Abstract
Background
It has been proposed that lncRNAs, widely transcribed from genomes, play pivotal regulatory roles in a variety of biological processes, but their function in regulating spermatogenesis in human males is rarely reported.
Methods
QRT‐PCR was adopted to detect HOTTIP expression level in testicular tissues from hypospermatogenesis (Hypo) patients or controls. The proliferation levels of NT2 and 293T were measured via CCK‐8 and EdU detection. Meanwhile, luciferase reporter gene assay and bioinformatics analysis were carried out to identify a target of HOTTIP. Additionally, the underlying mechanism of HOTTIP’s function was investigated using western blotting and RIP analysis.
Results
The research results manifested that the expression of HOTTIP in testicular tissues from Hypo patients was prominently reduced in comparison with that in control testicular tissues. Interestingly, it was noted that HOTTIP exhibited a high expression in testicular embryonal carcinoma cell line NT2 compared with that in normal control cell line 293T. It was denoted in cell function evaluation that cell proliferation was impeded by downregulated HOTTIP but evidently stimulated by overexpressed HOTTIP. Moreover, HOTTIP was capable of positively modulating HOXA13 expression via the competitive binding to miR‐128‐3p.
Conclusion
Therefore, HOTTIP acting as ceRNAs to promote testicular embryonal carcinoma cell proliferation.
Keywords: cell proliferation, HOTTIP, hypospermatogenesis, NT2, spermatogenesis
1. INTRODUCTION
As a global healthy issue in the reproductive system, infertility occurs in about one‐fifth couples planning for pregnancy (Szkodziak et al., 2016). Half infertility cases ascribe to male factors, 75% of which resulted from unknown factors due to the unclear molecular mechanisms of the defects (Chalyi, Akhvlediani, & Kharchilava, 2016). Male infertility is often clinically characterized by nonobstructive azoospermia (NOA) or severe oligozoospermia and uniform testicular maturation arrest (MA) (Esteves, 2015; Ferras et al., 2004; Oud et al., 2017), so unfolding the potential pathogenesis of hypospermatogenesis (Hypo) may be conducive to the therapeutic effects of these patients.
As a complicated development process, spermatogenesis sustains the production of spermatozoa and fertility during the whole life of an adult male. A complex transcriptional network strictly modulates the three main phases of spermatogenesis, namely mitotic proliferation of spermatogonia, haploid differentiation of spermatids, and meiosis of spermatocytes. During spermatogenesis, various ncRNAs, including microRNAs (miRNAs) (Kotaja, 2014), endogenous small interfering RNAs (siRNAs) (Neto, Bach, Najari, Li, & Goldstein, 2016), and long noncoding RNAs (lncRNAs) (Dianatpour & Ghafouri‐Fard, 2017) apart from protein‐coding messenger RNAs, also exert crucial regulatory effects on genes. However, how lncRNAs regulate spermatogenesis still remains elusive.
LncRNAs are a kind of RNAs with length exceeding 200 nucleotides, whose capacity to encode proteins is limited (Forrest & Khalil, 2017). Reports in recent years have pointed out that lncRNAs function in the pathogenesis of different diseases, such as reproductive system diseases, nervous system diseases, and different tumors (Kondo, Shinjo, & Katsushima, 2017; Quan, Zheng, & Qing, 2017; Shen & Zhong, 2015). According to studies, lncRNAs are capable of adjusting gene expression at transcriptional and posttranscriptional levels (Weidle, Birzele, Kollmorgen, & Ruger, 2017). HOTTIP (Gene ID: 100,316,868), an lncRNA on chromosome 7p15.2, can promote the proliferation ability of A, B, and C cancer cells (Deng, Zhao, Wu, & Song, 2017; Li et al., 2016; Yu, Nangia‐Makker, Farhana, & Majumdar, 2017). Nevertheless, the specific mechanism by which HOTTIP plays a role during spermatogenesis is not clear.
Through investigation, this research showed that HOTTIP was evidently decreased in testicular tissues of NOA patients with Hypo. Subsequently, a train of assays in vitro were performed to explore the underlying role of HOTTIP in the process of spermatogenesis. In general, the research results indicated that HOTTIP may participate in the mechanism of Hypo by influencing HOXA13 expression, which gives a new insight into studying Hypo from the perspective of lncRNA.
2. MATERIALS AND METHODS
2.1. Ethical compliance
The research was approved by the Ethics Committee of Jiangsu Provincial Hospital of Traditional Chinese Medicine.
2.2. Human testicular samples
In lncRNA microarray, testicular samples were obtained from Jiangsu Provincial Hospital of Traditional Chinese Medicine. In this study, 16 patients with Hypo and 16 control subjects receiving orchiectomy for prostate carcinoma were enrolled. The control samples were harvested from patients with normal spermatogenesis confirmed by histological examination and no meiotic defect history or infertility in the urology department. Informed consent was gained from all patients, and the Ethics Committee in Jiangsu Provincial Hospital of Traditional Chinese Medicine approved this study.
2.3. Cell culture
Tumor cell lines (NT2, NCCIT, TE1, A549, and SY5Y) and a human normal cell line (293T) were bought from ATCC (Manassas, VA, USA) and cultured with RPMI1640 or DMEM containing 10% FBS (Hyclone, UT, USA), 100 IU/ml penicillin and 100 μg/ml streptomycin (Invitrogen, USA) in the environment with humidity and 5% carbon dioxide at 37°C. According to the manufacturer’s protocol, all the cells were transfected with HOTTIP siRNAs, HOTTIP overexpression (HOTTIP OE) plasmids, miR‐128‐3p inhibitors, and miR‐128‐3p mimics synthesized by GenePharma (Shanghai, China) using Lipofectamine 3,000 (Invitrogen, CA, USA).
2.4. RNA separation and quantitative real‐time PCR (qRT‐PCR)
TRIzol reagent (Life Technologies, CA, USA) was applied to extract total RNAs, whose purity was determined after the measurement of A260 and A280 using a UV spectrophotometer. Then, the extracted total RNAs were stored in a −80‐C refrigerator for standby application. With reference to the instructions of the Reverse Transcription Kit (Takara, Tokyo, Japan), RNAs were subjected to reverse transcription into cDNAs and stored at −20°C for standby application. The primers used were diluted as required, and qRT‐PCR was performed to examine the expressions of lncRNAs, mRNAs and miRNAs on the ABI 7900HT (Applied Biosystems, CA, USA). HOXA13 primer sequences: F: 5'‐CTGCCCTATGGCTACTTCGG‐3', R: 5'‐CCGGCGGTATCCATGTACT‐3'. 2−△△Ct method was adopted for measuring the relative concentration of the samples to be tested. Each assay was repeated 3 times, followed by averaging.
2.5. Cell proliferation assessment
Following inoculation on 96‐well plates with DMEM free of serum, cells were incubated for required time, and each well was added with 10‐μl Cell Counting Kit‐8 (CCK‐8) reagents (Beyotime, Nantong, China). At 1 hr after incubation, TECAN infinite M200 multimode microplate reader (Tecan, Mechelen, Belgium) was applied to evaluate A450, and 5‐Ethynyl‐2'‐deoxyuridine (EdU) assay was carried out for evaluation of cell proliferation capacity. The same experiment was repeated 3 times at least.
2.6. Location of subcellular fractionation
Based on the protocol, RNAs in the cytoplasm and nucleus were separated using the PARIS Kit (Life Technologies, USA). Total RNAs separated from each fraction were determined via qRT‐RCR, with GAPDH as a cytoplasmic marker and U6 as a nuclear control transcript.
2.7. Dual‐luciferase reporter assay
The following fragments were constructed after the binding site between HOXA13 and HOTTIP was inserted into the KpnI and SacI sites of pGL3 promoter vectors (Realgene, Nanjing, China): pGL3‐HOXA13‐WT, pGL3‐HOXA13‐MUT, pGL3‐HOTTIP‐WT, and pGL3‐HOTTIP‐MUT. Then the cells were cultured on 24‐well plates until the cell density reached 50%–60% and co‐transfected with 50 nmol/L miR‐128‐3p mimics, 5 ng pRL‐SV40, 80 ng plasmids, and NC via Lipofectamine 2000 (Invitrogen, Shanghai, China). In line with the manufacturer’s protocol, the luciferase activity change at 36 hr after incubation was assessed through the dual‐Glo luciferase assay system (Promega, Madison, WI, USA). Each experiment was separately repeated 3 times.
2.8. RNA immunoprecipitation (RIP)
With reference to the protocol of the manufacturer, RIP assay was carried out using anti‐AGO2 (Abcam, ab32381, Cambridge, MA, USA) and the Magna Nuclear RIP™ (Native) Nuclear RNA‐Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA).
2.9. Western blotting
RIPA was conducted to extract total proteins, and the appropriate concentration of SDS‐PAGE gel was chosen in light of the molecular weight of target proteins. The proteins were transferred to a PVDF membrane and stained on the basis of normal immune staining after SDS‐PAGE. Anti‐HOXA13 and anti‐GAPDH antibodies (Abcam, Cambridge, MA, USA) were diluted at 1:500 and added for incubation at 4°C overnight. Diluted at 1:1,000, secondary antibodies were utilized for 2 hr at 37°C, followed by enhanced chemiluminescence. Finally, the color was fixed and pictures were taken.
2.10. Statistical analysis
Data were analyzed by means of GraphPad Prism 6.0 (GraphPad Software Inc., CA, USA) and SPSS 20.0 software (SPSS, Chicago, IL, USA) and expressed as mean ± SD. Statistical differences between data sets in line with normal distribution were investigated using the Student’s t‐test, whereas the differences were analyzed using nonparametric tests in case the data were not in line with normal distribution. The difference was considered to be statistically significant when p < .05.
3. RESULTS
3.1. HOTTIP expression changes in Hypo patients
QRT‐PCR analysis confirmed HOTTIP was downregulated in Hypo patients (Figure 1a). HOTTIP expression level was measured in tumor cell lines (NT2, NCCIT, TE1, A549, and SY5Y) and a human normal cell line (293T) by qRT‐PCR. It could be obviously seen from Figure 1b that compared with 293T, the HOTTIP level in testicular embryonal carcinoma cell line NT2 was markedly increased.
Figure 1.
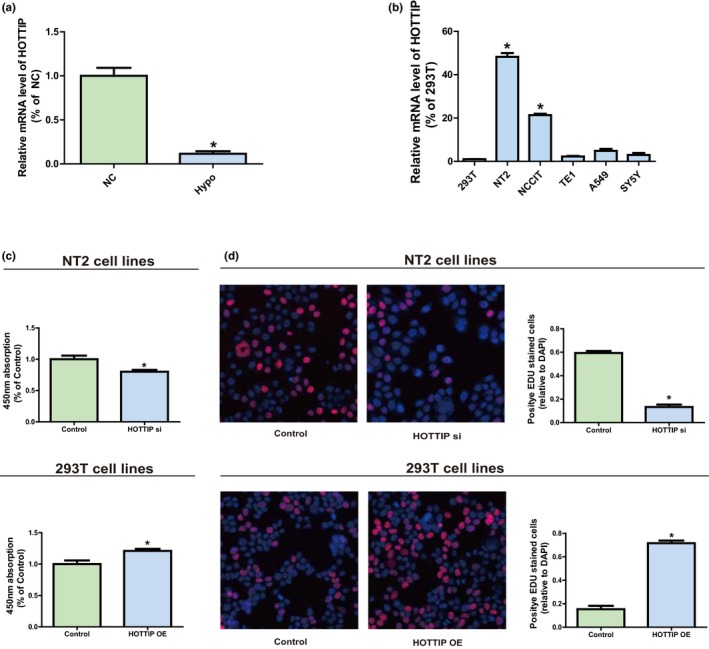
The role of HOTTIP in Hypo. (a) QRT‐PCR confirms HOTTIP is down‐regulated in Hypo patients. (b) Expression levels of HOTTIP in tumor cell lines (NT2, NCCIT, TE1, A549, and SY5Y) and a human normal cell line (293T). (c) Effect of transfection with HOTTIP siRNAs or OE plasmids on cell proliferation level of human cell lines NT2 and 293T observed via CCK‐8 assay. (d) Effect of transfection with HOTTIP siRNAs or OE plasmid on cell proliferation level of human cell lines NT2 and 293T observed via EdU assay. Data are presented as mean ± SD. *p ≤ .05, Student’s t‐test
3.2. The role of HOTTIP in NT2 and 293T
HOTTIP expression silencing was achieved through transfection of NT2 with HOTTIP siRNAs to figure out HOTTIP’s biological function in NT2 and 293T. Besides, 293T was transfected with HOTTIP OE plasmids so as to elevate HOTTIP expression. The results manifested that HOTTIP expression was substantially blocked after NT2 was transfected with HOTTIP siRNAs (Supplementary Figure S1a), and HOTTIP level was raised after 293T was transfected with HOTTIP OE plasmids (Supplementary Figure S1b). Subsequently, CCK‐8 assay and EdU assay showed that downregulation of HOTTIP could evidently reduce the proliferation ability of NT2. On the contrary, HOTTIP OE can significantly enhance the proliferation capacity of 293T (Figure 1c,d). In brief, it can be seen that HOTTIP may play a regulatory role in the proliferation of NT2 and 293T to certain extent.
3.3. Subcellular localization of HOTTIP
Subcellular localization of lncRNAs determines their biological functions. As such, cellular localization of HOTTIP was verified through isolation of NT2 and 293T into the nucleus and the cytoplasm, with U6 and GAPDH as controls. U6 is mainly found in the nucleus, while GAPDH is primarily distributed in the cytoplasm. As shown in qRT‐PCR results, HOTTIP was monitored in the cytoplasm of NT2 (45.4%) and 293T (49.1%) (Figure 2a). These findings imply that HOTTIP may be involved in the pathogenesis of Hypo through both transcriptional‐ and posttranscriptional‐level regulations.
Figure 2.
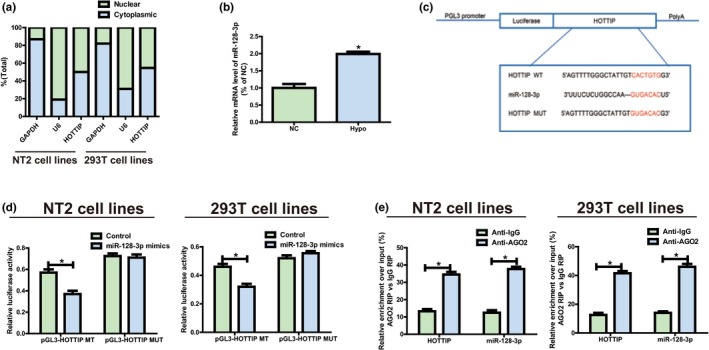
HOTTIP directly interacts with miR‐128‐3p. (a) Distribution characteristics of HOTTIP in NT2 and 293T detected via qRT‐PCR. (b) MiR‐128‐3p expression level is decreased in Hypo patients compared to the normal controls. (c) Bioinformatics evidence that miR‐128‐3p binds to HOTTIP’s 3’‐UTR. The speculated miRNAs recognition sites that are cloned into the luciferase gene downstream are named as pGL3‐HOTTIP‐WT. Bottom: Mutant HOTTIP sequences that produce the mutant luciferase reporter constructs gain the name of pGL3‐HOTTIP‐MUT. (d) Detection of luciferase activity in NT2 and 293T following co‐transfection with plasmids (pGL3‐HOTTIP‐WT or pGL3‐HOTTIP‐MUT) and miRNA mimics via dual‐luciferase reporter assay. (e) RIP assays are performed in NT2 and 293T to figure out whether HOTTIP is present in ribonucleoprotein complexes with miRNAs. QRT‐PCR is adopted to measure miR‐128‐3p and HOTTIP expression levels. Data are presented as mean ± SD. *p ≤ .05, Student’s t‐test
3.4. MiR‐128‐3p targets HOTTIP
The results showed that HOTTIP exhibited an obvious increase in NT2, and it was able to boost proliferation, but the specific mechanism remains not clear. It was hypothesized that in biological processes, HOTTIP might be considered as a ceRNA since HOTTIP is partially located in the cytoplasm. RegRNA 2.0, Starbase was applied for bioinformatics predictions, which unfolded that miR‐128‐3p was closely matched with the sequence in HOTTIP’s 3’‐UTR. MiR‐128‐3p expression level was lowered in normal controls compared to the Hypo patients (Figure 2b), which is opposite to the features of HOTTIP expression. HOTTIP fragments (pGL3‐HOTTIP‐WT and pGL3‐HOTTIP‐MUT) with predicted or mutated target sites were established into the firefly luciferase gene downstream to figure out the interaction between HOTTIP and miR‐128‐3p (Figure 2c). Treatment of NT2 and 293T with HOTTIP WT and miR‐128‐3p mimics remarkably weakened the intensity of luciferase compared to the control group, but the intensity did not change following co‐transfection with miR‐128‐3p mimics and HOTTIP MUT (Figure 2d). Moreover, to clarify whether HOTTIP was present in ribonucleoprotein complexes containing mRNAs, RIP assays were carried out in NT2 and 293T. Subsequently, compared with that in immunoglobulin G (IgG) controls, the relative RNA expression in immunoprecipitates was detected via qRT‐PCR, and it was discovered that anti‐AGO2 antibodies could enrich HOTTIP in NT2 and 293T, which was similar to the results of miR‐128‐3p (Figure 2e). The above findings suggest that miR‐128‐3p can bind to HOTTIP in vitro.
3.5. HOTTIP regulates miR‐128‐3p target gene, HOXA13
MiR‐128‐3p target genes were screened and subjected to intersection through bioinformatics prediction (TargetScan, Starbase, RegRNA) so as to study miR‐128‐3p molecular mechanism in its biological function. HOXA13 was ultimately selected for further research. To further prove the binding between miR‐128‐3p and HOXA13, dual‐luciferase reporter assay was conducted. pGL3‐HOXA13‐WT and pGL3‐HOXA13‐MUT with WT‐ or MUT‐binding sites were established and subjected to independent co‐transfection with NC or miR‐128‐3p mimics in NT2 and 293T (Figure 3a). It was confirmed that the luciferase activity of the groups co‐transfected with miR‐128‐3p WT plasmids and mimics was inhibited, while that in the groups co‐transfected miR‐128‐3p MUT plasmids and mimics was not blocked (Figure 3b), indicating that HOXA13 is an underlying miR‐128‐3p target gene. It was found in Figure 3c that the mRNA level of HOXA13 in Hypo patients was reduced remarkably. Additionally, western blotting was carried out to measure the protein level of HOXA13, and the results identical to those of qRT‐PCR were obtained (Figure 3d).
Figure 3.
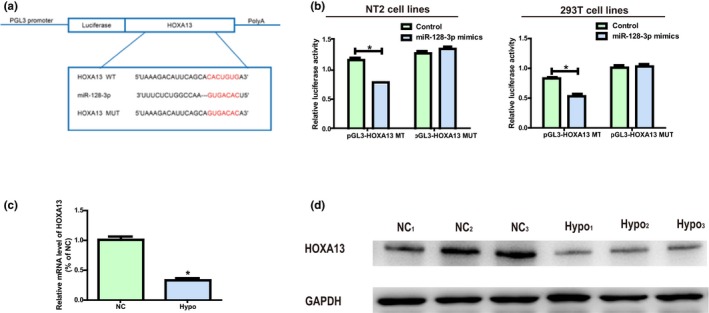
HOXA13 is a miR‐128‐3p direct target. (a) Prediction of binding site between HOXA13 and miR‐128‐3p and construction of HOXA13‐MUT plasmids. (b) Detection of the luciferase activity in NT2 and 293T after co‐transfection plasmid (pGL3‐HOXA13‐WT or pGL3‐HOXA13‐MUT) and miRNA mimics examined via dual‐luciferase reporter gene experiments. (c) The mRNA level of HOXA13 in Hypo patients is remarkably decreased. (d) The protein expression level of HOXA13 in Hypo patients is obviously decreased. Data are presented as mean ± SD. *p ≤ .05, Student’s t‐test
HOXA13 expression after transfection with HOTTIP and miR‐128‐3p was monitored via western blotting and qRT‐PCR to elucidate whether HOTTIP modulated HOXA13 expression level by binding to miR‐128‐3p. According to the results, miR‐128‐3p inhibitors markedly increased HOXA13 expression in NT2, while co‐transfection with HOTTIP siRNA could reverse this change (Figure 4a,b). Furthermore, miR‐128‐3p mimics evidently inhibited HOXA13 expression in 293T, whereas co‐transfection of HOTTIP OE plasmids could reverse this change (Figure 4c,d). The above findings generally confirm that HOTTIP elevates HOXA13 expression by directly binding to miR‐128‐3p.
Figure 4.
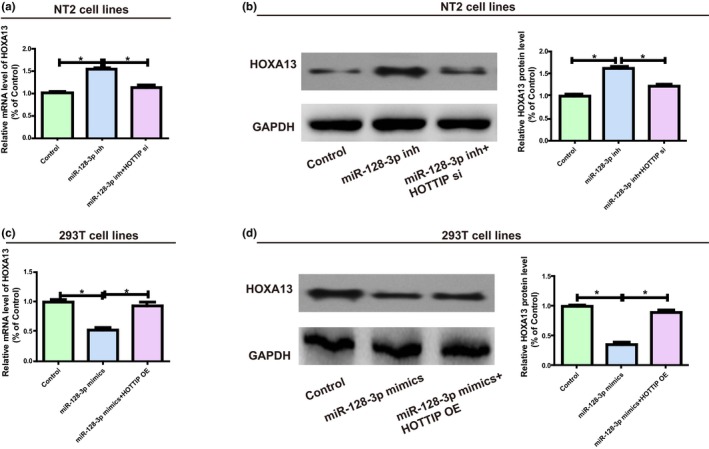
HOTTIP/miR‐128‐3p regulatory loop is pivotal for HOXA13 expression. (a) NT2 is transfected with miR‐128‐3p inhibitors (with or without HOTTIP siRNAs) and the mRNA level of HOXA13 is assessed via qRT‐PCR. (b) Following NT2 is treated with miR‐128‐3p inhibitors (with or without HOTTIP siRNAs), western blotting is carried out to detect the HOXA13 protein level, with GAPDH as a control. (c) 293T is transfected with miR‐128‐3p mimics (with or without HOTTIP OE plasmids), and the relative mRNA level of HOXA13 is detected using qRT‐PCR. (d) 293T is transfected with miR‐128‐3p mimics (with or without HOTTIP OE plasmids), and the relative protein level of HOXA13 is measured through qRT‐PCR. Data are presented as mean ± SD. *p ≤ .05, Student’s t‐test
3.6. HOTTIP/miR‐128‐3p regulatory loop is crucial for cell function
Then, whether miR‐128‐3p influenced proliferation of NT2 and 293T was determined. Determination of transfection efficiency proved that miR‐128‐3p inhibitors could impede the expression of miR‐128‐3p, while miR‐128‐3p mimics can improve the expression level of miRNA (Supplementary Figure S1c,d). Compared with NC, decreased miR‐128‐3p in NT2 obviously stimulated cell proliferation ability, and reduced HOTTIP could partially reverse the effect of miR‐128‐3p (Figure 5a,c). Besides, overexpressed miR‐128‐3p in 293T evidently impeded cell proliferation compared with NC, and upregulation of HOTTIP could reverse miR‐128‐3p’s effect to some extent (Figure 5b,d). In conclusion, all the research results reveal the interaction among HOTTIP, miR‐128‐3p and HOXA13.
Figure 5.
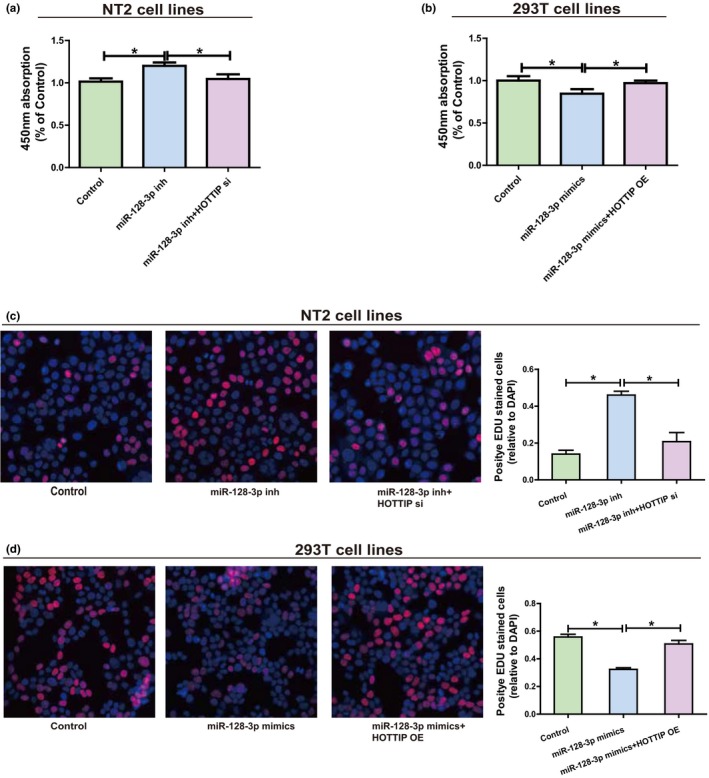
HOTTIP/miR‐128‐3p regulatory loop is important for cell function. (a) Following NT2 is transfected with miR‐128‐3p inhibitors (with or without HOTTIP siRNAs), the cell proliferation level is measured by CCK‐8. (b) After miR‐128‐3p mimics (with or without HOTTIP OE plasmids) are transfected into 293T, the cell proliferation level is determined by CCK‐8. (c) Following NT2 is transfected with miR‐128‐3p inhibitors (with or without HOTTIP siRNAs), the cell proliferation level is measured by EdU. (d) Following transfection of 293T with miR‐128‐3p mimics (with or without HOTTIP OE plasmids), the cell proliferation level is determined by EdU. Data are presented as mean ± SD. *p ≤ .05, Student’s t‐test
4. DISCUSSION
As a complicated and highly concerted process, spermatogenesis relies on the loss of germ cells in the processes of meiosis and spermiogenesis and the proliferation activity of spermatogonia (Nishimura & L’Hernault, 2017). Hence, any cause that affects the proliferation capacity of spermatogonia can lead to spermatogenesis process disorders. It has been proved that almost all cellular processes are modulated by lncRNAs, and the imbalance of these noncoding molecules appear to be associated with the development of a variety of diseases (Degirmenci & Lei, 2016). A study of Lu et al. confirmed that downregulation of miR‐320a/383 sponge‐like lncRNA NLC1‐C has correlation with male infertility and accelerates the proliferation of testicular embryonal carcinoma cells (Lu et al., 2015). As HOTTIP has been verified to exert a regulatory effect in cell proliferation in many diseases, it is hypothesized that HOTTIP may participate in the pathogenesis of infection.
The above study revealed that compared with the control testicular samples, HOTTIP was downregulated in Hypo patients. In the meantime, it was found that compared with the control normal cell line 293T, the HOTTIP level in testicular embryonal carcinoma cell line was evidently increased. These results suggest that HOTTIP may participate in the pathogenesis of Hypo by regulating the proliferation of NT2. In tumor‐related studies, HOTTIP has been proved to participate in the pathogenesis of many tumors including endometrial cancer (Guan, Zhang, Zhang, Liu, & Ren, 2018), ovarian cancer (Zou, Wang, Gao, & Liang, 2018), and renal cell carcinoma (Su et al., 2019) by promoting tumor cell proliferation level. To further verify our hypothesis, the changes in cell function of NT2 and 293T after knocking down and overexpressing HOTTIP in vitro were monitored. According to the results, NT2 proliferation was suppressed after HOTTIP knockdown, but HOTTIP OE sped up 293T proliferation. The above findings evidence that HOTTIP participates in the occurrence of Hypo.
By means of diverse mechanisms, lncRNAs are involved in human diseases. The subcellular localization of HOTTIP in NT2 and 293T was determined, the results of which displayed that HOTTIP showed primary expression in both the nucleus and cytoplasm, implying that HOTTIP is capable of modulating gene expression both at transcription and posttranscription levels. Recently, an enormous body of reports has shown that an lncRNA can be regarded as a sponge that adjusts the expression of miRNA targets and binds to miRNAs. For instance, Feng K et al. identified that the viability and invasion of papillary thyroid carcinoma cells can be enhanced by lncRNA PVT1, a ceRNA of miR‐30a, mediating IGF1 receptor expression (Feng et al., 2018). Yang et al. reported that lncRNA HOTTIP accelerates the migration and proliferation of prostate cancer cells by sponging miR‐216a‐5p (Xiong et al., 2018). After the intersection of bioinformatics prediction results, it was discovered that miR‐128‐3p was highly consistent with HOTTIP’s 3’UTR sequence. To further confirm the correlation between HOTTIP and miR‐128‐3p, dual‐luciferase reporter gene assay and RIP assay were conducted. The results manifested that miR‐128‐3p was capable of decreasing luciferase activity including pGL3‐HOTTIP‐WT sequence, while anti‐AGO2 antibodies decrease HOTTIP and miR‐128‐3p. The above results imply that HOTTIP can be directly combined with miR‐128‐3p and considered as a ceRNA. Numerous studies in recent years have confirmed that miR‐128‐3p can inhibit cell proliferation level (Huang et al., 2015; Huo et al., 2019; Zhou et al., 2018). The current research pointed out that miR‐128‐3p expression in testicular samples from Hypo patients was notably increased. In addition, the results displayed that miR‐128‐3p mimics impeded 293T proliferation, while miR‐128‐3p inhibitors exerted an opposite effect. In conclusion, these findings demonstrate that HOTTIP affects the proliferation of NT2 through interaction with miR‐128‐3p.
For further exploring the exact mechanism of miR‐128‐3p blocking cell proliferation, HOXA13 was proved to be a miR‐128‐3p target gene through dual‐luciferase reporter gene assay. Previously found in drosophila, the highly conservative HOX gene family encodes transcription factors that modulate cell differentiation and proliferation and exerts pivotal effects in the development of embryos (Mallo & Alonso, 2013). Abnormal differentiation and proliferation result from dysfunction of HOX proteins due to changed modes of HOX genes (Holland, 2013). In current studies, the expression of HOX gene, especially HOXA13, accelerating the proliferation of diverse tumor cells has been investigated (Lin et al., 2017; Qin et al., 2019). According to western blotting and qRT‐PCR, HOXA13 expression in testicular samples from Hypo patients was markedly elevated compared with control. However, whether there is interaction among HOXA13, HOTTIP, and miR‐128‐3p still needs further verification. It was also discovered that transfection with HOTTIP siRNAs can reverse the increase of HOXA13 caused by miR‐128‐3p inhibitors. As such, it is speculated that HOTTIP is involved in the pathogenesis of male infertility to adjust HOXA13 expression via the competitive binding to miR‐128‐3p.
5. CONCLUSION
The current research denotes that the expression level of HOTTIP in testicular samples from Hypo patients is decreased, and the downregulation of HOTTIP inhibits the cell proliferation ability of NT2. Furthermore, the above experiments eventually prove that HOXA13 expression can be adjusted by HOTTIP sponging miR‐128‐3p in NT2 as a ceRNA. In summary, this research demonstrates that lncRNA HOTTIP has correlation with male infertility and stimulates the proliferation of testicular embryonal carcinoma cells.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
None.
Su Y, Zhou L‐L, Zhang Y‐Q, Ni L‐Y. Long noncoding RNA HOTTIP is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Mol Genet Genomic Med. 2019;7:e870 10.1002/mgg3.870
Yang Su, Ling‐Ling Zhou, Yu‐Qing Zhang are contributed equally.
REFERENCES
- Chalyi, M. E. , Akhvlediani, N. D. , & Kharchilava, R. R. (2016). Male infertility. Urologiia, 1(Suppl 1), 2–16. https://www.ncbi.nlm.nih.gov/pubmed/28247742 [PubMed] [Google Scholar]
- Degirmenci, U. , & Lei, S. (2016). Role of lncRNAs in cellular aging. Frontiers in Endocrinology (Lausanne), 7, 151 10.3389/fendo.2016.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Zhao, Y. , Wu, X. , & Song, G. (2017). Upregulation of CCAT2 promotes cell proliferation by repressing the P15 in breast cancer. Biomedicine & Pharmacotherapy, 91, 1160–1166. 10.1016/j.biopha.2017.05.030 [DOI] [PubMed] [Google Scholar]
- Dianatpour, A. , & Ghafouri‐Fard, S. (2017). Long non coding RNA expression intersecting cancer and spermatogenesis: A systematic review. Asian Pacific Journal of Cancer Prevention, 18(10), 2601–2610. 10.22034/APJCP.2017.18.10.2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves, S. C. (2015). Clinical management of infertile men with nonobstructive azoospermia. Asian Journal of Andrology, 17(3), 459–470. 10.4103/1008-682X.148719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, K. , Liu, Y. , Xu, L. J. , Zhao, L. F. , Jia, C. W. , & Xu, M. Y. (2018). Long noncoding RNA PVT1 enhances the viability and invasion of papillary thyroid carcinoma cells by functioning as ceRNA of microRNA‐30a through mediating expression of insulin like growth factor 1 receptor. Biomedicine & Pharmacotherapy, 104, 686–698. 10.1016/j.biopha.2018.05.078 [DOI] [PubMed] [Google Scholar]
- Ferrás, C. , Fernandes, S. , Marques, C. J. , Carvalho, F. , Alves, C. , Silva, J. , … Barros, A. (2004). AZF and DAZ gene copy‐specific deletion analysis in maturation arrest and Sertoli cell‐only syndrome. Molecular Human Reproduction, 10(10), 755–761. 10.1093/molehr/gah104 [DOI] [PubMed] [Google Scholar]
- Forrest, M. E. , & Khalil, A. M. (2017). Review: Regulation of the cancer epigenome by long non‐coding RNAs. Cancer Letters, 407, 106–112. 10.1016/j.canlet.2017.03.040 [DOI] [PubMed] [Google Scholar]
- Guan, Q. , Zhang, Q. , Zhang, C. , Liu, Q. , & Ren, Q. L. (2018). HOTTIP regulates progression of endometrial cancer via activating PI3K/AKT pathway. European Review for Medical and Pharmacological Sciences, 22(12), 3727–3733. 10.26355/eurrev_201806_15252 [DOI] [PubMed] [Google Scholar]
- Holland, P. W. (2013). Evolution of homeobox genes. Wiley Interdisciplinary Reviews: Developmental Biology, 2(1), 31–45. 10.1002/wdev.78 [DOI] [PubMed] [Google Scholar]
- Huang, C.‐Y. , Huang, X.‐P. , Zhu, J.‐Y. , Chen, Z.‐G. , Li, X.‐J. , Zhang, X.‐H. , … Wu, G.‐B. (2015). miR‐128‐3p suppresses hepatocellular carcinoma proliferation by regulating PIK3R1 and is correlated with the prognosis of HCC patients. Oncology Reports, 33(6), 2889–2898. 10.3892/or.2015.3936 [DOI] [PubMed] [Google Scholar]
- Huo, L. , Wang, B. , Zheng, M. , Zhang, Y. , Xu, J. , Yang, G. , & Guan, Q. (2019). miR‐128‐3p inhibits glioma cell proliferation and differentiation by targeting NPTX1 through IRS‐1/PI3K/AKT signaling pathway. Experimental and Therapeutic Medicine, 17(4), 2921–2930. 10.3892/etm.2019.7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, Y. , Shinjo, K. , & Katsushima, K. (2017). Long non‐coding RNAs as an epigenetic regulator in human cancers. Cancer Science, 108(10), 1927–1933. 10.1111/cas.13342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja, N. (2014). MicroRNAs and spermatogenesis. Fertility and Sterility, 101(6), 1552–1562. 10.1016/j.fertnstert.2014.04.025 [DOI] [PubMed] [Google Scholar]
- Li, J. , Zhuang, C. , Liu, Y. , Chen, M. , Zhou, Q. , Chen, Z. , … Huang, W. (2016). shRNA targeting long non‐coding RNA CCAT2 controlled by tetracycline‐inducible system inhibits progression of bladder cancer cells. Oncotarget, 7(20), 28989–28997. 10.18632/oncotarget.8259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. , Wang, Y. , Wang, Y. , Zhang, S. , Yu, L. , Guo, C. , & Xu, H. (2017). Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene, 36(38), 5392–5406. 10.1038/onc.2017.133 [DOI] [PubMed] [Google Scholar]
- Lü, M. , Tian, H. , Cao, Y.‐X. , He, X. , Chen, L. , Song, X. , … Sun, F. (2015). Downregulation of miR‐320a/383‐sponge‐like long non‐coding RNA NLC1‐C (narcolepsy candidate‐region 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Cell Death & Disease, 6, e1960 10.1038/cddis.2015.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo, M. , & Alonso, C. R. (2013). The regulation of Hox gene expression during animal development. Development, 140(19), 3951–3963. 10.1242/dev.068346 [DOI] [PubMed] [Google Scholar]
- Neto, F. T. , Bach, P. V. , Najari, B. B. , Li, P. S. , & Goldstein, M. (2016). Spermatogenesis in humans and its affecting factors. Seminars in Cell & Developmental Biology, 59, 10–26. 10.1016/j.semcdb.2016.04.009 [DOI] [PubMed] [Google Scholar]
- Nishimura, H. , & L’Hernault, S. W. (2017). Spermatogenesis. Current Biology, 27(18), R988–R994. 10.1016/j.cub.2017.07.067 [DOI] [PubMed] [Google Scholar]
- Oud, M. S. , Ramos, L. , O’Bryan, M. K. , McLachlan, R. I. , Okutman, Ö. , Viville, S. , … Noordam, M. J. (2017). Validation and application of a novel integrated genetic screening method to a cohort of 1,112 men with idiopathic azoospermia or severe oligozoospermia. Human Mutation, 38(11), 1592–1605. 10.1002/humu.23312 [DOI] [PubMed] [Google Scholar]
- Qin, Z. , Chen, Z. , Weng, J. , Li, S. , Rong, Z. , & Zhou, C. (2019). Elevated HOXA13 expression promotes the proliferation and metastasis of gastric cancer partly via activating Erk1/2. OncoTargets and Therapy, 12, 1803–1813. 10.2147/OTT.S196986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan, Z. , Zheng, D. , & Qing, H. (2017). Regulatory roles of long non‐coding RNAs in the central nervous system and associated neurodegenerative diseases. Frontiers in Cellular Neuroscience, 11, 175 10.3389/fncel.2017.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C. , & Zhong, N. (2015). Long non‐coding RNAs: The epigenetic regulators involved in the pathogenesis of reproductive disorder. American Journal of Reproductive Immunology, 73(2), 95–108. 10.1111/aji.12315 [DOI] [PubMed] [Google Scholar]
- Su, Y. , Lu, J. , Chen, X. , Liang, C. , Luo, P. , Qin, C. , & Zhang, J. (2019). Long non‐coding RNA HOTTIP affects renal cell carcinoma progression by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway. Journal of Cancer Research and Clinical Oncology, 145(3), 573–588. 10.1007/s00432-018-2808-0 [DOI] [PubMed] [Google Scholar]
- Szkodziak, P. , Wozniak, S. , Czuczwar, P. , Wozniakowska, E. , Milart, P. , Mroczkowski, A. , & Paszkowski, T. (2016). Infertility in the light of new scientific reports ‐ focus on male factor. Annals of Agricultural and Environmental Medicine, 23(2), 227–230. 10.5604/12321966.1203881 [DOI] [PubMed] [Google Scholar]
- Weidle, U. H. , Birzele, F. , Kollmorgen, G. , & Ruger, R. (2017). Long non‐coding RNAs and their role in metastasis. Cancer Genomics & Proteomics, 14(3), 143–160. 10.21873/cgp.20027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, D.‐D. , Li, Z.‐Y. , Liang, L. U. , He, R.‐Q. , Ma, F.‐C. , Luo, D.‐Z. , … Chen, G. (2018). The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to Mir‐193a‐3p as a competitive endogenous RNA. Cellular Physiology and Biochemistry, 48(3), 905–918. 10.1159/000491958 [DOI] [PubMed] [Google Scholar]
- Yu, Y. , Nangia‐Makker, P. , Farhana, L. , & Majumdar, A. P. N. (2017). A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR‐145 expression by suppressing its maturation process in colon cancer cells. Molecular Cancer, 16(1), 155 10.1186/s12943-017-0725-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , He, Z. , Guo, L. E. , Zeng, J. , Liang, P. , Ren, L. , … Huang, X. (2018). MiR‐128‐3p directly targets VEGFC/VEGFR3 to modulate the proliferation of lymphatic endothelial cells through Ca(2+) signaling. International Journal of Biochemistry & Cell Biology, 102, 51–58. 10.1016/j.biocel.2018.05.006 [DOI] [PubMed] [Google Scholar]
- Zou, T. , Wang, P. L. , Gao, Y. , & Liang, W. T. (2018). Long noncoding RNA HOTTIP is a significant indicator of ovarian cancer prognosis and enhances cell proliferation and invasion. Cancer Biomarkers, 25(2), 133–139. 10.3233/CBM-181727 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


