Abstract
Background
Association between several single‐nucleotide polymorphisms (SNPs) and breast cancer risk has been identified through genome‐wide association studies (GWAS), but little is known about their significance in patients’ prognosis. We screened SNPs which were related to the prognosis of breast cancer in Henan Han population, analyzed relevant genes by bioinformatics in database, and further constructed the genetic regulatory network involved in the pathogenesis of breast cancer.
Methods
We evaluated five SNPs in 232 cases of breast cancer at the Affiliated Tumor Hospital of Zhengzhou University. Relationships between five SNPs, clinical prognostic indicators, and disease‐free survival (DFS) were evaluated by Kaplan–Meier analysis and Cox proportional hazards model. Gene ontology (GO) functional annotation and Kyoto Encyclopedia of genes and Genome (KEGG) analysis were carried out to preliminarily establish genetic regulation network model of breast cancer. Bayesian algorithm was used to optimize the model.
Results
The multivariate Cox proportional hazards model confirmed that SNP rs3803662 (TOX3/TNRC9) had correlation with DFS independently. In the multivariate Cox proportional hazards model, compared with GA/AA, GG increased the recurrent risk of breast cancer (p = .021, hazard ratio [HR] = 2.914). GO analysis showed that the function of TOX3/TNRC9 included biological_process, molecular_function, and cellular_component. According to KEGG signaling pathway database, the map of breast cancer‐related gene regulatory network was obtained. IGF‐IGF1R‐PI3K‐Akt‐mTOR‐S6K was the best possible pathway for the differentiation of breast cancer cells in this network and ER‐TOX3/TNRC9 was the best possible pathway for the survival of tumor cells in this network by Bayesian theorem optimization.
Conclusions
SNP rs3803662 (TOX3/TNRC9) is an independent prognostic factor for breast cancer in Henan Han Population. ER‐TOX3/TNRC9 is the best possible pathway involved in the pathogenesis of breast cancer.
Keywords: breast cancer, prognosis, regulatory network, SNPs
1. INTRODUCTION
Breast cancer is the most common malignant tumor in women worldwide (Bray et al., 2018). The occurrence of breast cancer is thought to be the result of the interaction of genetic and nongenetic factors. Recent studies have shown that different molecular subtypes of breast cancer have different genetic variables, which may indicate that different subtypes of breast cancer have different etiological pathways (Garcia‐Closas et al., 2008; Stacey et al., 2007). Traditional prognostic factors, such as tumor size, grade, and lymph node metastasis status, are still the most important prognostic factors for breast cancer. However, little interpretations of genetic information were included in the prognosis. As a relatively small allele variable, SNPs has a higher availability, and linkage disequilibrium exists among different populations and races, so it can be used as an important medium for studying different diseases or disease characteristics. With the development of GWAS, an increasing number of genetic variables have been confirmed to be associated with breast cancer. Studies about SNPs are currently focused on the correlation between clinicopathological features, DFS, overall survival (OS), and loci. Some researchers have even combined multiple gene locus models with clinicopathological features to forecast patients’ prognosis. However, their significances for patients or mechanism of pathogenesis remain unknown.
Our early work confirmed that SNP rs3803662 (TOX3/TNRC9) increased the risk of breast cancer in Henan Han population in a case–control study (He et al., 2016). This study was designed to analyze the correlation between five SNPs which were confirmed by GWAS (rs10069690 [TERT], rs2046210 [6q25.1], rs2981582 [FGFR2], rs889312 [MAP3K1], rs3803662[TOX3/TNRC9]) (Dai et al., 2012; Han et al., 2011; Hein et al., 2012; Mulligan et al., 2011; Palmer et al., 2013) and DFS of female breast cancer in Henan Han patients.
2. MATERIALS AND METHODS
2.1. Ethical compliance
The ethics committee of the Medical Ethics Committee of Henan Cancer Hospital approved the study, and informed consent was obtained from all patients prior to their enrollment in the study. The ethical review number is 2015ct072.
2.2. Subjects
In total, 232 female patients with invasive breast cancer who were treated in Affiliated Tumor Hospital of Zhengzhou University between 1 January 2014 and 31 May 2014, were enrolled in this study. Only subjects from Henan Han population were included in this study, furthermore, the subjects were required to have no family history of tumors or genetic diseases and to have received no neoadjuvant therapy. Among them, six patients were diagnosed as breast cancer with distant metastasis, and two patients were partially missing clinical information. All cases were confirmed by histopathology with information available about ER, PR, HER2, and Ki67. Details of patient characteristics were listed in Table 1.
Table 1.
Patient demographic and disease characteristics
| Characteristic | n (%) |
|---|---|
| No. of patients | 232 |
| Age | |
| <50 years | 128 (55.17) |
| ≥50 years | 103 (44.40) |
| Unknown | 1 (0.43) |
| Tumor size | |
| T1 | 88 (37.93) |
| T2 | 119 (51.29) |
| T3 | 17 (7.33) |
| T4 | 6 (2.59) |
| Unknown | 2 (0.86) |
| Number of metastatic lymph nodes | |
| N0 | 130 (56.03) |
| N1 | 54 (23.28) |
| N2 | 27 (11.64) |
| N3 | 19 (8.19) |
| Unknown | 2 (0.86) |
| ER and PR status | |
| Positive | 165 (71.12) |
| Negative | 67 (28.88) |
| HER2 status | |
| Negative | 161 (69.40) |
| Positive | 69 (29.74) |
| Unknown | 2 (0.86) |
| Menstrual statusa | |
| Premenopausal | 148 (63.79) |
| Postmenopausal | 82 (35.34) |
| Unknown | 2 (0.86) |
Abbreviations: T1, Tumor ≤20 mm or less in greatest dimension; T2, Tumor >20 mm but ≤50 mm in greatest dimension; T3, Tumor >50 mm in greatest dimension; T4, Tumor of any size with direct extension to the chest wall and/or to the skin (ulceration or skin nodules) N0, No regional lymph node metastasis histologically; N1, Metastases to movable ipsilateral level I, II axillary lymph node(s); N2, Metastases in ipsilateral level I, II axillary lymph nodes that are clinically fixed or matted; or in clinically detected ipsilateral internal mammary nodes in the absence of clinically evident axillary lymph node metastases; N3, Metastases in ipsilateral infraclavicular (level III axillary) lymph node(s) with or without level I, II axillary lymph node involventment; or in clinically detected ipsilateral interal mammary lymph node(s) with clinically evident level I, II axillary lymph node metastases in ipsilateral supraclavicular lymph node(s) with or without axillary or internal mammary lymph node involvment.
determining menopause includes any of the following: (a) prior bilateral oophorectomy; (b) age ≥60 years; (c) age <60 years and amenorrheic for 12 or more months in the absence of chemotherapy, tamoxifen, toremifene, or ovarian suppression and follicle‐stimulating hormone (FSH) and estradiol in the postmenopausal range.
2.3. Genotyping and quality control
In this study, we selected SNPs with minor allele frequency >5% from the Han Chinese population in Beijing (CHB) in the HapMap database (http://www.hapmap.org). Chinese keywords "single nucleotide polymorphisms", "breast cancer", "hormone receptor status", "molecular subtype", "prognosis", and English keywords "SNPs", "breast cancer", "ER", "PR", "subtypes of breast cancer", "prognosis", "GWAS" were used as keywords to search in PubMed, Embase, China knowledge Network, cqvip, Wanfang Database, and Chinese Biomedical Literature Database for Chinese and foreign literature. Eventually, we selected five SNPs (rs10069690 [TERT], rs2046210 [6q25.1], rs2981582 [EGFR2], rs889312 [MAP3K1], and rs3803662 [TOX3]/[TNRC9]). All DNA samples were blindly duplicated to assess the reproducibility of genotypes. An average reproducibility of 100% was obtained. The mean call rate in the final data set was 99% for SNPs; the quality evaluation is listed in Table 2 (2016). DNA was isolated from peripheral blood samples at the Central Laboratory of Henan Tumor Hospital using whole blood genomic DNA extraction kits (TianGen, M2023). DNA samples were stored at −80°C before genotyping. The genotyping of SNPs was carried out by Shanghai Genesky Bio‐Tech Co., Ltd. (http://biotech.geneskies.com/index.html) using the improved multiplex ligase detection reaction (iMLDR) method; the primers are listed in Table 3.
Table 2.
Quality evaluation
| SNP | Call rate (%) | Test for HWE (p value) | MAF (the study) | MAF (Hapmap‐HCB) | ||
|---|---|---|---|---|---|---|
| rs10069690 | 99.5 | .36 | 0.192 | T | 0.202 | T |
| rs2046210 | 99.82 | .18 | 0.393 | A | 0.380 | A |
| rs2981582 | 99.83 | .13 | 0.368 | A | 0.336 | A |
| rs3803662 | 99.83 | .38 | 0.314 | G | 0.347 | G |
| rs889312 | 99.83 | .20 | 0.494 | C | 0.500 | A |
Abbreviations: SNPs, single‐nucleotide polymorphisms; HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency.
Table 3.
SNPs and PCR primer
| SNPs | Chr | Chromosome position | Gene | PCR primer |
|---|---|---|---|---|
| rs10069690 | 5 | 1279790 | TERT | rs10069690F: CCCAGCTTCCTCAGACCCTGTT |
| rs10069690R: CTGGATCCGTGTCCTGCTGTG | ||||
| rs2046210 | 6 | 151948366 | – | rs2046210F: GAGGTGTGACCACTGCCATCGT |
| rs2046210R: GAAACCATCAGGGTGCCTCAAC | ||||
| rs2981582 | 10 | 123352317 | FGFR2 | rs2981582F: GAGGCTGGGCTCTCTGTCCTCT |
| rs2981582R: GAACCTCTCTCCCAGCCCTTTG | ||||
| rs3803662 | 16 | 52586341 | LOC643714 | rs3803662F: GGTGGGGGTCAGTCCACAGTTT |
| rs3803662R: TGCTGCTAGTCCTTGGCTGTTC | ||||
| rs889312 | 5 | 56031884 | – | rs889312F: TTCCAGTCTGGGGTGGCTTGTA |
| rs889312R: TGGGAAGGAGTCGTTGAGTTTTCA |
Abbreviations: SNPs, single‐nucleotide polymorphisms; PCR, polymerase chain reaction; F, forward; R, reverse.
2.4. Statistical analysis and outcome measures
Disease‐free survival(DFS) was defined as the elapsed time between the date of initial treatment (surgery) and the first date of documented disease recurrence or death due to breast cancer. Follow‐up information was obtained by telephone and outpatient data. The information included patient's status (alive; death; local recurrence; distant metastasis), results of clinical physical examinations, and imaging data (color Doppler ultrasound, CT, whole body bone imaging or MRI). Clinical prognostic indicators included the age of onset, tumor size, lymph node metastasis status, and subtypes of breast cancer. Kaplan–Meier analysis and the log‐rank test were used to identify the correlation between DFS and clinical prognostic indicators as well as five SNPs. The factors which were significant in univariate analysis were then analyzed in the multivariate Cox proportional hazards model to determine the independent prognostic factor. All statistical procedures were performed with SPSS (version 20.0; SPSS Company, Chicago, IL). All p values reported were two‐sided and were calculated at a significance level of .05.
2.5. Construction and optimization of regulatory network
Gene ontology functional annotation and KEGG analysis of selected genes were carried out. In GO analysis, the screening conditions of AmiGO (http://geneontology.org) were “name of selected genes” and”Homo sapiens”. GO functional annotation of selected genes was preliminarily analyzed. For KEGG analysis of breast cancer‐related genes, the first step was that entering the homepage of KEGG signaling pathway database (http://www.kegg.jp/kegg/pathway.html). Screening condition organism was "hsa", keywords were "names of selected genes" and "Breast cancer". Queried the regulatory network of breast cancer related genes. Bayesian algorithm was used to optimize the regulatory network model to find out the most possible pathway of gene regulation of breast cancer cell proliferation by probability.
3. RESULTS
The final analysis included 209 patients with breast invasive cancer. The follow‐up deadline was 31 December 2017 and the median follow‐up length was 44.5 months. There were 23 (11.0%) DFS events. Four patients had regional recurrence, 19 had distant metastasis (five had regional recurrence synchronously accompanied by distant metastasis). No patient died due to breast cancer. There were 15 (7.18%) cases who were not interviewed (listed in Table 4). The survival curve is shown in Figure 1.
Table 4.
Events of all patients
| Events | Number of events |
|---|---|
| Only regional recurrence | 4 |
| Distance recurrence | 19 |
| Distance recurrence and regional recurrence | 5 |
| Only distance recurrence | 14 |
| Death caused by any recurrence of disease | 0 |
| Total | 23 |
Figure 1.
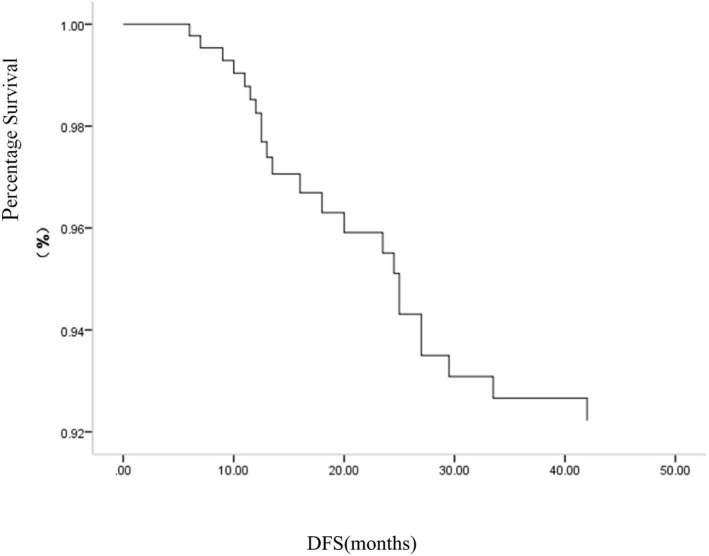
Kaplan–Meier curves of DFS for 209 patients
The results of the Kaplan–Meier analysis showed that tumor size, lymph node metastasis status, and subtypes of breast cancer were significantly associated with DFS (p = .024, .000, .028) (Table 5). There were no associations between SNPs rs10069690 (TERT), rs2046210 (6q25.1), rs2981582 (FGFR2) and rs889312 (MAP3K1) and DFS (p = .202, .096, .686, .172), but SNP rs3803662 (TOX3/TNRC9) had association with DFS (p = .010) (Table 6). The multivariate Cox proportional hazards model showed that lymph node metastasis status and SNP rs3803662 (TOX3/TNRC9) were correlated with DFS. Compared with GA/AA, GG increased the recurrent risk of breast cancer (p = .021, HR = 2.914) (Table 7). The results of GO analysis showed that gene TOX3/TNRC9 had three functions: biological_process, molecular_function, and cellular_component (Table 8). According to KEGG signaling pathway database, the map of breast cancer‐related gene regulatory network was obtained (Figure 2). The optimal pathway of breast cancer cell regulatory network in which TOX3/TNRC9 was involved was selected by Bayesian theorem optimization (Figure 3).
Table 5.
Survival analysis by Kaplan–Meier analysis and log‐rank test between clinicopathological factors and DFS
| Clinicopathological factors | No. of patients | Events (%) | Log Rank (Mantel‐Cox) | |
|---|---|---|---|---|
| χ 2 | p | |||
| Age | ||||
| <50 years | 120 | 15 (12.5) | 0.606 | .436 |
| ≥50 years | 89 | 8 (9.0) | ||
| Tumor size | ||||
| T1 + T2 | 189 | 18 (9.5) | 5.113 | .024 |
| T3 + T4 | 20 | 5 (25.0) | ||
| No. of metastatic lymph nodes | ||||
| N0 + N1 | 173 | 11 (6.4) | 55.425 | 0.000 |
| N2 | 22 | 4 (18.2) | ||
| N3 | 14 | 8 (57.1) | ||
| Subtypes | ||||
| L‐A | 20 | 0 (0.0) | 4.846 | 0.028 |
| L‐B | 97 | 8 (8.2) | ||
| L‐H | 31 | 4 (12.9) | ||
| HER2 | 32 | 7 (21.8) | ||
| TNBC | 29 | 4 (13.8) | ||
Abbreviations: T1, Tumor ≤20 mm or less in greatest dimension; T2, Tumor >20 mm but ≤50 mm in greatest dimension; T3, Tumor >50 mm in greatest dimension; T4, Tumor of any size with direct extension to the chest wall and/or to the skin (ulceration or skin nodules) N0, No regional lymph node metastasis histologically; N1, Metastases to movable ipsilateral level I, II axillary lymph node(s); N2, Metastases in ipsilateral level I, II axillary lymph nodes that are clinically fixed or matted; or in clinically detected ipsilateral internal mammary nodes in the absence of clinically evident axillary lymph node metastases; N3, Metastases in ipsilateral infraclavicular (level III axillary) lymph node(s) with or without level I, II axillary lymph node involventment; or in clinically detected ipsilateral interal mammary lymph node(s) with clinically evident level I, II axillary lymph node metastases in ipsilateral supraclavicular lymph node(s) with or without axillary or internal mammary lymph node involvment; L‐A, Luminal A; L‐B, Luminal B (not contains) L‐H, Luminal‐HER2; HER2:HER2‐enrich; TNBC:triple‐negative breast cancer.
Table 6.
Survival analysis by Kaplan–Meier analysis and log‐rank test between SNPs and DFS
| SNPs | No. of patients | Events (%) | Log Rank (Mantel–Cox) | |
|---|---|---|---|---|
| χ2 | p | |||
| rs10069690 | ||||
| TT | 8 | 2 (25.0) | 1.629 | .202 |
| CT + CC | 201 | 21 (10.4) | ||
| rs2046210 | ||||
| GG | 70 | 11 (15.7) | 2.765 | .096 |
| GA + GG | 139 | 12 (8.6) | ||
| rs2981582 | ||||
| AA | 34 | 3 (8.8) | 0.163 | .686 |
| GA + GG | 175 | 20 (11.4) | ||
| rs3803662 | ||||
| GG | 29 | 7 (24.1) | 6.703 | .010 |
| GA + AA | 180 | 16 (8.8) | ||
| rs889312 | ||||
| CC | 49 | 8 (16.3) | 1.866 | .172 |
| CA + AA | 160 | 15 (9.4) | ||
Abbreviation: SNPs, single‐nucleotide polymorphisms.
Table 7.
Multivariate Cox proportional hazards model for DFS in the study cohort
| Variables | HR | 95% CI | p |
|---|---|---|---|
| Age: ≥50 years versus <50 years (reference) | 1.304 | 0.535–3.175 | .559 |
| T: T3 + T4 versus T1 + T2 (reference) | 1.897 | 0.679–5.302 | .222 |
| No. of metastatic lymph nodes | |||
| N2 versus N0 + N1 (reference) | 3.505 | 1.099–11.182 | .034 |
| N3 versus N0 + N1 (reference) | 18.277 | 7.254–46.053 | .000 |
| Subtypes | |||
| L‐HER2 + HER2 versus L‐A + L‐B (reference) | 1.506 | 0.568–3.991 | .411 |
| TNBC versus L‐A + L‐B (reference) | 2.038 | 0.594–6.993 | .258 |
| rs3803662: GG versus GA + AA (reference) | 2.914 | 1.1733–7.239 | .021 |
Abbreviations: CI, confidence interval; HR, hazard ratio; T1, Tumor ≤20 mm or less in greatest dimension; T2, Tumor >20 mm but ≤50 mm in greatest dimension; T3, Tumor > 50 mm in greatest dimension; T4, Tumor of any size with direct extension to the chest wall and/or to the skin (ulceration or skin nodules) N0, No regional lymph node metastasis histologically; N1, Metastases to movable ipsilateral level I, II axillary lymph node(s); N2, Metastases in ipsilateral level I, II axillary lymph nodes that are clinically fixed or matted; or in clinically detected ipsilateral internal mammary nodes in the absence of clinically evident axillary lymph node metastases; N3, Metastases in ipsilateral infraclavicular (level III axillary) lymph node(s) with or without level I, II axillary lymph node involventment; or in clinically detected ipsilateral interal mammary lymph node(s) with clinically evident level I, II axillary lymph node metastases in ipsilateral supraclavicular lymph node(s) with or without axillary or internal mammary lymph node involvment; L‐A, Luminal A; L‐B, Luminal B (not contains) L‐H, Luminal‐HER2; HER2, HER2‐enrich; TNBC, triple‐negative breast cancer.
Table 8.
The result of GO Analysis about Gene TOX3/TNRC9
| Genes | Name | Gene ontology | GO number |
|---|---|---|---|
| TOX3/TNRC9 | Estrogen response element binding | Biological_process | GO:0000107 |
| DNA binding | Biological_process | GO:0000104 | |
| DNA‐binding transcription factor activity, RNA polymerase II‐specific | Molecular_function | GO:0000113 | |
| Nucleus | Cellular_component | GO:0000039 | |
| Regulation Of transcription by RNA polymerase II | MOLECULAR_function | GO:0000981 | |
| Apoptotic process | Biological_process | GO:0007177 | |
| Protein homodimerization activity | Molecular_function | GO:1901185 | |
| Regulation of apoptotic process | Biological_process | GO:0008185 | |
| Positive regulation of transcription, DNA‐templated | Biological_process | GO:0038095 |
Figure 2.
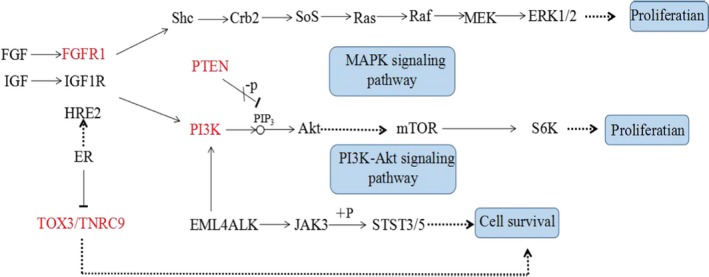
Regulation Network of Breast Cancer‐related genes contained GeneTOX3/TNRC9
Figure 3.
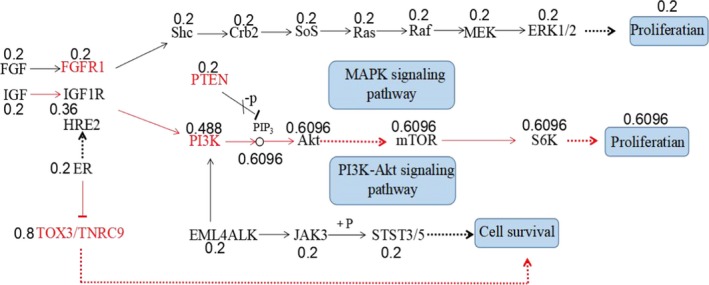
Optimized breast cancer cell regulatory network contained Gene TOX3/TNRC9 by Bayesian networks
4. DISCUSSION
Breast cancer is the most common malignant tumor in Chinese women (Li et al., 2018). In recent years, with the improvement of comprehensive treatment, the survival of breast cancer patients has greatly improved, but 20%–30% of patients still have recurrence and metastasis (Cameron et al., 2017; Pan et al., 2017). Currently, clinicopathological factors determining the prognosis of patients mainly include lymph node staging, tumor size, molecular subtype, and so on, but these factors still cannot accurately evaluate the prognosis of patients. There are many studies on correlations between SNPs and prognosis of breast cancer, but conclusions are different. Some researchers believed that rs88931 (MAP3K1) was highly correlated with distant disease‐free survival (DDFS), DFS, and OS of hormone receptor‐positive breast cancer (Kuo et al., 2017). Yamamoto‐Ibusuki et al. (2015) confirmed that homozygous alleles of rs2046210 showed worse relapse‐free survival. Hein et al. (2017) showed that rs2981582 (FGFR2), rs889312 (MAP3K1), and rs3803662 (TOX3) did not affect the overall survival and progression‐free survival in breast cancer patients. Similar finding was noted in the current study, but the role of rs3803662 in the prognosis of breast cancer patients in Han population was rarely analyzed. Our study explored genetic factors related to the prognosis of female breast cancer patients in Henan by means of SNPs (a third‐generation genetic marker) which have regional and ethnic differences. We identified SNPs rs10069690 (TERT), rs2046210 (6q25.1), rs2981582 (FGFR2), and rs889312 (MAP3K1) were not associated with DFS, while rs3803662 (TOX3/TNRC9) was associated with DFS. Genotype GG of rs3803662 (TOX3/TNRC9) was associated with a worse prognosis and increased the recurrent risk of breast cancer nearly threefold. This locus was an independent prognostic factor in this study population, which was consistent with foreign studies (Fasching et al., 2012).
At the same time, combined with our previous research results (He et al., 2016), we believe that gene TOX3 plays a certain role in the occurrence and development of breast cancer in Henan Han women. TOX3 gene in malignant tumors has been reported to be mainly involved in the transcription process (Dittmer et al., 2010; Yahata et al., 2001; Yuan, Qiu, & Ghosh, 2009). Studies about breast cancer showed that TOX3 was an anti‐oncogene (Cowper‐Sal·lari et al., 2012), and was expressed more in luminal tumors (Han, Zhang, Zheng, Huo, & Olopade, 2016). These studies confirmed the roles of TOX3 in breast cancer, but how to regulates is complex and unknown (Cowper‐Sal·lari et al., 2012; Han et al., 2016; Yu & Li, 2015).
Recently, with the continuous development of bioinformatics, a large number of multifunctional bioinformatics softwares have emerged and they have greatly accelerated the integration and utilization of existing biomedical data. Bioinformatics research helps us to find the most reasonable and effective methods or approaches for treatment and prevention of diseases (Ethier, Desautels, Templeton, Shah, & Amir, 2017). We used tools of bioinformatics, such as GO, KEGG, and Bayesian networks to analyze TOX3. We found TOX3/TNRC9 had three functions: molecular_function, cellular_component, and biological_process by GO analysis. IGF‐IGF1R‐PI3K‐Akt‐mTOR‐S6K was the best possible pathway for the differentiation of breast cancer cells by KEGG analysis and ER‐TOX3/TNRC9 was the best possible pathway for the survival of tumor cells by Bayesian networks. These results provide a theoretical basis for targeted therapy of breast cancer, and further lay the theoretical foundation for studies of mechanisms of TOX3 gene in breast cancer.
Several limitations of this study should be acknowledged. First, the number of SNPs tested was limited to five, which is not a sufficient evaluation of the correlation between the GWAS‐identified SNPs and prognosis of breast cancer. Second, the sample size was too small to definitively evaluate outcomes of breast cancer, and the study needs to be replicated in a larger sample. We need to further verify the mechanism or the regulatory network in the real world (cells, animals, or populations).
In summary, these findings provide further evidence that some genetic factors are strongly associated with breast cancer prognosis among the Han Population. These differences may be due to racial differences. The main purpose of the gene regulatory network is to obtain a network of interactions between genes, and to reveal the overall or partial network characteristics related to life processes. Understanding the mechanism of life activity at the molecular level is conducive to the study of cell function and life process, thus providing directions for exploring the causes of human diseases.
5. CONCLUSIONS
SNP rs3803662 (TOX3/TNRC9) is an independent prognostic factor for breast cancer in Henan Han Population. ER‐TOX3/TNRC9 is the best possible pathway involved in the pathogenesis of breast cancer.
6. DECLARATIONS
Ethics approval and consent to participate: The Medical Ethics Committee of Henan Cancer Hospital approved the study, and informed consent was obtained from all patients prior to their enrollment in the study. The ethical review number is 2015ct072.
Consent for publication: Not applicable.
Availability of data and material: Not applicable.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Conception/Design: Yaning He, Hui Liu and Suxia Luo. Collection of data and extraction of DNA: Qi Chen and Yingbo Shao. Data analysis and interpretation: Yaning He and Qi Chen. Manuscript writing: Yaning He.
ACKNOWLEDGMENTS
We thank all subjects for providing the DNA and information necessary for our study. We also thank the Central Laboratory of Henan Tumor Hospital and Shanghai Genesky Bio‐Tech Co., Ltd. (http://biotech.geneskies.com/index.html) for their valuable help with the isolation of DNA and the genotyping of the SNPs.
He Y, Liu H, Chen Q, Shao Y, Luo S. Relationships between SNPs and prognosis of breast cancer and pathogenic mechanism. Mol Genet Genomic Med. 2019;7:e871 10.1002/mgg3.871
Funding Information
This study was funded by the scientific and technological project of the Science and Technology Department of Henan Province. The award number is 142102310129. The grant recipient was Hui Liu.
REFERENCES
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. [DOI] [PubMed] [Google Scholar]
- Cameron, D. , Piccart‐Gebhart, M. J. , Gelber, R. D. , Procter, M. , Goldhirsch, A. , de Azambuja, E. , … Jackisch, C. (2017). 11 years' follow‐up of trastuzumab after adjuvant chemotherapy in HER2‐positive early breast cancer: Final analysis of the HERceptin Adjuvant (HERA) trial. The Lancet, 389(10075), 1195–1205. 10.1016/S0140-6736(16)32616-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper‐Sal·lari, R. , Zhang, X. , Wright, J. B. , Bailey, S. D. , Cole, M. D. , Eeckhoute, J. , … Lupien, M. (2012). Breast cancer risk‐associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nature Genetics, 44(11), 1191–1198. 10.1038/ng.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, J. , Hu, Z. , Jiang, Y. , Shen, H. , Dong, J. , Ma, H. , & Shen, H. (2012). Breast cancer risk assessment with five independent genetic variants and two risk factors in Chinese women. Breast Cancer Research, 14, R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer, S. , Kovacs, Z. , Yuan, S. H. , Siszler, G. , Kögl, M. , Summer, H. , … Methner, A. (2010). TOX3 is a neuronal survival factor that induces transcription depending on the presence of CITED1 or phosphorylated CREB in the transcriptionally active complex. Journal of Cell Science, 124(Pt 2), 252–260. 10.1242/jcs.068759 [DOI] [PubMed] [Google Scholar]
- Ethier, J. L. , Desautels, D. , Templeton, A. , Shah, P. S. , & Amir, E. (2017). Prognostic role of neutrophil‐to‐lymphocyte ratio in breast cancer: A systematic review and meta‐analysis. Breast Cancer Research, 19(1), 2 10.1186/s13058-016-0794-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching, P. A. , Pharoah, P. D. P. , Cox, A. , Nevanlinna, H. , Bojesen, S. E. , Karn, T. , … Schmidt, M. K. (2012). The role of genetic breast cancer susceptibility variants as prognostic factors. Human Molecular Genetics, 21(17), 3926–3939. 10.1093/hmg/dds159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Closas, M. , Hall, P. , Nevanlinna, H. , Pooley, K. , Morrison, J. , Richesson, D. A. , … Milne, R. L. (2008). Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genetics, 4, e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, W. , Woo, J. H. , Yu, J. H. , Lee, M. J. , Moon, H. G. , Kang, D. , & Noh, D. Y. (2011). Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer Epidemiology and Prevention Biomarkers, 20, 793–798. [DOI] [PubMed] [Google Scholar]
- Han, Y.‐J. , Zhang, J. , Zheng, Y. , Huo, D. , & Olopade, O. I. (2016). Genetic and epigenetic regulation of TOX3 expression in breast cancer. PLoS ONE, 11(11), e0165559 10.1371/journal.pone.0165559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Liu, H. , Chen, Q. , Sun, X. , Liu, C. , & Shao, Y. (2016). Relationship between five GWAS‐identified single nucleotide polymorphisms and female breast cancer in the Chinese Han population. Tumor Biology 37(7), 9739–9744. 10.1007/s13277-016-4795-6 [DOI] [PubMed] [Google Scholar]
- Hein, A. , Rack, B. , Li, L. , Ekici, A. , Reis, A. , Lux, M. , … Häberle, L. (2017). Genetic breast cancer susceptibility variants and prognosis in the prospectively randomized success a study. Geburtshilfe und Frauenheilkunde, 77(6), 651–659. 10.1055/s-0042-113189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein, R. , Maranian, M. , Hopper, J. L. , Kapuscinski, M. K. , Southey, M. C. , Park, D. J. , … Muir, K. R. (2012). Comparison of 6q25 breast cancer hits from Asian and European genome wide association studies in the Breast Cancer Association Consortium (BCAC). PLoS ONE, 7(8), e42380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, S. H. , Yang, S. Y. , You, S. L. , Lien, H. C. , Lin, C. H. , Lin, P. H. , & Huang, C. S. (2017). Polymorphisms of ESR1, UGT1A1, HCN1, MAP3K1 and CYP2B6 are associated with the prognosis of hormone receptor‐positive early breast cancer. Oncotarget, 8(13), 20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Zheng, R. S. , Zhang, S. W. , Zeng, H. M. , Sun, K. X. , Xia, C. F. , … He, J. (2018). Incidence and mortality of female breast cancer in China, 2014. Chinese Journal of Oncology, 40(3), 166–171. [DOI] [PubMed] [Google Scholar]
- Mulligan, A. M. , Couch, F. J. , Barrowdale, D. , Domchek, S. M. , Eccles, D. , Nevanlinna, H. , … Wappenschmidt, B. (2011). Common breast cancer susceptibility alleles are associated with tumor subtypes in BRCA1 and BRCA2 mutation carriers: Results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Research, 13, R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J. R. , Ruiz‐Narvaez, E. A. , Rotimi, C. N. , Cupples, L. A. , Cozier, Y. C. , Adams‐Campbell, L. L. , & Rosenberg, L. (2013). Genetic susceptibility loci for subtypes of breast cancer in an African American population. Cancer Epidemiology and Prevention Biomarkers, 22(1), 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, H. , Gray, R. , Braybrooke, J. , Davies, C. , Taylor, C. , McGale, P. , … Hayes, D. F. (2017). 20‐year risks of breast‐cancer recurrence after stopping endocrine therapy at 5 years. New England Journal of Medicine, 377(19), 1836–1846. 10.1056/NEJMoa1701830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, S. N. , Manolescu, A. , Sulem, P. , Rafnar, T. , Gudmundsson, J. , Gudjonsson, S. A. , … Aben, K. K. (2007). Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor–positive breast cancer. Nature Genetics, 39(7), 865–869. [DOI] [PubMed] [Google Scholar]
- Yu, X. , & Li, Z. (2015). TOX gene: A novel target for human cancer gene therapy. American Journal of Cancer Research, 5(12), 3516–3524. www.ajcr.us/ISSN:2156–6976/ajcr0005402 [PMC free article] [PubMed] [Google Scholar]
- Yahata, T. , Shao, W. , Endoh, H. , Hur, J. , Coser, K. R. , Sun, H. , … Shioda, T. (2001). Selective coactivation of estrogen‐dependent transcription by CITED1 CBP/p300‐binding protein. Genes & Development, 15(19), 2598–2612. 10.1101/gad.906301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto‐Ibusuki, M. , Yamamoto, Y. , Fujiwara, S. , Sueta, A. , Yamamoto, S. , Hayashi, M. , … Iwase, H. (2015). C6ORF97‐ESR1 breast cancer susceptibility locus: Influence on progression and survival in breast cancer patients. European Journal of Human Genetics, 23(7), 949 10.1038/ejhg.2014.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, S. H. , Qiu, Z. , & Ghosh, A. (2009). TOX3 regulates calcium‐dependent transcription in neurons. Proceedings of the National Academy of Sciences, 106(8), 2909–2914. 10.1073/pnas.0805555106 [DOI] [PMC free article] [PubMed] [Google Scholar]


