Abstract
Introduction
Comorbidity with metabolic diseases indicates that lipid metabolism plays a role in the etiology of Alzheimer's disease (AD). Comprehensive lipidomic analysis can provide new insights into the altered lipid metabolism in AD.
Method
In this study, a total 349 serum lipids were measured in 806 participants enrolled in the Alzheimer's Disease Neuroimaging Initiative Phase 1 cohort and analyzed using lipid-set enrichment statistics, a data mining method to find coregulated lipid sets.
Results
We found that sets of blood lipids were associated with current AD biomarkers and with AD clinical symptoms. AD diagnosis was associated with 7 of 28 lipid sets of which four also correlated with cognitive decline, including polyunsaturated fatty acids. Cerebrospinal fluid amyloid beta (Aβ1-42) correlated with glucosylceramides, lysophosphatidylcholines and unsaturated triacylglycerides; cerebrospinal fluid total tau and brain atrophy correlated with monounsaturated sphingomyelins and ceramides, in addition to EPA-containing lipids.
Discussion
AD-associated lipid sets indicated that lipid desaturation, elongation, and acyl chain remodeling processes are disturbed in AD subjects. Monounsaturated lipid metabolism was important in early stages of AD, whereas the polyunsaturated lipid metabolism was associated with later stages of AD. Our study provides several new hypotheses for studying the role of lipid metabolism in AD.
Keywords: Alzheimer's disease, Lipidomics, Dyslipidemias, Lipid biochemistry, Mass spectrometry
Highlights
-
•
Twenty eight coregulatory lipid sets are identified in Alzheimer's Disease Neuroimaging Initiative lipidomics data.
-
•
Distinct lipid pathways are associated with cerebrospinal fluid biomarkers, brain atrophy, and Alzheimer's disease (AD) diagnosis.
-
•
Monounsaturated lipids play a role in the AD initiation and early neurodegeneration.
-
•
Polyunsaturated lipids are associated with AD diagnosis.
1. Introduction
Alzheimer's disease (AD) is often presented with diabetes comorbidity and a wide range of metabolic perturbations occurring early in the disease process [1]. APOE-ε4 is by far the strongest single gene variant contributing to increased AD risk and plays key roles in lipid transport and metabolism. Presence of the APOE-ε4 variant is correlated with higher cholesterol levels in the blood [2]. More than twenty additional genes have recently been implicated in AD with functional roles in lipid-processing, immune regulation, and phagocytosis. Hence, both comorbidities and known gene variants support the idea that metabolic dysfunctions may contribute to AD onset and progression. Comprehensive biochemical profiling of biofluids can provide new insights into AD biology and expand the battery of hypotheses to find new therapeutic options for AD.
Lipidomics methods using liquid chromatography and mass spectrometry yield reliable measurements of hundreds of lipids in biological samples. Liquid chromatography and mass spectrometry methods have been used in AD studies in attempts to define possible risk factors [3], [4], [5], [6], [7], diagnostic markers [8], and for highlighting novel drug targets [9], [10], [11]. Perturbations in ceramides and related sphingomyelin (SM) metabolism [4], [7] were noted in many of these studies pointing toward a possible role of these lipids in aberrant signaling pathways, membrane remodeling, and apoptotic cascades during AD progression. Changes in phosphatidylcholines were observed in several studies [11], [12], [13] pointing to a possible role for phospholipid metabolism in AD pathogenesis. Yet, AD risk prediction failed to replicate using a phosphatidylcholine (PC) biomarker panel [11], [14]. Partial correlation network analysis indicated early AD biomarker Aβ1-42 was associated with PC and SM [11]. These studies support the hypothesis that distinct lipid biochemical pathways were dysregulated in early and late phase of AD.
Here, we used LC-MS/MS-based serum lipidomics analysis measured in the Alzheimer's Disease Neuroimaging Initiative Phase 1 (ADNI1) cohort to define the lipid coregulatory network of AD phenotypes, a statistical analysis tool that previously has been successfully used in the analysis of transcriptomic data [15]. We investigated correlation of lipid sets with (1) disease diagnosis, (2) CSF markers of disease Aβ1-42, CSF total tau, and (3) cognitive decline and brain atrophy.
2. Materials and methods
2.1. Study cohort
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by the Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging, positron emission tomography, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment, and early AD. For up-to-date information, see www.adni-info.org.
The ADNI cohort information was downloaded from the ADNI data repository (http://adni.loni.ucla.edu/), supplying all the demographic information, neuropsychological and clinical assessment data, and diagnostic information that was previously published [11]. Prior Institutional Review Board approval was obtained at each participating institution and written informed consent was obtained for all participants. Information about the ADNI project is provided on http://www.adni-info.org/ and the associated publication [16].
2.2. Pathology, clinical and lipidomics data
Untargeted lipidomics, AD diagnosis, CSF biomarkers including total Tau (t-tau) and amyloid beta (Aβ1-42), cognitive decline (ADAScog13), and brain atrophy represented by Spatial Pattern of Abnormality for Recognition of Early Alzheimer's disease (SPARE-AD) [16] data were obtained from the ADNI repository (http://www.adni-info.org/). Generation and quality control of lipidomics data have been described in the study of Barupal et al [17].
2.3. Detection of sets of coregulated lipids
A pairwise Spearman-rank correlation matrix was generated for lipids using the R function cor.test. The matrix was converted to a hierarchical tree model using the hclust function in R with the ward linkage method. The resulting tree model was divided into clusters using the tree cutting algorithm dynamicTreeCut [18]. We used a minimum cluster size of three and a split depth of four in the tree-cutting method.
2.4. Association modeling
Linear regression models were established for association of serum lipid abundances and CSF biomarkers and indices for cognitive decline and brain atrophy. No confounding variables were included in the regression models. Logistic regression models were calculated to associate serum lipids with AD diagnosis. Lipid abundances were scaled by the mean subtracting approach. All models were unadjusted to identify all the lipid coregulatory sets that were associated specifically with only AD or with AD and other demographics or confounding factors. Data from all ADNI1 participants were included in the analysis.
2.5. Lipid-set enrichment analysis
Coregulatory lipid sets detected by the dynamicTreeCut method [18] were used as an input for cluster enrichment analysis using the Kolmogorov–Smirnov test as described in the ChemRICH method [19]. In this test, the distribution of P-values was assumed to be uniform under a null hypothesis for a lipid cluster. Raw P-values obtained from the linear and logistic models were used as input for computing the enrichment statistics by comparing the experimental P-values with the uniform distribution. Set level P-values were adjusted for false discovery rate using the Benjamini–Hochberg method.
2.6. Source code
All statistical analyses were performed in R programming language version 3.4.0. The R-script is available at http://github.com/barupal/ADNI.
3. Results
The main objective of the study was to identify lipid coregulatory modules that were associated with AD diagnosis and its clinical and pathological features. Coregulation of lipids can indicate biochemical mechanisms that can be explored for new therapeutic options for AD. In this direction, we first computed univariate association models and obtained raw P-values for each lipid. Next, we identified lipid coregulatory modules, which were then used as set definitions for a lipid-set enrichment analysis using the univariate P-values for lipids.
3.1. Subject cohort and lipidomics details
Supplementary Table 1 summarizes the details for the 806 ADNI participants at the baseline included in the present study. The baseline ADNI1 serum lipidomics data set contained 16 different lipid chemical classes summarizing 349 annotated lipids (Table 1). ADNI study collects blood samples at the fasted state. In our analysis, 737 (91%) samples were from subjects who were fasting. Few subjects (9%) donated blood in the nonfasting state. Key findings did not change by exclusion of these nonfasting samples.
Table 1.
Lipid classes and subclasses in the ADNI serum lipidomics untargeted data set
| Class | Subclass | Count |
|---|---|---|
| Acylcarnitine (AC) | Acylcarnitine | 9 |
| Free fatty acid (FA) | Fatty acid | 29 |
| Sterol lipids | Cholesterol | 1 |
| Cholesteryl ester (CE) | 8 | |
| Phospholipid | Lysophosphatidylcholine (LPC) | 22 |
| Lysophosphatidylethanolamine (LPE) | 4 | |
| Phosphatidylcholine (PC) | 53 | |
| Phosphatidylethanolamine (PE) | 11 | |
| Phosphatidylinositol (PI) | 11 | |
| Plasmalogen phosphatidylcholine (p-PC) | 28 | |
| Plasmalogen phosphatidylethanolamine (p-PE) | 15 | |
| Sphingolipid | Ceramide (CER) | 19 |
| Glucosylceramide (GluCer) | 8 | |
| Sphingomyelin (SM) | 34 | |
| Acylglycerols | Diacylglycerol (DG) | 13 |
| Triacylglycerol (TG) | 84 |
Abbreviation: ADNI, Alzheimer's Disease Neuroimaging Initiative.
3.2. Regression models for individual lipids
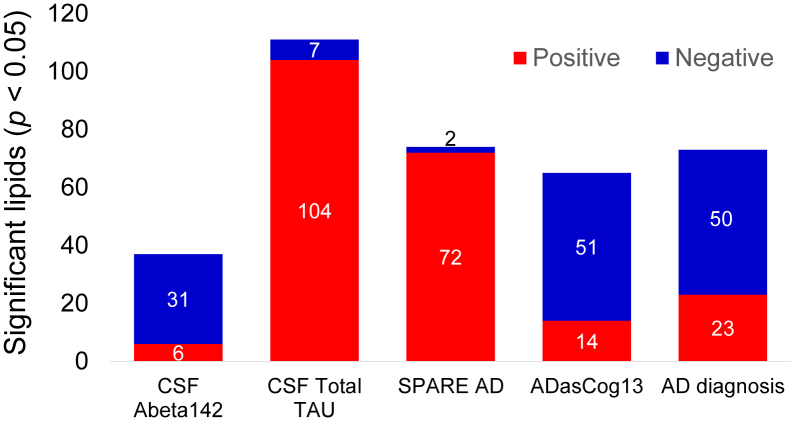
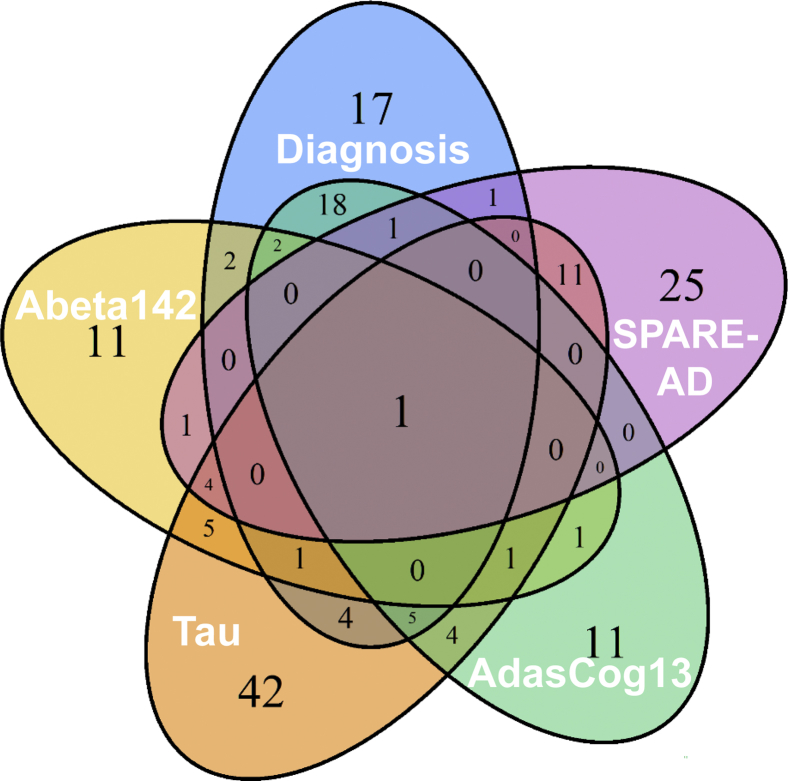
We first tested all individual lipids for their association with both early and late AD pathogenic markers and cognitive changes (Supplementary Table 2). Raw P-values from these association models will be used as an input for the lipid-set enrichment analysis in the following section. Fig. 1 shows the number of significantly associated lipids in these regression models. A total of 168 lipids were found to be significant in at least one regression model (raw P-value < .05), making it difficult to biologically interpret them on individual lipid level. Two AD phenotypes showed strong positive associations with individual serum lipids, CSF total tau and SPARE-AD. Conversely, three phenotypes were mostly negatively associated with individual blood lipids, including the two related phenotypes AD diagnosis and its major contributor, ADASCog13. Overall, AD diagnosis was associated with a decline in many lipid levels which could point to lower metabolic activity in specific lipid metabolic pathways. When analyzing all individual lipids that were associated with at least one AD phenotype, we found a very high specificity of lipid associations with a particular AD phenotype (Fig. 2 and Supplementary Table 3). Forty eight percentage of (168/349) of all lipids were associated with at least one AD phenotype (Supplementary Table 3). Specifically, for known lipids, 63% of all AD phenotype–associated lipids were specific to only one phenotype and not shared with others (Fig. 3). Twenty eight percentages of the detected associations of known lipids were shared between two phenotypes, driven by lipids shared between the two related phenotypes AD diagnosis and its major contributor, ADASCog13, in addition to lipids shared between total tau and SPARE-AD. Conversely, Abeta142 showed few shared lipids. Importantly, there was no identified lipid that was shared between four phenotypes, and only one lipid that was associated with all AD-phenotypes (arachidonyl-lysophosphatidylethanolamine; LPE C20:4). Many lipids are coregulated by the activity of specific lipases or other enzymes involved in lipid remodeling. Identifying commonalities of biochemical mechanisms may lead to underlying genetic drivers or environmental factors implicated in AD etiology. Therefore, we next focused on identifying sets of coregulated lipids associated with AD pathophysiology rather than interpreting individual lipids.
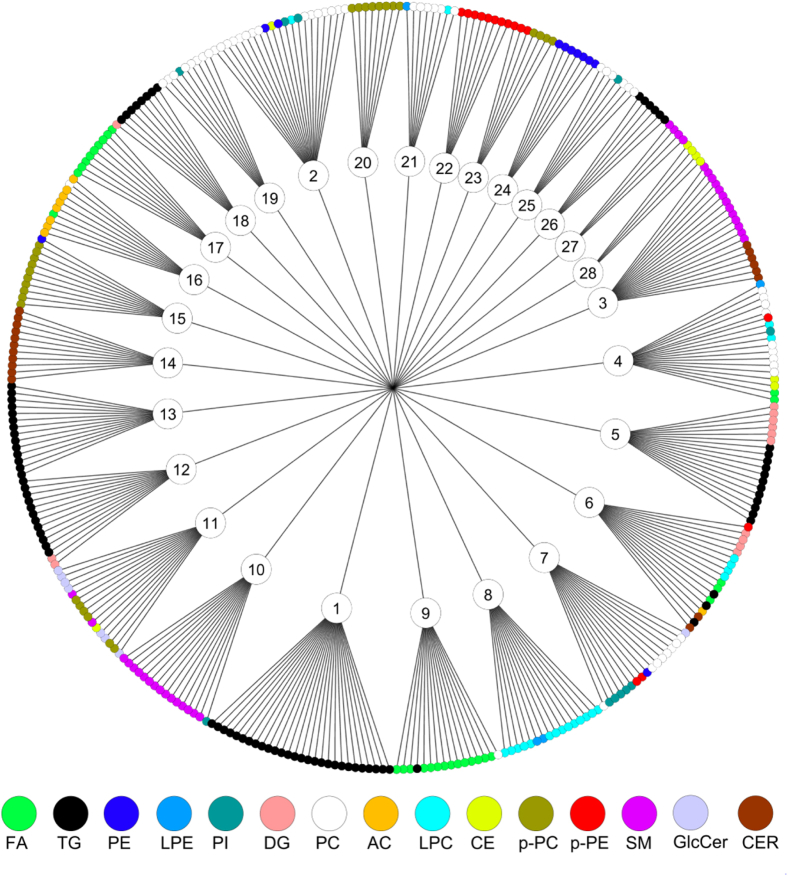
Fig. 1.
Coregulated sets of serum lipids in the ADNI lipidomics data set. Sets were detected by the dynamicTreeCut algorithm (see method). Node colors show different chemical classes. Abbreviations: FA, fatty acids; AC, acyl carnitines; PC, phosphatidylcholines; CE, cholesteryl esters; SM, sphingomyelins; Cer, ceramides; PE, phosphatidylethanolamines; TG, triacylglycerols; PI, phosphatidylinositols; DG, diacylglycerols; LPC, lysophosphatidylcholines.
Fig. 2.
Number of significantly different lipids with AD phenotypes in univariate statistics. Directions of beta coefficients in regression models are given by colors as blue (negative) and red (positive) associations using uncorrected P < .05 values. Abbreviations: CN, cognitively normal; LMCI, late mild cognitive impairment; AD, Alzheimer's disease. DIAG, linear models for diagnosis; tau, linear model for tau; Aβ1-42, linear model for amyloid beta peptide 42; SPARE-AD, linear model for Spatial Pattern of Abnormality for Recognition of Early Alzheimer's disease index; ADASCog13, Cognitive Subscale of the Alzheimer's Disease Assessment Scale index.
Fig. 3.
Number of significantly associated lipids across AD phenotypes. Uncorrected P < .05 values for five AD phenotypes. Abbreviations: DIAG, linear models for diagnosis; tau, linear model for tau; ABETA142, linear model for amyloid beta peptide 42; SPARE-AD, linear model for Spatial Pattern of Abnormality for Recognition of Early Alzheimer's disease index; ADASCog13, Cognitive Subscale of the Alzheimer's Disease Assessment Scale index.
3.3. Identifying sets of coregulated lipids
Lipid classification often uses chemical structure as the only criterion. To specify the biochemical relationships between circulating blood lipids, we instead used correlation analysis to determine sets of lipids. A pairwise Spearman correlation matrix followed by hierarchical clustering with the DynamicTreeCut dendrogram cutting method [18] yielded a total of 28 coregulated lipid sets in the Alzheimer's Disease Neuroimaging Initiative Phase 1 data set (Fig. 3). The mean size was 12.5 lipids per set, ranging from 4 to 28 members. These lipid sets (LM) were named LM1 to LM28. The average Spearman correlation coefficient rho across sets was 0.63 with a range of 0.19 < ρ < 0.82. Fig. 3 and Supplementary Table 2 show that some lipid coregulatory sets were composed of lipids from the same chemical classes (such as set 17 for free fatty acids, set 1 for triacylglycerides, and set 14 for ceramides) whereas other sets were heterogeneous (such as set 3 consisting of ceramides and SMs, or set 7 that includes phosphatidylinositols and phosphatidylcholines). Interestingly, several classes of lipids were found with distinct coregulation within each class. For example, triacylglycerides were not found as one large group of coregulated species, but clustered in three specific sets, and similarly, free fatty acids were found in two different sets, set 9 consisting only of saturated fatty acids and set 17 comprised only of unsaturated fatty acids. Similarly, other lipid classes were distributed across different sets, too. For example, phosphatidylcholines were found in five sets and SMs were coregulated in three sets, indicating downstream regulation of lipid biochemistry by specific elongases, desaturases, lipases, and acyl-transferases within each lipid class (Fig. 2).
3.4. Associating lipid sets with AD phenotypes
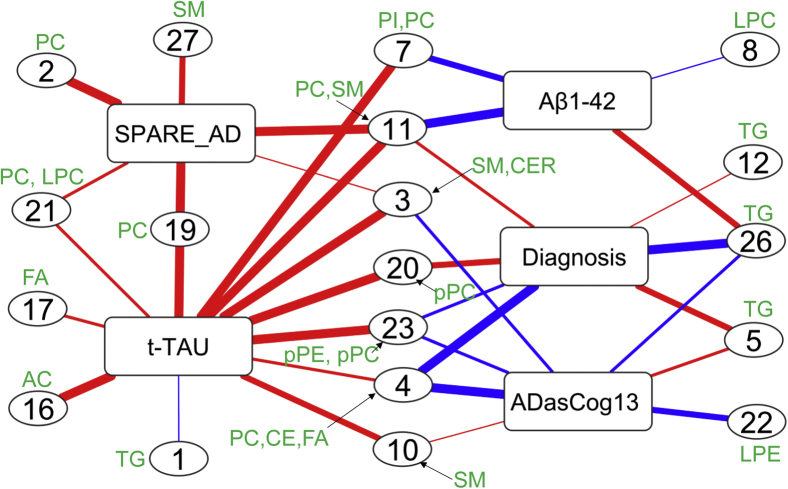
These lipid groups served as input for a lipid-set enrichment analysis [19] along with the P-value and beta coefficient results from the regression models. Overall, 19 of 28 lipid sets were significantly associated with at least one AD phenotype (Fig. 4, Supplementary Table 3) using the Kolmogorov-Smirnoff statistical test with false discovery rate (FDR) corrections. Eight sets were uniquely associated with only one specific AD phenotype, but only one set was associated with four phenotypes, set 11, consisting primarily of ceramides and phosphatidylcholines. Set 11 did not include polyunsaturated acyl chains with three or more double bonds. Six sets were associated with two AD phenotypes and four sets were correlated with three AD phenotypes, but no set correlated with all five phenotypes.
Fig. 4.
Coregulated sets of lipids significantly associated with AD phenotypes. Direction of associations is given by red (positive) and blue (negative) edge colors. Line thickness indicates the significance of associations (see Supplementary Table 4 for details). Lipid compositions for each set are shown in Fig. 1 and Supplementary Table 1. Abbreviations: DIAG, linear models for diagnosis; tau, linear model for tau; ABETA142, linear model for amyloid beta peptide 42; SPARE-AD, linear model for Spatial Pattern of Abnormality for Recognition of Early Alzheimer's disease index; ADASCog13, Cognitive Subscale of the Alzheimer's Disease Assessment Scale index.
More than two-thirds of all associations were positively correlated between lipid sets and phenotypes, mostly driven by the t-tau phenotype that also had the highest number of correlated lipid sets. Conversely, ADASCog13 showed the highest number of negative associations with lipid sets. We therefore investigated the individual phenotypes with respect to the composition of their associated lipid sets.
3.4.1. Lipid sets associated with AD diagnosis
AD diagnosis was significantly associated with seven distinct lipid sets (Fig. 4) after FDR correction. Specifically, the phenotype was highly significantly negatively correlated with lipid set 26 and set 4. Both sets comprised acyl chains with at least one polyunsaturated fatty acyl chain (PUFA) (see Supplementary Table 2), either eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA), or arachidonic acid (AA). Set 26 consisted exclusively of triacylglycerides that also contained either EPA or DHA, but not a single saturated fatty acid. Set 4 was mixed between different phospholipid head groups, cholesteryl esters and free fatty acids, indicating that the coregulation mechanisms focused on the modulation and incorporation of acyl chains irrespective of the lipid class. Set 23 was also negatively correlated with AD diagnosis and comprised DHA-containing choline- and ethanolamine-plasmalogens. Conversely, two other sets of lipids were positively associated with AD diagnosis, most significantly for set 5 and set 20, and less significantly with set 11 and set 12. Set-5 contained coregulated di- and triacylglycerols with acyl groups that did not contain any PUFA with four or more double bonds, and only one single lipid with linolenic acid (C18:3). Set-20 was exclusively composed of unsaturated choline-plasmalogens, but not containing any EPA or DHA acyl chain.
3.4.2. Lipid sets associated with CSF Aβ1-42
CSF Aβ1-42 was significantly associated with four lipid sets (Fig. 4). Three sets were negatively correlated, set 11, set 7, and set 8. Set-7 was the only lipid set that contained phosphatidylinositols, in addition to phosphatidylcholines. Acyl chains were primarily saturated or mono- and di-unsaturated. Similarly, set 8 consisted mostly of desaturated acyl groups with less than four double bonds, exclusively found as lysophosphatidylcholines. In the same way, no PUFA acyl chains were found in set 11, a heterogenous set of ceramides and choline-plasmalogens. Importantly, the only positive association of a lipid set with CSF Aβ1-42 was set 26 that was completely made of PUFA triacylglycerides.
3.4.3. Lipid sets associated with CSF tau
CSF total tau correlated with 12 lipid sets, the highest number of associated lipid sets among all phenotypes (Fig. 4). All sets except set 1 were positively correlated with CSF-total tau. Three unique sets were not shared with other phenotypes. Set-16 was composed of acylcarnitines with increasing degree of desaturation, and set 17 was a set of monounsaturated fatty acids. Set-1 was less significant in comparison with other set associations. Four sets were shared with brain atrophy, four sets were shared with AD diagnosis, two sets with Aβ and four lipid sets were shared with cognitive decline. Notably, set 19 was also associated with brain atrophy and contained mostly EPA-side chain phosphatidylcholines. Set-3 was composed of SMs and ceramides that were not associated with diagnosis or Aβ, but instead was also linked with cognitive decline and SPARE-AD.
3.4.4. Lipid sets associated with brain atrophy (SPARE-AD)
Brain atrophy was most significantly associated with set 27, 19, 11, and 2 (Fig. 4). Three sets (set 2, set 11, and set 27) were void of PUFA side chains with either phospholipid or sphingolipid head groups. Conversely, set 19 contained mostly EPA–side chain phosphatidylcholines and was further associated with CSF total tau. Similarly, set 21 was associated with both phenotypes, containing phospholipids and their lyso-forms with the PUFA acyl chain arachidonic acid.
3.4.5. Lipid sets associated with cognitive functions
Most of the lipid sets associated with cognitive decline were also associated with AD diagnosis (Fig. 4). In addition, it was negatively associated with set set 22 which consisted of ethanolamine plasmalogens.
4. Discussion
We here focused on associations between blood lipids and five AD phenotypes guided by known contributions of lipids and metabolic comorbidities to AD. We systematically tested both the association of individual lipids and the association with sets of coregulated lipids. This approach showed an important advantages over previous “feature” based lipidomics AD studies [9], [20] that did not focus on specific lipid groups, their side chains, or their biological regulation. Without clear lipid identification, feature-based associations miss biological insights and have a high risk of not being validated in subsequent studies because each individual lipid may cause more than one feature in LC/MS based lipidomics [20], [21]. Instead we used the largest published AD lipidomics data set [17] to date with 349 identified lipids belonging to 13 major lipid classes, identified by extensive MS/MS fragmentation analysis [22] and enabling analyses reaching to the level of acyl chains. A second difference to previous efforts was a focus on summarizing lipids by statistical coregulation instead of only relying on univariate analysis or grouping by lipid head groups. This expansion of classic statistical analysis was critical to extend from diagnostic biomarkers (that need correction for multiple testing using FDR adjustments) to revealing underlying biological mechanisms. The axiom of univariate analyses, the mutual independence of variables, is untrue in lipid biology. Moreover, stringent FDR corrections lead to an increased number of false negative results and compromise the statistical power to investigate the biological mechanisms and pathways. As lipids are poorly presented in biochemical pathway databases [19], classic metabolic pathway enrichment analysis [23] ignores most detected lipids and is unsuitable for lipidomics. Instead, Spearman-rank correlation–based matrices yielded specific clusters of lipids associated with AD phenotypes by using the robust Kolmogorov–Smirnov test for P-value distributions. These lipid sets showed very distinct metabolic features that we identified as preferential use of specific polyunsaturated fatty acids that replaced saturated or monounsaturated fatty acids for distinct lipid classes. These mechanisms of lipid desaturation, elongation, and acyl-chain remodeling were disturbed in early and later stages of AD. A minimal overlap among lipid sets was observed (Fig. 3) with respect to statistical associations with AD phenotypes, indicating that quite distinct lipid biochemical processes were involved in the early and later stages of AD. Lipid metabolic pathways associated with the early stage, in particular, may provide new therapeutic targets to stop AD progression. MUFA-containing lipids were positively associated with the brain atrophy and tau accumulation, whereas PUFA-containing lipids were negatively associated with AD diagnosis and cognitive decline. Therapeutic strategies targeting MUFA lipid pathways at the early stages of AD could therefore be potentially more effective in delaying the progression of the disease.
4.1. Lipids linked to the amyloid beta clearance pathway
A decrease in the CSF Aβ1-42 peptide marker is indicating a poor clearance of the peptide in the brain, leading to its extraneuronal accumulation. In our study, poor clearance was indicated by negative associations with lipids sets, including sets that contained phosphatidylinositols, lysoPCs, ceramides and choline-plasmalogens and PUFA TGs. The Aβ peptide is known to cause mitochondrial dysfunction [24] which can lead to neurodegeneration via autophagic cascades [25], [26]. The associated lipids, specifically ceramides and phosphatidylinositols and lysoPCs have been linked with cell death and may also contribute in the Aβ–mediated toxicity in neurons [27], [28], [29]. Higher levels of ceramides containing oleic acid (C18:1) have been shown to increase AD risk [4], [5]. We validate this finding in our study and also observed lower levels of phosphoinositols containing polyunsaturated fatty acids to correlate with poor Aβ clearance. An alternative explanation to our data is an impaired Aβ clearance in the liver [30] that subsequently leads to dysregulation of lipid metabolism in the liver. Overall, our data suggest that these lipid sets can serve as serum biomarkers for disturbed Aβ pathway regulation in brain and can complement Aβ imaging assays.
4.2. Cerebrospinal fluid total tau
CSF tau is a marker for accumulating tau tangles in the brain tissues, causing neurodegeneration. We found that the total CSF tau marker was significantly associated with lipid sets enriched in monounsaturated fatty acids, acylcarnitines, ceramides, SMs, and EPA-containing phosphatidylcholines. Increased fatty acids and acylcarnitines are known markers of impaired fatty acid beta oxidation in mitochondria [31], specially during metabolic diseases such as diabetes and obesity [32], [33]. We found free fatty acids and acylcarnitines to be positively correlated with total tau, supporting the notion of tau mediated neurodegeneration and mitochondrial dysfunctions. Mitochondrial impairment was further evidenced by positive associations of total tau with sets of ceramides because accumulating ceramides are known to induce cell death and to increase the AD risk in normal subjects [5]. Higher ceramide levels were also reported for early-stage AD [34], [35], [36]. These findings indicate that these lipids may be involved in early neurodegenerative pathways, and their underlying pathways might lead to candidates for new therapeutic strategies.
4.3. Lipid sets linked with brain atrophy
SPARE-AD is a composite index of brain atrophy and indicates the neurodegeneration magnitude. We found a high overlap of lipid sets that were associated with both SPARE-AD and total tau, reinforcing the usability of these serum lipids as biomarkers for neurodegeneration. These lipid sets included phosphatidylcholines and sphingolipids that were enriched in PUFA eicosapentaenoic acid and arachidonic acid (EPA, AA). These fatty acids are main components of brain lipids [37], [38]. The loss of brain tissue may cause an increase in levels of serum lipids that include EPA and AA as acyl groups through lipid remodeling [39], [40]. We here identify lipid pathways associated with tau-mediated brain atrophy that eventually contributes to AD.
4.4. AD diagnosis and cognitive decline
Previous publications reported that lower levels of PUFA in AD subjects across multiple lipid classes, along with higher levels of monounsaturated lipids [4], [8], [9], [41], [42], [43], [44], [45]. We found numerous, very significant associations of omega-3 and omega-6 containing complex lipids with AD diagnosis and cognitive functions. Our analysis is consistent with these results as shown by AD-associated lipids in set 4, set 20, and set 23 (Fig. 4). We here specify that the major difference is not related to total levels of mono- or polyunsaturated fatty acids, but the extent at which these fatty acids are incorporated into different complex lipids. Clear differences in circulating PUFA phospholipid and PUFA triacylglycerol levels in AD subjects in comparison with normal subjects were observed, likely due to dysregulation of biosynthesis in liver. Here, substrate preference of MGAT and DGAT enzymes in the liver may play an important role, but the exact specificities of acyltransferase enzymes (and their corresponding lipase enzymes) are not well studied. Levels of anti-inflammatory plasmalogens [46], important lipids for brain membrane functions [44], [47], were decreased in AD patients in comparison with cognitively normal subjects. Lower levels of plasmalogens have been linked to AD [44]. However, in clinical trials, EPA and DHA supplementation do not improve the cognitive function of AD subjects [48]. Nutritional intervention trials such as the European LipiDiDiet have failed to show any cognitive improvement in AD subjects. A broad-spectrum effect of fish oil supplements on additional lipid pathways may explain the failure of this trial and warrants further lipidomics studies for serum specimens of this trial's participants. It was observed that the incorporation of omega-6 fatty acids was increased in AD subjects. These fatty acids are well-known precursors to proinflammatory molecules such as leukotrienes. Further studies are needed to test if postmortem brain tissues of AD subjects show similar disruption in fatty acid incorporation and if these patterns correlate with AD severity or other AD phenotypes.
Coregulatory lipid sets and their associations with AD biomarkers will be further tested in reference to a range of covariates including age, BMI, sex, race, diet, and comorbidities in larger cohorts and diverse population settings.
5. Conclusions
Using coregulated sets of lipids enabled us to find significant associations of lipids with AD that led to biochemical mechanisms. Across the spectrum of AD progression, pathways were dysregulated that pointed to lipid desaturation, elongation and remodeling. These pathways provide new targets as well candidate biomarkers for the population screening for AD prevention. Future studies are needed to tease out the roles of genetic variations, drug, and diet the metabolism of MUFA and PUFAs and their complex lipids and their roles in AD.
Research in Context.
-
1.
Systematic review: Genes such as APOE-ε4 as well as comorbidity with metabolic diseases such as type 2 diabetes support the concept that lipid metabolism is critical in the etiology of Alzheimer's disease (AD). Yet, current understanding of altered lipid biochemistry in AD remains sparse with respect to both brain metabolism and the relationship to circulating lipids.
-
2.
Interpretation: Sets of blood lipids were associated with CSF AD biomarkers and with clinical diagnosis of AD or mild cognitive impairment. Our study provides several new hypotheses for studying the role of lipid metabolism in AD.
-
3.
Future directions: The association of specific blood lipids with AD phenotypes raises new hypotheses to be tested in a subsequent analyses and studies. (A) The coregulation of human blood lipids in triacylglycerol and phospholipids classes needs to be tested and replicated with respect to universality or specificity for age-dependent cohorts. (B) The remodeling of acyl chains needs to be studied in relation to genetic variants and environmental factors. (C) Specifically, the impact of dietary supplements and drugs on sets of coregulated lipids and their associations with AD phenotypes must be investigated.
Acknowledgments
This work was funded through NIH awards U54 AI138370, U19 AG023122 and U2C ES030158. National Institute on Aging (R01AG046171, RF1AG051550, and RF1AG057452 and 3U01AG024904-09S4) supported the Alzheimer Disease Metabolomics Consortium which is a part of NIA national initiatives AMP-AD and M2OVE AD. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Authors' contributions: D.K.B. and O.F. generated the lipidomics data set. D.K.B., R.B., R.K.D., and O.F. designed the study. D.K.B. and S.F. performed the statistical analyses. A.J.S., P.J.M., M.A., and K.N. contributed in data interpretation. All author contributed in the manuscript writing.
Footnotes
R B. reported receiving a salary from Rosa & Co outside the submitted work. A.J.S. reported receiving grants from the NIH during the conduct of the study; receiving nonfinancial support from Avid Radiopharmaceuticals; receiving investigator-initiated research support from Eli Lilly unrelated to the work reported here; receiving consulting fees and travel expenses from Eli Lilly and Siemens Healthcare; serving as a consultant to Arkley BioTek; and receiving support from Springer-Nature publishing as Editor-In-Chief of Brain Imaging and Behavior. R.K.-D. reported being an inventor on key patents in the field of metabolomics including applications for Alzheimer disease. P.J.M. has licensed biomarker intellectual property to Zora Biosciences and holds other relevant patents (Au2018901220). No other disclosures were reported.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2019.07.002.
Contributor Information
Oliver Fiehn, Email: ofiehn@ucdavis.edu.
Rima Kaddurah-Daouk, Email: kaddu001@mc.duke.edu.
Supplementary Data
References
- 1.Scheltens P., Blennow K., Breteler M.M., de Strooper B., Frisoni G.B., Salloway S. Alzheimer's disease. Lancet. 2016;388:505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 2.Liu C.C., Liu C.C., Kanekiyo T., Xu H., Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressler J., Yu B., Mosley T.H., Knopman D.S., Gottesman R.F., Alonso A. Metabolomics and cognition in African American adults in midlife: the atherosclerosis risk in communities study. Translational psychiatry. 2017;7:e1173. doi: 10.1038/tp.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han X., Rozen S., Boyle S.H., Hellegers C., Cheng H., Burke J.R. Metabolomics in early Alzheimer's disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PloS one. 2011;6:e21643. doi: 10.1371/journal.pone.0021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mielke M.M., Bandaru V.V., Haughey N.J., Xia J., Fried L.P., Yasar S. Serum ceramides increase the risk of Alzheimer disease: the Women's Health and Aging Study II. Neurology. 2012;79:633–641. doi: 10.1212/WNL.0b013e318264e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schilling S., Tzourio C., Soumare A., Kaffashian S., Dartigues J.F., Ancelin M.L. Differential associations of plasma lipids with incident dementia and dementia subtypes in the 3C Study: A longitudinal, population-based prospective cohort study. PLoS Med. 2017;14:e1002265. doi: 10.1371/journal.pmed.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Lee S.J., Teunissen C.E., Pool R., Shipley M.J., Teumer A., Chouraki V. Circulating metabolites and general cognitive ability and dementia: Evidence from 11 cohort studies. Alzheimer's Dement. 2018;14:707–722. doi: 10.1016/j.jalz.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Proitsi P., Kim M., Whiley L., Simmons A., Sattlecker M., Velayudhan L. Association of blood lipids with Alzheimer's disease: A comprehensive lipidomics analysis. Alzheimers Dement. 2017;13:140–151. doi: 10.1016/j.jalz.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Proitsi P., Kim M., Whiley L., Pritchard M., Leung R., Soininen H. Plasma lipidomics analysis finds long chain cholesteryl esters to be associated with Alzheimer's disease. Translational Psychiatry. 2015;5:e494. doi: 10.1038/tp.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varma V.R., Oommen A.M., Varma S., Casanova R., An Y., Andrews R.M. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: A targeted metabolomics study. PLoS Med. 2018;15:e1002482. doi: 10.1371/journal.pmed.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo J.B., Arnold M., Kastenmuller G., Chang R., Baillie R.A., Han X., I. Alzheimer's Disease Neuroimaging. C. the Alzheimer Disease Metabolomics Metabolic network failures in Alzheimer's disease: A biochemical road map. Alzheimers Dement. 2017;13:965–984. doi: 10.1016/j.jalz.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mapstone M., Cheema A.K., Fiandaca M.S., Zhong X., Mhyre T.R., MacArthur L.H. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiandaca M.S., Zhong X., Cheema A.K., Orquiza M.H., Chidambaram S., Tan M.T. Plasma 24-metabolite Panel Predicts Preclinical Transition to Clinical Stages of Alzheimer's Disease. Front Neurol. 2015;6:237. doi: 10.3389/fneur.2015.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casanova R., Varma S., Simpson B., Kim M., An Y., Saldana S. Blood metabolite markers of preclinical Alzheimer's disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement. 2016;12:815–822. doi: 10.1016/j.jalz.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu M., Zhang D.F., Luo R., Wu Y., Zhou H., Kong L.L. A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer's disease. Alzheimers Dement. 2018;14:215–229. doi: 10.1016/j.jalz.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Toledo J.B., Da X., Bhatt P., Wolk D.A., Arnold S.E., Shaw L.M. Relationship between plasma analytes and SPARE-AD defined brain atrophy patterns in ADNI. PloS one. 2013;8:e55531. doi: 10.1371/journal.pone.0055531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barupal D.K., Fan S., Wancewicz B., Cajka T., Sa M., Showalter M.R. Generation and quality control of lipidomics data for the Alzheimer's disease neuroimaging initiative cohort. Sci Data. 2018;5:180263. doi: 10.1038/sdata.2018.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langfelder P., Zhang B., Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 19.Barupal D.K., Fiehn O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci Rep. 2017;7:14567. doi: 10.1038/s41598-017-15231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahieu N.G., Patti G.J. Systems-Level Annotation of a Metabolomics Data Set Reduces 25000 Features to Fewer than 1000 Unique Metabolites. Anal Chem. 2017;89:10397–10406. doi: 10.1021/acs.analchem.7b02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden J.A., Heckert A., Ulmer C.Z., Jones C.M., Koelmel J.P., Abdullah L. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in Frozen Human Plasma. J lipid Res. 2017;58:2275–2288. doi: 10.1194/jlr.M079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kind T., Liu K.H., Lee D.Y., DeFelice B., Meissen J.K., Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia J., Wishart D.S. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 24.Mossmann D., Vogtle F.N., Taskin A.A., Teixeira P.F., Ring J., Burkhart J.M. Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab. 2014;20:662–669. doi: 10.1016/j.cmet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 26.Suomalainen A., Battersby B.J. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cel Biol. 2017;19:77–92. doi: 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]
- 27.Elliott M.R., Ravichandran K.S. The Dynamics of Apoptotic Cell Clearance. Dev Cell. 2016;38:147–160. doi: 10.1016/j.devcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magtanong L., Ko P.J., Dixon S.J. Emerging roles for lipids in non-apoptotic cell death. Cell Death Differ. 2016;23:1099–1109. doi: 10.1038/cdd.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong M.W., Braidy N., Poljak A., Pickford R., Thambisetty M., Sachdev P.S. Dysregulation of lipids in Alzheimer's disease and their role as potential biomarkers. Alzheimers Dement. 2017;13:810–827. doi: 10.1016/j.jalz.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Maarouf C.L., Walker J.E., Sue L.I., Dugger B.N., Beach T.G., Serrano G.E. Impaired hepatic amyloid-beta degradation in Alzheimer's disease. PloS one. 2018;13:e0203659. doi: 10.1371/journal.pone.0203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciavardelli D., Piras F., Consalvo A., Rossi C., Zucchelli M., Di Ilio C. Medium-chain plasma acylcarnitines, ketone levels, cognition, and gray matter volumes in healthy elderly, mildly cognitively impaired, or Alzheimer's disease subjects. Neurobiol Aging. 2016;43:1–12. doi: 10.1016/j.neurobiolaging.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Adams S.H., Hoppel C.L., Lok K.H., Zhao L., Wong S.W., Minkler P.E. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139:1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serra D., Mera P., Malandrino M.I., Mir J.F., Herrero L. Mitochondrial fatty acid oxidation in obesity. Antioxid Redox Signal. 2013;19:269–284. doi: 10.1089/ars.2012.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han X., M Holtzman D., McKeel D.W., Jr., Kelley J., Morris J.C. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer's disease: potential role in disease pathogenesis. J Neurochem. 2002;82:809–818. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- 35.He X., Huang Y., Li B., Gong C.X., Schuchman E.H. Deregulation of sphingolipid metabolism in Alzheimer's disease. Neurobiol Aging. 2010;31:398–408. doi: 10.1016/j.neurobiolaging.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cutler R.G., Kelly J., Storie K., Pedersen W.A., Tammara A., Hatanpaa K. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer's disease. Proc Natl Acad Sci U S A. 2004;101:2070–2075. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyall S.C. Long-chain omega-3 fatty acids and the brain: a review of the independent and shared effects of EPA, DPA and DHA. Front Aging Neurosci. 2015;7:52. doi: 10.3389/fnagi.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin V., Fabelo N., Santpere G., Puig B., Marin R., Ferrer I. Lipid alterations in lipid rafts from Alzheimer's disease human brain cortex. J Alzheimers Dis. 2010;19:489–502. doi: 10.3233/JAD-2010-1242. [DOI] [PubMed] [Google Scholar]
- 39.Hashidate-Yoshida T., Harayama T., Hishikawa D., Morimoto R., Hamano F., Tokuoka S.M. Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. Elife. 2015;4 doi: 10.7554/eLife.06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang B., Rong X., Duerr M.A., Hermanson D.J., Hedde P.N., Wong J.S. Intestinal Phospholipid Remodeling Is Required for Dietary-Lipid Uptake and Survival on a High-Fat Diet. Cell Metab. 2016;23:492–504. doi: 10.1016/j.cmet.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iuliano L., Pacelli A., Ciacciarelli M., Zerbinati C., Fagioli S., Piras F. Plasma fatty acid lipidomics in amnestic mild cognitive impairment and Alzheimer's disease. J Alzheimers Dis. 2013;36:545–553. doi: 10.3233/JAD-122224. [DOI] [PubMed] [Google Scholar]
- 42.Cunnane S.C., Schneider J.A., Tangney C., Tremblay-Mercier J., Fortier M., Bennett D.A. Plasma and brain fatty acid profiles in mild cognitive impairment and Alzheimer's disease. J Alzheimer's Dis : JAD. 2012;29:691–697. doi: 10.3233/JAD-2012-110629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conquer J.A., Tierney M.C., Zecevic J., Bettger W.J., Fisher R.H. Fatty acid analysis of blood plasma of patients with Alzheimer's disease, other types of dementia, and cognitive impairment. Lipids. 2000;35:1305–1312. doi: 10.1007/s11745-000-0646-3. [DOI] [PubMed] [Google Scholar]
- 44.Wood P.L., Mankidy R., Ritchie S., Heath D., Wood J.A., Flax J. Circulating plasmalogen levels and Alzheimer Disease Assessment Scale-Cognitive scores in Alzheimer patients. J Psychiatry Neurosci. 2010;35:59–62. doi: 10.1503/jpn.090059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mielke M.M., Haughey N.J., Bandaru V.V., Schech S., Carrick R., Carlson M.C. Plasma ceramides are altered in mild cognitive impairment and predict cognitive decline and hippocampal volume loss. Alzheimers Dement. 2010;6:378–385. doi: 10.1016/j.jalz.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmiena A.A., Barlow C.K., Stefanovic N., Huynh K., Tan R., Sharma A. Plasmalogen modulation attenuates atherosclerosis in ApoE- and ApoE/GPx1-deficient mice. Atherosclerosis. 2015;243:598–608. doi: 10.1016/j.atherosclerosis.2015.10.096. [DOI] [PubMed] [Google Scholar]
- 47.Scott B.L., Bazan N.G. Membrane docosahexaenoate is supplied to the developing brain and retina by the liver. Proc Natl Acad Sci U S A. 1989;86:2903–2907. doi: 10.1073/pnas.86.8.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazereeuw G., Lanctot K.L., Chau S.A., Swardfager W., Herrmann N. Effects of omega-3 fatty acids on cognitive performance: a meta-analysis. Neurobiol Aging. 2012;33:1482.e1417–1482.e1429. doi: 10.1016/j.neurobiolaging.2011.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.