St. Louis, MO). HPLC grade methanol, 12.1 N hydrochloric acid (HCl), 50% w/w sodium hydroxide solution (NaOH), glacial acetic acid (CH3COOH), sodium borohydride (NaBH4), disodium orthophosphate heptahydrate (Na2HPO4•7H2O), monobasic sodium phosphate ACS grade (NaH2PO4•H2O), sodium chloride (NaCl), bovine serum albumin (BSA) lyophilized powder, pyridine, and sodium acetate trihydrate (CH3COONa•3H2O) were purchased from Thermo Fisher Scientific (Bellefonte, PA). The disaccharides ΔUA-GalNAc, ΔUA-GalNAc6S, and ΔUA-GalNAc4S were purchased from Iduron Ltd. (Manchester, UK). The disaccharides ΔUA2S-GlcNAc; ΔUA2S-GlcNAc6S; ΔUA-GlcNAc6S; and ΔUA-GlcNAc and monosaccharides D-glucosamine-2-N, 3-O-disulfate (GlcNS3S); D-glucosamine-2-N, 6-O-disulfate (GlcNS6S); and D-glucoamine-2-N, 3-O, 6-O-trisulfate (GlcNS3S6S) were purchased from Dextra Laboratories Ltd. (Reading, UK). Sodium hyaluronate (HA) was purchased from Spectrum Chemical MFG Corp. (New Brunswick, NJ). Chitin disaccharide (N-acetyl-glucosamine)2 was purchased from Qingdao BZ Oligos Biotech Co., LTD (Qingdao, China). Koptec Pure Ethanol (200 proof) was purchased from Decon Labs, Inc. (King of Prussia, PA). Deuterium oxide (D2O, 99.9% D), 40% w/w sodium deuteroxide (NaOD, 99.5% D), deuterated dimethyl sulfoxide (DMSO-d6, 99.9% D), and the low temperature methanol standard were purchased from Cambridge Isotope Laboratories (Andover, MA). Ultrapure deionized water (18 MΩ/cm) was prepared using a Simplicity UV water purification system from Millipore (Billerica, MA). Buffers at pH 2.00, 4.00, 7.00, and 10.00 for calibration of the pH meter were purchased from Fisher Scientific (Nazareth, PA). All pH measurements were performed at room temperature using an AB 15 Accumet Basic pH meter and a 9110DJWP double-junction Ag/AgCl micro pH electrode, both from Thermo Scientific (Chelmsford, MA). All pH values measured in deuterated solutions are reported as pD with correction for the deuterium isotope effect.35,36 NMR NE-H5/3-Br tubes were purchased from New Era Enterprises (Vineland, NJ).
NMR Measurements
Variable temperature experiments.
Milligram quantities of each mono- or oligosaccharide were dissolved in 300 μL of an aqueous 10 mM phosphate buffer at pH 7.48 containing 0.5 mM DSS-d6. Actual weights for each solution are given in the Supporting Information (SI). The pH of each solution was adjusted to 7.4 with NaOH and HCl, lyophilized and stored at −20°C. Prior to the NMR experiments, the solid samples were reconstituted in 270 μL of H2O and, if necessary, the pH was re-adjusted to 7.4. Then, 30 μL of D2O was added to provide a lock signal and 200 μL aliquots were transferred to NE-H5/3-Br NMR tubes. Experimental details for the isolation and/or chemical modification and structure elucidation of CS dp4-ol, CS dp6-ol, HA dp3, HA dp4, HA dp5, HA dp6, HA dp6-ol, GlcNAc3S, and N-acetylated Arixtra (NAcA) are described in detail in the SI.
Variable temperature experiments were performed over the range 283.2 K to 338.2 K using a Bruker Avance spectrometer operating at 599.88 MHz and equipped with a 5 mm TBI probe. One-dimensional spectra were acquired by coadding 128 scans using a 90° pulse length between 9.5 and 10.6 μs with solvent suppression provided by excitation sculpting. A spectral window of 6887 Hz and a relaxation delay of 2.0 s were used. FIDs were acquired into 32,768 complex points, zero filled to 65,536 points and apodized by multiplication with an exponential function equivalent to a 0.3 Hz line broadening prior to Fourier transformation. At least 7 min was allowed for sample thermal equilibration at each temperature. Sample temperatures were calibrated using an external low temperature methanol standard obtained from Cambridge Isotope Labs (Andover, MA) and calculated using the Van Geet equation.37 The spectra were referenced to the 1H resonance of DSS-d6 (0.000 ppm) and all spectra were analyzed using MestReNova-12.0.1.
Temperature coefficient (Δδ/ΔT) measurements.
To characterize the temperature dependence of the amide proton resonances and explore the possibility of hydrogen bonds involving the amide protons, 1H NMR spectra of each oligosaccharide shown in Figure 1 were acquired at pH 7.40 over the temperature range 283.2 – 338.2 K. Temperature coefficients were determined by plotting chemical shift vs temperature as illustrated in Figure S4 and were calculated from the absolute value of the slopes of the lines obtained by plotting amide proton chemical shifts as a function of the calibrated temperature.
Line-shape analysis and determination of exchange energy barriers (ΔG‡).
Resonance line widths were measured for the amide protons by nonlinear least squares fitting of a Lorentzian peak shape to the experimental spectra using MestReNova-12.0.1 as shown in Figure S5a. The extracted line widths at half-height were plotted as a function of the calibrated temperatures as shown in Figure S5b, followed by fitting with the Eyring-Polanyi equation (eq 1) to calculate the energy barrier, ΔG‡, associated with amide proton solvent exchange. A representative example showing the calculation of ΔG‡ is given in Figure S5.
pH titrations.
The samples of GlcNAc, GlcNAc3S, GlcNAc6S, chitin disaccharide, NAcA, and GalNAc used for the 1H NMR temperature experiments were also titrated with NaOH and HCl solutions prepared in 90% H2O/10% D2O. After adjustment to the desired pH, the solutions were transferred to NE-H5/3-Br NMR tubes and the spectra recorded at 298.2 K using a Bruker Avance spectrometer operating at 599.88 MHz and equipped with a 5 mm TBI probe. Spectra were acquired as a function of pH using the same acquisition and processing parameters used for the variable temperature experiments.
For experiments with the N-sulfo-glucosamines, 1.5 mg GlcNS3S6S, 0.9 mg GlcNS, 1.3 mg GlcNS3S, and 0.9 mg GlcNS were each dissolved in 800 μL of a 90% H2O/10% D2O 25 mM phosphate buffer at pH 6.09 containing mM DSS-d6. The samples were titrated with NaOH and HCl in 90% H2O/10% D2O and aliquots of the solution at each pH were transferred to NE-H5/3-Br NMR tubes. 1H NMR spectra were acquired as a function of pH using a Bruker Avance spectrometer operating at 1H frequency of 599.52 MHz and equipped with a 5 mm BBI probe. NMR spectra were at 298.2 K using WATERGATE-W5 solvent suppression.38 The acquisition and processing parameters were as described for the variable temperature experiments.
MD Simulations.
The AmberTools 16 module xleap39 was used to model the structures of the HA dp6(α), CS dp6(α), and NAcA. These saccharides were modeled to generate the parameter, topology, and coordinate files for molecular dynamics simulations using a GLYCAM 06g force field.40 The xleap module was used to add sodium ions (ff99SB parameters) to oligosaccharide structures to make the system’s overall charge neutral (addions function) and to construct a cubic water box with each face a distance 12 Å from the solute. 37,40–42 Minimization, heating, equilibration, and molecular dynamics simulation production runs were performed using NAMD 2.11.43 To remove possible bad contacts, conjugate-gradient minimization was performed separately in the following order: solute, solvent, and the entire system. Periodic boundary conditions were applied throughout. The particle mesh Ewald algorithm was used to treat long range electrostatic interactions with the grid spacing set to 1 Å.44 Non-bonded interactions were cut off at 12 Å and a smooth switching function was applied to 10 Å. The VDW and electrostatic 1–4 scaling factors (SCNB and SCEE, respectively) were set to unity for consistency with the GLYCAM 06g parameters.40 The SETTLE algorithm was used to hold rigid hydrogen-oxygen bonds within water molecules.45 Slow system heating from 0 K to 298 K was performed in 398 ps, including a 100 ps hold at 298 K using a constant-NVE ensemble. A constant-NPT ensemble with a pressure of 1 atm and a temperature of 298 K was used for system equilibration and the production run. Temperature control was performed using the Langevin thermostat with a damping coefficient of 5 ps−1, while control over pressure was performed by using NAMD’s Nosé-Hoover Langevin piston method.43 Trajectory outputs of 11 ns simulations were saved every 500 fs with a time step of simulation integration in 1 fs. Monitoring of temperature, pressure, and energy, in addition to trajectory visualization, was performed in VMD 1.9.2.46 The hydrogen bonding analysis, including a 3.5 Å heavy-atom cutoff distance and a 120° angle cutoff, was performed using the AmberTools 16 module cpptraj.39
RESULTS AND DISCUSSION
To experimentally explore amide proton solvent exchange properties, we generated a library of amide-containing saccharides found in GAG polysaccharides, structures shown in Figure 1 with descriptions provided in Table 1. Figures 1(c), (q) also highlight two chemically modified structures that do not occur naturally: 3-O-sulfo-N-acetyl-glucosamine (GlcNAc3S) and N-acetylated Arixtra (NAcA). The variations in length, composition, sulfation position and extent, overall oligosaccharide charge, and glycosidic linkage of the compounds listed in Figure 1 allow the evaluation of the effect of these structural features on amide proton solvent exchange rates.
Table 1.
Description of oligosaccharide structures shown in Figure 1.
| Letter code | Name and abbreviations |
Oligosaccharide sequence |
|---|---|---|
| a | N-acetyl-D-glucosamine | GlcNAc |
| b | N-acetyl-D-glucosamine-6-O-sulfate | GlcNAc6S |
| c | N-acetyl-D-glucosamine-3-O-sulfate | GlcNAc3S |
| d | N-acetyl-D-galactosamine | GalNAc |
| e | HEP disaccharide IVA | ΔUA(I)(β1→4)GlcNAc(II) |
| f | Chitin disaccharide | GlcNAc(I)(β1→4)GlcNAc(II) |
| g | HEP disaccharide IIA | ΔUA(I)(β1→4)GlcNAc6S(II) |
| h | HEP disaccharide IIIA | ΔUA2S(I)(β1→4)GlcNAc(II) |
| i | HEP disaccharide IA | ΔUA2S(I)(β1→4)GlcNAc6S(II) |
| j | CS/DS disaccharide | ΔUA(I)(β1→3)GalNAc(II) |
| k | CS-A disaccharide | ΔUA(I)(β1→3)GalNAc4S(II) |
| l | CS-C disaccharide | ΔUA(I)(β1→3)GalNAc6S(II) |
| m | unsaturated HA trisaccharide, abbr. HA dp3 | ΔUA(I)(β1→3)GlcNAc(II)(β1→4)GlcA(III) |
| n | unsaturated HA tetrasaccharide, abbr. HA dp4 | ΔUA(I)(β1→3)GlcNAc(II)(β1→4)GlcA(III)(β1→3)GlcNAc(IV) |
| o | unsaturated reduced CS-A tetrasaccharide, abbr. CS dp4-ol | ΔUA(I)(β1→3)GalNAc4S(II)(β1→4)GlcA(III)(β1→3)GalNAc4S(IV-ol) |
| p | unsaturated HA pentasaccharide, abbr. HA dp5 | ΔUA(I)(β1→3)GlcNAc(II)(β1→4)GlcA(III)(β1→3)GlcNAc(IV) (β1→4)GlcA(V) |
| q | N-acetylated Arixtra, abbr. NAcA | GlcNAc6S(I)(α1→4)GlcA(II)(β1→4)GlcNAc3S6S(III)(α1→4)IdoA2S(IV)(α1→4)GlcNAc6S(V)-CH3 |
| r | unsaturated HA hexasaccharide, abbr. HA dp6 | ΔUA(I)(β1→3)GlcNAc(II)(β1→4)GlcA(III)(β1→3)GlcNAc(IV)(β1→4)GlcA(V)(β1→3)GlcNAc(VI) |
| s | unsaturated reduced HA hexasaccharide, abbr. HA dp6-ol | ΔUA(I)(β1→3)GlcNAc(II)(β1→4)GlcA(III)(β1→3)GlcNAc(IV)(β1→4)GlcA(V)(β1→3)GlcNAc(VI-ol) |
| t | unsaturated reduced CS-A hexasaccharide, abbr. CS dp6-ol | ΔUA(I)(β1→3)GalNAc4S(II)(β1→4)GlcA(III)(β1→3)GalNAc4S(IV)(β1→4)GlcA(V)(β1→3)GalNAc4S(VI-ol) |
Amide Proton Temperature Coefficients (Δδ/ΔT).
Calculated temperature coefficients (presented as absolute value of Δδ/ΔT throughout) for the oligosaccharide amide protons are presented in Table S6. None of the investigated structures produced an amide proton temperature coefficient that suggests participation in an intramolecular hydrogen bond. To visualize the data shown in Table S6, the results are depicted in a graphical form in Figure 2 with the Δδ/ΔT values plotted as a function of the corresponding residue. Because the α conformers at the reducing end of oligosaccharides are generally more energetically favorable and therefore the dominant species in aqueous solutions, they are the focus of the following discussion.
Figure 2.
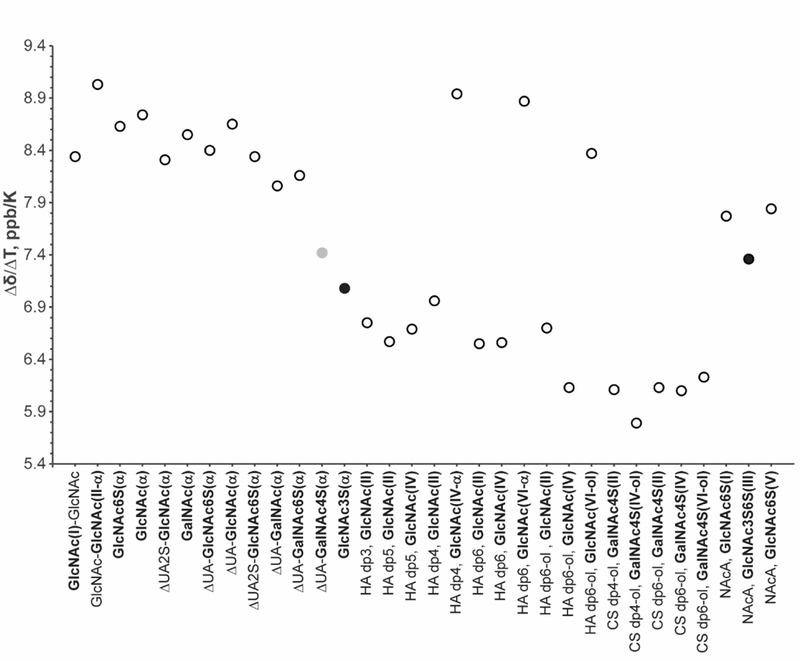
Graphical depiction of the absolute value of the amide proton temperature coefficients presented in Table S6. The graph does not include the temperature coefficient of β conformers. The 3-O-sulfated residues are indicated by solid black circles and the 4-O-sulfated GalNAc residue is highlighted by a solid grey circle.
The GlcNAc3S monosaccharide and NAcA pentasaccharide are structurally unique and were prepared to facilitate evaluation the effect of a 3-O-sulfo group on an adjacent amide moiety. The earlier work of Langeslay et al.20 identified that the same 3-O-sulfo group in Arixtra was involved in a hydrogen bond with the adjacent sulfamate group proton. In this work, the N-sulfo groups of Arixtra were chemically replaced by N-acetyl groups to evaluate possible hydrogen bond formation involving an amide proton and an adjacent 3-O-sulfo group. To our surprise, all three amide groups of NAcA gave virtually identical temperature coefficients, a result that does not support the formation of a persistent hydrogen bond between the 3-O-sulfo group and the adjacent amide group of the internal glucosamine residue. This is in contrast to what was reported for the N-sulfated glucosamine of Arixtra.20 Moreover, the temperature coefficients measured for NAcA were slightly higher than those of the GlcNAc3S monosaccharide and opposed to the general trend formed by the other larger oligosaccharides.
Interestingly, the amide proton of the GlcNAc3S monosaccharide containing a 3-O-sulfo group showed the most reduced temperature coefficient of all the mono- and disaccharides examined. A slightly higher Δδ/ΔT value was observed for ΔUA(I) (β1→3) GalNAc4S(II). This disaccharide has three distinctive features that distinguish it from the GlcNAc3S monosaccharide, including a 4-O-sulfated GalNAc residue (GalNAc4S), and an unsulfated unsaturated uronic acid residue (ΔUA) connected to GalNAc4S by a β1→3 glycosidic linkage. The temperature coefficients for GalNAc and GlcNAc are not significantly different, so the effect of sugar type can be excluded. Replacement of the GalNAc 4-O-sulfo group with a 6-O-sulfo group in ΔUA(I) (β1→3) GalNAc6S(II) led to increased Δδ/ΔT relative to ΔUA(I) (β1→3) GalNAc6S(II). Also, there is no difference in the Δδ/ΔT values for ΔUA(I) (β1→3) GalNAc6S(II) and the unsubstituted ΔUA(I) (β1→3) GalNAc(II), indicating that the 6-O-sulfo substituent is too distant to affect the amide proton.
Comparison of the effect of different glycosidic linkages for ΔUA(I) (β1→3) GalNAc(II) and ΔUA(I) (β1→4) GlcNAc(II) indicates that slightly higher temperature coefficients are associated with the β1→4 linkage. Addition of the 2-O-sulfo group to the unsaturated uronic acid residue of ΔUA2S(I) (β1→4) GlcNAc(II) produced a slight decrease in temperature coefficient compared with ΔUA(I) (β1→4) GlcNAc(II). Identical results were obtained for ΔUA2S(I) (β1→4) GlcNAc(II) and ΔUA2S(I) (β1→4) GlcNAc6S(II), as with GalNAc, no difference was observed between the unsulfated and 6-O-sulfated GlcNAc monosaccharides. Interestingly, however, replacement of the ΔUA with a GlcNAc residue produced a larger temperature coefficient for the chitin disaccharide GlcNAc(I) (β1→4) GlcNAc(II).
Comparison of the amide proton temperature coefficients within monosaccharides and disaccharides revealed the following trends: (1) no difference is observed between types of unsulfated amino sugars; (2) the amide protons in disaccharides with a β1→3 glycosidic linkage have slightly reduced temperature coefficients relative to the β1→4 linkage; (3) 6-O-sulfation had very little effect on temperature coefficient; (4) negatively charged unsaturated uronic acid residues decreased the temperature coefficient and 2-O-sulfo substituted ΔUA residues contributed to further decreases; and (5) 4-O-sulfated and 3-O-sulfated amino sugars had significantly reduced temperature coefficients compared to other mono- and disaccharides.
The monosaccharide GlcNAc3S gave temperature coefficients comparable to the larger oligosaccharides. Across the larger oligosaccharides, the most reduced Δδ/ΔT values were observed for the reduced CS dp4-ol and CS dp6-ol derived from CS-A, and HA dp6. The temperature coefficients of amide protons of the reducing end residues were consistent with the values measured for the mono- and disaccharides. The amides of the CS-A oligosaccharides had lower temperature coefficients compared to the unsulfated HA oligosaccharides, further supporting the effect of 4-O-sulfation in reducing the magnitude of the temperature coefficients.
Another important observation is that the presence of glucuronic acid (GlcA) reduces the temperature coefficient of the amide protons in adjacent residues, especially when the amide-containing residue is situated between two GlcA residues. For disaccharides with an unsaturated uronic acid at the non-reducing end, the presence of the ΔUA residue appears to slightly reduce the temperature coefficient of the adjacent amide proton. In larger oligosaccharides, when the amide group is preceded by an unsaturated uronic acid and followed by a GlcA residue, the amide proton temperature coefficient is reduced more substantially, as can be observed for the HA dp3. Furthermore, the GlcNAc6S(I) and GlcNAc6S(V) residues of NAcA, which are linked to only one uronic acid, showed higher temperature coefficients than the amide of GlcNAc3S6S(III) which is situated between GlcA(II) and IdoA2S(IV) residues.
Because the concentrations of the saccharide samples studied varied due to the limited availability of some compounds, we evaluated the effect of sugar and phosphate buffer concentrations on our measurements. Temperature coefficients of 10 mM GlcNAc in 10 mM phosphate buffer (α = 8.7 ppb/K, β = 7.8 ppb/K), 100 mM GlcNAc in 10 mM phosphate buffer (α = 8.6 ppb/K, β = 7.6 ppb/K), and 10 mM GlcNAc in 50 mM phosphate buffer (α = 8.6 ppb/K, β = 7.5 ppb/K) showed no significant deviations that can be ascribed to either the saccharide or buffer concentration. Similarly, 10 mM GalNAc in 10 mM phosphate buffer (α = 8.6 ppb/K, β = 7.5 ppb/K), 100 mM GalNAc in 10 mM phosphate buffer (α = 8.5 ppb/K, β = 7.5 ppb/K), and 10 mM GalNAc in 50 mM phosphate buffer (α = 8.7 ppb/K, β = 7.6 ppb/K) also showed no large deviations due to the concentration of the monosaccharide or the buffer. Errors in the experimental values of temperature coefficients are estimated to be around ± 0.1 ppb/K due to differences in sample preparation, shimming, temperature fluctuations and error in the temperature calibration using the external methanol standard.
Line-Shape Analysis and Exchange Energy Barriers (ΔG‡).
Resonance line widths are related to the rate constant (kex) of the solvent exchange reaction and can be used to calculate the energy barrier for solvent exchange (ΔG‡). Therefore, the variable temperature experiments used for the calculation of temperature coefficients can also provide information about the solvent exchange of the labile protons. The calculated values of ΔG‡ are summarized in Table S7 of the supplemental information. As with the temperature coefficient results, the data presented in Table S7 can be more easily visualized as a graph of ΔG‡ plotted as a function of the corresponding amide-containing residue. Figure 3 plots the measured ΔG‡ values using the same saccharide order as in Figure 2. Comparison of Figures 2 and 3 reveals that the trend of the ΔG‡ values is reciprocal to that for the Δδ/ΔT values. In other words, amide protons with reduced temperature coefficients have generally higher activation energy barriers, corresponding to slower rates of exchange with water.
Figure 3.
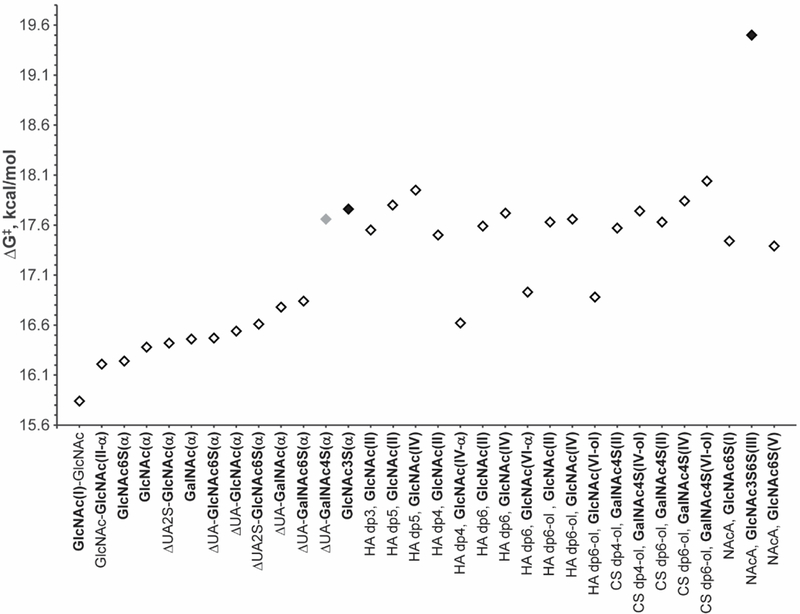
Graphical depiction of the amide proton ΔG‡ results from Table S7 with the results for the β conformers omitted. The saccharide order is the same as in Figure 2. The 3-O-sulfated GlcNAc residues are highlighted with solid black diamonds and the 4-O-sulfated GalNAc residue is indicated with a solid grey diamond.
Most interesting is the high energy barrier calculated for the internal GlcNAc3S6S(III) residue of NAcA. The hydrogen bond identified by Langeslay et al. involving the internal Arixtra GlcNS3S6S(III) sulfamate proton was supported by both a reduced temperature coefficient and a high energy barrier for solvent exchange.20 In contrast, despite its higher ΔG‡ value, the absence of a reduced temperature coefficient does not support the participation of the NAcA GlcNAc3S6S(III) amide proton in a hydrogen bond.
Distinctively different ΔG‡ values were observed for the 3-O- and 4-O-sulfated residues of GlcNAc3S and ΔUA(I) (β1→3) GalNAc4S(II), respectively. Their ΔG‡ values are comparable to those measured for the larger oligosaccharides. Furthermore, the energy barrier for amide proton solvent exchange in the chitin disaccharide, which lacks a linkage to an uronic acid residue, was the lowest value of all the saccharides examined.
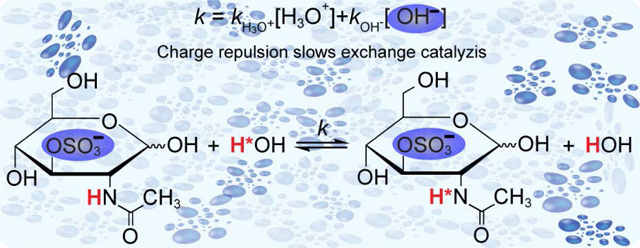
These observations suggest that charge plays an important role in determining the energy barriers for amide proton exchange with water in aqueous solutions. The pH of solution is important experimental parameter because the chemical exchange reaction between the labile protons and water is catalyzed by H3O+ and OH- ions. This exchange occurs in solution with large excess of the water and therefore is governed by the pseudo first-order kinetics as described by eq 2.50
| (2) |
According to eq 2, the overall rate constant (kobs) of the proton exchange reaction is the sum of two terms, kH3O+[H3O+] and kOH-[OH-], representing the exchange rates of the base and acid catalyzed reactions, respectively. Exchange rate constants for the acid (kH3O+) and base (kOH-) catalyzed reactions are not equal and depend on the chemical system. In each given system, concentrations of H3O+ and OH- ions might also play important role in catalysis of the exchange of labile protons with solvent, as described by eq 2. Therefore, the proximity of centers of negative charge close to an amide proton would be expected to slow exchange by repulsing OH- ions, which would be reflected by higher values of ΔG‡. To examine the effects of negatively charged groups on amide proton exchange, pH titrations were performed for a set of saccharides with unique structural differences as described in the following section.
Effects of pH on Labile Proton Resonance Line Widths.
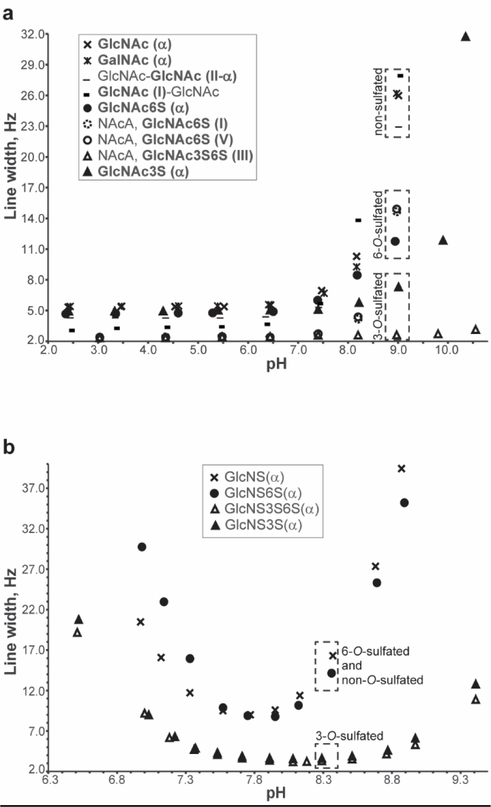
1H NMR spectra were acquired as a function of pH for GalNAc, GlcNAc, GlcNAc6S, GlcNAc3S, the chitin disaccharide and NAcA. The line widths of the amide proton resonances of these saccharides are plotted in Figure 4a as a function of pH. It was previously reported that amide proton exchange with water occurs through an acid-base catalysis mechanism.50,51 From Figure 4a, it is apparent that in the acidic region there are no changes in the amide proton line widths even at the lowest pH tested (~2.5). In the resonance structure of amide groups, the carbonyl oxygen atom is partially negatively charged, while the amide nitrogen carries partial positive charge. Consequently, charge repulsion between the partially positively charged nitrogen and hydronium ions at low pH prevents significant acid catalyzed exchange. As the pH is raised, the concentration of hydroxide increases and the base catalyzed exchange rate also increases producing line broadening. Closer examination of the basic region of Figure 4a reveals interesting trends correlated to the saccharide structures. At pH 9, the uncharged saccharides have the broadest resonances reflecting faster solvent exchange. In contrast, for the sulfated sugars, where the bulky negatively charged sulfate groups may repel the negatively charged hydroxide, reducing base catalyzed solvent exchange and producing less line broadening. Because of the close spatial proximity of the 3-O-sulfo and amide groups (discussed in the Molecular Dynamics Simulation section), base catalyzed exchange may be less effective for the 3-O-sulfated saccharide residues. Though increased line broadening is observed for the GlcNAc3S monosaccharide above pH 9, the NAcA GlcNAc3S6S(III) amide proton is unaffected by increases in pH. In addition to the 3-O and 6-O sulfo groups of this residue, the additional negative charges from the adjacent GlcA(II) and IdoA2S(IV) residues contribute to the charge density in the neighborhood of the amide proton, essentially shutting down the base catalyzed exchange mechanism.
Figure 4.
Dependence of NH resonance line widths on solution pH at 298.2K. (a) Results for the uncharged N-acetylated saccharides GalNAc, GlcNAc, chitin disaccharide, and the charged N-acetylated saccharides GlcNAc6S, GlcNAc3S, and NAcA. (b) Results for a family of N-sulfated glucosamine monosaccharides: D-glucosamine-2-N-sulfate (GlcNS), D-glucosamine-2-N,3-O-disulfate (GlcNS3S), D-glucosamine-2-N,3-O,6-O-trisulfate (GlcNS3S6S), and D-glucosamine-2- N,6-O-disulfate (GlcNS3S6S).
Closer examination of the basic region of Figure 4a reveals interesting trends correlated to the saccharide structures. At pH 9, the uncharged saccharides have the broadest resonances reflecting faster solvent exchange. In contrast, for the sulfated sugars, the bulky negatively charged sulfate groups repel the negatively charged hydroxyl ions, reducing base catalyzed solvent exchange and producing less line broadening. Because of the close spatial proximity of the 3-O-sulfo and amide groups, base catalyzed exchange is least effective for the 3-O-sulfated saccharide residues. Though increased line broadening is observed for the GlcNAc3S monosaccharide above pH 9, the NAcA GlcNAc3S6S(III) amide proton is unaffected by increases in pH. In addition to the 3-O and 6-O sulfo groups of this residue, the additional negative charges from the adjacent GlcA(II) and IdoA2S(IV) residues contribute to the negative charge density in the neighborhood of the amide proton, essentially shutting down the base catalyzed exchange mechanism.
To further evaluate the effect of structure and charge repulsion on the solvent exchange rates of labile protons, we performed a similar NMR pH titration for the N-sulfated monosaccharides GlcNS, GlcNS3S, GlcNS6S, and GlcNS3S6S (Figure 4b). Because the sulfamate nitrogen does not have a resonance structure similar to the amide group and the charge on nitrogen is neutral, acid and base catalysis of the sulfamate proton exchange is observed in Figure 4b as expected. However, based on the results for the N-acetylated saccharides (Figure 4a), monosaccharides containing a 3-O-sulfo group should repel hydroxide ions and reduce the contributions from base catalyzed exchange. Among the monosaccharides examined, only GlcNS has no O-sulfo groups and therefore should experience the greatest base catalyzed exchange as observed in Figure 4b. The pH dependent line broadening experienced by the 6-O-sulfated GlcNS sulfamate proton is only slightly less than that of GlcNS because the 6-O-sulfo group is located far from the sulfamate proton and only weakly repels hydroxide. Finally, the sulfamate line widths of the GlcNS3S and GlcNS3S6S monosaccharides are least affected by increasing pH because the adjacent bulky negatively charged 3-O-sulfate group effectively repels hydroxide. A similar pH titration curve was reported for the sulfamate group of the Arixtra GlcNS3S6S(III) residue by Langeslay et al. with narrow line widths observed even at pH values near 11.20 Though Langeslay et al. attributed this unusual pH stability solely to the participation of the sulfamate proton in a hydrogen bond with the 3-O-sulfo group, the results obtained for the N-sulfo glucosamine monosaccharides in Figure 4b and the similar pH dependence of the NAcA titration curve (Figure 4a) suggest that the combination of hydrogen bonding and less effective base catalysis due to charge repulsion of the hydroxide ions both contribute to the lack of line broadening of the Arixtra GlcNS3S6S(III) sulfamate proton in basic solution.
In another study, Beecher et al. identified three hydrogen bonds involving the hydroxyl protons of Arixtra by measurements of the hydroxyl proton solvent exchange rates and temperature coefficients.21 The reduced exchange rates measured for two of the hydroxyl protons involved in hydrogen bonds were supported by reduced temperature coefficients. However, there was a discrepancy between the lower solvent exchange rate of the IdoA2S(IV) hydroxyl proton (OH3) and its large temperature coefficient. Because of the proximity of the IdoA2S 2-O-sulfo group, the reduced exchange rate observed for OH3 likely reflects charge repulsion of hydroxide ions and a reduction in base catalyzed solvent exchange rate. These results confirm that in aqueous solutions of GAGs temperature coefficients may be reliable indicators of hydrogen bonding, and that while exchange rates can provide important insights, their interpretation must include an understanding of the effects of local structure on the acid-base catalyzed solvent exchange mechanism.
Molecular Dynamics Simulations.
To visualize the oligosaccharide structures and estimate the distances separating amide protons from the oxygens of O-sulfo groups and uronic acid carboxylates, we selected three oligosaccharides with variable length and sulfation patterns. MD results were obtained for NAcA, the HA dp6(α), and CS dp6(α). Representative structures of each oligosaccharide taken from one of the simulation frames are shown in Figure 5.
Figure 5.
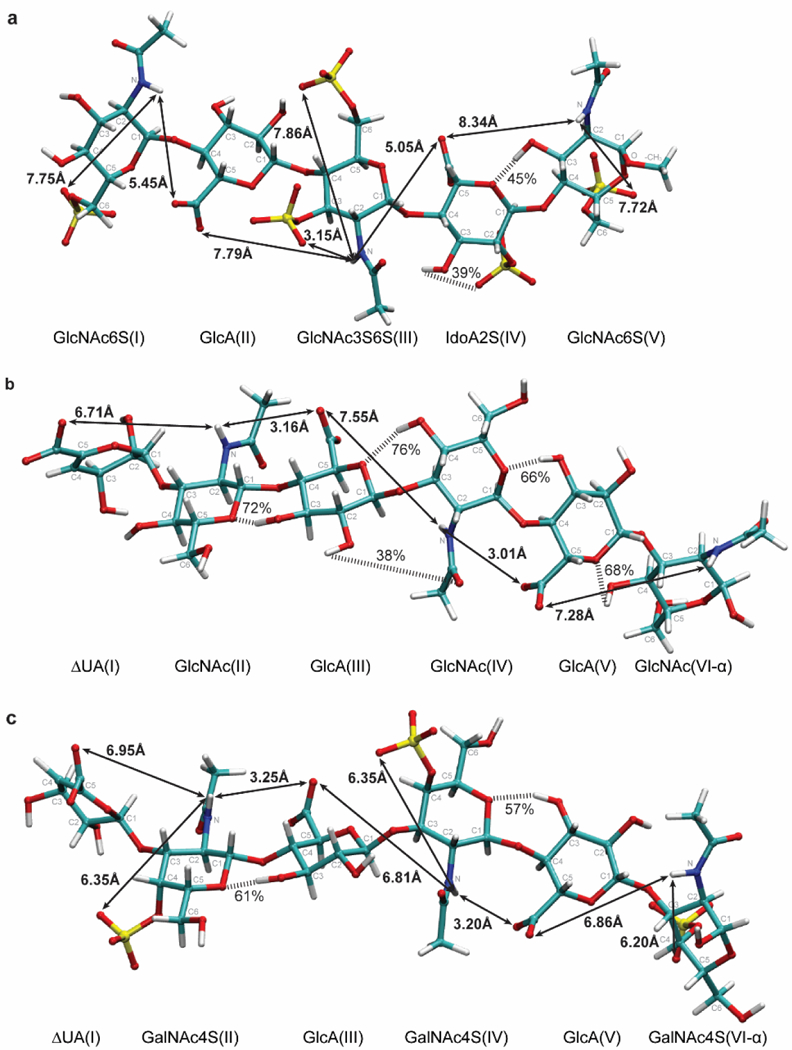
3D representation of the penta- and hexasaccharides (a) N-acetylated Arixtra (NAcA), (b) unsaturated HA hexasaccharide (HA dp6 (α)), and (c) unsaturated 4-O-sulfated chondroitin sulfate hexasaccharide (CS dp6 (α)) calculated by MD simulation. Each carbon of the sugar ring is labeled according to its position relative to anomeric carbon (C1). Measured shortest average distances separating amide proton and the O-sulfo groups are shown. In addition, distances separating amide protons from the uronic acid carboxylate oxygens are also reported, and hydrogen bonds, shown by striped bars, with occupancies greater than 25% are indicated.
We can see that among O-sulfo groups the 3-O-sulfo group in GlcNAc3S6S(III) (Figure 5a) is indeed closest to the amide group separated by 3.15 Å. Furthermore, the distance for the 4-O-sulfo group of the GalNAc4S residues (Figure 5c) is 3.20 Å longer than that of the 3-O-sulfo group, which explains why the amide proton had the second highest activation energy of the measured mono- and disaccharides.
The experimentally observed effect of glucuronic acid on the adjacent amide-containing residue is reflected by the proximity of the negatively charged carboxylate oxygens to the amide protons. For example, for the HA hexasaccharide, shown in Figure 5c, the amide proton of the GlcNAc(II) residue is only 3.16 Å from the GlcA(III) carboxylate oxygens. Similarly, the amide proton of the GlcNAc(IV) residue is only 3.01 Å from the carboxylate oxygens of the GlcA(V) residue. In contrast, the distance between the amide proton in GlcNAc(IV) and carboxylate oxygens of GlcA(III) was almost two times greater, approaching 7.55 Å. Also, the longer distance separating the carboxylate of the ΔUA(I) residue and the amide proton of GlcNAc(II) residue (~ 6.71 Å) explains the smaller effect of the ΔUA residue on solvent exchange rates. These observations are also applicable to the GlcA residues of the CS dp6 and NAcA, shown in Figure 5c and 5a, respectively. In summary, the calculated distances support the interpretation of the experimentally determined exchange rates through the effects of charge repulsion of hydroxide by neighboring negatively charged groups.
Finally, analysis of the trajectories of the MD simulations for the structures shown in Figure 5 predicted hydrogen bonds, however, none of the amide protons are involved in hydrogen bonds, consistent with experimental temperature coefficient data. Hydrogen bond donor-acceptor pairs predicted by MD with occupancies greater than 25% lifetime of frames are highlighted with black dashed lines. The hydrogen bond predicted between the 2-O-sulfo group and OH3 hydroxyl proton in IdoA2S(IV) (39%) was consistent with what was previously reported in Arixtra (32%) when IdoA2S residue adopts a 2S0 chair conformation. Another hydrogen bond between hydroxyl proton OH3 in GlcNAc6S(V) and ring oxygen of IdoA2S(IV) (45%) was also previously reported for Arixtra (32% when IdoA2S residue adopts 2S0, 60% when IdoA2S residue adopts 1C4, and 43% overall). For the HA hexasaccharide, MD predicted hydrogen bonds involving all ring oxygens in internal residues with hydroxyl protons in adjacent residues. However, as can be seen from Figure 5c for the CS hexasaccharide, disruption of hydrogen bonds was observed between GalNAc4S(VI) and GlcA(V) and between GalNAc4S(IV) and GlcA(III) when the 4-O-sulfo group was present in GalNAc residues, perhaps due to steric effects.
CONCLUSIONS
By generating a library of N-acetylated saccharides, the effects of structural features, including length, sulfation, and glycosidic linkage on amide proton solvent exchange could be evaluated. Through variable temperature experiments carried out at pH 7.4, one-dimensional NMR spectra were acquired from which amide proton temperature coefficients and energy barriers for solvent exchange were calculated. None of the saccharides were identified as having temperature coefficients that indicate amide protons involved in hydrogen bonds, however, 3-O-sulfated amide-containing residues showed very distinctive and high energy barriers associated with reduced exchange rates. By comparison of the activation energy barriers for the saccharide library, elements of structure that slow the rate of solvent exchange through charge repulsion of hydroxide were identified. Negatively charged sulfate and carboxylate groups in close spatial proximity to amide protons reduce their solvent exchange rate by decreasing base catalysis. These experimental observations were supported by distances calculated in MD simulations. The results of this work indicate that reduced proton exchange rates or very high activation energies need to be interpreted within the context of a given chemical environment, as it is the change in rate for this environment that is indicative of participation in a hydrogen bond. Slow exchange rates cannot alone be used to identify hydrogen bonds involving the labile protons of oligosaccharides and should be interpreted in concert with additional evidence supporting hydrogen bond participation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the Analytical Chemistry Instrumentation Facility (ACIF) and support by NSF CHE-1626673 for the NMR instrumentation used for some of our experiments. LJM gratefully acknowledges support by NIH Grant R01GM097569 and KL acknowledges support by the China Scholarship Council (201504910405).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The following files are available free of charge at the ACS Publications website: http://pubs.acs.org:
Methods for the preparation and structure elucidation of oligosaccharides; Tables of carbon-bound and amide proton NMR resonance assignments; Additional experimental methods for the variable temperature experiments; Representative plot of the determination of amide proton temperature coefficients; Table of amide proton temperature coefficients; Representative line shape analysis results and example plot for the determination of activation energy barriers, ΔG‡, for the GlcNAc α anomer.
REFERENCES
- (1).Pomin VH; Mulloy B Glycosaminoglycans and Proteoglycans. Pharmaceuticals (Basel) 2018, 11, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Deepa SS; Yamada S; Fukui S; Sugahara K Structural Determination of Novel Sulfated Octasaccharides Isolated from Chondroitin Sulfate of Shark Cartilage and Their Application for Characterizing Monoclonal Antibody Epitopes. Glycobiology 2007, 17, 631–45. [DOI] [PubMed] [Google Scholar]
- (3).Pothacharoen P; Kalayanamitra K; Deepa SS; Fukui S; Hattori T; Fukushima N; Hardingham T; Kongtawelert P; Sugahara K Two Related but Distinct Chondroitin Sulfate Mimetope Octasaccharide Sequences Recognized by Monoclonal Antibody WF6. J. Biol. Chem. 2007, 282, 35232–46. [DOI] [PubMed] [Google Scholar]
- (4).Li JP; Vlodavsky I Heparin, Heparan Sulfate and Heparanase in Inflammatory Reactions. Thromb. Haemost. 2009, 102, 823–8. [DOI] [PubMed] [Google Scholar]
- (5).Mousa SA Heparin, Low Molecular Weight Heparin, and Derivatives in Thrombosis, Angiogenesis, and Inflammation: Emerging Links. Semin. Thromb. Hemost. 2007, 33, 524–33. [DOI] [PubMed] [Google Scholar]
- (6).Parish CR The Role of Heparan Sulphate in Inflammation. Nat. Rev. Immunol. 2006, 6, 633–43. [DOI] [PubMed] [Google Scholar]
- (7).Tyrrell DJ; Kilfeather S; Page CP Therapeutic Uses of Heparin Beyond Its Traditional Role as an Anticoagulant. Trends Pharmacol. Sci. 1995, 16, 198–204. [DOI] [PubMed] [Google Scholar]
- (8).Nairn AV; Kinoshita-Toyoda A; Toyoda H; Xie J; Harris K; Dalton S; Kulik M; Pierce JM; Toida T; Moremen KW; Linhardt RJ Glycomics of Proteoglycan Biosynthesis in Murine Embryonic Stem Cell Differentiation. J. Proteome. Res. 2007, 6, 4374–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kresse H; Schonherr E Proteoglycans of the Extracellular Matrix and Growth Control. J. Cell. Physiol. 2001, 189, 266–74. [DOI] [PubMed] [Google Scholar]
- (10).Whitelock JM; Iozzo RV Heparan Sulfate: A Complex Polymer Charged with Biological Activity. Chem. Rev. 2005, 105, 2745–64. [DOI] [PubMed] [Google Scholar]
- (11).Raman R; Sasisekharan V; Sasisekharan R Structural Insights Into Biological Roles of Protein-Glycosaminoglycan Interactions. Chem. Biol. 2005, 12, 267–77. [DOI] [PubMed] [Google Scholar]
- (12).Sasisekharan R; Shriver Z; Venkataraman G; Narayanasami U Roles of Heparan-Sulphate Glycosaminoglycans in Cancer. Nat. Rev. Cancer 2002, 2, 521–8. [DOI] [PubMed] [Google Scholar]
- (13).Vlodavsky I; Beckhove P; Lerner I; Pisano C; Meirovitz A; Ilan N; Elkin M Significance of Heparanase in Cancer and Inflammation. Cancer Microenviron. 2012, 5, 115–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bartlett AH; Park PW Proteoglycans in Host-Pathogen Interactions: Molecular Mechanisms and Therapeutic Implications. Expert. Rev. Mol. Med. 2010, 12, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shukla D; Liu J; Blaiklock P; Shworak NW; Bai X; Esko JD; Cohen GH; Eisenberg RJ; Rosenberg RD; Spear PG A Novel Role for 3-O-Sulfated Heparan Sulfate in Herpes Simplex Virus 1 Entry. Cell 1999, 99, 13–22. [DOI] [PubMed] [Google Scholar]
- (16).Tiwari V; Clement C; Xu D; Valyi-Nagy T; Yue BY; Liu J; Shukla D Role for 3-O-sulfated Heparan Sulfate as the Receptor for Herpes Simplex Virus Type 1 Entry Into Primary Human Corneal Fibroblasts. J. Virol. 2006, 80, 8970–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Garcia B; Merayo-Lloves J; Martin C; Alcalde I; Quiros LM; Vazquez F Surface Proteoglycans as Mediators in Bacterial Pathogens Infections. Front. Microbiol. 2016, 7, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cierpicki T; Otlewski J Amide Proton Temperature Coefficients as Hydrogen Bond Indicators in Proteins. J. Biomol. Nmr 2001, 21, 249–261. [DOI] [PubMed] [Google Scholar]
- (19).Wagner G; Pardi A; Wuethrich K Hydrogen Bond Length and Proton NMR Chemical Shifts in Proteins. J. Am. Chem. Soc. 1983, 105, 5948–5949. [Google Scholar]
- (20).Langeslay DJ; Young RP; Beni S; Beecher CN; Mueller LJ; Larive CK Sulfamate Proton Solvent Exchange in Heparin Oligosaccharides: Evidence for a Persistent Hydrogen Bond in the Antithrombin-Binding Pentasaccharide Arixtra. Glycobiology 2012, 22, 1173–82. [DOI] [PubMed] [Google Scholar]
- (21).Beecher CN; Young RP; Langeslay DJ; Mueller LJ; Larive CK Hydroxyl-Proton Hydrogen Bonding in the Heparin Oligosaccharide Arixtra in Aqueous Solution. J. Phys. Chem. B 2014, 118, 482–91. [DOI] [PubMed] [Google Scholar]
- (22).Beecher CN; Larive CK (1)H and (15)N NMR Characterization of the Amine Groups of Heparan Sulfate Related Glucosamine Monosaccharides in Aqueous Solution. Anal. Chem. 2015, 87, 6842–8. [DOI] [PubMed] [Google Scholar]
- (23).Battistel MD; Pendrill R; Widmalm G; Freedberg DI Direct Evidence for Hydrogen Bonding in Glycans: A Combined NMR and Molecular Dynamics Study. J. Phys. Chem. B 2013, 117, 4860–9. [DOI] [PubMed] [Google Scholar]
- (24).Eyring H The Activated Complex in Chemical Reactions. J. Chem. Phys. 1935, 3, 107–115. [Google Scholar]
- (25).Pechukas P Transition-State Theory. Annu. Rev. Phys. Chem. 1981, 32, 159–177. [Google Scholar]
- (26).Bain AD Chemical Exchange in NMR. Prog. Nucl. Mag. Res. Sp. 2003, 43, 63–103. [Google Scholar]
- (27).Sandstrom C; Baumann H; Kenne L NMR Spectroscopy of Hydroxy Protons of 3,4-Disubstituted Methyl Alpha-D-Galactopyranosides in Aqueous Solution. J. Chem. Soc. Perk. T 2 1998, 809–815. [Google Scholar]
- (28).Dobson CM; Lian LY; Redfield C; Topping KD Measurement of Hydrogen-Exchange Rates Using 2d Nmr-Spectroscopy. J. Magn. Reson. 1986, 69, 201–209. [Google Scholar]
- (29).Chen JH; Mao XA Measurement of Chemical Exchange Rate Constants with Solvent Protons Using Radiation Damping. J. Magn. Reson. 1998, 131, 358–361. [DOI] [PubMed] [Google Scholar]
- (30).Chen JH; Mao XA Radiation Damping Transfer in Nuclear Magnetic Resonance Experiments Via Chemical Exchange. J. Chem. Phys. 1997, 107, 7120–7126. [Google Scholar]
- (31).Baxter NJ; Williamson MP Temperature Dependence of H-1 Chemical Shifts in Proteins. J. Biomol. Nmr 1997, 9, 359–369. [DOI] [PubMed] [Google Scholar]
- (32).Adams B; Lerner L Observation of Hydroxyl Protons of Sucrose in Aqueous-Solution - No Evidence for Persistent Intramolecular Hydrogen-Bonds. J. Am. Chem. Soc. 1992, 114, 4827–4829. [Google Scholar]
- (33).Kindahl L; Sandstrom C; Norberg T; Kenne L H-1 NMR Studies of Hydroxy Protons of the V[beta-Gal(1 -> 3)-Alpha-GalNAc(1 -> O)THPGY Glycopeptide. Carbohyd. Res. 2001, 336, 319–323. [DOI] [PubMed] [Google Scholar]
- (34).Kindahl L; Sandstrom C; Norberg T; Kenne L H-1 NMR Studies of Hydroxy Protons of Asn- and Ser-Linked Disaccharides in Aqueous Solution. J. Carbohyd. Chem. 2000, 19, 1291–1303. [Google Scholar]
- (35).Glasoe PK; Long FA Use of Glass Electrodes to Measure Acidities in Deuterium Oxide. J. Phys. Chem. 1960, 64, 188–190. [Google Scholar]
- (36).Mazák K; Beecher CN; Kraszni M; Larive CK The Interaction of Enoxaparin and Fondaparinux with Calcium. Carbohyd. Res. 2014, 384, 13–19. [DOI] [PubMed] [Google Scholar]
- (37).Van Geet AL Calibration of the Methanol and Glycol Nuclear Magnetic Resonance Thermometers with a Static Thermistor Probe. Anal. Chem. 1968, 40, 2227–2229. [Google Scholar]
- (38).Liu ML; Mao XA; Ye CH; Huang H; Nicholson JK; Lindon JC Improved WATERGATE Pulse Sequences for Solvent Suppression in NMR Spectroscopy. J. Magn. Reson. 1998, 132, 125–129. [Google Scholar]
- (39).Case DA, B. R M, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li S, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L, Kollman PA AMBER 16. University of California; San Francisco. [Google Scholar]
- (40).Kirschner KN; Yongye AB; Tschampel SM; Gonzalez-Outeirino J; Daniels CR; Foley BL; Woods RJ GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hornak V; Abel R; Okur A; Strockbine B; Roitberg A; Simmerling C Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins 2006, 65, 712–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- (43).Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Darden T; York D; Pedersen L Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar]
- (45).Miyamoto S; Kollman PA Settle - an Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar]
- (46).Humphrey W; Dalke A; Schulten K VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–8, 27–8. [DOI] [PubMed] [Google Scholar]
- (47).Andersen NH; Neidigh JW; Harris SM; Lee GM; Liu ZH; Tong H Extracting Information From the Temperature Gradients of Polypeptide NH Chemical Shifts .1. The Importance of Conformational Averaging. J. Am. Chem. Soc. 1997, 119, 8547–8561. [Google Scholar]
- (48).Blundell CD; Almond A Temperature Dependencies of Amide 1H- and 15N- Chemical Shifts in Hyaluronan Oligosaccharides. Magn. Reson. Chem. 2007, 45, 430–3. [DOI] [PubMed] [Google Scholar]
- (49).Blundell CD; Deangelis PL; Almond A Hyaluronan: The Absence of Amide-Carboxylate Hydrogen Bonds and the Chain Conformation in Aqueous Solution are Incompatible with Stable Secondary and Tertiary Structure Models. Biochem. J. 2006, 396, 487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Englander SW; Kallenbach NR Hydrogen Exchange and Structural Dynamics of Proteins and Nucleic Acids. Q. Rev. Biophys. 1983, 16, 521–655. [DOI] [PubMed] [Google Scholar]
- (51).Woodward CK; Hilton BD Hydrogen Isotope Exchange Kinetics of Single Protons in Bovine Pancreatic Trypsin Inhibitor. Biophys. J. 1980, 32, 561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.