Abstract
Probiotics are live microorganisms which when consumed in large number together with a food promote the health of the consumer. The aim of this study was to evaluate in vitro probiotic properties of lactic acid bacteria (LAB) isolated from traditional Ethiopian fermented Teff injera dough, Ergo, and Kocho products. A total of 90 LAB were isolated, of which 4 (4.44%) isolates showed 45.35–97.11% and 38.40–90.49% survival rates at pH values (2, 2.5, and 3) for 3 and 6 h, in that order. The four acid-tolerant isolates were found tolerant to 0.3% bile salt for 24 h with 91.37 to 97.22% rate of survival. The acid-and-bile salt-tolerant LAB isolates were found inhibiting some food-borne test pathogenic bacteria to varying degrees. All acid-and-bile-tolerant isolates displayed varying sensitivity to different antibiotics. The in vitro adherence to stainless steel plates of the 4 screened probiotic LAB isolates were ranged from 32.75 to 36.30% adhesion rate. The four efficient probiotic LAB isolates that belonged to Lactobacillus species were identified to the strain level using 16S rDNA gene sequence comparisons and, namely, were Lactobacillus plantarum strain CIP 103151, Lactobacillus paracasei subsp. tolerans strain NBRC 15906, Lactobacillus paracasei strain NBRC 15889, and Lactobacillus plantarum strain JCM 1149. The four Lactobacillus strains were found to be potentially useful to produce probiotic products.
1. Introduction
Worldwide, a variety of fermented food products are produced, which contribute significantly to the diets of many people [1]. Fermented food products are used to describe a special class of the food products characterized by various kinds of carbohydrate breakdowns in the presence of probiotic microorganisms, but seldom is carbohydrate the only constituent acted upon [2]. Fermented food and beverage products have emerged as not only the source of nutrition but also as functional and probiotic foods, which besides nutritional value have health effects or provide protection against food-borne diseases.
The problem of food-borne diseases (FBD) is multifactorial, and their prevention and control require multidisciplinary approaches that involve human beneficial live microbes (probiotics) in order to combat these pathogens and their associated health risks [3]. Several in vitro studies indicate that the growth of food-borne pathogenic microbes is inhibited by probiotic lactic acid bacteria [4–6]. The consumption of a large number of probiotic live microorganisms together with a food fundamentally promotes the health of the consumers [7]. Lactic acid bacteria (LAB) are a diverse group of microorganisms consisting of Gram-positive, aerotolerant, acid-tolerant, usually nonsporulating and nonrespiring rod or cocci microorganisms, and play an important role in the process of fermentation of food by inhibiting spoilage/pathogenic bacteria and by producing excellent flavor, aroma, and texture of fermented foods [8, 9]. Some LAB are probiotics, and others may be potential probiotics or just fermentation cultures that are widely distributed in nature and can be used in the food industry [10].
LAB could be isolated from many kinds of sources such as milk products, fermented foods, animal intestines or freshwater fishes, soil samples, sugar cane plants, and poultry farms [11]. The most common types of probiotic LAB include different Lactobacillus spp. (Lb. acidophilus, Lb. johnsonii, Lb. casei, Lb. rhamnosus, Lb. gasseri, and Lb. reuteri) and genus Bifidobacteria (Bf. bifidum, Bf. animalis subsp. lactis, Bf. longum subsp. longum, and Bf. longum subsp. infantis) [12, 13]. LAB are also useful in the treatment of various diseases caused by drug-resistant pathogenic microbes [14]. Probiotic microbes may provide nutrients, enhance growth, produce enzymes, inhibit pathogens, and enhance immune responses [15].
In Ethiopia, traditionally fermented food products are prepared at the household level and the consumption of these fermented food products is commonly practiced. Therefore, some studies on isolation and screening of antibacterial producing lactic acid bacteria from traditionally fermented foods were undertaken by many workers [16, 17]. Likewise, Tesfaye et al. [4] have revealed the antagonistic effect of lactic acid bacterial strains either as pure or defined mixed cultures against some food-borne pathogens during fermentation and storage of fermented milk. However, there are still few research data available on the characterization of probiotic LAB. Most of the traditionally fermented products of Ethiopia are consumed without further heat processing which can be considered as ideal vehicles to carry probiotic bacteria into the human gastrointestinal tract.
Probiotic strains isolated from traditionally fermented foods and drinks could have application as a starter culture for large-scale production of the traditional product and have a desirable functional property for their application as probiotics against food-borne pathogens. Therefore, this study attempts to evaluate the in vitro probiotic properties of LAB isolated from three traditionally fermented Ethiopian food products such as Teff dough, Ergo, and Kocho with respect to their potential probiotic properties against some food-borne pathogens.
2. Materials and Methods
2.1. Sample Collection
Traditionally fermented food products (Teff dough, Kocho, and Ergo) were obtained from Addis Ababa and its surroundings, Ethiopia. Each sample (200 g) was aseptically collected by using sterilized containers. The samples were brought to the laboratory with an ice box and stored in a refrigerator at +4°C until further analysis was carried out. Ergo is a locally fermented milk product. Teff dough is made by fermenting teff (Eragrostis tef) flour which is used to prepare a thin pancake-like product with many eyes known as injera. Kocho is a product which is prepared from decorticated and pounded pulp of enset plant (Ensete ventricosum), which is further mixed and kneaded into a mash and fermented in a pit.
2.2. Isolation and Purification of LAB from Traditional Fermented Foods
For isolation of LAB, 25 ml or 25 g of each sample of traditionally fermented foods (Teff dough, Kocho and Ergo) was mixed with 225 ml of separate sterile peptone water (0.1% W/V). Then, a sequential decimal dilution of the homogenate was obtained. From the appropriate dilutions, 0.1 ml aliquots were spread plated on duplicate predried surfaces of MRS (de Man, Rogosa, and Sharp) agar (Oxoid, Basingstok, Hampshire, England) plates. The inoculated plates were incubated under anaerobic condition using an anaerobic jar (BBL, Gas Pak Anaerobic Systems) at 37°C for 48 hours. Then, 10–20 distinct colonies were randomly picked from countable MRS plates for further purification. The isolated colonies of LAB were transferred into about 5 ml MRS broth (Oxoid) and purified by repeated streaking on MRS agar. Pure cultures of LAB were then streaked onto slants of MRS agar and stored at +4°C for further characterization [18].
2.3. Confirmation Tests of LAB Isolates
2.3.1. KOH Test
The KOH test was used to determine the gram reaction of LAB isolates. LAB cultures were grown on MRS agar at 37°C for 24 h under anaerobic conditions. A drop of 3% aqueous KOH was placed on a clean slide. Using a sterile loop, visible cells from fresh cultures were transferred to the drop of 3% KOH. The cells and KOH were mixed thoroughly on the slide and stirred constantly over an area about 1-2 cm2. The isolates, which did not give a viscid product, were selected since lactic acid bacteria (LAB) are known as Gram-positive cells [19].
2.3.2. Catalase Test
Overnight cultures of isolates were grown on MRS agar at +37°C for 24 h under anaerobic conditions. The catalase test was conducted by dripping two drops of hydrogen peroxide (3%) on 24 h-old cultures on a glass slide. The catalase test showed positive reaction characterized by the formation of oxygen bubbles that indicate the production of catalase enzyme by the test bacterium. Therefore, the isolates, which did not give gas bubbles, were selected for subsequent activities.
2.3.3. Spore Staining
Gram-positive and catalase-negative isolates were grown on MRS agar at +37°C for 24 h under anaerobic conditions. The spore-staining procedure was applied [20]. After the spore-staining technique, the endospore formulation was examined under light microscopy using oil immersion objectives. The isolates which did not form endospores were selected for further analysis.
2.4. In Vitro Characterization of Probiotic Properties
The common methods for in vitro analysis of probiotic properties include tolerance to low pH, tolerance against bile salt, antibiotic susceptibility, antimicrobial activity, and bacterial adherence to stain steel plates.
2.4.1. Tolerance to Low pH
The isolates were grown separately overnight in 5 ml MRS broth at +37°C under anaerobic conditions. A volume of 1 ml of log 7 CFU/ml of each overnight-grown culture was inoculated into 10 ml of MRS broth to give an initial inoculum level of log 6 CFU/ml. The culture was then centrifuged at 5000 rpm for 10 min at +4°C. The pellets were washed twice in phosphate buffer (pH 7.2). The pellets were resuspended in 5 ml sterile MRS broth which was adjusted to pH values of 2.0, 2.5, and 3.0 using 1 N·HCl to simulate the gastric environment. The test tubes were incubated for 3 and 6 hours at 37°C. After an appropriate incubation period, 1 ml of the culture was diluted in sterile 9 ml phosphate buffer (Sigma, St. Louis, MO USA) prepared according to the manufacturer's instruction (0.1 M, pH 6.2) in order to neutralize the medium acidity. Briefly, a 100 μl aliquot of the culture and its 10-fold serial dilutions were plated on the MRS agar medium. The inoculated plates were incubated at 37°C for 24 to 48 h under anaerobic condition using an anaerobic jar (BBL, Gas Pack System). The grown LAB colonies were expressed as colony-forming units per milliliter (CFU/ml). A positive control consisting of regular MRS broth inoculated with the culture was used [21]. The survival rate was calculated as the percentage of LAB colonies grown on MRS agar compared to the initial bacterial concentration:
| (1) |
where N1 is the viable count of isolates after incubation and N0 is the initial viable count.
2.4.2. Tolerance to Bile Salts
To estimate bile tolerance of acid-tolerant LAB (those only were grown in pH 2.0, 2.5, and/or 3.0), the isolates were separately grown overnight in MRS broth at 37°C under anaerobic conditions [21]. Each culture with the initial concentration of 106 CFU/ml was then centrifuged at 5000 rpm for 10 min at 4°C. The pellets were washed twice in the phosphate-saline buffer (PBS at pH 7.2). Cell pellets were resuspended in sterile MRS broth supplemented with 0.3% (w/v) bile salt (Oxgall, USA). Samples were taken at 24 h from the onset of incubation to determine the survivability of cells as described previously [21]. A positive control consisting of plain MRS broth without bile salts inoculated with each separate culture was simultaneously set up. After appropriate incubation, 1 ml of each separate culture was diluted separately in sterile 9 ml phosphate buffer (Sigma, St. Louis, USA) prepared according to the manufacturer's instruction (0.1 M, pH 6.2) in order to neutralize the medium. Concisely, a 100 μl aliquot of the culture and its 10-fold serial dilutions were plated on MRS agar medium. Plates were incubated at 37°C for 24 to 48 h under anaerobic condition using an anaerobic jar (BBL, Gas Pack System). LAB counts were expressed in colony-forming units per milliliter (CFU/ml). The survival rate was calculated as the percentage of LAB colonies grown on MRS agar compared to the initial bacterial concentration:
| (2) |
where N1 is the viable count of isolates after incubation and N0 is the initial viable count.
2.4.3. Antimicrobial Activity
Antibacterial activity of the acid-bile-tolerant LAB strains against some food-borne pathogens was determined using the agar-well diffusion method with some modifications of the protocol indicated by Fontana et al. [22]. The test organisms (Staphylococcus aureus ATCC 25923, Listeria monocytogenes (clinical isolate), Salmonella enterica Typhimurium, and Escherichia coli ATCC 25922) were obtained from the Ethiopian Public Health Institute (EPHI), Addis Ababa, Ethiopia.
The selected acid-bile-tolerant LAB isolates were inoculated from slants to fresh MRS broth containing 1% glucose and incubated overnight at 37°C. The overnight active culture broth of each isolate was centrifuged separately at 5000 rpm for 10 min at 4°C. The cell-free supernatant from each separate culture was collected as a crude extract for the antagonistic study against selected food-borne pathogens. The pure cultures of food-borne pathogens were inoculated from slants to brain heart infusion broth. After 24-hour incubation at 37°C, a volume of 100 μl of inoculum of each indicator bacteria was swabbed evenly over the surface of nutrient agar plates with a sterile cotton swab. The plates were allowed to dry, and a sterile cork borer (diameter 5 mm) was used to cut uniform wells in the agar. Each well was filled with 100 μl culture-free filtrate obtained from each of the acid-bile-tolerant LAB isolates. After incubation at 37°C for 24 to 48 hours, the plates were observed for a zone of inhibition (ZOI) around the well. The diameter of the inhibition zone was measured by calipers in millimeters, and a clear zone of 1 mm or more was considered positive inhibition [23, 24]. The experiment was carried out in triplicates, and the activity was reported as the diameter of ZOI ± SD.
2.4.4. Antibiotic Susceptibility Tests
Each of acid-bile-tolerant and antagonistic lactic acid bacteria isolates was assessed for its antibiotic resistance by the disc diffusion method as described by Zhang et al. [25] against some antibiotics that included ampicillin (10 μg/ml), erythromycin (15 μg/ml), streptomycin (10 μg/ml), kanamycin (25 μg/ml), and tetracycline (30 μg/ml). Thus, a volume of 100 μl of actively growing cultures of each acid-bile-tolerant and antagonistic lactic acid bacteria was swabbed evenly over the surface of nutrient agar plates with a sterile cotton swab. After drying, the antibiotic discs were placed on the solidified agar surface, and the plates were left aside for 30 min at 4°C for the diffusion of antibiotics and then anaerobically incubated at 37°C for 24 to 48 h. Resistance was defined according to the disc diffusion method by using the above antibiotic discs, and the diameters of inhibition zones were measured using calipers [26]; the zone of inhibition (diameter in mm) for each antibiotic was measured and expressed as susceptible, S (≥21 mm); intermediate, I (16–20 mm), and resistance, R (≤15 mm).
2.4.5. Bacterial Adhesion to Stainless Steel Plates
The adherence assay of acid-bile-tolerant, antagonistic, and antibiotic-sensitive lactic acid bacterial isolates was determined on stainless steel plates with some modifications of the protocol given by El-Jeni et al. [27]. Briefly, LAB were cultured in sterile MRS broth. Thereafter, the overnight bacterial culture (500 μl) was deposited in a test tube, which was then filled with 450 μl of MRS broth, wherein the sterile stainless steel plate was deposited, and the test tubes were then incubated for 24 h at 37°C. The stainless steel plate was removed under aseptic conditions, washed with 10 ml of sterile 1% peptone water, and left for 5 min in a sterile 1% peptone water tube. The plate was then washed again in the same conditions and vortexed for 3 min in a sterile 1% peptone water tube (6 ml) consecutively to detach the bacterial cells adhering to the steel plate surface. The cell number was determined by counting on MRS agar after 24 h of incubation at 37°C. Simultaneously, the total initial cell numbers were estimated to calculate the percentage of adhered bacterial cells for each LAB.
2.5. Morphological, Biochemical, and Physiological Tests
The probiotic LAB isolates were identified according to their morphological, physiological, and biochemical characteristics based on Bergey's Manual [28].
2.5.1. Cell Morphology
Overnight cultures were wet mounted on microscopic slides and examined under a light microscope using oil immersion objectives. Cellular morphological criteria considered during the examination were cell shape and cell arrangements.
2.5.2. Growth at Different Temperatures
Each of the overnight LAB cultures of 50 μl was transferred into four separate tubes that contained 5 ml medium (modified MRS broth) containing bromecresol purple indicator at a concentration of 0.12 g/l. After inoculation, two of the inoculated test tubes were incubated for 7 days either at 15°C and the other two test tubes at 45°C. During this incubation time, growth at any temperature was observed by the change of the growth medium (cultures) from purple to yellow.
2.5.3. Growth at Different NaCl Concentrations
LAB isolates were tested for their tolerance to different NaCl concentrations. For this purpose, 4% and 6.5% NaCl concentrations were used for testing. Similarly, test tubes with 5 ml of modified MRS broth containing bromecresol purple indicator were prepared according to the appropriate concentrations. Four test tubes with 4% NaCl and the other four test tubes with 6.5% NaCl were inoculated separately with 50 μl of 1% of each overnight culture of LAB and incubated at +37°C for 7 days. The change of the color from purple to yellow was considered as proof of cell growth.
2.5.4. Arginine Hydrolysis Test
Arginine containing MRS medium and Nessler's reagent (HgI4K2), a 0.09 mol/L solution of potassium tetraiodomercurate (II) (K2 [HgI4]) in 2.5 mol/L potassium hydroxide, were used in order to test ammonia production from arginine. MRS containing 0.3% L-arginine hydrochloride was transferred into duplicate tubes with 5 ml amount and inoculated with each of 1% overnight cultures of LAB. Thereafter, tubes were incubated at +37°C for 24 h. A 100 µl of cultures were transferred onto a white background. Thereafter, the same amount of Nessler's reagent was pipetted to the cultures. The change in color to bright orange indicates a positive reaction, while the yellow color indicates the negative reaction. A negative control, which does not contain arginine, was used as a negative control.
2.5.5. Gas Production from Glucose
In order to determine the homofermentative and heterofermentative characteristics of LAB isolates, CO2 production from glucose was determined in modified MRS broth containing inverted Durham tubes with 1% glucose. MRS broth (8 ml) in separate tubes containing 1% glucose with inverted Durham tubes was prepared and inoculated separately with 50 µl of 1% overnight fresh LAB culture. Then the test tubes were incubated at +37°C for 5 days. The presence of gas in Durham tubes during 5 days of observation indicates CO2 production from glucose.
2.6. Identification of Probiotic LAB Isolates Using 16S rRNA Gene Sequencing
2.6.1. Genomic DNA Extraction
Genomic DNA was extracted from pure cultures (n = 4) of potential probiotic LAB. Separately, 1 ml of each pure liquid culture was centrifuged for 3 min at 10000 rpm. The supernatant was removed, and the cells were resuspended in 300 µl buffer (10 mM Tris-HCl, pH 8.0; 50 mM glucose, and 10 mM EDTA). To the suspension, 3 µl lysozyme (10 mg/ml) was added, and cells were lysed at 37°C for 60 min under occasional stirring of the tube content by overturning it. Lysing buffer (20 mM Tris-HCl, pH 8.0; 75 mM NaCl; 1% SDS; 10 mM EDTA) of 300 µl and 3 µl RNAse (10 mg/ml) was added to the mixture. The mixture was incubated at 37°C for 30 min and then cooled on ice for 1 min. Then, 100 µl solution of ammonium acetate (7.5 M) was added to the mixture, mixed on a vortex for 20 seconds and was centrifuged at 13000 rpm for 5 min. The supernatant was transferred into clean 1.5 ml tubes, and 300 μl isopropanol was added. Thereafter, the mixture was mixed by overturning for 1 min and stored at −20°C for 30 min. The mixture was centrifuged at 13000 rpm for 5 min. The supernatant was accurately decanted, and the tubes were placed overturned on a clean filter. Then, 400 µl of 70% ethanol was added and mixed several times by overturning to wash the DNA sediment. Finally, the sediment was dried at 37°C for 15 min till ethanol drops disappeared completely. The dried sediment was dissolved in 30 µl TE buffer.
2.6.2. PCR Amplification of 16S rDNA
For the amplification of the 16S rDNA gene, the specific primers AMP_F 5′- GAG AGT TTG ATY CTG GCT CAG -3′ and AMP_R 5′- AAG GAG GTG ATC CAR CCG CA -3′ were used. PCR reaction mixture was prepared by mixing 25 µl of the Tag 2x Master mix (buffer, polymerase and dNTPs), forward primer 1 µl, reverse primer 1 µl, and UPH2O 22 µl. Then, 49 µl of the mixtures was added to a sterile PCR tube, and 1 µl of the gDNA was used as a template; the amplification reactions were carried out in a thermal cycler (Bio-Rad Mycycler).
2.6.3. DNA Electrophoresis
PCR products were separated in a 1% agarose gel and stained with ethidium bromide followed by examination on a UV illuminator, and images were captured by a digital camera.
2.6.4. Sequencing of the PCR Products
A 16S rRNA PCR amplification and sequencing was performed by Eurofins, Novogene (Hong Kong). The V4 hypervariable region of the 16S rRNA was amplified using specific primers 515F and 806R. All the PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). The libraries generated with TruSeq DNA PCR-Free Sample Preparation Kit were sequenced using paired-end Illumina sequencing (2 × 250 bp) on the HiSeq2500 platform (Illumina, USA).
2.6.5. Phylogenetic Analysis
Forward and reverse sequences were assembled and edited using BioEdit Sequence Alignment Editor Version 5.0.9. Sequence similarity was estimated by searching the homology in the Genbank DNA database using BLAST. Finally, the isolate was then identified based upon the sequence.
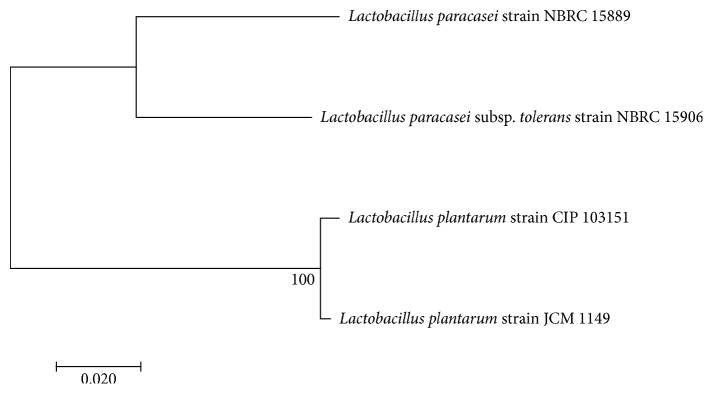
The evolutionary history was inferred using the neighbor-joining method [29]. The optimal tree with the sum of branch length = 0.19972900 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 891 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.
2.6.6. Statistical Analysis
All the measurements were performed in triplicate, and the results were expressed as mean standard deviation (SD). Data were analyzed by the one-way ANOVA plus post hoc Duncan's test by SAS software (ver. 9.2, Raleigh, NC). The phylogenetic tree was prepared using MEGA7 (version 7.0). Statistical significance was determined at p < 0.05.
3. Results
3.1. Isolation of Potentially Probiotic Lactic Acid Bacteria from Traditional Fermented Foods
A total of 90 (30 from each sample) lactic acid bacteria were isolated from three different traditionally fermented Ethiopian food products (Teff dough, Ergo, and Kocho). Among them, 56 (62.22%) isolates were found Gram-positive, endospore-negative, and catalase-negative.
3.2. Morphological, Biochemical, and Physiological Characterization
The 4 acid-bile-tolerant LAB isolates were identified through their morphological, biochemical, and physiological features (Table 1). All the isolates were straight rod-shaped and able to grow at 4% NaCl salt concentration. From the 4 acid-bile-tolerant LAB isolates, 3 of the isolates were found growing at 6.5% NaCl salt concentration. Testing the abilities of isolates growing at 15°C and 45°C indicated that 3 of the isolates were able to grow at both 15°C and 45°C (Table 1). On the contrary, isolate E031 was not found growing at 45°C.
Table 1.
Physiological, morphological, and biochemical characteristics of the isolates.
| Isolate | KOH test | Catalase test | Shape | Spore staining | Temperature (°C) | Salt concentration (%) | Production | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 45 | 4 | 6.5 | CO2 | NH3 | |||||
| E052 | −ve | −ve | Rod | − | + | + | + | − | − | − |
| E031 | −ve | −ve | Rod | − | + | − | + | + | − | + |
| T035 | −ve | −ve | Rod | − | + | + | + | + | + | + |
| K011 | −ve | −ve | Rod | − | + | + | + | + | + | + |
Out of the 4 acid-bile-tolerant isolates, 2 isolates (T035 and K011) produced gas from glucose (Table 1). Thus, the four isolates were found to be equally divided into homofermentative and heterofermentative types (50 : 50) (Table 1). Regarding the arginine hydrolysis test, isolates E031, T035, and K011 were found positive for arginine hydrolysis, whereas E052 was not able to hydrolyze (Table 1).
3.3. In vitro Characterization of Probiotic Properties
3.3.1. Tolerance to Low pH
Out of 56 isolates, 4 isolates (7.14%), 4 isolates (7.14%), and 9 isolates (16.07%) tolerated pH values of 2, 2.5, and 3 for 3 h, respectively (Table 2). Upon further extension of the incubation period to 6 h, the 8 isolates (each 4 tested at pH 2.0 and 2.5) survived, whereas only 5 survived from 9 with the extension of incubation period to 6 h at pH 3.0 (Table 2).
Table 2.
Acid tolerance patterns of probiotic LAB at different pH values after 3 and 6 h exposure.
| Source | No. of isolates | No. of survived isolates (%) | |||||
|---|---|---|---|---|---|---|---|
| 3 h | 6 h | ||||||
| pH 2 | pH 2.5 | pH 3 | pH 2 | pH 2.5 | pH 3 | ||
| Ergo | 21 | 2 (9.5%) | 2 (9.5%) | 4 (19.1%) | 2 (9.5%) | 2 (9.5%) | 3 (14.3%) |
| Teff dough | 19 | 1 (5.3%) | 1 (5.3%) | 3 (15.8%) | 1 (5.3%) | 1 (5.3%) | 1 (5.3%) |
| Kocho | 16 | 1 (6.3%) | 1 (6.3%) | 2 (12.5%) | 1 (6.3%) | 1 (6.3%) | 1 (6.3%) |
| Total | 56 | 4 (7.14%) | 4 (7.14%) | 9 (16.1%) | 4 (7.14%) | 4 (7.14%) | 5 (8.93%) |
Therefore, out of the total 56 LAB isolates, 4 (7.14%) isolates survived pH 2.0, 2.5, and 3.0 upon exposure for 3 and 6 hours, and the mean value of the treatments was significantly different at p < 0.05 (Table 3). Among them, 1 (25%) was isolated from each Teff dough and Kocho sample. And also, 2 (50%) LAB cultures were isolated from Ergo samples. In general, the survival rate of the isolates was ranged from 38.40 to 97.11% at different pH values for 3 and 6 h incubation periods (Table 3). Isolates, like E052, K011, and T035 were found highly tolerant and persisted above 50% for both 3 and 6 hours. On the contrary, isolate K031 was not able to grow above 50% at pH 2.0 upon exposure for 3 and 6 h (Table 3). Though the survival rate of the isolates was markedly reduced at pH 2.0 for 6 h exposure, all the isolates were taken to the next experiments.
Table 3.
Percentage survival of probiotic LAB at different pH levels and 0.3% bile salt.
| Isolates | pH tolerance | Bile tolerance | |||||
|---|---|---|---|---|---|---|---|
| 3 h | 6 h | 24 h | |||||
| pH 2 | pH 2.5 | pH 3 | pH 2 | pH 2.5 | pH 3 | 0.3% | |
| E052 | 57.36 ± 1.63c | 80.31 ± 1.17b | 93.02 ± 1.40bc | 54.68 ± 1.63a | 65.58 ± 1.68b | 88.68 ± 1.88a | 93.62 ± 2.19ab |
| E031 | 45.35 ± 1.08d | 79.39 ± 1.59b | 94.00 ± 1.62ab | 38.40 ± 1.24b | 71.50 ± 1.08a | 83.39 ± 1.27b | 97.22 ± 0.35a |
| K011 | 73.29 ± 1.44a | 90.13 ± 1.10a | 97.11 ± 2.17a | 49.34 ± 1.27a | 70.52 ± 1.99a | 90.49 ± 1.46a | 93.38 ± 0.70b |
| T035 | 60.15 ± 0.74b | 77.98 ± 1.19b | 90.28 ± 2.03c | 51.41 ± 0.77a | 70.11 ± 1.11a | 83.89 ± 0.93b | 91.37 ± 2.66b |
a–dMeans within a column with different superscripts are significantly different (p < 0.05). Results expressed as average (n = 3) ± SD (standard deviation).
3.3.2. Tolerance to Bile Salts
All of the 4 LAB isolates (E052, E031, K011 and T035) were able to survive above 90% in the presence of 0.3% of bile salt (Table 3). Isolate E031 was the most tolerant with 97.22% survival rate followed by isolates E052 and K031 with 93.62% and 93.38% survival rates, respectively. However, isolate T035 showed 91.37% survival rate (Table 3).
3.3.3. Antimicrobial Activities
The diameter of inhibition zones showed that crude extracts from each isolate had antimicrobial effect against each tested food-borne pathogen (Table 4). The average zones of inhibition by which the crude extracts inhibited the growth of the test food-borne pathogens were found ranging between 17 to 21 mm. Isolate T035 displayed highest antagonistic activity against Staphylococcus aureus ATCC 25923, Listeria monocytogenes (clinical isolate), E. coli ATCC 25922, and Salmonella enterica Typhimurium with the inhibition zone ranged from 19.33 to 21 mm in diameters. However, E052 showed a minimum inhibition zone of diameter ranging from 17 to 19 mm against the indicator microorganisms (Table 4).
Table 4.
Antimicrobial activities of LAB against some food-borne pathogens and adhesion of LAB to stainless steel plate.
| Isolates | Diameter of inhibition zone (mm) | Adherence (%) | |||
|---|---|---|---|---|---|
| L. monocytogenes | S. aureus | E. coli | S. typhimurium | ||
| E052 | 19.00 ± 1.00b | 17.00 ± 1.00b | 17.33 ± 0.58b | 18.67 ± 0.58ab | 33.17 ± 1.45b |
| E031 | 19.67 ± 0.58ab | 20.33 ± 0.58a | 20.00 ± 1.00a | 18.33 ± 0.58b | 32.75 ± 2.11b |
| T035 | 20.67 ± 0.58a | 21.00 ± 1.00a | 19.33 ± 1.15a | 20.00 ± 1.00a | 33.48 ± 1.05b |
| K011 | 19.67 ± 0.58ab | 20.33 ± 1.15a | 20.67 ± 0.58a | 19.67 ± 0.58ab | 36.30 ± 1.36a |
a,bMeans within a column with different superscripts are significantly different (p < 0.05). Results expressed as average (n = 3) ± SD (standard deviation).
3.3.4. Antibiotic Susceptibility Test
The adherence ability of the potential probiotic LAB isolates was found ranging between 32.75 and 36.30% (Table 4). Isolate K011 showed the highest (36.30%) adherence rate followed by isolates T035 and E052 with 33.48% and 33.17% adherence rates, respectively. However, isolate E031 showed the least (32.75%) adherence rate (Table 4).
3.3.5. Bacterial Adhesion to Stainless Steel Plates
The antibiotic susceptibility test of the selected LAB isolates to some common antibiotics showed sensitivity to tetracycline, ampicillin, and erythromycin. However, all the 4 potential probiotic LAB isolates displayed resistance to kanamycin and streptomycin (Table 5).
Table 5.
Antibiotic susceptibility profile of probiotic LAB isolates.
| Isolates | Diameter of inhibition zone | ||||
|---|---|---|---|---|---|
| Kanamycin | Streptomycin | Tetracycline | Ampicillin | Erythromycin | |
| E052 | R | R | S | S | S |
| E031 | R | R | S | S | S |
| T035 | R | R | S | S | S |
| K011 | R | R | S | S | S |
Zone of inhibition (diameter in mm) for each antibiotic was measured and expressed as susceptible, S (≥21 mm); intermediate, I (16–20 mm); and resistance, R (≤15 mm).
3.3.6. Identification of Probiotic LAB Isolates by 16S rRNA Gene Sequencing
The 16S rRNA gene sequences of the 4 LAB isolates with the best potential probiotic properties showed the highest homology to the known species of bacteria in the database (Figure 1). Accordingly, E052 showed 99% match with Lactobacillus plantarum strain JCM 1149, T035 showed 99% homology with Lactobacillus plantarum strain CIP 103151, K011 showed 95% similarity with Lactobacillus paracasei subsp. tolerans strain NBRC 15906, and E031 showed 99% homology with Lactobacillus paracasei strain NBRC 15889 (Figure 1).
Figure 1.
Evolutionary relationships of the isolation to the known strains.
4. Discussion
The study of probiotic activities, for application in preservation of food products and human health, remains to be of great interest. Currently, interest in antagonistic feature of probiotic LAB against food-borne pathogens has indicated their potential to be likely alternatives to chemical drugs [30]. Therefore, a significant effort has been made to select lactic acid bacteria originating from the traditional Ethiopian fermented food products on the basis of the most important technological, functional, and safety criteria in order to obtain potentially probiotic lactic acid bacteria.
All the selected four potential probiotic bacteria were identified as LAB based on their morphological, biochemical, and physiological characteristics that were found to be equally divided into homofermentative and heterofermentative types. This finding is in accordance with Mamo et al. [31] who isolated Lactobacillus species from Ergo and found that they were grouped as homofermentative and heterofermentative types. Akalu et al. [17] have also reported that the probiotic LAB isolated from Shamita and Kocho were identified morphologically, biochemically, and physiologically and comprised of both heterofermentative and homofermentative types. Azadnia and Khan Nazer [32] reported that the lactic acid bacteria isolated from traditional drinking yoghurt in tribes of Fars Province were able to grow at 4% NaCl concentration but not at 6.5% NaCl concentration.
In the present study, out of 56 probiotic LAB isolates tested to pH 2 for 3 and 6h, only 4 (7.14%) isolates showed tolerance to pH 2 for 6 h (Table 1). Thus, the tolerance of the four Lactobacillus spp. to pH 2.0 for 6 h showed a survival rate of 38.40 to 73.29%. Similar to this study, Mourad and Nour-Eddine [33] have demonstrated that Lactobacillus plantarum OL12, L. plantarum OL9, L. plantarum OL15, and L. plantarum OL33 isolated from fermented olives showed survival percentages of 55%, 49%, 65% and 57%, respectively, when exposed to pH 2.0 for 2 h. However, these results are not in accordance with those reported by Akalu et al. [17] and Rajoka et al. [34] who have indicated that most strains of Lb. plantarum isolated from different sources showed a survival rate above 80% at pH 2 for 3 h. Other reports revealed that 5 acid-tolerant Lactobacillus strains showed above 89% survival rate after exposure to pH 2 for 3 h [35]. Akalu et al. [17] verified that 14/17 Lactobacillus strains isolated from traditionally fermented Shamita and Kocho were found tolerant to pH 2 when incubated for 6 h with a survival rate of 74–89%. However, incubation at low pH resulted in a significant decrease in the survival rate of all LAB isolates as noted in another study by Guo et al. [36]. The authors noted that the viable counts of all lactic acid bacteria were significantly affected by low acidity, especially at pH 2.
In this study, out of the 56 probiotic LAB isolates, only 4 (7.14%) of the isolates were tolerant to pH 2.5 for 3 and 6 h (Table 1). Similarly, out of 56 LAB isolates, 9 isolates (16.1%) and 5 isolates (8.93%) survived at pH 3 for 3 and 6 h, respectively. Thus, the survival rate of the four Lactobacillus strains at pH 2.5 and 3 for 3 and 6 h was found to be tolerant with a different survival rate (77.98–97.11%) and (65.58–90.49%), respectively (Table 2). Similarly, Akalu et al. [17] have reported that out of 30 LAB isolated from traditionally fermented Ethiopian beverage and food (Shamita and Kocho), 17 lactobacilli showed 82 to 97% and 81 to 91% survival rate at pH 2.5 and 3 for 3 and 6 h, respectively. This is also similar to the report of Oh and Jung [35] who have shown a better survival rate (above 88%) of Lactobacillus species in pH 2.5 and 3 for 3 h that were isolated from Omegisool, a traditionally fermented millet alcoholic beverage of North Korea. As reported by Haghshenas et al. [37], Lactobacillus strains isolated from Iranian fermented dairy products survived (71% to 76%) at pH 2.5 for 3 h. From the previous investigations, it has generally been accepted that an isolate with full tolerance to pH 3.0 for 3 h can be considered as high-acid-resistant strain with promising probiotic properties [36, 38]. Very recently, Tang et al. [39] have reported that all the 9 Lactobacillus plantarum strains recovered from the faeces of breast-feeding piglets were found to be highly tolerant to pH 3 for 3 h.
The present results are in contrast with those of Mamo et al. [31] who have found low to high survival rates (1.03–100%) for the six Lactobacillus species at pH 2.5 and 3.0 for 3 h. The same authors indicated that the maximum survival rate of the six strains were 22.5% at pH 3 for 6 h. Nevertheless, complete loss of viability of lactobacilli isolated from traditional Ethiopian ergo was recorded at pH 2.5 for 6 h [31]. In addition, the current survival rate was higher than that of previously reported strains such as Lactobacillus plantarum at pH 3 for 2 and 6 h exposure [33]. The 4 acid-tolerant LAB isolates showed high tolerance to bile salt conditions (91.37% to 97.22%) (Table 3). Bile salt tolerance is also considered as an important selection criterion for probiotic isolates in order to survive the conditions in the small intestine. Moreover, tolerance to a high bile salt condition is also straining specific as demonstrated earlier where different Lactobacillus species isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea showed considerable tolerance to bile salt [35]. Similar to the present findings, the results in other studies have revealed that all the isolated strains displayed high tolerance to bile salt conditions and the survival rates of Lactobacillus strains ranged from 88% to 92% [37]. In a related study, Akalu et al. [17] have also shown that out of the 30 tested LAB isolates, 17 Lactobacillus isolates obtained from Ethiopian traditionally fermented Shamita and Kocho showed remarkably high tolerance to an environment containing 0.3% bile salt. On the contrary, Boke et al. [40] have indicated that Lactobacillus strains B3, G12, A13, and 22 exhibited low level of tolerance to 0.3% bile salts with survival rates of 36%, 33%, 3%, and 3%, respectively. In line with this, Handa [41] has reported that all of the LAB isolates demonstrated a low level of tolerance to bile salts by displaying surviving percentage less than 50% when exposed to 0.3% bile salts after 24 h at 37°C.
The selected four potential probiotic lactic acid bacterial strains (Lactobacillus plantarum strain JCM 1149, Lactobacillus plantarum strain CIP 103151, Lactobacillus paracasei subsp. tolerans strain NBRC 15906, and Lactobacillus paracasei strain NBRC 15889) exhibited varying degree of antagonism against Staphylococcus aureus, Listeria monocytogenes, Salmonella Typimurium, and Escherichia coli (Table 4). According to Handa (2012), isolates having clearance zones ≤9 mm and ≥12 mm diameter against the test pathogens indicated poor and strong antimicrobial activity, respectively. Accordingly, all the selected potential probiotic LAB strains (n = 4) exhibited strong antimicrobial activity against the food-borne pathogens, where T035 (Lactobacillus plantarum strain CIP 103151) displayed the highest antagonistic activity against Staphylococcus aureus, Listeria monocytogenes, E. coli, and S. typhimurium with the inhibition zone ranged from 19.33 to 21 mm in diameters.
In accordance to the current study, Bassyouni et al. [42] have demonstrated that all of the Lactobacillus isolates obtained from Egyptian dairy product had a strong antibacterial effect against E. coli and Salmonella typhimurium. However, among the results that were revealed by the same author, 3 isolates had the most potent antimicrobial activity against the tested pathogenic microorganisms with the inhibition zone ranged from 17 to 21 mm in diameters. In agreement to this study, Tadesse et al. [5] have verified that all the LAB isolates (n = 118) originated from Borde and Shamita belonging to the genera Lactobacillus, Lactococcus, Leuconostoc, and Streptococcus were found to inhibit the growth of the test strains such as S. aureus, Salmonella spp., and E. coli O157 : H7 with inhibition zones that ranged from 15 to 17 mm in diameters. In line with this, Choi et al. [43] have reported that out of the 4 strains of LAB, the Lactobacillus strain has completely inhibited the growth of food-borne pathogens, E. coli O157 : H7 ATCC 35150, Salmonella enteritidis KCCM 12021, Salmonella typhimurium KCTC 1925, and S. aureus. Tigu et al. [16] have also revealed that out of the 11 probiotic LAB isolated from traditional Ethiopian fermented condiments, namely, Datta and Awaze, 2 Lactobacillus isolates inhibited the growth of Salmonella typimurium and Escherichia coli with inhibition zones ranging from 10.3 to 14.3 mm in diameters. In line with this, Haghshenas et al. [37] have reported that among the selected 8 LAB isolated from Iranian fermented dairy products, Lactobacillus species, particularly Lb. plantarum 15HN, showed the most efficient antagonistic activity against Staphylococcus aureus, Listeria monocytogenes, Salmonella typimurium, and Escherichia coli with inhibition zones of 11.7, 13.7, 12.3, and 12.3 mm diameters, respectively. Likewise, Rajoka et al. [34] have verified that all the Lactobacillus rhamnosus isolated from human milk inhibited the growth of Staphylococcus aureus, Salmonella typimurium, and Escherichia coli using the agar-well diffusion method with variable diameters (6 mm to 14 mm).
Disparity in the antagonism activity against different pathogens indicates that probiotic strains are highly pathogen-specific and a prerequisite for probiotic potential. In general, the antimicrobial activity of probiotics might be caused by the production of antimicrobial compounds such as organic acids, ethanol, carbon dioxide, hydrogen peroxide, short-chain fatty acids, and bacteriocins. Therefore, by producing these antimicrobial compounds, probiotic microorganisms gain an advantage over other microorganisms to survive in the adverse conditions of the gastrointestinal tract [41].
All of the tested four Lactobacillus strains were found to be resistant to streptomycin and kanamycin but sensitive towards tetracycline, ampicillin, and erythromycin (Table 5). These results were found in agreement with the results that were obtained using Lactobacillus species [34]. Tigu et al. [16] have also reported that all of the LAB isolates obtained from traditional fermented condiments such as Datta and Awaze were susceptible to ampicillin, erythromycin, and tetracycline. Similarly, according to Amraii et al. [44], all of the selected LAB isolates were sensitive towards ampicillin, erythromycin, and tetracycline. On the contrary, Sukmarini et al. [45] have reported that out of 120 isolates of LAB from four different Indonesian traditional fermented foods, 16 isolates were resistant to erythromycin. In line with this, Pan et al. [46] observed that among the 12 Lactobacillus species obtained from Chinese fermented foods, 5 isolates were sensitive to kanamycin, 7 resistant to erythromycin, 9 resistant to ampicillin, and 8 isolates resistant to tetracycline.
Among the main important characteristics of probiotic bacteria, adhesion to the intestinal mucosa is required. The current results showed that the screened probiotic LAB isolates possessed in vitro adherence property to stainless steel plates with the adhesion rate ranging from 32.75 to 36.30% (Table 4). In agreement with this study, El-Jeni et al. [27] have reported that the adhesion rate of lactic acid bacteria to stainless steel plates ranged from 32 to 35%. Winkelströter et al. [47] have also revealed that pure culture of Lb. sakei ATCC 15521 showed strong adherence to stainless steel surface. Generally, this suggests that our potential probiotic LAB isolates may have a potential capacity to colonize the gastrointestinal (GI) tract mucosa.
The phylogenetic analysis and the 16S rDNA sequencing assigned all the LAB isolates with probiotic properties to genus Lactobacillus, and the identified species were Lactobacillus plantarum strain JCM 1149, Lactobacillus paracasei strain NBRC 15889, Lactobacillus plantarum strain CIP 103151, and Lactobacillus paracasei subsp. tolerans strain NBRC 15906 (Figure 1). According to Shokryazdan et al. [48], the results of comparative 16S rRNA gene analysis showed LAB isolates belonged to L. acidophilus, L. fermentum, L. buchneri, and L. casei. In addition, Cho, Lee, and Hahm [49] have identified Lactobacillus strains with potential probiotic properties from the faeces of breast-feeding piglets using 16S rRNA genes analysis. Similarly, other recent studies (Dowarah et al. [6] have revealed the strain level identification of diverse lactic acid bacteria with potent probiotic properties isolated from some substrates using phylogenetic estimation of 16S rDNA genes.
5. Conclusion
In the present study, four Lactobacillus isolates from Ergo, Kocho, and Teff dough were found to have potentially probiotic characteristics. It is suggested that these strains can be good candidates for food industries as prospective probiotic cultures with other human health benefits. However, further research work is needed to evaluate the in vivo probiotic characteristics of these potential lactic acid bacteria.
Acknowledgments
The authors acknowledge Microbial, Cellular and Molecular Biology Department of Addis Ababa University and Department of Food and Environmental Sciences, Helsinki University, Finland, for provision of all laboratory facilities, Department of Biology, College of Natural and Computational Sciences, Aksum University, Ethiopia, for financial support, and Ethiopian Public Health Institute (EPHI), Addis Ababa, Ethiopia, for providing test organisms. The authors are also thankful to Dr. Wesley Morovic, Scientist, Strain Discovery and Microbiome Science, DuPont Nutrition and Biosciences, Finland, for his support in 16S rDNA analysis.
Data Availability
The data used to support the findings of this study are included within the article and are available (the SAS data) from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Three (Teff dough, Ergo, and Kocho) traditionally fermented Ethiopian food products were used as the isolation source. The isolates were designated with E for Ergo, T for Teff dough, and K for Kocho, followed with different numbers. In general, the abovementioned traditionally fermented food products are prepared at the household level and their consumption is commonly practiced throughout the country.
References
- 1.Nath A., Gupta A., Neopany B., et al. Biotechnology and Traditional Fermented foods. Indigenous Fermented Foods of South East Asia. Boca Raton, FL, USA: CRC Press; 2016. [Google Scholar]
- 2.Hasan M. N., Sultan M. Z., Mar-E-Um M. Significance of fermented food in nutrition and food science. Journal of Scientific Research. 2014;6(2) doi: 10.3329/jsr.v6i2.16530. [DOI] [Google Scholar]
- 3.Sivapalasingam S., Friedman C. R., Cohen L., Tauxe R. V. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. Journal of Food Protection. 2004;67(10):2342–2353. doi: 10.4315/0362-028x-67.10.2342. [DOI] [PubMed] [Google Scholar]
- 4.Tesfaye A., Mehari T., Ashenafi M. Evaluation of the in vitro and in vivo probiotic qualities of lactic acid bacteria (LAB) recovered from locally fermented products. International Journal of Probiotics & Prebiotics. 2011;6(2) [Google Scholar]
- 5.Tadesse G., Ephraim E., Ashenafi M. Assessment of the antimicrobial activity of lactic acid bacteria isolated from Borde and Shamita, traditional Ethiopian fermented beverages, on some foodborne pathogens and effect of growth medium on the inhibitory activity. Internet Journal of Food Safety. 2005;5:13–20. [Google Scholar]
- 6.Dowarah R., Verma A. K., Agarwal N., Singh P., Singh B. R. Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PloS One. 2018;13(3) doi: 10.1371/journal.pone.0192978.e0192978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill C., Guarner F., Reid G., et al. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Reviews Gastroenterology & Hepatology. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 8.Akkoç N., Ghamat A., Akçelik M. Optimisation of bacteriocin production of lactococcus lactis subsp: lactis MA23, a strain isolated from Boza. International Journal of Dairy Technology. 2011;64(3):425–432. doi: 10.1111/j.1471-0307.2011.00671.x. [DOI] [Google Scholar]
- 9.O’Bryan C., Crandall P., Ricke S., Ndahetuye J. Handbook of Natural Antimicrobials for Food Safety and Quality. Cambridge, UK: Woodhead Publishing; 2015. 7-Lactic acid bacteria (LAB) as antimicrobials in food products: analytical methods and applications; pp. 137–151. [Google Scholar]
- 10.Ricci A., Cirlini M., Maoloni A., et al. Use of dairy and plant-derived lactobacilli as starters for cherry juice fermentation. Nutrients. 2019;11(2):p. 213. doi: 10.3390/nu11020213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Rab G. D., Barakat O. S., Ibrahim G., Tawfik N., El-Kholy W. Identification and probiotic characteristics of Lactobacillus strains isolated from traditional Domiati cheese. International Journal of Microbiology Research. 2011;3(1):p. 59. doi: 10.9735/0975-5276.3.1.59-66. [DOI] [Google Scholar]
- 12.Ouwehand A. C., Salminen S., Isolauri E. Probiotics: an overview of benefical effects. Antonie Van Leeuwenhoek. 2002;82(1–4):279–289. [PubMed] [Google Scholar]
- 13.Chassard C., Grattepanche F., Lacroix C. Probiotics and Health Claims. Hoboken, NJ, USA: Wiley; 2011. Probiotics and health claims: challenges for tailoring their efficacy; pp. 49–74. [Google Scholar]
- 14.Marco M. L., Heeney D., Binda S., et al. Health benefits of fermented foods: microbiota and beyond. Current Opinion in Biotechnology. 2017;44:94–102. doi: 10.1016/j.copbio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Manhar A. K., Bashir Y., Saikia D., et al. Cellulolytic potential of probiotic bacillus subtilis AMS6 isolated from traditional fermented soybean (churpi): an in-vitro study with regards to application as an animal feed additive. Microbiological Research. 2016;186-187:62–70. doi: 10.1016/j.micres.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Tigu F., Assefa F., Mehari T., Ashenafi M. Probiotic property of lactic acid bacteria from traditional fermented condiments: Datta and Awaze. International Food Research Journal. 2016;23(2):p. 770. [Google Scholar]
- 17.Akalu N., Assefa F., Dessalegn A. In vitro evaluation of lactic acid bacteria isolated from traditional fermented Shamita and Kocho for their desirable characteristics as probiotics. African Journal of Biotechnology. 2017;16(12):594–606. doi: 10.5897/AJB2016.15307. [DOI] [Google Scholar]
- 18.Patil M. M., Pal A., Anand T., Ramana K. Isolation and characterization of lactic acid bacteria from curd and cucumber. Indian Journal of Biotechnology. 2010;9:166–172. [Google Scholar]
- 19.Powers E. M. Efficacy of the Ryu nonstaining KOH technique for rapidly determining gram reactions of food-borne and waterborne bacteria and yeasts. Applied and Environmental Microbiology. 1995;61(10):3756–3758. doi: 10.1128/aem.61.10.3756-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescott L. M., Harley J. P., Klein D. A. Laboratory Exercises in Microbiology. NY, USA: McGraw-Hill Companies; 2002. [Google Scholar]
- 21.Grosu-Tudor S. S., Zamfir M. Probiotic potential of some lactic acid bacteria isolated from Romanian fermented vegetables. Annals of the Romanian Society for Cell Biology. 2012;17(1) [Google Scholar]
- 22.Fontana C., Cocconcelli P. S., Vignolo G., Saavedra L. Occurrence of antilisterial structural bacteriocins genes in meat borne lactic acid bacteria. Food Control. 2015;47:53–59. doi: 10.1016/j.foodcont.2014.06.021. [DOI] [Google Scholar]
- 23.del Mar Lleo M., Tafi M. C., Canepari P. Nonculturable Enterococcus faecalis cells are metabolically active and capable of resuming active growth. Systematic and Applied Microbiology. 1998;21(3):333–339. doi: 10.1016/s0723-2020(98)80041-6. [DOI] [PubMed] [Google Scholar]
- 24.Mir-Hoseini M. Study of Effect of Nisin and Producer Bacteria of Nisin on Listeria Monocytogenes and Bacillus Cereus. Isfahan, Iran: University of Isfahan; 2004. [Google Scholar]
- 25.Zhang B., Wang Y., Tan Z., Li Z., Jiao Z., Huang Q. Screening of probiotic activities of lactobacilli strains isolated from traditional Tibetan Qula, a raw yak milk cheese. Asian-Australasian Journal of Animal Sciences. 2016;29(10):1490–1499. doi: 10.5713/ajas.15.0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vlková E., Rada V., Popelářová P., Trojanová I., Killer J. Antimicrobial susceptibility of bifidobacteria isolated from gastrointestinal tract of calves. Livestock Science. 2006;105(1–3):253–259. doi: 10.1016/j.livsci.2006.04.011. [DOI] [Google Scholar]
- 27.El-Jeni R., El Bour M., Calo-Mata P., et al. In vitro probiotic profiling of novel Enterococcus faecium and Leuconostoc mesenteroides from Tunisian freshwater fishes. Canadian Journal of Microbiology. 2015;62(1):60–71. doi: 10.1139/cjm-2015-0481. [DOI] [PubMed] [Google Scholar]
- 28.Nair P. S., Surendran P. K. Biochemical characterization of lactic acid bacteria isolated from fish and prawn. Journal of Culture Collections. 2005;4(1):48–52. [Google Scholar]
- 29.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Rather I. A., Choi K.-H., Bajpai V. K., Park Y.-H. Antiviral mode of action of Lactobacillus plantarum YML009 on Influenza virus H1N1. Bangladesh Journal of Pharmacology. 2015;10(2):475–482. doi: 10.3329/bjp.v10i2.23068. [DOI] [Google Scholar]
- 31.Mamo N. J., Assefa T. F., Tesfaye T. A. Evaluation of the antagonistic effect of six mixed cultures of lactic acid bacteria, isolated from the Ethiopian fermented milk ergo, against some foodborne pathogens inoculated into the Ethiopian cottage cheese ayib. African Journal of Microbiology Research. 2015;9(29):1789–1797. doi: 10.5897/AJMR2015.7504. [DOI] [Google Scholar]
- 32.Azadnia P., Khan Nazer A. Identification of lactic acid bacteria isolated from traditional drinking yoghurt in tribes of Fars province. Iranian Journal of Veterinary Research. 2009;10(3):235–240. doi: 10.22099/ijvr.2009.1698. [DOI] [Google Scholar]
- 33.Mourad K., Nour-Eddine K. In vitro preselection criteria for probiotic Lactobacillus plantarum strains of fermented olives origin. International Journal of Probiotics and Prebiotics. 2006;1(1):p. 27. [Google Scholar]
- 34.Rajoka M. S. R., Mehwish H. M., Siddiq M., et al. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT-food Science and Technology. 2017;84:271–280. doi: 10.1016/j.lwt.2017.05.055. [DOI] [Google Scholar]
- 35.Oh Y. J., Jung D. S. Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT—Food Science and Technology. 2015;63(1):437–444. doi: 10.1016/j.lwt.2015.03.005. [DOI] [Google Scholar]
- 36.Guo X.-H., Kim J.-M., Nam H.-M., Park S.-Y., Kim J.-M. Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe. 2010;16(4):321–326. doi: 10.1016/j.anaerobe.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Haghshenas B., Nami Y., Almasi A., et al. Isolation and characterization of probiotics from dairies. Iranian Journal of Microbiology. 2017;9(4):234–243. [PMC free article] [PubMed] [Google Scholar]
- 38.Argyri A. A., Zoumpopoulou G., Karatzas K.-A. G., et al. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiology. 2013;33(2):282–291. doi: 10.1016/j.fm.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Tang H., Qian B., Xia B., et al. Screening of lactic acid bacteria isolated from fermented Cornus officinalis fruits for probiotic potential. Journal of Food Safety. 2018;38(6) doi: 10.1111/jfs.12565.e12565 [DOI] [Google Scholar]
- 40.Boke H., Aslim B., Alp G. The role of resistance to bile salts and acid tolerance of exopolysaccharides (EPSS) produced by yogurt starter bacteria. Archives of Biological Sciences. 2010;62(2):323–328. doi: 10.2298/abs1002323b. [DOI] [Google Scholar]
- 41.Handa S. New Delhi, India: KrishiKosh; 2012. Isolation of lactic acid bacteria and to study their potential as probiotics. M.SC thesis. [Google Scholar]
- 42.Bassyouni R. H., Abdel-all W. S., Fad M. G., Abdel-all S., Kamel Z. Characterization of lactic acid bacteria isolated from dairy products in Egypt as a probiotic. Life Science Journal. 2012;9(4) [Google Scholar]
- 43.Choi A.-R., Patra J. K., Kim W. J., Kang S.-S. Antagonistic activities and probiotic potential of lactic acid bacteria derived from a plant-based fermented food. Frontiers in Microbiology. 2018;9 doi: 10.3389/fmicb.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amraii H. N., Abtahi H., Jafari P., Mohajerani H. R., Fakhroleslam M. R., Akbari N. In vitro study of potentially probiotic lactic acid bacteria strains isolated from traditional dairy products. Jundishapur Journal of Microbiology. 2014;7(6) doi: 10.5812/jjm.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sukmarini L., Mustopa A. Z., Normawati M., Muzdalifah I. Identification of antibiotic-resistance genes from lactic acid bacteria in Indonesian fermented foods. HAYATI Journal of Biosciences. 2014;21(3):144–150. doi: 10.4308/hjb.21.3.144. [DOI] [Google Scholar]
- 46.Pan L., Hu X., Wang X. Assessment of antibiotic resistance of lactic acid bacteria in Chinese fermented foods. Food Control. 2011;22(8):1316–1321. doi: 10.1016/j.foodcont.2011.02.006. [DOI] [Google Scholar]
- 47.Winkelströter L. K., Gomes B. C., Thomaz M. R. S., Souza V. M., De Martinis E. C. P. Lactobacillus sakei 1 and its bacteriocin influence adhesion of Listeria monocytogenes on stainless steel surface. Food Control. 2011;22(8):1404–1407. doi: 10.1016/j.foodcont.2011.02.021. [DOI] [Google Scholar]
- 48.Shokryazdan P., Sieo C. C., Kalavathy R., et al. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. BioMed Research International. 2014;2014:16. doi: 10.1155/2014/927268.927268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho I. J., Lee N. K., Hahm Y. T. Characterization of Lactobacillus spp. isolated from the feces of breast-feeding piglets. Journal of Bioscience and Bioengineering. 2009;108(3):194–198. doi: 10.1016/j.jbiosc.2009.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three (Teff dough, Ergo, and Kocho) traditionally fermented Ethiopian food products were used as the isolation source. The isolates were designated with E for Ergo, T for Teff dough, and K for Kocho, followed with different numbers. In general, the abovementioned traditionally fermented food products are prepared at the household level and their consumption is commonly practiced throughout the country.
Data Availability Statement
The data used to support the findings of this study are included within the article and are available (the SAS data) from the corresponding author upon request.