Abstract
Background: The liver-specific microRNA-122 (miR-122) has been demonstrated as a powerful and promising biomarker of hepatic diseases. However, the researches on the accuracy of miR122 detection in chronic viral hepatitis have been inconsistent, leading us to conduct this meta-analysis to systematically summarize the diagnostic value of circulating miR-122 in patients with hepatitis B virus (HBV) and/or hepatitis C virus (HCV)-associated chronic viral hepatitis.
Methods: A comprehensive literature search (updated to January 30, 2019) in PubMed, Cochrane library, EMBASE, CNKI, Wanfang, and CQVIP databases was performed to identify eligible studies. The sensitivity (SEN), specificity (SPE), positive and negative likelihood ratios (PLR and NLR), diagnostic odds ratio (DOR), and area under the curve (AUC) were pooled to explore the diagnostic performance of circulating miR-122. Subgroup and threshold effect analysis were further carried out to explore the heterogeneity.
Results: Overall, 15 studies were finally included in this meta-analysis according to the exclusion and inclusion criteria. The pooled estimates indicated a moderately high diagnostic accuracy for circulating miR-122, with a sensitivity of 0.92 [95% confidence interval (CI), 0.86–0.95], a specificity of 0.84 (95% CI, 0.78–0.89), a PLR of 5.7 (95% CI, 4.7–8.1), a NLR of 0.1 (95% CI, 0.06–0.18), a DOR of 57 (95% CI 25-129), and an AUC of 0.93 (95% CI, 0.91–0.95). The subgroup analysis demonstrated that diagnostic accuracy was better for HCV-associated chronic viral hepatitis patients and non-Chinese compared with other subgroups. In addition, we found that serum might be a more promising matrix for detecting the expression of miR-122 than plasma.
Conclusions: Our results demonstrated that circulating miR-122 have a relatively high diagnostic value for chronic viral hepatitis detection, especially in the patients with HCV-associated chronic viral hepatitis. However, further large cohort studies are still required to confirm our findings.
Keywords: chronic viral hepatitis, diagnosis, meta-analysis, miR-122
Introduction
Hepatitis usually refers to inflammation of the liver tissue, which may result from both infectious (e.g. viral and bacterial) and noninfectious causes (e.g. alcohol, certain medications, and toxins). Severe liver disease usually contributes to persistent inflammation and necrosis, of which the two primary adverse outcomes are cirrhosis and hepatocellular carcinoma, consequently may cause liver-related death. Here, this article focuses on viral hepatitis because viruses mainly including hepatitis B virus (HBV), and hepatitis C virus (HCV), which have already been proved to be the most common cause of hepatitis worldwide. Furthermore, about a million of patients die from chronic viral hepatitis, most of which occur indirectly from liver scarring or liver cancer [1]. Therefore, early diagnosis of chronic viral hepatitis not only plays a significant role in hepatitis treatment and prevention but also allows for inhibiting disease progression, and decreasing transmission to others to a large extent [1,2].
Nowadays, the laboratory diagnostic tests for hepatitis are mainly based on blood tests, imaging, and liver biopsy [3]. Blood tests include antigens, antibodies, liver-associated enzymes, nucleic acid testing (e.g. hepatitis virus DNA/RNA). Furthermore, hepatic ultrasound, computed tomography, and magnetic resonance imaging are the procedures which can identify steatosis of the liver tissue and nodularity of the liver surface suggestive of cirrhosis. Liver biopsy is still the gold standard for assessing the precise extent and pattern of inflammation and fibrosis of the liver. However, some of these diagnostic approaches are invasive while some fail in early detection of disease due to the limited sensitivity which can only be used in advanced cases [4]. Since the delayed diagnosis may result in poor prognosis, it is necessary to find minimally invasive and cost-effective biomarkers to expand the diagnosis range for liver diseases. Accumulating evidence has witnessed the potential role of circulating miRNAs, as a part of “liquid biopsy”, in the diagnostic value for viral hepatitis [5]. miRNAs, a family of highly conserved single-stranded RNA molecules (19–22 nucleotides), have been proved to participated in multiple biological processes mainly including cell cycle, cell proliferation, differentiation, and apoptosis through binding to the complimentary 3′UTR of their target mRNAs and degrading the mRNAs [6,7]. Several research groups have demonstrated that circulating miRNAs, deriving from intracellular miRNAs and secreted out of the cell via exosomes and microvesicles during the process of cell death, can be stably detected in the biofluid like serum, plasma, urine, and cerebrospinal fluid (CSF) from patients making them relatively non-invasive biomarker for infectious diseases [8–10]. As a major miRNA in liver, miR-122 accounts for approximately 70% of the total liver miRNAs and has been widely reported to suffer from dysregulation in HCV and HBV infection. Besides, the level of miR-122 has been shown to correlate with the severity and stage of infection and helps to evaluate the treatment response [11–13]. In order to verify the hypothesis and assess the diagnostic value of miR-122 in chronic viral hepatitis, we systematically reviewed the literature and conducted this meta-analysis.
Materials and methods
Search strategy
In order to retrieve all the articles analyzing the diagnostic value of miR-122 in patients with HBV and/or HCV chronic viral hepatitis, a comprehensive literature search (updated to January 30, 2019) in PubMed, Cochrane library, EMBASE, and CNKI Wanfang and CQVIP databases was performed without language restrictions. The medical subject heading (MeSH) terms (“microRNA-122” or “miRNA-122” or “miR-122” or “hsa-miR-122”) and (“diagnostic value” or “diagnoses” or “receiver operating characteristics curve” or “ROC curve” or “sensitivity and specificity”) and (“HCV” or “Chronic hepatitis C” or “CHC” or “HBV” or “Chronic hepatitis B” or “CHB”) were used to identify all the relevant articles. Besides, we examined the reference lists of review articles and selected papers manually to identify whether there are any other eligible articles.
Inclusion and exclusion criteria
Studies were considered eligible for inclusion in this meta-analysis had to fulfill the following criteria: (1) evaluate the diagnostic value of miR-122 in patients with HBV and/or HCV-associated chronic viral hepatitis; (2) the patients with HBV-associated chronic viral hepatitis should be positive for HBV surface antigen, positive for HBV DNA, while the diagnosis of HCV-associated chronic viral hepatitis was based on the detection of anti-HCV antibodies and consistent detection of HCV RNA, for at least 6 months; (3) each study involved both experimental and control groups; (4) the miR-122 expression was measured in serum or plasma samples; (5) articles provided sufficient data. Exclusion criteria were as follows: (1) studies with duplicate data; (2) meta-analysis, letters, reviews, case reports; (3) studies without sufficient data or with 20 patients or less.
Data extraction and quality assessment
Two investigators (XHZH and SHQF) are responsible for assessing all the publications. Any disagreements were resolved through discussion with a third reviewer (MW). The following information was extracted from eligible studies: first author’s name, year and country of the publication, characteristics of participants (ethnicity, total number of cases and controls, mean/median age), sample types, methods of miR-122 testing, and the data (true-positive (TP), false-positive (FP), false-negative (FN), and true-negative (TN), sensitivity, and specificity). In addition, the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) was performed to assess the methodological qualities of each included article.
Statistical analysis
Stata 14.0 (STATA Corp, College Station, TX, U.S.A.) and Meta-DiSc version 1.4 software were used to perform all the statistical analysis and P-value less than 0.05 was considered statistically significant. We calculated the pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and corresponding 95% confidence intervals (CIs) for the miR-122 studied using the bivariate random-effect regression model [14]. In addition, we determined the sensitivity and specificity of the miRNAs in each study using a bivariate summary receiver operating characteristic (SROC) curve. We calculated the area under the curve (AUC) and the maximum point of intersection between sensitivity and specificity [15]. Moreover, subgroup and sensitivity analysis were carried out to explore potential sources of between-study heterogeneity. And publication bias was assessed by Deeks’ funnel plot asymmetry test.
Results
Selection process and characteristics of the eligible studies
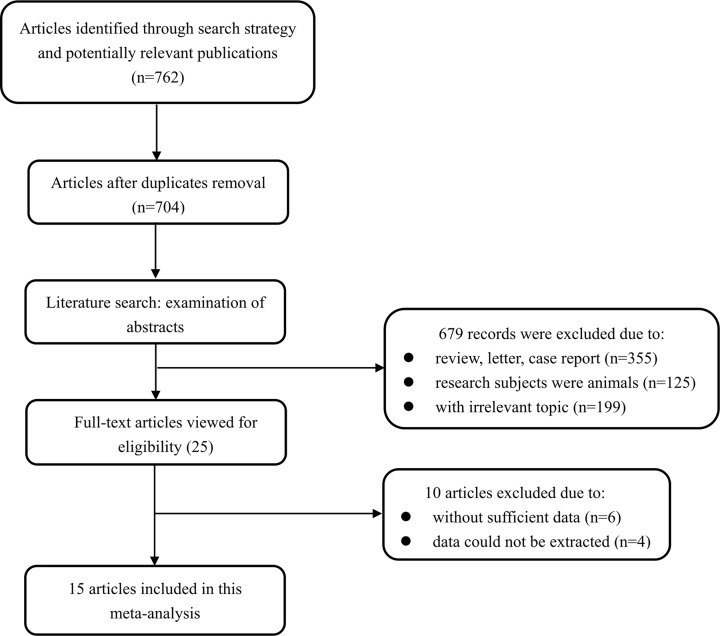
As presented in Figure 1, a total of 762 articles were initially identified from the primary literature search strategy. 704 articles were left for screening after 65 duplicates were removed through Endnote X7 software. After reviewing their abstracts and titles, 679 articles were removed due to unfit literary forms, research subjects of animal model and irrelevant research topic. Subsequently, the full-texts of the remaining 25 articles were read to assess the eligibility and 10 articles were further excluded. As a result, 15 articles were included in the current meta-analysis [13,16–29].
Figure 1. Flowchart of the articles selection process in this meta-analysis.
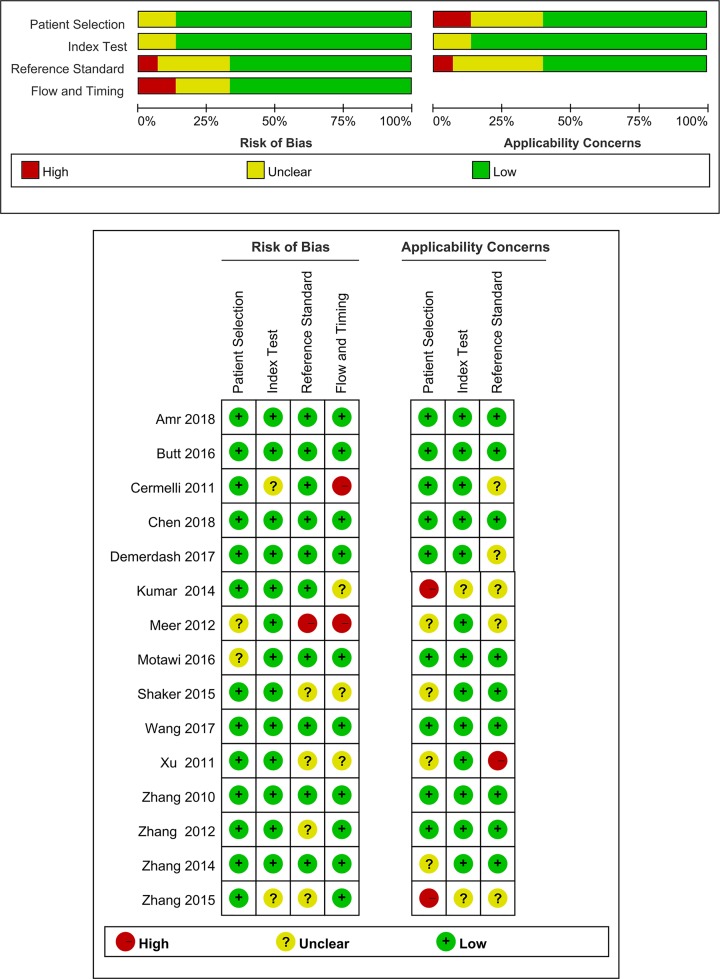
The main characteristics, along with the QUADAS-2 scores of the 15 included articles are summarized in Table 1, in an order by the publication year, ranging from 2010 to 2018. In all studies, quantitative real time polymerase chain reaction (qRT-PCR) assays were performed to detect the expression levels of miRNAs in either plasma (n = 4) or serum (n = 11). Seven articles of these selected studies were conducted in China, while the rest eight articles did research in foreign countries. As for the virus types, six articles focus on HBV-associated chronic viral hepatitis, and nine articles focus on HCV-associated chronic viral hepatitis. In addition, according to the 14-item QUADAS assessment tool, the quality assessments for each included study are presented in Figure 2.
Table 1. The summary characteristics and quality assessment of diagnostic clinical trials included in this meta-analysis.
| Included studies | Country | Detection method | Internal reference | Source of the virus | Sample size | Mean age (year) | Sensitivity | Specificity | Specimen | QUADAS-2 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case age | Control age | |||||||||
| Zhang, 2010 | China | SYBR PCR | U6 snRNA | HBV | 83 | 40 | 40.2 ± 13.1 | 39.1 ± 13.4 | 98% | 93% | Plasma | 9 |
| Xu, 2011 | China | SYBR PCR | U6 snRNA | HBV | 48 | 89 | NA | NA | 80% | 96% | Serum | 6 |
| Cermelli, 2011 | Egypt | SYBR PCR | miR-238 | HCV | 18 | 19 | NA | NA | 94% | 83% | Serum | 7 |
| Meer, 2012 | Netherlands | Taqman PCR | NA | HCV | 102 | 25 | 48.65 ± 10.34 | 35.3 ± 11.5 | 95% | 92% | Serum | 5 |
| Zhang, 2012 | China | SYBR PCR | NA | HBV | 24 | 24 | 37.6 ± 9.0 | 35.6 ± 10.2 | 88% | 100% | Serum | 6 |
| Kumar, 2014 | India | Taqman PCR | U6 snRNA | HCV | 25 | 25 | 38.08 ± 10.81 | 32.53 ± 9.63 | 92% | 84% | Serum | 6 |
| Zhang, 2014 | China | SYBR PCR | U6 snRNA | HBV (active) | 112 | 22 | NA | NA | 86% | 63% | Plasma | 7 |
| China | SYBR PCR | U6 snRNA | HBV (indolent) | 19 | 22 | NA | NA | 84% | 37% | Plasma | ||
| Zhang, 2015 | China | SYBR PCR | U6 snRNA | HCV | 39 | 29 | 49.0 ± 14.3 | 45.0 ± 16.1 | 92% | 79% | Serum | 6 |
| Shaker, 2015 | Egypt | SYBR PCR | SNORD68 | HCV | 30 | 55 | 60.27 ± 8.2 | 55.88 ± 15.91 | 90% | 100% | Serum | 7 |
| Motawi, 2016 | Egypt | SYBR PCR | SNORD68 | HCV | 40 | 30 | 42.95 ± 11.21 | 49.9 ± 14.9 | 93% | 100% | Serum | 7 |
| Butt, 2016 | Egypt | SYBR PCR | U6 snRNA | HCV (abnormal ALT) | 80 | 60 | 32.7 ± 9.9 | 39.2 ± 12.9 | 87% | 97% | Serum | 8 |
| Egypt | SYBR PCR | U6 snRNA | HCV (normal ALT) | 43 | 60 | 32.7 ± 9.9 | 39.2 ± 12.9 | 65% | 93% | Serum | ||
| Demerdash, 2017 | Egypt | SYBR PCR | SNORD68 | HCV | 60 | 60 | 35.1 ± 6.7 | 33.9 ± 8.64 | 80% | 88% | Plasma | 7 |
| Wang, 2017 | China | Taqman PCR | cel-miR-39 | HBV (occult) | 119 | 117 | 42.30 ± 13.60 | 45.58 ± 13.08 | 79% | 55% | Serum | 9 |
| China | Taqman PCR | cel-miR-39 | HBV (active) | 115 | 117 | 42.30 ± 13.60 | 44.40 ± 13.10 | 86% | 83% | Serum | ||
| Amr, 2018 | Egypt | SYBR PCR | SNORD68 | HCV | 50 | 20 | 41.5 | 41.7 | 72% | 85% | Serum | 8 |
| Chen, 2018 | China | Taqman PCR | Hsa-miR-25-3p | HBV | 30 | 30 | 42.7 ± 10.3 | 37.6 ± 12.8 | 80% | 83% | Plasma | 8 |
NA, not available.
Figure 2. Overall methodological quality assessments of the included 15 articles based on QUADAS-2 tool.
Diagnostic accuracy of miR-122 in chronic viral hepatitis
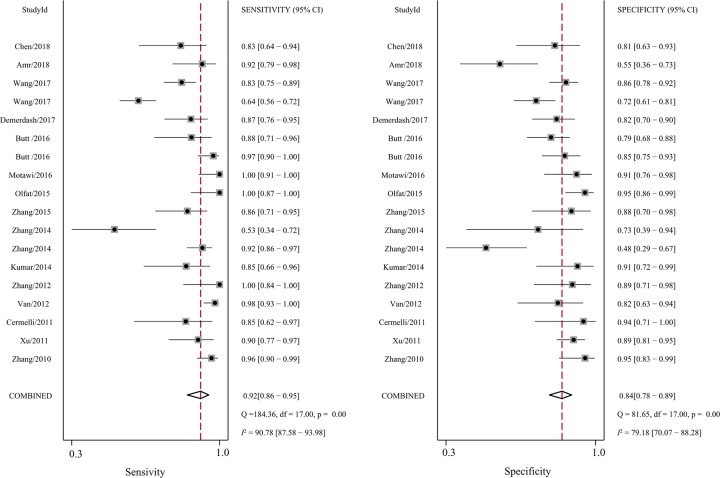
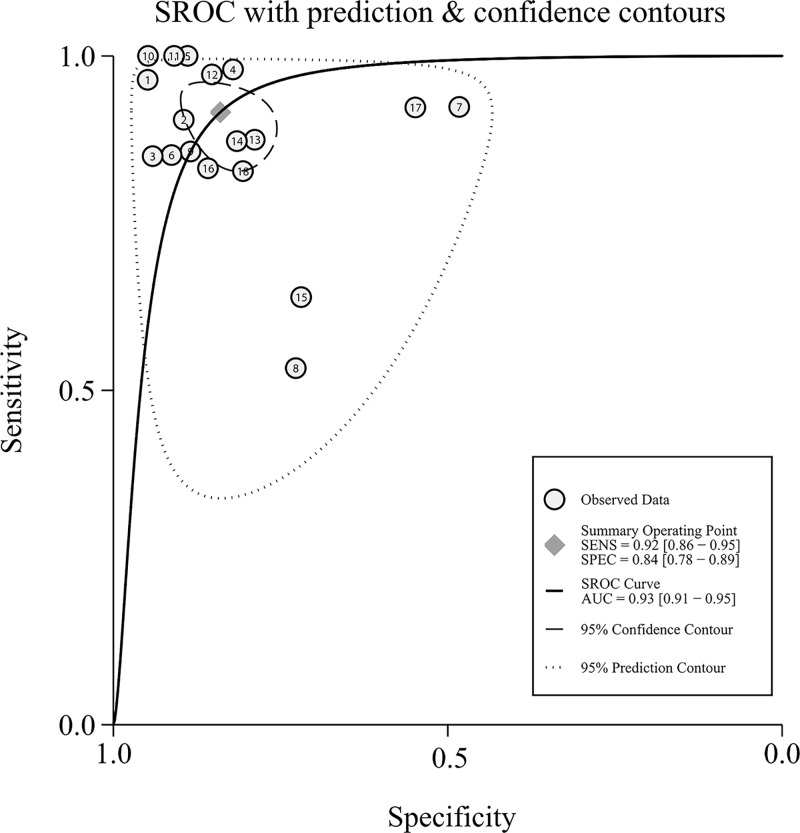
First, significant heterogeneity was found in our meta-analysis, as demonstrated by the results (I2 = 90.78% for sensitivity and I2 = 79.18% for specificity, respectively). Thus, the random-effect model was selected for the next calculation. Forest plots of the sensitivity and specificity results are shown in Figure 3. Overall, as shown in Table 2, the pooled sensitivity was 0.92 (95%CI: 0.86–0.95), specificity was 0.84 (95% CI: 0.78–0.89), PLR was 5.7 (95%CI: 4.7–8.1), NLR was 0.10 (95%CI: 0.06–0.18), DOR was 57 (95%CI: 25–129), and AUC was 0.93 (95%CI: 0.91–0.95). The SROC curve for the overall results is shown in Figure 4. The above results both suggest that miR-122 can be served as an adjuvant tool for the diagnosis of HBV- and/or HCV-associated chronic viral hepatitis.
Figure 3. Forest plots of summary sensitivities and specificity of circulating miR-122 in the diagnosis of HBV- and HCV-associated chronic viral hepatitis.
Table 2. Summary diagnostic accuracy of circulating miR-122 for HBV and/or HCV.
| Analysis | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) |
|---|---|---|---|---|---|---|
| Virus types | ||||||
| HBV | 0.87 (0.75–0.94) | 0.81 (0.75–0.87) | 4.7 (3.3–6.7) | 0.16 (0.07–0.33) | 30 (11–79) | 0.88 (0.85–0.91) |
| HCV | 0.94 (0.89–0.97) | 0.85 (0.78–0.90) | 6.6 (4.4–10.0) | 0.07 (0.04–0.14) | 89 (36–217) | 0.95 (0.93–0.97) |
| Sample types | ||||||
| Serum | 0.93 (0.86–0.97) | 0.86 (0.80–0.90) | 6.4 (4.5–9.2) | 0.08 (0.04–0.17) | 79 (30–207) | 0.94 (0.91–0.96) |
| Plasma | 0.87 (0.72–0.95) | 0.79 (0.61–0.90) | 4.1 (2.0–8.5) | 0.16 (0.07–0.40) | 25 (6–109) | 0.90 (0.87–0.92) |
| Ethnicity | ||||||
| Chinese | 0.87 (0.76–0.93) | 0.83 (0.73–0.89) | 5.1 (3.0–8.5) | 0.16 (0.08–0.31) | 32 (11–95) | 0.91 (0.88–0.93) |
| Non-Chinese | 0.95 (0.89–0.97) | 0.85 (0.77–0.91) | 6.6 (4.4–10.0) | 0.06 (0.03–0.13) | 100 (36–279) | 0.96 (0.94–0.97) |
| Overall | 0.92 (0.86–0.95) | 0.84 (0.78–0.89) | 5.7 (4.7–8.1) | 0.10 (0.06–0.18) | 57 (25–129) | 0.93 (0.91–0.95) |
| Outliers excluded | 0.93 (0.87–0.95) | 0.86 (0.82–0.89) | 6.6 (4.9–8.9) | 0.09 (0.05–0.19) | 66 (27–160) | 0.94 (0.90–0.96) |
Figure 4. Summary ROC curves for miR-122 in the diagnosis of HBV- and HCV-associated chronic viral hepatitis.
Threshold effect and subgroup analysis
Since threshold effect has been reported to be major cause of between-study heterogeneity and occurs when differences in sensitivities and specificities arise; this effect can be assessed with the spearman correlation coefficient [30]. And a value of −0.298 (P=0.229; P>0.05) indicated the absence of the threshold effect in our study. Afterwards, subgroup analyses based on ethnicity, sample types and virus types were also performed. The pooled sensitivity, specificity, PLR, NLR, DOR, and AUC for each subgroup are listed in Table 2. As for virus types, the HCV-associated chronic viral hepatitis group showed a higher accuracy than HBV-associated chronic viral hepatitis group with sensitivity of 0.94 versus 0.87, specificity of 0.85 versus 0.81, PLR of 6.6 versus 4.7, NLR of 0.07 versus 0.16, DOR of 89 versus 30 and AUC of 0.95 versus 0.88, respectively. Furthermore, a comparison of miR-122 expression profile in serum and plasma showed that the sensitivity (0.93 versus 0.87), specificity (0.86 versus 0.79), and AUC (0.94 versus 0.90) were higher in serum-based test than in plasma, providing additional evidence for the use of serum as a better matrix for diagnostic profiling of miR-122 in patients with HBV- and/or HCV-associated chronic viral hepatitis. Afterwards, the analysis based on ethnicity demonstrated the non-Chinese populations yield a better diagnosis accuracy than their Chinese counterparts. Specifically, for the non-Chinese population group, the pooled sensitivity, specificity, PLR, NLR, DOR, and AUC were 0.95, 0.85, 6.6, 0.06, 100, and 0.96, while the results for the Chinese population group were 0.97, 0.83, 5.1, 0.16, 32, and 0.91, respectively.
Sensitivity analysis and publication bias
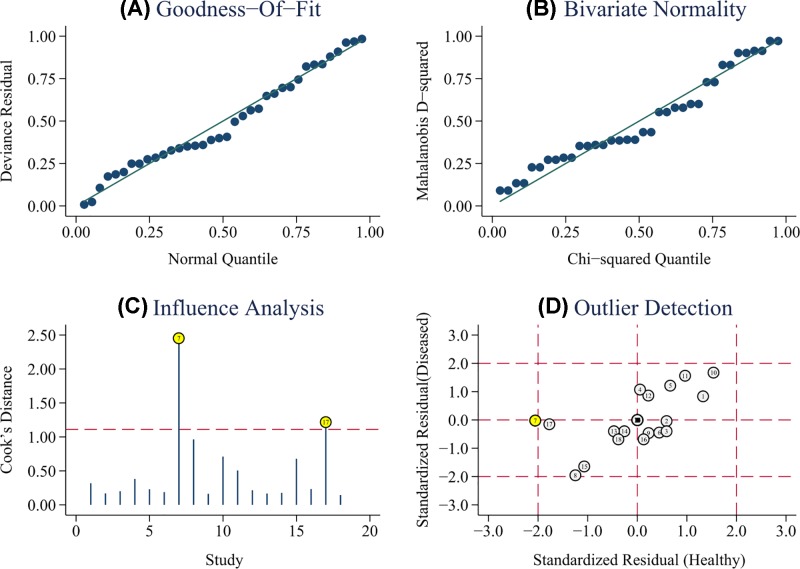
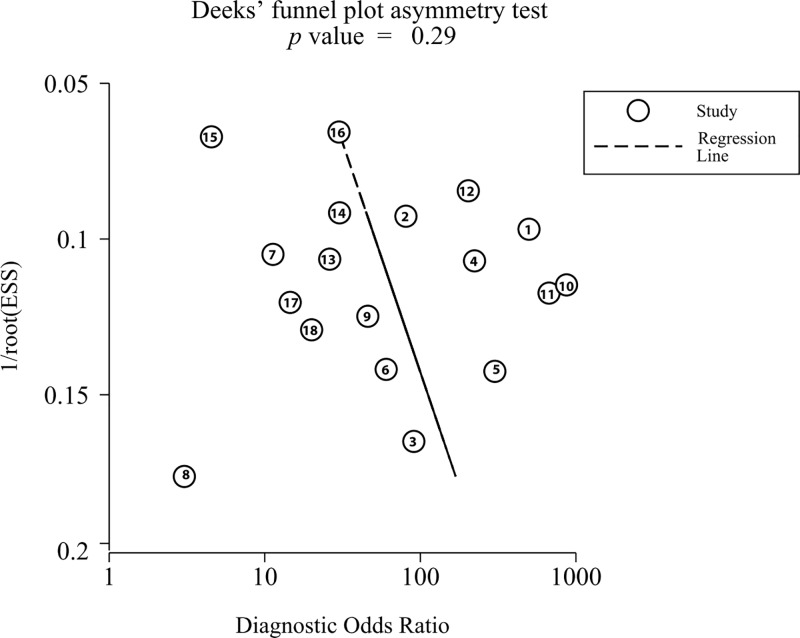
As shown in Figure 5A,B, the results of goodness of fit and bivariate normality analysis suggested that the random-effect model was suitable for subsequent calculation of the pooled estimates. Afterwards, influence analysis and outlier detection (Figure 5C,D) identified two outlier researches. After excluding these outliers, sensitivity increased from 0.92 to 0.93, specificity increased from 0.84 to 0.86, PLR increased from 5.7 to 6.6, NLR decreased from 0.10 to 0.09, DOR increased from 57 to 66, and AUC increased from 0.93 to 0.94. In addition, heterogeneity increased from 90.78% to 91.19% for sensitivity and from 79.18% to 62.02% for specificity. Furthermore, Deeks’ funnel plot asymmetry test was carried out to explore the potential publication bias of the included studies. As demonstrated in Figure 6, an obtained P value of 0.29 indicated the absence of publication bias in this meta-analysis.
Figure 5. Sensitivity analysis: graphical depiction of goodness of fit and bivariate normality analysis (A,B), influence and outlier detection analysis (C,D), respectively.
Figure 6. Deeks’ funnel plot asymmetry test for the assessment of potential publication bias.
Discussion
Though significant progress has been occurred in diagnostic techniques over the years, the accurate and convenient diagnosis of HBV- and/or HCV-associated chronic viral hepatitis remains a clinical challenge. Subsequently, the application of miRNAs, which can control gene expression in multiple biological processes including growth, cell proliferation, differentiation, apoptosis, and carcinogenesis through RNA interference (RNAi) [31,32], has gained much attention. As a part of miRNAs, miR-122 has already been demonstrated to be the most frequent miRNA in adult human liver accounting up to 70% of the total hepatic miRNAs, and a central player in liver development, differentiation, and homeostasis as well as in metabolic functions [33]. There is a great deal of researches into the use of miR-122 as a biomarker for HBV and HCV. In the aspect of HBV regulation, Chen et al. [34] have demonstrated that miR122 can bind to the highly conserved region of a bicistronic mRNA called HBV pregenomic RNA, which reported to encode the HBV polymerase and core protein, thereby ultimately leading to inhibition of HBV gene expression and replication. Furthermore, it has also been confirmed that increase in miR122 can attenuated the replication of HBV by regulating cyclin G(1) -modulated P53 activity [35]. As for the aspect of HCV regulation, liver-specific miR122 can stabilize HCV viral RNA through a process involving protecting highly conserved 5′ untranslated region the HCV genome from degradation by the host exonuclease, Xrn1 or from host innate immune responses [36–39]. A phase 2a study has shown that miravirsen, an antisense oligonucleotide, exhibited remarkable prolonged dose-dependent reductions in HCV RNA levels in patients with chronic HCV genotype 1 infection [40]. All the above researches both suggest that miR122 is a significant regulator of HBV and HCV replication by either directly affecting viral RNA or modulating host gene expression.
However, due to the different study designs and study subjects, some studies dispute its diagnostic efficacy [41]. Thus, the present study was carried out to summarize the results of individual studies. As the present results show, circulating miR-122 achieved a pooled sensitivity of 0.92, specificity of 0.84, and AUC of 0.93, indicating an overall moderate test performance for the diagnosis of HBV- and HCV-associated chronic viral hepatitis. Furthermore, the PLR value of 5.7 suggested that HBV- and HCV-associated chronic viral hepatitis patients had an almost six-fold higher chance of being miR-122 test positive than other individuals without the disease, and a NLR value of 0.1 implied that in a negative result from the miR-122 test, only 10% is likely to be false-negative. Obviously, the DOR value, which combines the strengths of both sensitivity and specificity, was 57 in our meta-analysis, indicating a high level of discriminating accuracy for clinical practice [42].
Furthermore, to explore the potential sources of heterogeneity, subgroup analysis based on virus types, sample types and ethnicity were subsequently performed. The results of subgroup analysis based on virus types suggest that miRNA-122 yielded an overall higher diagnostic accuracy in HCV-associated chronic viral hepatitis patients with a sensitivity of 0.94, specificity of 0.85, PLR of 6.6, NLR of 0.07, DOR of 89, and AUC of 0.95. Besides, with regard to sample types, miRNA expression profiles have been reported to be considerably affected by the coagulation process in the blood [43]. In our study, serum turned out to be a better matrix for diagnostic profiling of miR-122 in HBV- and HCV-associated chronic viral hepatitis than plasma: the pooled sensitivity was 0.93 versus 0.87, specificity was 0.86 versus 0.79, PLR was 6.4 versus 4.1, NLR was 0.08 versus 0.16, DOR was 79 versus 25, and AUC was 0.94 versus 0.90. However, as only four studies were included in the plasma specimen group, thus large-scale prospective researches are still needed to consolidate the results. In the subgroup analysis based on ethnicity, we found compared with Chinese populations, miR-122 assay may be more accurate in non-Chinese populations, with the DOR value hiked from 32 to 100, and AUC increased from 0.91 to 0.96.
Admittedly, heterogeneity still exists when interpreting the results of any meta-analysis. In our study, heterogeneity does not come from the threshold effect. However, in the sensitivity analysis, after excluding the outlier, the overall pooled sensitivity, sensitivity, PLR, DOR, and AUC all increased while NLR decreased suggesting that the outlier is probably a source of heterogeneity. In addition, as the P value of I2 for the overall study altered inconspicuously, substantial heterogeneity from non-threshold effect still exists among studies to some extent. Although we have made every effort to avoid bias during the process of our study, there were still some limitations in this meta-analysis. In the first place, several valuable studies may be missed in spite of the comprehensive search strategy during our literature search. What’s more, diagnostic performance maybe affected as the majority of healthy people were randomly selected as controls and were not blind designed.
Conclusions
Taken together, for the first time, our meta-analysis focuses on the diagnostic performance of circulating miRNA-122 in HBV- and/or HCV-associated chronic viral hepatitis detection, and it is concluded that circulating miRNA-122 has a relatively high diagnostic value for chronic viral hepatitis detection, especially in patients of HCV-associated chronic viral hepatitis. However, in the future, well-designed, large-scale and accurate researches are still needed to consolidate the results of this meta-analysis in clinical practice. The ultimate purpose is to combine clinical variables with those available in public databases to open avenues for prospective trials of a non-invasive diagnostic test for enhancing the early detection of patients with chronic viral hepatitis.
Abbreviations
- AUC
area under the curve
- CI
confidence interval
- DOR
diagnostic odds ratio
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- NLR
negative likelihood ratio
- PLR
positive likelihood ratio
- QUADAS
Quality Assessment of Diagnostic Accuracy Studies
- SROC
summary receiver operating characteristic
Author Contribution
Study design: Xinhao Zhou, Shiqiang Fang and Mian Wang. Literature search and data collection: Mian Wang, Ali Xiong and Chao Zheng. Statistical analysis: Xinhao Zhou and Shiqiang Fang. Manuscript preparation and revision: Xinhao Zhou, Jiulong Wang, Changqing Yin. All authors agreed to be accountable for all aspects of the work and approved of the final version.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 31500729].
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Weinbaum C.M., Mast E.E. and Ward J.W. (2009) Recommendations for identification and public health management of persons with chronic hepatitis B virus infection. Hepatology 49, S35–S44 10.1002/hep.22882 [DOI] [PubMed] [Google Scholar]
- 2.Chou R., Dana T., Bougatsos C., Blazina I., Zakher B. and Khangura J. (2014), Screening for Hepatitis B Virus Infection in Nonpregnant Adolescents and Adults: Systematic Review to Update the 2004 U.S. Preventive Services Task Force Recommendation, Agency for Healthcare Research and Quality (US), Rockville (MD) [PubMed] [Google Scholar]
- 3.Eguchi A., Wree A. and Feldstein A.E. (2014) Biomarkers of liver cell death. J. Hepatol. 60, 1063–1074 10.1016/j.jhep.2013.12.026 [DOI] [PubMed] [Google Scholar]
- 4.Gougelet A. and Colnot S. (2016) Hepatocellular carcinoma diagnosis: circulating microRNAs emerge as robust biomarkers. Clin. Res. Hepatol. Gastroenterol. 40, 367–369 10.1016/j.clinre.2015.12.010 [DOI] [PubMed] [Google Scholar]
- 5.Musaddaq G., Shahzad N., Ashraf M.A. and Arshad M.I. (2018) Circulating liver-specific microRNAs as noninvasive diagnostic biomarkers of hepatic diseases in human. Biomarker 24, 103–109 [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A. and Slack F.J. (2006) Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 7.Hunter M.P., Ismail N., Zhang X., Aguda B.D., Lee E.J., Yu L.. et al. (2008) Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 3, e3694 10.1371/journal.pone.0003694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L.. et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 105, 10513–10518 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferracin M., Veronese A. and Negrini M. (2010) Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 10, 297–308 10.1586/erm.10.11 [DOI] [PubMed] [Google Scholar]
- 10.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F.. et al. (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 108, 5003–5008 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koberle V., Waidmann O., Kronenberger B., Andrei A., Susser S., Fuller C.. et al. (2013) Serum microRNA-122 kinetics in patients with chronic hepatitis C virus infection during antiviral therapy. J. Viral Hepat. 20, 530–535 10.1111/jvh.12075 [DOI] [PubMed] [Google Scholar]
- 12.Dubin P.H., Yuan H., Devine R.K., Hynan L.S., Jain M.K., Lee W.M.. et al. (2014) Micro-RNA-122 levels in acute liver failure and chronic hepatitis C. J. Med. Virol. 86, 1507–1514 10.1002/jmv.23987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X., Zhang Z., Dai F., Shi B., Chen L., Zhang X.. et al. (2014) Comparison of circulating, hepatocyte specific messenger RNA and microRNA as biomarkers for chronic hepatitis B and C. PLoS One 9, e92112 10.1371/journal.pone.0092112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deeks J.J. (2001) Systematic reviews in health care: systematic reviews of evaluations of diagnostic and screening tests. BMJ 323, 157–162 10.1136/bmj.323.7305.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moses L.E., Shapiro D. and Littenberg B. (1993) Combining independent studies of a diagnostic test into a summary roc curve: data-analytic approaches and some additional considerations. Stat. Med. 12, 1293–1316 10.1002/sim.4780121403 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Jia Y., Zheng R., Guo Y., Wang Y., Guo H.. et al. (2010) Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 56, 1830–1838 10.1373/clinchem.2010.147850 [DOI] [PubMed] [Google Scholar]
- 17.Xu J., Wu C., Che X., Wang L., Yu D., Zhang T.. et al. (2011) Circulating microRNAs, mir-21, mir-122, and mir-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 50, 136–142 10.1002/mc.20712 [DOI] [PubMed] [Google Scholar]
- 18.Cermelli S., Ruggieri A., Marrero J.A., Ioannou G.N. and Beretta L. (2011) Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One 6, e23937 10.1371/journal.pone.0023937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Meer A.J., Farid W.R., Sonneveld M.J., de Ruiter P.E., Boonstra A., van Vuuren A.J.. et al. (2013) Sensitive detection of hepatocellular injury in chronic hepatitis C patients with circulating hepatocyte-derived microRNA-122. J. Viral Hepat. 20, 158–166 10.1111/jvh.12001 [DOI] [PubMed] [Google Scholar]
- 20.Zhang H., Li Q.Y., Guo Z.Z., Guan Y., Du J., Lu Y.Y.. et al. (2012) Serum levels of microRNAs can specifically predict liver injury of chronic hepatitis B. World J. Gastroenterol. 18, 5188–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S., Chawla Y.K., Ghosh S. and Chakraborti A. (2014) Severity of hepatitis C virus (genotype-3) infection positively correlates with circulating microRNA-122 in patients sera. Dis. Markers 2014, 435476 10.1155/2014/435476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S., Ouyang X., Jiang X., Gu D., Lin Y., Kong S.K.. et al. (2015) Dysregulated serum microRNA expression profile and potential biomarkers in hepatitis C virus-infected patients. Int. J. Med. Sci. 12, 590–598 10.7150/ijms.11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaker O.G., Elshehaby A.R., Zahra A.A., Abdelaleem O.O. and Yousry A. (2015) Dysregulated serum microRNA expression profile and potential biomarkers in hepatitis C virus-infected patients. Med. J. Cairo Univ. 80, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motawi T.M., Sadik N.A., Shaker O.G. and Ghaleb M.H. (2016) Elevated serum microRNA-122/222 levels are potential diagnostic biomarkers in egyptian patients with chronic hepatitis C but not hepatic cancer. Tumour Biol. 37, 9865–9874 10.1007/s13277-016-4884-6 [DOI] [PubMed] [Google Scholar]
- 25.Butt A.M., Raja A.J., Siddique S., Khan J.S., Shahid M., Tayyab G.U.. et al. (2016) Parallel expression profiling of hepatic and serum microRNA-122 associated with clinical features and treatment responses in chronic hepatitis C patients. Sci. Rep. 6, 21510 10.1038/srep21510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demerdash H.M., Hussien H.M., Hassouna E. and Arida E.A. (2017) Detection of microRNA in hepatic cirrhosis and hepatocellular carcinoma in hepatitis C genotype-4 in Egyptian patients. BioMed. Res. Int. 2017, 1806069 10.1155/2017/1806069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Zhu P., Qiu J., Wang J., Zhu H., Zhu Y.. et al. (2017) Identification and characterization of interferon signaling-related microRNAs in occult hepatitis B virus infection. Clin. Epigenetics 9, 101 10.1186/s13148-017-0404-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amr K.S., Ezzat W.M., Elbatae H., Atia H., Reda E. and Elhosary Y. (2018) Assessment of serum level of miRNAs before and after treatment with sofosbuvir in Egyptian patients with chronic HCV infection. Meta Gene 18, 93–99 10.1016/j.mgene.2018.08.005 [DOI] [Google Scholar]
- 29.Cheng J.L., Zhao H., Yang S.G., Chen E.M., Chen W.Q. and Li L.J. (2018) Plasma miRNA-122-5p and miRNA-151a-3p identified as potential biomarkers for liver injury among CHB patients with PNALT. Hepatol. Int. 12, 277–287 10.1007/s12072-018-9871-0 [DOI] [PubMed] [Google Scholar]
- 30.Zamora J., Abraira V., Muriel A., Khan K. and Coomarasamy A. (2006) Meta-disc: a software for meta-analysis of test accuracy data. BMC Med. Res. Method 6, 31 10.1186/1471-2288-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond S.M., Bernstein E., Beach D. and Hannon G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296 10.1038/35005107 [DOI] [PubMed] [Google Scholar]
- 32.Martinez J., Patkaniowska A., Urlaub H., Luhrmann R. and Tuschl T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574 10.1016/S0092-8674(02)00908-X [DOI] [PubMed] [Google Scholar]
- 33.Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A.. et al. (2004) Mir-122, a mammalian liver-specific microRNA, is processed from HCR mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1, 106–113 10.4161/rna.1.2.1066 [DOI] [PubMed] [Google Scholar]
- 34.Chen Y., Shen A., Rider P.J., Yu Y., Wu K., Mu Y.. et al. (2011) A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 25, 4511–4521 10.1096/fj.11-187781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S., Qiu L., Yan X., Jin W., Wang Y., Chen L.. et al. (2012) Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin g(1)-modulated p53 activity. Hepatology 55, 730–741 10.1002/hep.24809 [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Masaki T., Yamane D., McGivern D.R. and Lemon S.M. (2013) Competing and noncompeting activities of mir-122 and the 5′ exonuclease XRN1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 110, 1881–1886 10.1073/pnas.1213515110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M. and Sarnow P. (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309, 1577–1581 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 38.Machlin E.S., Sarnow P. and Sagan S.M. (2011) Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. PNAS 108, 3193–3198 10.1073/pnas.1012464108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimakami T., Yamane D., Jangra R.K., Kempf B.J., Spaniel C., Barton D.J.. et al. (2012) Stabilization of hepatitis C virus RNA by an Ago2-mir-122 complex. Proc. Natl. Acad. Sci. USA 109, 941–946 10.1073/pnas.1112263109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K.. et al. (2013) Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- 41.Morita K., Taketomi A., Shirabe K., Umeda K., Kayashima H., Ninomiya M.. et al. (2011) Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 31, 474–484 10.1111/j.1478-3231.2010.02433.x [DOI] [PubMed] [Google Scholar]
- 42.Glas A.S., Lijmer J.G., Prins M.H., Bonsel G.J. and Bossuyt P.M. (2003) The diagnostic odds ratio: a single indicator of test performance. J. Clin. Epidemiol. 56, 1129–1135 10.1016/S0895-4356(03)00177-X [DOI] [PubMed] [Google Scholar]
- 43.Wang K., Yuan Y., Cho J.H., McClarty S., Baxter D. and Galas D.J. (2012) Comparing the microRNA spectrum between serum and plasma. PLoS One 7, e41561 10.1371/journal.pone.0041561 [DOI] [PMC free article] [PubMed] [Google Scholar]