Abstract
Alzheimer’s disease (AD) is an age- and gender-associated brain disorder. Long noncoding RNAs (lncRNAs) have emerged as key regulators of brain development, homeostasis, and pathologies. Here we utilized gene array datasets and bioinformatics analysis to identify differentially-expressed age- and gender-associated lncRNAs in human AD brains. We found that the expressions of 16 age-associated and 13 gender-associated lncRNAs were dysregulated in AD brains. Notably, the expressions of age-associated lncRNAs - SNHG19 and LINC00672, were significantly correlated with Braak stage of AD, positively and negatively, respectively, while the expressions of gender-associated lncRNAs - RNF144A-AS1, LY86- AS1, and LINC00639 were negatively correlated with Braak stage of AD. Functional analysis suggests that the pathways involved in neurodegenerative diseases, synaptic vesicle cycle and endocytosis were overly represented within age- and gender-associated lncRNA-correlating genes. The identification of age- and gender-associated lncRNAs and their differential expressions in human AD brain provide potential targets for further experimental validation and mechanistic investigation, which could, in turn, pave the way for developing age- and gender-specific prevention and adjunctive therapeutic options for AD patients.
Keywords: long noncoding RNA, aging, Alzheimer’s disease, brain, gender differences
Introduction
Alzheimer's disease (AD) is a slowly progressive brain disorder characterized by cognitive decline, irreversible memory loss, disorientation, and language impairment (Masters et al., 2015). Aging is one of the major risk factors for AD. In the U.S., more than ten percent of people aged 65 years and older are living with AD (Masters et al., 2015). Cognitive decline in aging and AD is associated with altered expression of genes involved in synaptic function, energy metabolism (including mitochondrial function) and protein synthesis in the brain (Grimm and Eckert, 2017; Saura et al., 2015). The alteration of age-related genes is accelerated in the brain of AD patients (Saetre et al., 2011); suggesting aberrant regulation of aging-associated transcripts in the human brain could contribute to AD pathogenesis.
The overall incidence of AD in females is up to twice that of males in the U.S. (Chene et al., 2015; Drew, 2018). Gender differences in brain development, structure, and function are considered as important implications for mechanistic investigation of psychiatric diseases and neurodegenerative disorders including AD (Mazure and Swendsen, 2016). Studies have demonstrated that sex steroids can modulate brain development and are crucial for the development of brain regions, such as the hippocampus and parietal lobe that are significantly affected in AD (Compton et al., 2001; Fitch and Denenberg, 1998; Murphy et al., 1996). For example, plasma levels of circulating sex steroids have been shown positively correlated with cognitive performance in women who undergo bilateral oophorectomy (Sherwin, 1988). Subsequent studies have further demonstrated that the administration of estrogens affects brain organization for memory and improves cognitive ability in postmenopausal women (Shaywitz et al., 1999; Sherwin, 1988). Additionally, Barnes et al. (Barnes et al., 2005) have demonstrated a stronger relationship between brain pathology (such as neuritic plaques, diffuse plaques, and neurofibrillary tangles) and clinical AD in female than in male, indicating gender differences in the brain could be a confounding factor for the development of AD.
Gender-specific gene expression related to brain function has also been implicated in AD pathogenesis. For example, Köglsberger et al. have demonstrated a male-specific reduction of sex-linked ubiquitin-specific peptidase 9 (USP9) in AD (Koglsberger et al., 2017). Mechanistic studies have demonstrated that USP9 is a positive regulator of an AD-associated protein - MAPT (microtubule-associated protein tau). Another elegant study by Bangasser et al. has demonstrated that overexpression of CRF (Corticotropin-releasing factor) in females is associated with increased tau phosphorylation - a critical step in the formation of fibrillary tangles that are often observed in AD brains (Bangasser et al., 2017).
Recent compelling evidence has indicated that long noncoding RNAs (lncRNAs) are key mediators in the development and progression of various brain disorders, including AD. A group of differentially expressed lncRNAs, including n341006, and AD-linc1 were identified in human AD brains by Zhou and Xu (Zhou and Xu, 2015) and Magistri et al. (Magistri et al., 2015). Subsequent analysis has revealed a brain-region-specific expression pattern of lncRNAs that is age-dependent and AD-associated (Zhou et al., 2018).
Herein, we investigated the expression patterns of lncRNAs in the human brain during aging and between genders as well as their differential expressions in the AD brains. We found that 16 age-associated and 13 gender-associated lncRNAs were dysregulated in AD brains. The functional analysis suggests that these lncRNAs could be pivotal regulators of neuronal functions.
Materials and methods
Microarray data acquisition
Five sets of microarray gene expression data (GSE53890, GSE48350, GSE5281, GSE84422, and GSE66333) were obtained from the Gene Expression Omnibus (GEO) database of NCBI (http://www.ncbi.nlm.nih.gov/geo/) (Barrett and Edgar, 2006; Barrett et al., 2013). The statistics and description of the datasets are shown in Table 1. Of note, microarray dataset GSE66333 contains expression profiles of neurons isolated from eight cases (four high and four low oxidative damage and an associated DNA damage response [DDR]) by laser capture microdissection (Simpson et al., 2016). High and low DDRs are defined base on the activation of DNA-protein kinase catalytic subunit (DNA-PKcs) and the phosphorylation of the histone H2AX (γH2AX) as reported earlier (Shrivastav et al., 2008; Simpson et al., 2015; Simpson et al., 2016). The raw CEL data were normalized with the Robust Multichip Average (RMA) method using the R software 'limma' package.
Table 1:
The statistics and description of the datasets used in this study
| Datasets (GEO ID) $ | Data | Sample Type/Source |
References |
|---|---|---|---|
| GSE53890 | 41 adult human brain (aged 24-106 years) | Frontal cortical regions | (Lu et al., 2014) |
| GSE48350 | 48 normal controls (aged 20-99 years); 21 AD cases (aged 74-95 years) |
Superior frontal gyrus | (Astarita et al., 2010; Berchtold et al., 2013; Berchtold et al., 2008; Blair et al., 2013; Cribbs et al., 2012; Sarvari et al., 2012) |
| GSE5281 | 23 AD brains (aged 68-95 years); 11 normal controls (aged 63-102years) |
laser-capture microdissected neurons from superior frontal gyrus | (Liang et al., 2007; Liang et al.,2008; Readhead et al., 2018) |
| GSE84422 | 102 patients (aged 62-102 years; clinical dementia rating:0-5) |
Amygdala and nucleus accumbens | (Wang et al., 2016) |
| GSE66333 | Four high DNA damage response [DDR]). Four low DDR |
Frontal cortex pyramidal neurons | (Simpson et al., 2016) |
All datasets used in this study were generated on the microarray platform GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array.
Reannotation of the Affymetrix microarray and identification of differentially expressed IncRNAs
The probe sets of the Affymetrix Human Genome U133 Plus 2.0 array were first reannotated using the method developed by Van Grembergen et al. (Van Grembergen et al., 2016). Briefly, sequences of all probe sets were locally mapped against a reference transcriptome in LNCipedia database - a database dedicated to lncRNAs and Ensembl 84 transcriptome. Probes that target lncRNAs in the LNCipedia database remain while probes that are discordant between LNCipedia and Ensembl were discarded from the future analysis. Probe sets that target multiple genes were also excluded unless the targets are duplicated lncRNAs. lncRNA transcripts were defined as duplicated if more than 95% of one sequence is identical to the other. Finally, 3,053 lncRNAs represented by 3,668 probe sets were reannotated.
The GEO2R (Barrett et al., 2013) web tool (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to identify differentially expressed genes between two given groups of samples in a GEO profile. lncRNAs with p ≤ 0.05 and ∣log Fold Change∣ ≥ 0.5 were selected for further analysis. The age- and gender-associated lncRNAs were further subjected to coding potential analysis using CPC (coding potential calculator) (Kong et al., 2007), and CPAT (Coding-Potential Assessment Tool) (Wang et al., 2013). Transcripts with the CPC score between −1 and 1 are marked as ‘weak noncoding’ or ‘weak coding’. Transcripts with CPC score < −1 or CPAT score < 364 were considered as non-coding RNAs.
Functional enrichment analysis of lncRNAs based on their correlated mRNAs
Pathway enrichment analysis on lncRNA-correlating genes was performed using the R2 KEGG Pathway Finder by Gene correlation (R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl)) using the dataset GSE48350 (173 normal and 80 AD brain samples). Genes with a p-value < 0.05 were considered as lncRNA-correlating genes. Pathways with P-value <=0.01 (cutoff 0.01) were considered as having significant over-representation in the dataset and were ranked by the sum of the negative log10 p-value of each lncRNA for each pathway.
Statistical analysis
P-values were calculated using a two-tailed unpaired t-test or two-way analysis of variance (ANOVA) for differential expression as indicated in the figure legends. The linear regression equations were derived based on the averages expression of lncRNAs in each age group (20: [age ranging 20-29, n=7]; 30: [30-39, n=5]; 40 [40-49, n=9]; 60: [52-69, n=5]; 70: [70-79, n=8]; 80: [80-89, n=6]; and 90 [90-99, n=10]) for the normal brains, where n is the age group index. The Pearson correlation coefficients were calculated using the R package and GraphPad Prism version 6.01 for Window (GraphPad Software, San Diego, CA, USA).
Data Availability Statement
The original data (GSE53890, GSE48350, GSE5281, GSE84422, and GSE66333) used for this study are available at the Gene Expression Omnibus (GEO) database of NCBI (http://www.ncbi.nlm.nih.gov/geo/). Anonymized data will be shared upon request from qualified investigators.
Results
Common lncRNA expression patterns identified in aged and AD brains
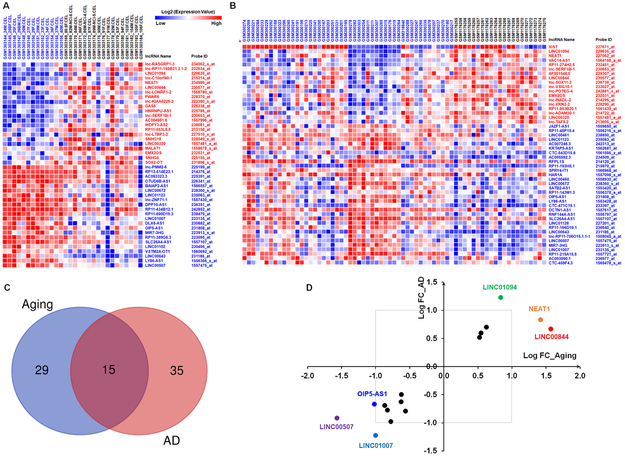
To explore the expression pattern of lncRNAs in aged and AD brains, we first investigated differentially expressed lncRNAs in the prefrontal cortex of young (≤ 40 years) and aged (≥ 80 years) individuals without AD (from dataset GSE53890). As shown in supplementary table 1, the expressions of 22 and 22 lncRNAs were upregulated and downregulated, respectively, in the aged brains compared with young adult brains. The dysregulated lncRNAs in aged brains are shown in Figure 1A. Analysis of dataset GSE48350 demonstrated that the expressions of 18 and 32 lncRNAs were significantly upregulated and downregulated respectively in AD brains compared with normal brains (supplementary table 2). The dysregulated lncRNAs in AD brains are shown in Figure 1B. We next compared the significantly regulated lncRNAs in aged brains and in AD brains. As shown in Figure 1C, the expressions of 15 lncRNAs were commonly altered in both aged and AD brains compared with normal brains. Interestingly, all of these lncRNAs were either commonly upregulated (6 lncRNAs) or downregulated (9 lncRNAs) in both aged and AD brains compared with normal brains. As shown in Figure 1D, the expressions of LINC01094, NEAT1 and LINC00844 were significantly increased in both aged and AD brains with a log2 fold change greater than 1, and the expressions of LINC01007; LINC00507 and OIP5-AS1 were significantly decreased in both aged and AD brains with a log2 fold change less than −1.
Figure 1. Common lncRNA expression patterns in the frontal cortices of aged and AD individuals.
A Heat map showing microarray data (GSE53890) for the up- and down-regulated lncRNAs in the frontal cortices of aged individuals (age ≥ 80; n = 17) compared with that in young individuals (age ≤ 40; n = 13). B Heat map showing microarray data (GSE48350) for the up- and down-regulated lncRNAs in the frontal cortices of individuals with AD (n = 21) compared with that in individuals with no disease (nd; n = 48). C Venn diagram illustrating the overlap of 15 lncRNAs that were altered (∣log2 (Fold Change)∣ > 0.5; p < 0.05) by both aging and AD. D Scatter plot illustrating the 15 overlapping lncRNA genes showing log2 of fold change by AD (x-axis) against log2 of fold change with aging (y-axis). The upper right quadrant represents 6 lncRNAs that were increased with both aging and AD. Conversely, the lower left quadrant illustrates 9 lncRNAs that were decreased by both aging and AD.
Abnormal alteration of age-associated lncRNAs in AD brains
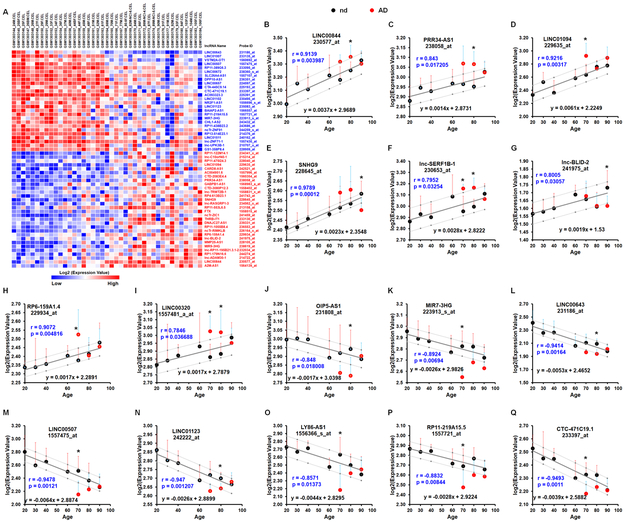
Next, we sought to determine whether there is an abnormal alteration in the expression level of age-correlated lncRNAs in AD brains. For this, we retrieved microarray expression profiles of the human frontal cortex (GSE53890) and performed Pearson’s correlation analysis between age and log2 expression value. As shown in Figure 2A and supplementary table 3, the expressions of 23 lncRNAs were negatively correlated with age and the expressions of 29 lncRNAs were positively correlated with age (all ∣r∣ > 0.5; p< 0.05). To further confirm that the expressions of these lncRNAs were correlated with age and to investigate their alterations in AD brains, we first, using data retrieved from GSE48350, performed Pearson’s correlation analysis between the average log2 expression value of each lncRNA in each age group (normal brain with no AD) and age index; next, we conducted a 95% prediction interval for the regression slope for each lncRNA followed by expression comparison between AD and normal normal brains within each age group. As shown in Figure 2B-2Q, the expressions of 16 lncRNAs were strongly correlated with age in individuals with no diseases (all ∣r∣ > 0.75; p< 0.05), either positively or negatively; while their expressions in AD brains fell outside of their prediction intervals calculated based on their expression in normal aging brains; indicating the expressions of these 16 lncRNAs were abnormally alternated in AD brains during aging. For example, the expressions of the positively-age-associated lncRNAs- LINC00844, PRR34-AS1, and LINC01094 were abnormally increased in AD brains compared with age-matched normal brains (Figures 2B-2D). In contrast, the expressions of the negatively-age-associated lncRNAs-MIR7-3HG, LINC00643, and LINC00507 were significantly decreased in AD brains compared with age-matched normal brains (Figure 2K-2M).
Figure 2. Abnormal alteration of age-associated lncRNAs in the frontal cortices of AD patients.
A Heat map representation of microarray data (GSE53890) for age correlated lncRNAs (∣r∣ > 0.5; p < 0.05; Pearson’s test). Blue and black colors indicate lncRNAs that are negatively and positively associated with age, respectively. B - Q Relative alterations of indicated lncRNAs in the frontal cortices of individuals with no disease (nd; black dots) and AD (red dots) during aging. Bars indicate SD; r and p values were obtained by Pearson’s correlation and are shown within the graphs. The black dots with error bars denote means for expression levels of indicated lncRNAs in the frontal cortices of individuals with nd in each age group (20: [age ranging 20-29, n=7]; 30: [30-39, n=5]; 40 [40-49, n=9]; 60: [52-69, n=5]; 70: [70-79, n=8]; 80: [80-89, n=6]; and 90 [90-99, n=10]). The red dots with error bars denote means for expression levels of indicated lncRNAs in the frontal cortices of individuals with AD in each age group (70: [70-79, n=3]; 80: [80-89, n=8]; and 90: [90-99], n=8). The solid line shows the expression trend of indicated lncRNA in the frontal cortices during normal aging. Pearson correlation coefficients (r) and p values are shown in the graphs. The dotted lines show the 95% prediction interval of linear regression analysis calculated from the data (GSE48350). P-values were calculated using Two-tailed Unpaired t-test where: *P < 0.05 vs the same age group.
Dysregulated gender-associated lncRNAs in AD brains
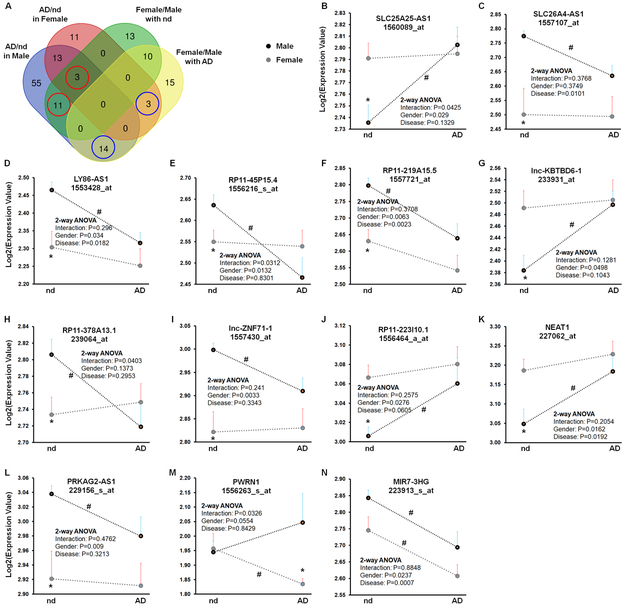
To determine gender-associated lncRNAs and their alterations in AD brains, we first compared differentially expressed lncRNAs in four comparison pairs: (1) Female (n = 24) vs. Male (n = 24) in individuals with no diseases; (2) Female (n =12) vs. Male (n = 7) in AD patients; (3) Female with AD vs. Female with no diseases; and (4) Male with AD vs. Male with no diseases (Data from dataset GSE48350). As shown in Figure 3A, a total of 37 (13+10+11+3) lncRNAs were differentially expressed (∣log2 (Fold Change)∣ > 0.5; p < 0.05) in female versus male normal brains. Among them, 3 and 11 lncRNAs were also differentially expressed in female AD brains and male AD brains compared with female and male normal brains, respectively (red circles in Figure 3A). A total of 42 lncRNAs were differentially expressed in female and male AD brains. Among them, 3 and 14 of them were differentially expressed in female and male AD brains compared with female and male normal brains, respectively (blue circles in Figure 3A). Next, to avoid age bias, data from a total of 43 individuals, age ranging from 70 to 99, (9 males and 13 females with no disease; 7 males and 14 females with AD), were selected for further analysis. As shown in Figures 3B-3N and Supplementary Table 4, the expressions of 13 gender-associated lncRNAs were significantly dysregulated in AD brains compared with normal brains in either females or males. For example, SLC25A25-AS1 shows lower levels in normal male brains compared with normal female brains but was significantly upregulated in male AD brains specifically compared with normal male brains (Figure 3B). PWRN1 was expressed at similar levels in normal female and male brains, while the expression of PWRN1 was significantly decreased in female AD brains specifically (Figure 3M). Interestingly, two-way ANOVA (gender × AD) demonstrated a significant interaction and significant main effects of gender on the expressions of SLC25A25-AS1 and RP11-45P15.4 (Figures 3B and 3E), indicating that AD affects the expression of the gender-associated lncRNAs in a gender-dependent manner.
Figure 3. Dysregulated gender-associated lncRNAs in the frontal cortices of AD patients.
A Venn graph summarizes the number of common and unique lncRNAs (dataset GSE48350) identified in the four comparisons: Female (n = 24) vs. Male (n = 24) in individuals with no disease (nd), Female (n =12) vs. Male (n = 7) in AD patients, Female with AD vs. Female with nd and Male with AD vs. Male with no disease. B - N Relative expression of indicated lncRNAs in the frontal cortices of male (black dots) and female (gray dots) individuals with and without AD (GSE48350). To avoid age bias, a total of 43 subjects, age ranging from 70 to 99, (9 males and 13 females with nd; 7 males and 14 females with AD), were selected for this analysis. Bars indicate SEM. P-values were calculated using Two-tailed Unpaired t-test where: *P < 0.05 vs male in the same disease status; # P < 0.05 vs nd in the same gender. Results of Two-Way ANOVA are given within the graphs.
Next, we sought to determine the coding potential of the age- and gender-associated lncRNAs. For this, CPC and CPAT tools were applied to evaluate the coding potential of the age- and gender-associated lncRNAs. As shown in Supplementary Table 5, all of the 16 age-associated and 13 gender-associated lncRNAs that were dysregulated in AD brains are noncoding or have weak potential coding RNAs.
Correlation of the age- and gender-associated lncRNAs with Braak stage of AD and their host genes
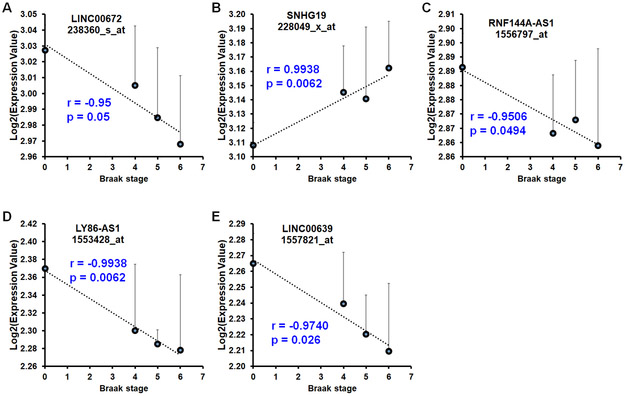
We next sought to determine whether the expressions of these lncRNAs are correlated with disease stage using age-matched samples in the dataset GSE48350 (Supplementary Table 6). To this end, we performed Pearson’s correlation analysis between the average log2 expression value of each lncRNA in each Braak stage and Braak stage of AD (normal was considered as 0). As shown in Figure 4A, expression of age-associated lncRNA - LINC00672, was negatively correlated with Braak stage of AD. As shown in Figures 4B-4E, expression of gender-associated lncRNA -SNHG19, was positively correlated with Braak stage of AD, while the expressions of gender-associated lncRNAs (RNF144A-AS1, LY86-AS1, and LINC00639) were negatively correlated with Braak stage of AD.
Figure 4. Expression of aging- and gender-associated lncRNAs was correlated with Braak stage of AD in the anterior prefrontal cortex (PFC).
A - E Expression of age-associated lncRNAs - LINC00672 (A) and SNHG19 (B), was positively and negatively correlated with Braak stage of AD, respectively, while the expression of gender-associated lncRNAs - RNF144A-AS1 (C), LY86-AS1 (D), and LINC00639 (E) was negatively correlated with Braak stage of AD. (Dataset GSE48350; n=22 for nd age ranging from 70-99; n = 6 for braak stage of IV; n = 4 for braak stage of V; n = 5 for braak stage of VI).
We next asked whether expressions of the age- and gender-associated lncRNAs are correlated with neuritic plaque density which is one of the major histopathological lesions of AD. To this end, we computed correlations between the log2 expression value of the age- and gender- associated lncRNAs and average neuritic plaque density using data in the dataset GSE84422 which was generated on the GPL507 microarray platform. Interestingly, as shown in Supplementary Figure 4, the expressions of 4 lncRNAs, such as RP11-223I10.1, LINC01094, OIP5-AS1, and LINC01007, showed a moderate linear correlation with average neuritic plaque density (∣r∣ > 0.3 and p < 0.05, n =102).
Additionally, given the fact that expression of lncRNA often correlates with the expression of its host or neighbor genes due to shared regulatory elements and cis-regulatory role of lncRNA, we, therefore, calculated the Pearson correlation coefficient between each lncRNA-host mRNA pair using the expression values in the dataset GSE48350 (173 normal and 80 AD brain samples). We found that the expressions of 9 lncRNAs have positive correlations with the expressions of their intersecting mRNAs, the expressions of 2 lncRNAs have positive correlations with the expressions of their intersecting genes (∣r∣ > 0.5, p < 0.0001; Supplementary Figure 5).
Dysregulation of age- and gender-associated lncRNAs in neurons of AD patients
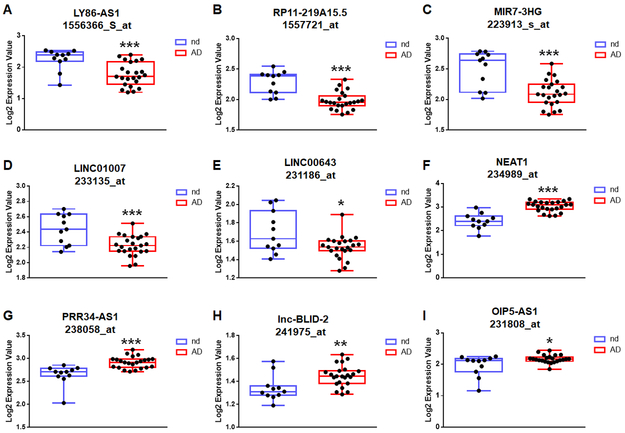
To determine whether the alteration of the age- and gender-associated lncRNAs is neuron-specific, we assessed dataset GSE5281 that contains gene expression profile in neurons from superior frontal gyrus of AD cases and normal controls (Liang et al., 2008). Interestingly, most of the age- and gender associated lncRNAs showed similar alteration in the neurons from AD brains compared with normal brains (Figure 5 and Supplementary Figures 1 and 2). For example, age negative correlated lncRNAs- LY86-AS1, RP11-219A15.5, MIR7-3HG, LINC01007 and LINC00643 were significantly downregulated in the neurons of AD patients compared with controls (Figures 5A-5E), while age positively correlated lncRNAs- NEAT1, PRR34-AS1, and lnc-BLID-2 were significantly upregulated in the neurons of AD patients compared with controls (Figures 5F-5H). However, as shown in Figure 5I, the expression of OIP5-AS1 (age negatively correlated) was significantly upregulated in the neurons of AD patients compared with controls, indicating a distinct alteration pattern of OIP5-AS1 in various CNS cells, such as increased in the AD neurons but likely decreased in the AD glial cells.
Figure 5. Age-associated lncRNAs are dysregulated in neurons of AD patients.
Boxplots of expression levels of age-associated lncRNAs in frontal neurons of individuals with no disease (n=11) and AD (n=23) based on GSE5281 dataset. Boxplots show median, interquartile range, sample minimum, and maximum. P-values were calculated using Two-tailed Unpaired t-test where: *P<0.05, **P<0.01, and *** P<0.001.
Oxidative damage and an associated DNA damage response (DDR) occur at the earliest stages of Alzheimer pathology (Simpson et al., 2010; Simpson et al., 2016) and could be reflected by AD-associated lncRNAs. We thus sought to assess the expression of the AD-associated lncRNAs in neurons isolated from brains with a high and low neuronal DDR from dataset GSE66333 (Simpson et al., 2016). Interestingly, as shown in Supplementary Figure 3 and Supplementary Table 4, the downregulated lncRNAs in AD brains were expressed at higher levels in neurons isolated from high DDR cases than from low DDR cases, which further suggests that high levels of oxidative stress and an associated neuronal DDR are very early events in the progression of AD and the downregulation of the AD-associated lncRNAs in AD neurons could be triggered by these events.
Functional analysis of age- and gender-associated lncRNAs
Finally, to investigate the potential functions of the age- and gender-associated lncRNAs in AD brains, pathway enrichment analysis on lncRNA-correlating genes was performed using the R2 KEGG Pathway Finder. Data used for the correlation analyses were from the GSE48350 dataset (173 normal and 80 AD brain samples) in the R2 platform. Supplementary table 7 shows the KEGG pathways that were significantly over-represented within the age- and gender-associated lncRNA-correlating genes. The pathways were then ranked by the sum of the negative log10 p-value of each lncRNA for each pathway. Top 30 pathways are shown in Figure 6. Interestingly, KEGG pathways involved in neurodegenerative diseases such as AD, HD, and PD (Huntington’s, and Parkinson’s disease), were over-represented within both age- and gender-associated lncRNA- correlating genes. Endocytosis and Prostate_cancer KEGG pathways were significantly correlated with AD-associated age- and gender-lncRNAs (Figures 6A and 6B). Notably, lysosome and TNF_signaling pathways were in the top 30 enriched pathways among age- and AD-associated lncRNA-correlating genes (Figure 6A), while Glioma and FoxO_signaling pathways were in the top 30 significantly over-represented pathways of gender- and AD-associated lncRNA-correlating genes(Figure 6B). Furthermore, Proteasome and Long_term_potentiation KEGG pathways were commonly correlated with both age- and gender-lncRNAs in normal brains (Figures 6C and 6D).
Figure 6. Top 30 KEGG pathways significantly correlated with the expression of lncRNAs associated with age (A) and gender (B) in human AD brains and (C) and gender (D) in normal brains.
Each column corresponds to a single lncRNA, and each row corresponds to a KEGG pathway with an overrepresentation of genes correlating with lncRNA. The pathways were ranked by the sum of the negative log10 p-value of each lncRNA for each pathway. Top 30 pathways are shown.
Discussion
In the current study, we investigated the expression patterns of lncRNAs in the human brain during aging and between genders as well as their differential alterations in human AD brains. We found that the expressions of 18 age-associated and 13 gender-associated lncRNAs were dysregulated in AD brains compared with normal brains. Moreover, the expressions of age-associated lncRNAs - SNHG19 and LINC00672 - were positively and negatively correlated with Braak stage of AD, respectively; while the expressions of gender-associated lncRNAs - RNF144A-AS1, LY86-AS1 and LINC00639 were negatively correlated with Braak stage of AD. Further functional analysis suggests that these lncRNAs could function as key players in a broad array of essential signaling pathways, including ribosome, endocytosis, synaptic vesicle cycle, and axon guidance, that are critical for AD pathogenesis.
Independent lines of evidence suggest that lncRNAs are a new class of players involved in the development and progression of brain aging, cognitive decline, and AD. For example, studies by Faghihi et al. (Faghihi et al., 2008) have demonstrated that the BACE1-antisense transcript (BACE1-AS) is concordantly expressed with BACE1 (β-site amyloid precursor protein cleaving enzyme 1, also known as β-secretase - an enzyme central to the pathology of AD (Querfurth and LaFerla, 2010)) and acts as a feed-forward positive regulator of BACE1. Moreover, Mus et al. (Mus et al., 2007) have demonstrated a steady decrease of a brain-specific lncRNA -BC200 with aging. However, the level of BC200 is significantly upregulated in AD brains compared with age-matched normal brains (Mus et al., 2007). Indeed, studies have shown that BC200 is involved in regulating neuronal protein translation that subsequently could contribute to amyloid plaque formation and AD pathogenesis (Muddashetty et al., 2002; Roberts et al., 2014; Tiedge et al., 1993). Additionally, comprehensive studies have identified a group of lncRNAs with aberrant expression in human AD brains (Magistri et al., 2015; Zhou and Xu, 2015) and aged brains (Kim et al., 2016; Kour and Rath, 2016). By analyzing two microarray datasets, we here identified a total of 23 lncRNAs that were negatively correlated with age and 29 lncRNAs that were positively correlated with age (∣r∣ > 0.5; p < 0.05). Furthermore, the expressions of 18 age-correlated lncRNAs, including LINC00844, LINC01094, LINC00507, and OIP5-AS1, were abnormally alternated in AD brains compared with age-matched normal brains. The functioning of the age-associated lncRNAs, however, remains largely unknown. Our functional analysis suggests that the pathways involved in protein synthesis and neuronal functioning such as ribosome, synaptic vesicle cycle, and lysosome were over-represented within the age-associated lncRNA-correlating genes. It is well documented that lysosomal dysfunction in the brain results in a failure to clear accumulated protein aggregates which in turn contributes to the process of aging and pathogenesis of various neurodegenerative diseases (Fraldi et al., 2016; Kaur et al., 2017; Leeman et al., 2018). In the current study, we found the alterations of 3 age-associated lncRNAs (LINC00844, LINC00643, and OIP5-AS1) were abnormally alternated in AD brains, and lysosome was one of the significantly over-represented pathways of the lncRNA-correlating genes, indicating these lncRNAs could serve as master regulators. The underlying mechanisms(s) by which these lncRNAs regulate lysosomal function in aging and AD, however, warrant further investigation.
Females are at higher risk of developing AD than males. Studies have demonstrated that gender differences in genetic risk factors, including Apolipoprotein E, known as APOE, is associated with an increased risk for AD. Females with APOE4 are more at risk to develop AD than males with APOE4 (Altmann et al., 2014; Farrer et al., 1997). Recent studies have also demonstrated that single nucleotide polymorphisms (SNPs), including sex-specific SNPs, are associated with AD pathology (Gusareva et al., 2018; Wachinger et al., 2018). Whether gender-associated lncRNAs are differentially expressed in AD brains as well as their functions in AD pathogenesis however are uncertain. Herein, we identified 13 gender- and AD-associated lncRNAs, including SLC25A25-AS1, PWRN1, and LY86-AS1 that were significantly dysregulated in AD brains compared with age-matched normal brains in either females or males. Functional analysis suggests that FoxO_signaling and Glioma pathways were over-represented within the gender-associated lncRNA-correlating genes. Further investigations, however, are needed to better understand the role of these gender-associated lncRNAs in AD pathology.
The frontal cortex, located at the very front of the brain, controls cognitive functions in humans (Fuster, 2002). The superior frontal gyrus is the upper part of the frontal cortex and primarily involved in higher cognitive functions and working memory (du Boisgueheneuc et al., 2006). Structural and gene expression changes in the superior frontal gyrus have been demonstrated in both normal aging and AD (Bakkour et al., 2013). In the current study, we first identified age-associated lncRNAs, such as LINC00844, in the frontal cortex during normal aging. We then confirmed that the expressions of some of the age-associated lncRNAs were also age-correlated in the superior frontal gyrus during normal aging but is abnormally alternated in the superior frontal gyrus of AD patients, such as LINC00507.
Interestingly, a recent study has demonstrated that NEAT1 is significantly upregulated in the temporal cortex and hippocampus of AD patients (Spreafico et al., 2018). The authors have further demonstrated that NEAT1 plays a neuroprotective role through regulating cyclin-dependent kinase 5 regulatory subunit 1 (CDK5R1), an AD-associated gene. In addition, LINC00507 has been recently implicated as an age-dependent lncRNA that is expressed in a cortex-specific manner in both non-human primates and humans (Mills et al., 2016). In another study, Kang et al. have demonstrated an increase in BACE1AS levels in the superior temporal gyrus of AD patients (Kang et al., 2014). Increased BACE1AS can stabilize BACE1 mRNA and promote BACE1 expression (Faghihi et al., 2008) which, in turn, promote the cleavage of APP the accumulation of toxic aggregates of Aβ peptide and ultimately contribute to AD pathogenesis (Kang et al., 2014). Strikingly, Zhou et al. (Zhou et al., 2018) have recently performed a comparative study on the expression pattern of lncRNAs in four brain regions of aged or AD patients and demonstrated that the alteration of age- and AD-associated lncRNAs in the brain is region-dependent. The function of these age- AD- and brain-region associate lncRNA, however, warrants further, in-depth, investigations.
Although the major findings of this study were reproducible in two independent datasets, there are several limitations. First, for the analysis of the correlation between the expression of lncRNAs and braak stage of AD, there were only four and five samples for stage V and VI, respectively. Larger sample size will lead to more reliable results. To this end, datasets generated from different platforms and/or studies may be integrated which will allow us to perform a large-scale meta-analysis. Second, experimental validation will provide more insight into the differential alterations of the age- and gender-associated lncRNAs in AD brains as well as their actual functions. Third, other clinical factors, such as treatments, body mass index (BMI), smoking status, and sample heterogeneity which could dysregulate the expression of lncRNAs, were not considered in this study.
Taken together, the identification of age- and gender-associated lncRNAs and their differential alterations in human AD brain provide potential targets for further mechanistic investigation which could, in turn, pave the way for developing age- and gender-specific diagnosis, prevention and individualized treatment options for AD patients.
Supplementary Material
Highlights.
The expressions of 16 age-associated lncRNAs are dysregulated in AD brains.
The expressions of 13 gender-associated lncRNAs are alternated in AD brains.
The expressions of age- and gender-associated lncRNAs are correlated with Braak stage of AD and neuritic plaque density.
Acknowledgments
This work was supported by grants DA042704, DA046831, MH112848, DA043138 (to G.H.) and 1P20GM104320 and 1R01DK107264 (to J.C.) from the National Institutes of Health (NIH) and grant 31270175 (to J.Z.) from the National Natural Science Foundation of China. The support of the Nebraska Center for Substance Abuse Research is acknowledged. The project described was also supported by the NIH, National Institute of Mental Health, grant 2P30MH062261. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosure
Dr. Hu was supported by NIH grants (DA042704 and DA046831). Dr. Zhao was supported by the National Natural Science Foundation of China (31270175); Dr. Cui is funded by NIH-funded COBRE [1P20GM104320], NIH/NIFA [1R01DK107264/2016-67001-06314], and UNL Food for Health seed grant. Drs. Cao and Li report no disclosures.
Study funding: Supported by the NIH and the National Natural Science Foundation of China.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All the authors concur with the submission, and these findings have neither been reported before nor have been submitted elsewhere for publication, in whole or in part, and there is no conflicting financial interest.
References
- Altmann A, Tian L, Henderson VW, Greicius MD, Alzheimer's Disease Neuroimaging Initiative, I., 2014. Sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol 75(4), 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarita G, Jung KM, Berchtold NC, Nguyen VQ, Gillen DL, Head E, Cotman CW, Piomelli D, 2010. Deficient liver biosynthesis of docosahexaenoic acid correlates with cognitive impairment in Alzheimer's disease. PLoS One 5(9), e12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkour A, Morris JC, Wolk DA, Dickerson BC, 2013. The effects of aging and Alzheimer's disease on cerebral cortical anatomy: specificity and differential relationships with cognition. Neuroimage 76, 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangasser DA, Dong H, Carroll J, Plona Z, Ding H, Rodriguez L, McKennan C, Csernansky JG, Seeholzer SH, Valentino RJ, 2017. Corticotropin-releasing factor overexpression gives rise to sex differences in Alzheimer's disease-related signaling. Mol Psychiatry 22(8), 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes LL, Wilson RS, Bienias JL, Schneider JA, Evans DA, Bennett DA, 2005. Sex differences in the clinical manifestations of Alzheimer disease pathology. Arch Gen Psychiatry 62(6), 685–691. [DOI] [PubMed] [Google Scholar]
- Barrett T, Edgar R, 2006. Mining microarray data at NCBI's Gene Expression Omnibus (GEO)*. Methods Mol Biol 338, 175–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A, 2013. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41(Database issue), D991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Coleman PD, Cribbs DH, Rogers J, Gillen DL, Cotman CW, 2013. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer's disease. Neurobiol Aging 34(6), 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW, 2008. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A 105(40), 15605–15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LJ, Nordhues BA, Hill SE, Scaglione KM, O'Leary JC 3rd, Fontaine SN, Breydo L, Zhang B, Li P, Wang L, Cotman C, Paulson HL, Muschol M, Uversky VN, Klengel T, Binder EB, Kayed R, Golde TE, Berchtold N, Dickey CA, 2013. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J Clin Invest 123(10), 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chene G, Beiser A, Au R, Preis SR, Wolf PA, Dufouil C, Seshadri S, 2015. Gender and incidence of dementia in the Framingham Heart Study from mid-adult life. Alzheimers Dement 11(3), 310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J, van Amelsvoort T, Murphy D, 2001. HRT and its effect on normal ageing of the brain and dementia. Br J Clin Pharmacol 52(6), 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs DH, Berchtold NC, Perreau V, Coleman PD, Rogers J, Tenner AJ, Cotman CW, 2012. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J Neuroinflammation 9, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew L, 2018. An age-old story of dementia. Nature 559(7715), S2–S3. [DOI] [PubMed] [Google Scholar]
- du Boisgueheneuc F, Levy R, Volle E, Seassau M, Duffau H, Kinkingnehun S, Samson Y, Zhang S, Dubois B, 2006. Functions of the left superior frontal gyrus in humans: a lesion study. Brain : a journal of neurology 129(Pt 12), 3315–3328. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C, 2008. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14(7), 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM, 1997. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278(16), 1349–1356. [PubMed] [Google Scholar]
- Fitch RH, Denenberg VH, 1998. A role for ovarian hormones in sexual differentiation of the brain. Behav Brain Sci 21(3), 311–327; discussion 327-352. [DOI] [PubMed] [Google Scholar]
- Fraldi A, Klein AD, Medina DL, Settembre C, 2016. Brain Disorders Due to Lysosomal Dysfunction. Annu Rev Neurosci 39, 277–295. [DOI] [PubMed] [Google Scholar]
- Fuster JM, 2002. Frontal lobe and cognitive development. J Neurocytol 31(3-5), 373–385. [DOI] [PubMed] [Google Scholar]
- Grimm A, Eckert A, 2017. Brain aging and neurodegeneration: from a mitochondrial point of view. J Neurochem 143(4), 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusareva ES, Twizere JC, Sleegers K, Dourlen P, Abisambra JF, Meier S, Cloyd R, Weiss B, Dermaut B, Bessonov K, van der Lee SJ, Carrasquillo MM, Katsumata Y, Cherkaoui M, Asselbergh B, Ikram MA, Mayeux R, Farrer LA, Haines JL, Pericak-Vance MA, Schellenberg GD, Genetic, Environmental Risk in Alzheimer's Disease, c., Alzheimer's Disease Genetics, C., European Alzheimer Disease Initiative, I., Sims R, Williams J, Amouyel P, van Duijn CM, Ertekin-Taner N, Van Broeckhoven C, Dequiedt F, Fardo DW, Lambert JC, Van Steen K, 2018. Male-specific epistasis between WWC1 and TLN2 genes is associated with Alzheimer's disease. Neurobiol Aging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Abdelmohsen K, Hutchison ER, Mitchell SJ, Grammatikakis I, Guo R, Noh JH, Martindale JL, Yang X, Lee EK, Faghihi MA, Wahlestedt C, Troncoso JC, Pletnikova O, Perrone-Bizzozero N, Resnick SM, de Cabo R, Mattson MP, Gorospe M, 2014. HuD regulates coding and noncoding RNA to induce APP-->Abeta processing. Cell Rep 7(5), 1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Pawlik M, Gandy SE, Ehrlich ME, Smiley JF, Levy E, 2017. Lysosomal dysfunction in the brain of a mouse model with intraneuronal accumulation of carboxyl terminal fragments of the amyloid precursor protein. Mol Psychiatry 22(7), 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim KM, Noh JH, Yoon JH, Abdelmohsen K, Gorospe M, 2016. Long noncoding RNAs in diseases of aging. Biochim Biophys Acta 1859(1), 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koglsberger S, Cordero-Maldonado ML, Antony P, Forster JI, Garcia P, Buttini M, Crawford A, Glaab E, 2017. Gender-Specific Expression of Ubiquitin-Specific Peptidase 9 Modulates Tau Expression and Phosphorylation: Possible Implications for Tauopathies. Mol Neurobiol 54(10), 7979–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G, 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35(Web Server issue), W345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kour S, Rath PC, 2016. Long noncoding RNAs in aging and age-related diseases. Ageing Res Rev 26, 1–21. [DOI] [PubMed] [Google Scholar]
- Leeman DS, Hebestreit K, Ruetz T, Webb AE, McKay A, Pollina EA, Dulken BW, Zhao X, Yeo RW, Ho TT, Mahmoudi S, Devarajan K, Passegue E, Rando TA, Frydman J, Brunet A, 2018. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359(6381), 1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette C, Schmechel D, Alexander GE, Reiman EM, Rogers J, Stephan DA, 2007. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics 28(3), 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA, 2008. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A 105(11), 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, Bennett DA, Colaiacovo MP, Yankner BA, 2014. REST and stress resistance in ageing and Alzheimer's disease. Nature 507(7493), 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M, Velmeshev D, Makhmutova M, Faghihi MA, 2015. Transcriptomics Profiling of Alzheimer's Disease Reveal Neurovascular Defects, Altered Amyloid-beta Homeostasis, and Deregulated Expression of Long Noncoding RNAs. J Alzheimers Dis 48(3), 647–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL, 2015. Alzheimer's disease. Nat Rev Dis Primers 1, 15056. [DOI] [PubMed] [Google Scholar]
- Mazure CM, Swendsen J, 2016. Sex differences in Alzheimer's disease and other dementias. Lancet Neurol 15(5), 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JD, Ward M, Chen BJ, Iyer AM, Aronica E, Janitz M, 2016. LINC00507 Is Specifically Expressed in the Primate Cortex and Has Age-Dependent Expression Patterns. J Mol Neurosci 59(4), 431–439. [DOI] [PubMed] [Google Scholar]
- Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Huttenhofer A, Tiedge H, Brosius J, 2002. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol 321(3), 433–445. [DOI] [PubMed] [Google Scholar]
- Murphy DG, DeCarli C, McIntosh AR, Daly E, Mentis MJ, Pietrini P, Szczepanik J, Schapiro MB, Grady CL, Horwitz B, Rapoport SI, 1996. Sex differences in human brain morphometry and metabolism: an in vivo quantitative magnetic resonance imaging and positron emission tomography study on the effect of aging. Arch Gen Psychiatry 53(7), 585–594. [DOI] [PubMed] [Google Scholar]
- Mus E, Hof PR, Tiedge H, 2007. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci U S A 104(25), 10679–10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM, 2010. Alzheimer's disease. N Engl J Med 362(4), 329–344. [DOI] [PubMed] [Google Scholar]
- Readhead B, Haure-Mirande JV, Funk CC, Richards MA, Shannon P, Haroutunian V, Sano M, Liang WS, Beckmann ND, Price ND, Reiman EM, Schadt EE, Ehrlich ME, Gandy S, Dudley JT, 2018. Multiscale Analysis of Independent Alzheimer's Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus. Neuron 99(1), 64–82 e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC, Morris KV, Wood MJ, 2014. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci 369(1652). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetre P, Jazin E, Emilsson L, 2011. Age-related changes in gene expression are accelerated in Alzheimer's disease. Synapse 65(9), 971–974. [DOI] [PubMed] [Google Scholar]
- Sarvari M, Hrabovszky E, Kallo I, Solymosi N, Liko I, Berchtold N, Cotman C, Liposits Z, 2012. Menopause leads to elevated expression of macrophage-associated genes in the aging frontal cortex: rat and human studies identify strikingly similar changes. J Neuroinflammation 9, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Parra-Damas A, Enriquez-Barreto L, 2015. Gene expression parallels synaptic excitability and plasticity changes in Alzheimer's disease. Front Cell Neurosci 9, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz SE, Shaywitz BA, Pugh KR, Fulbright RK, Skudlarski P, Mencl WE, Constable RT, Naftolin F, Palter SF, Marchione KE, Katz L, Shankweiler DP, Fletcher JM, Lacadie C, Keltz M, Gore JC, 1999. Effect of estrogen on brain activation patterns in postmenopausal women during working memory tasks. JAMA 281(13), 1197–1202. [DOI] [PubMed] [Google Scholar]
- Sherwin BB, 1988. Estrogen and/or androgen replacement therapy and cognitive functioning in surgically menopausal women. Psychoneuroendocrinology 13(4), 345–357. [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA, 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res 18(1), 134–147. [DOI] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Haynes LJ, Theaker R, Gelsthorpe C, Baxter L, Forster G, Lace GL, Shaw PJ, Matthews FE, Savva GM, Brayne C, Wharton SB, Function M.R.C.C., Ageing Neuropathology Study, G., 2010. Population variation in oxidative stress and astrocyte DNA damage in relation to Alzheimer-type pathology in the ageing brain. Neuropathol Appl Neurobiol 36(1), 25–40. [DOI] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Matthews FE, Shaw PJ, Heath PR, Brayne C, Garwood C, Higginbottom A, Wharton SB, Function M.R.C.C., Ageing Neuropathology Study, G., 2015. A neuronal DNA damage response is detected at the earliest stages of Alzheimer's neuropathology and correlates with cognitive impairment in the Medical Research Council's Cognitive Function and Ageing Study ageing brain cohort. Neuropathol Appl Neurobiol 41(4), 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Minett T, Matthews FE, Heath PR, Shaw PJ, Goodall E, Garwood CJ, Ratcliffe LE, Brayne C, Rattray M, Wharton SB, Function M.R.C.C., Ageing Neuropathology Study, G., 2016. Neuronal DNA damage response-associated dysregulation of signalling pathways and cholesterol metabolism at the earliest stages of Alzheimer-type pathology. Neuropathol Appl Neurobiol 42(2), 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreafico M, Grillo B, Rusconi F, Battaglioli E, Venturin M, 2018. Multiple Layers of CDK5R1 Regulation in Alzheimer's Disease Implicate Long Non-Coding RNAs. Int J Mol Sci 19(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Chen W, Brosius J, 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci 13(6), 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Grembergen O, Bizet M, de Bony EJ, Calonne E, Putmans P, Brohee S, Olsen C, Guo M, Bontempi G, Sotiriou C, Defrance M, Fuks F, 2016. Portraying breast cancers with long noncoding RNAs. Sci Adv 2(9), e1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachinger C, Nho K, Saykin AJ, Reuter M, Rieckmann A, Alzheimer's Disease Neuroimaging, I., 2018. A Longitudinal Imaging Genetics Study of Neuroanatomical Asymmetry in Alzheimer's Disease. Biol Psychiatry 84(7), 522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W, 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41(6), e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Roussos P, McKenzie A, Zhou X, Kajiwara Y, Brennand KJ, De Luca GC, Crary JF, Casaccia P, Buxbaum JD, Ehrlich M, Gandy S, Goate A, Katsel P, Schadt E, Haroutunian V, Zhang B, 2016. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer's disease. Genome Med 8(1), 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Zhao H, Wang X, Sun J, Su J, 2018. Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer's disease. Brief Bioinform. [DOI] [PubMed] [Google Scholar]
- Zhou X, Xu J, 2015. Identification of Alzheimer's disease-associated long noncoding RNAs. Neurobiol Aging 36(11), 2925–2931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data (GSE53890, GSE48350, GSE5281, GSE84422, and GSE66333) used for this study are available at the Gene Expression Omnibus (GEO) database of NCBI (http://www.ncbi.nlm.nih.gov/geo/). Anonymized data will be shared upon request from qualified investigators.