Abstract
The dataset presented in this article supports “Selected persistent organic pollutants associated with the risk of primary ovarian insufficiency in women” (Pan et al., 2019). The supplementary data were as follows: (1) Detailed information regarding pretreatment methods, instrumental analysis and methods validation of quantification of serum concentrations of persistent organic pollutants (POPs). (2) The total dioxin equivalents (TEQs) levels of dioxin-like PCBs (DL-PCBs) in primary ovarian insufficiency (POI) cases and controls, as well as the association of TEQ levels with the risk of POI. (3) The results of principal components analyses (PCA) about 20 POPs that were detected in >40% samples.
Keywords: Persistent organic pollutants, Polychlorinated biphenyls, Organochlorine pesticides, Primary ovarian insufficiency, Pretreatment methods
Specifications Table
| Subject area | Chemistry |
| More specific subject area | Analytical chemistry |
| Type of data | Tables and figures |
| How data was acquired | Gas chromatography-triple quadrupole mass spectrometer (GC-MS/MS) (Agilent 7890B GC/7000C) |
| Data format | Raw and Analyzed |
| Experimental factors | Spiked 0.3 mL of serum sample in a centrifuge tube with internal standards [PCB 209, tetrachloro-m-xylene (TCMX), isotopically labeled standards of PBDEs]. After three times of liquid-liquid extraction by extractant of n-hexane and dichloromethane (DCM) (1:1, v/v), evaporating the extracts to about 1 mL, and cleaned by a column filled with activated silica gel and Na2SO4. The elution was evaporated to dryness and redissolved in 50 μL of n-decane. |
| Experimental features | Recruited 157 primary ovarian insufficiency (POI) cases and 217 healthy controls. Serum concentrations of polychlorinated biphenyls (PCBs), organochlorine pesticides (OCPs), polybrominated diphenyl ethers (PBDEs) were measured. |
| Data source location | Zhejiang, China |
| Data accessibility | The data are given in this article |
| Related research article | Pan, W.; Ye, X.; Yin, S.; Ma, X.; Li, C.; Zhou, J.; Liu, W.; Liu, J. Selected persistent organic pollutants associated with the risk of primary ovarian insufficiency in women. Environment international. 129 (2019) 51–58[1] |
Value of the data
|
1. Data
The data reported here constitute the basis for the article by Pan et al. [1] The detailed information about sample pretreatment method, instrumental analysis and method validation for determination of persistent organic pollutants (POPs) in serum samples were presented in Table 1, Table 2, Table 3, Table 4, Table 5 and Fig. 1, Fig. 2. Table 6 and Table 7 showed the total dioxin equivalents (TEQs) levels of dioxin-like PCBs (DL-PCBs) in primary ovarian insufficiency (POI) cases and heathy controls, as well as the association of TEQ levels with the risk of POI. Principal components analyses results about 20 POPs that were detected in >40% samples were summarized in Table 8 and Table 9. The raw data of Table 2, Table 3, Table 6 were available in the file Supplementary Table 1, 2 and 4, respectively. The raw data of Fig. 1, Fig. 2 were available in the file Supplementary Table 3.
Table 1.
Instrumental and quantification methods.
| Type | Compound | IS | Quantifier |
Qualifier |
RT |
||||
|---|---|---|---|---|---|---|---|---|---|
| P | F | CE | P | F | CE | (min) | |||
| IS | TCMX | 244 | 209 | 15 | 136 | 75.2 | 15 | 10.8 | |
| AT | PCB8 | TCMX | 222 | 152 | 30 | 152.1 | 151 | 5 | 11.7 |
| AT | α-HCH | TCMX | 216.9 | 181 | 5 | 181 | 145 | 5 | 11.7 |
| AT | HCB | TCMX | 283.9 | 213.9 | 35 | 283.9 | 248.8 | 25 | 11.9 |
| AT | β-HCH | TCMX | 216.9 | 181 | 5 | 181 | 145 | 5 | 12.3 |
| AT | γ-HCH | TCMX | 216.9 | 181 | 5 | 181 | 145 | 5 | 12.4 |
| AT | PCB18 | TCMX | 256 | 186 | 30 | 258 | 186.1 | 30 | 12.6 |
| AT | δ-HCH | TCMX | 216.9 | 181 | 5 | 181 | 145 | 5 | 12.9 |
| AT | PCB28 | TCMX | 256 | 186 | 30 | 258 | 186.1 | 30 | 13.6 |
| AT | Heptachlor | TCMX | 272 | 237 | 25 | 272 | 117 | 40 | 13.9 |
| AT | PCB52 | TCMX | 292 | 220 | 35 | 222 | 150 | 35 | 14.3 |
| AT | Aldrin | TCMX | 262.9 | 192.9 | 40 | 262.9 | 190.9 | 40 | 14.6 |
| AT | PCB44 | TCMX | 292 | 220 | 35 | 222 | 150 | 35 | 14.7 |
| AT | HCEX | TCMX | 352.9 | 262.8 | 25 | 352.9 | 281.9 | 20 | 15.5 |
| AT | PCB66 | TCMX | 292 | 220 | 35 | 220 | 150 | 35 | 15.6 |
| AT | o,p'-DDE | TCMX | 246 | 176.2 | 30 | 248 | 176.2 | 30 | 16.1 |
| AT | PCB101 | TCMX | 326 | 255.9 | 35 | 256 | 183.9 | 35 | 16.2 |
| AT | PCB81 | TCMX | 292 | 220 | 35 | 222 | 150 | 35 | 16.8 |
| AT | p,p'-DDE | TCMX | 246 | 176.2 | 30 | 248 | 176.2 | 30 | 16.8 |
| AT | PCB77 | TCMX | 292 | 220 | 35 | 222 | 150 | 35 | 17.0 |
| AT | o,p'-DDD | TCMX | 235 | 165.2 | 30 | 237 | 165.2 | 20 | 17.0 |
| AT | Endrin | TCMX | 262.9 | 192.9 | 35 | 262.9 | 190.9 | 35 | 17.4 |
| AT | PCB123 | TCMX | 326 | 255.9 | 35 | 256 | 183.9 | 35 | 17.5 |
| AT | PCB118 | TCMX | 326 | 255.9 | 35 | 256 | 183.9 | 35 | 17.6 |
| AT | p,p'-DDD | TCMX | 235 | 165.2 | 30 | 237 | 165.2 | 20 | 17.8 |
| AT | PCB114 | TCMX | 326 | 255.9 | 35 | 256 | 183.9 | 35 | 17.9 |
| AT | o,p'-DDT | TCMX | 235 | 165.2 | 30 | 237 | 165.2 | 20 | 17.9 |
| AT | PCB153 | PCB209 | 360 | 289.9 | 30 | 290 | 218 | 30 | 18.2 |
| AT | PCB105 | TCMX | 326 | 255.9 | 35 | 256 | 183.9 | 35 | 18.3 |
| AT | p,p'-DDT | TCMX | 235 | 165.2 | 30 | 237 | 165.2 | 20 | 18.8 |
| AT | PCB138 | PCB209 | 360 | 289.9 | 30 | 290 | 218 | 30 | 18.9 |
| AT | PCB126 | TCMX | 326 | 255.9 | 35 | 256 | 183.9 | 35 | 19.2 |
| AT | PCB187 | PCB209 | 394 | 324 | 30 | 324 | 254 | 30 | 19.4 |
| AT | PCB167 | PCB209 | 360 | 289.9 | 30 | 290 | 218 | 30 | 19.7 |
| AT | PCB156 | PCB209 | 360 | 289.9 | 30 | 290 | 218 | 30 | 20.3 |
| AT | PCB157 | PCB209 | 360 | 289.9 | 30 | 290 | 218 | 30 | 20.5 |
| AT | PCB170 | PCB209 | 394 | 324 | 30 | 324 | 254 | 30 | 20.8 |
| IS | BDE47 | 498 | 338 | 20 | 496 | 336 | 30 | 20.9 | |
| AT | BDE47 | BDE47 | 486 | 326 | 20 | 484 | 324 | 30 | 20.9 |
| AT | PCB169 | PCB209 | 360 | 290 | 30 | 290 | 218 | 30 | 21.4 |
| AT | PCB180 | PCB209 | 394 | 324 | 30 | 324 | 254 | 30 | 21.7 |
| AT | PCB189 | PCB209 | 394 | 324 | 30 | 324 | 254 | 30 | 22.5 |
| AT | PCB195 | PCB209 | 430 | 360 | 30 | 358 | 288 | 30 | 22.9 |
| IS | BDE99 | 576 | 416 | 20 | 576 | 418 | 20 | 23.5 | |
| AT | BDE99 | BDE99 | 564 | 404 | 20 | 564 | 406 | 20 | 23.5 |
| IS | BDE100 | 576 | 416 | 20 | 576 | 418 | 20 | 24.3 | |
| AT | BDE100 | BDE100 | 564 | 404 | 20 | 564 | 406 | 20 | 24.3 |
| AT | PCB206 | PCB209 | 464 | 392 | 25 | 392 | 322 | 35 | 24.7 |
| IS | PCB209 | 498 | 427 | 30 | 214 | 178 | 20 | 25.6 | |
| IS | BDE153 | 656 | 496 | 20 | 496 | 387 | 40 | 26.4 | |
| AT | BDE153 | BDE153 | 644 | 484 | 20 | 484 | 375 | 40 | 26.4 |
| IS | BDE154 | 656 | 496 | 20 | 496 | 387 | 40 | 27.5 | |
| AT | BDE154 | BDE154 | 644 | 484 | 20 | 484 | 375 | 40 | 27.5 |
AT: Analytical Target compound, IS: internal standard. RT: retention time, P: Parent ion (m/z). F: Fragment ion (m/z).
CE: Collision Energy (eV).
Table 2.
The accuracy and precision methods of PCBs.
| Compound | Spiking levels | Blank Matrix |
Within-run precision for serum from random donors (n = 3, RSD %) | |||||
|---|---|---|---|---|---|---|---|---|
| Accuracy (Bias %) |
Precision (RSD %) |
|||||||
| Within-run | Between-run | Within-run | Between-run | |||||
| PCB8 | Low | 1.3% | 4.4% | 1.2% | 3.8% | 11.70% | 4.65% | 2.40% |
| High | 0.2% | 2.7% | 4.8% | 10.5% | ||||
| PCB18 | Low | 3.4% | 11.4% | 6.9% | 8.6% | 6.00% | 8.10% | 2.10% |
| High | 1.9% | 6.5% | 2.5% | 11.3% | ||||
| PCB28 | Low | 1.1% | 6.8% | 6.9% | 11.6% | 7.50% | 0.90% | 2.25% |
| High | 1.8% | 3.0% | 0.3% | 11.0% | ||||
| PCB44 | Low | 6.4% | 7.2% | 7.1% | 15.6% | 8.10% | 8.10% | 6.15% |
| High | 4.6% | 5.6% | 6.8% | 10.7% | ||||
| PCB52 | Low | 2.3% | 4.2% | 3.8% | 6.9% | 5.12% | 4.56% | 7.33% |
| High | 1.2% | 5.0% | 2.9% | 4.1% | ||||
| PCB66 | Low | 8.0% | 11.4% | 9.5% | 10.4% | 4.95% | 3.90% | 8.55% |
| High | 5.1% | 9.0% | 6.0% | 8.1% | ||||
| PCB101 | Low | 2.2% | 15.0% | 4.2% | 5.4% | 1.50% | 1.20% | 2.70% |
| High | 6.7% | 8.0% | 5.9% | 9.9% | ||||
| PCB81 | Low | 9.2% | 10.0% | 2.1% | 2.4% | 4.80% | 5.40% | 6.15% |
| High | 6.8% | 11.1% | 2.8% | 8.4% | ||||
| PCB77 | Low | 9.9% | 10.0% | 3.0% | 7.0% | 10.95% | 11.70% | 8.85% |
| High | 2.1% | 5.0% | 3.8% | 8.7% | ||||
| PCB123 | Low | 0.3% | 7.0% | 0.5% | 1.4% | 3.00% | 8.40% | 5.10% |
| High | 9.6% | 10.5% | 0.3% | 5.1% | ||||
| PCB118 | Low | 11.3% | 13.4% | 0.3% | 2.2% | 9.90% | 10.65% | 7.50% |
| High | 0.2% | 6.8% | 5.6% | 10.1% | ||||
| PCB114 | Low | 5.4% | 9.6% | 0.5% | 4.6% | 9.00% | 5.25% | 2.40% |
| High | 3.1% | 3.6% | 10.5% | 11.6% | ||||
| PCB153 | Low | 1.3% | 14.2% | 3.6% | 10.2% | 4.20% | 11.70% | 6.45% |
| High | 0.9% | 11.7% | 5.3% | 10.7% | ||||
| PCB105 | Low | 1.7% | 11.2% | 4.8% | 9.8% | 10.35% | 4.50% | 10.65% |
| High | 5.7% | 6.8% | 4.1% | 5.1% | ||||
| PCB138 | Low | 0.7% | 1.4% | 2.1% | 12.4% | 8.70% | 9.75% | 4.20% |
| High | 0.8% | 1.4% | 5.1% | 7.5% | ||||
| PCB126 | Low | 7.2% | 9.4% | 7.6% | 13.2% | 6.90% | 4.20% | 1.95% |
| High | 1.5% | 2.1% | 4.1% | 5.0% | ||||
| PCB187 | Low | 7.2% | 14.6% | 9.8% | 13.2% | 4.20% | 8.70% | 1.80% |
| High | 4.4% | 4.5% | 2.4% | 7.4% | ||||
| PCB167 | Low | 4.3% | 8.2% | 1.1% | 9.4% | 5.10% | 10.65% | 6.90% |
| High | 4.1% | 10.5% | 6.0% | 6.3% | ||||
| PCB156 | Low | 0.1% | 3.2% | 1.7% | 6.4% | 0.75% | 3.75% | 8.70% |
| High | 0.9% | 6.9% | 0.2% | 6.3% | ||||
| PCB157 | Low | 7.7% | 10.6% | 6.6% | 11.0% | 5.70% | 6.90% | 6.15% |
| High | 6.5% | 8.3% | 1.8% | 8.0% | ||||
| PCB170 | Low | 6.0% | 10.4% | 1.7% | 2.0% | 3.60% | 5.10% | 2.70% |
| High | 0.4% | 3.6% | 2.1% | 9.6% | ||||
| PCB169 | Low | 2.1% | 8.4% | 4.8% | 7.6% | 8.25% | 7.05% | 7.65% |
| High | 1.7% | 9.3% | 2.6% | 11.0% | ||||
| PCB180 | Low | 1.9% | 2.0% | 5.2% | 12.8% | 7.80% | 6.30% | 7.05% |
| High | 3.2% | 6.8% | 1.4% | 3.6% | ||||
| PCB189 | Low | 3.6% | 3.8% | 5.8% | 13.4% | 2.70% | 0.90% | 3.75% |
| High | 2.2% | 2.6% | 6.8% | 10.4% | ||||
| PCB195 | Low | 3.1% | 4.0% | 3.5% | 8.6% | 9.75% | 10.80% | 11.40% |
| High | 3.4% | 9.9% | 2.7% | 5.1% | ||||
| PCB206 | Low | 0.1% | 12.4% | 1.8% | 4.8% | 6.75% | 4.50% | 10.80% |
| High | 0.0% | 3.3% | 4.8% | 5.9% | ||||
Table 3.
The accuracy and precision methods of OCPs and PBDEs.
| Compound | Spiking levels | Blank Matrix |
Within-run precision for serum from random donors (n = 3, RSD%) | |||||
|---|---|---|---|---|---|---|---|---|
| Accuracy (Bias%) |
Precision (RSD%) |
|||||||
| Within-run | Between-run | Within-run | Between-run | |||||
| α-HCH | Low | 0.6% | 4.4% | 4.0% | 15.2% | 6.60% | 8.25% | 6.30% |
| High | 1.7% | 2.1% | 5.4% | 6.0% | ||||
| HCB | Low | 2.0% | 12.4% | 3.0% | 7.8% | 9.00% | 3.45% | 11.55% |
| High | 5.9% | 11.3% | 2.9% | 11.4% | ||||
| β-HCH | Low | 13.8% | 15.4% | 1.6% | 13.0% | 7.05% | 9.90% | 1.95% |
| High | 0.3% | 3.6% | 3.3% | 3.6% | ||||
| γ-HCH | Low | 2.5% | 10.2% | 4.4% | 6.8% | 4.95% | 8.40% | 4.95% |
| High | 0.9% | 1.4% | 5.2% | 11.1% | ||||
| δ-HCH | Low | 2.1% | 11.8% | 4.2% | 12.4% | 5.68% | 8.63% | 4.58% |
| High | 1.5% | 7.9% | 3.1% | 5.7% | ||||
| Heptachlor | Low | 8.0% | 11.2% | 4.2% | 9.4% | 6.75% | 1.20% | 10.95% |
| High | 1.7% | 5.3% | 3.3% | 6.9% | ||||
| Aldrin | Low | 8.2% | 15.6% | 3.5% | 6.6% | 3.75% | 3.15% | 2.55% |
| High | 0.7% | 8.3% | 0.9% | 1.4% | ||||
| HCEX | Low | 8.7% | 14.6% | 5.2% | 8.6% | 9.00% | 10.95% | 7.35% |
| High | 4.9% | 11.7% | 1.0% | 6.6% | ||||
| o,p'-DDE | Low | 4.3% | 10.6% | 3.3% | 6.8% | 4.95% | 11.25% | 9.15% |
| High | 0.3% | 1.5% | 3.6% | 10.5% | ||||
| p,p'-DDE | Low | 1.5% | 6.0% | 3.4% | 6.6% | 2.40% | 4.05% | 10.80% |
| High | 6.9% | 9.6% | 1.5% | 3.3% | ||||
| o,p'-DDD | Low | 13.5% | 14.0% | 1.2% | 2.4% | 4.50% | 3.75% | 4.50% |
| High | 3.6% | 9.8% | 4.7% | 10.7% | ||||
| Endrin | Low | 0.4% | 3.0% | 3.9% | 12.8% | 10.05% | 2.55% | 2.85% |
| High | 4.8% | 6.2% | 5.2% | 11.4% | ||||
| p,p'-DDD | Low | 6.4% | 15.2% | 8.7% | 14.6% | 9.30% | 5.85% | 6.45% |
| High | 7.0% | 7.4% | 0.2% | 4.5% | ||||
| o,p'-DDT | Low | 0.9% | 1.4% | 1.8% | 2.0% | 0.75% | 3.75% | 11.40% |
| High | 1.4% | 5.4% | 2.4% | 11.3% | ||||
| p,p'-DDT | Low | 1.3% | 4.2% | 2.3% | 3.2% | 3.15% | 1.20% | 6.75% |
| High | 5.3% | 8.9% | 5.0% | 11.7% | ||||
| BDE47 | Low | 3.6% | 14.6% | 0.7% | 1.0% | 11.40% | 8.10% | 10.05% |
| High | 3.1% | 6.0% | 0.4% | 3.5% | ||||
| BDE99 | Low | 2.1% | 2.6% | 0.8% | 1.8% | 7.95% | 9.30% | 8.10% |
| High | 0.1% | 1.7% | 0.6% | 6.6% | ||||
| BDE100 | Low | 1.2% | 7.4% | 4.1% | 13.8% | 4.20% | 6.15% | 1.50% |
| High | 6.4% | 8.7% | 5.1% | 5.7% | ||||
| BDE153 | Low | 3.9% | 7.2% | 2.1% | 4.8% | 10.50% | 3.90% | 11.70% |
| High | 3.0% | 3.8% | 1.3% | 4.7% | ||||
| BDE154 | Low | 9.3% | 10.8% | 1.4% | 2.6% | 1.35% | 5.10% | 11.40% |
| High | 1.8% | 4.1% | 8.8% | 9.0% | ||||
Table 4.
The calibration of POPs.
| Compound | Calibration Curve | R2 | Compound | Calibration Curve | R2 |
|---|---|---|---|---|---|
| TCMX | y = 5134.48x | 0.9996 | o,p'-DDT | y = 11368.51x | 0.9990 |
| PCB8 | y = 21697.78x | 0.9995 | PCB153 | y = 11887.47x | 0.9992 |
| α-HCH | y = 3236.73x | 0.9998 | PCB105 | y = 7448.54x | 0.9956 |
| HCB | y = 6203.90x | 0.9996 | p,p'-DDT | y = 8156.91x | 0.9971 |
| β-HCH | y = 2133.68x | 0.9993 | PCB138 | y = 5233.74x | 0.9987 |
| γ-HCH | y = 2585.35x | 0.9976 | PCB126 | y = 6742.35x | 0.9985 |
| PCB18 | y = 14785.66x | 0.9995 | PCB187 | y = 4682.77x | 0.9996 |
| δ-HCH | y = 2006.67x | 0.9975 | PCB167 | y = 11514.99x | 0.9986 |
| PCB28 | y = 20311.56x | 0.9994 | PCB156 | y = 5974.90x | 0.9990 |
| Heptachlor | y = 4481.43x | 0.9994 | PCB157 | y = 6378.60x | 0.9992 |
| PCB52 | y = 6847.47x | 0.9994 | PCB170 | y = 4395.06x | 0.9987 |
| Aldrin | y = 1769.68x | 0.9998 | BDE47 (IS) | y = 2094.29x | 0.9985 |
| PCB44 | y = 6168.34x | 0.9987 | BDE47 | y = 2252.61x | 0.9982 |
| HCEX | y = 951.55x | 0.9997 | PCB169 | y = 4938.22x | 0.9997 |
| PCB66 | y = 9201.21x | 0.9993 | PCB180 | y = 3991.75x | 0.9992 |
| o,p'-DDE | y = 11969.61x | 0.9996 | PCB189 | y = 4780.02x | 0.9987 |
| PCB101 | y = 6792.92x | 0.9993 | PCB195 | y = 2569.60x | 0.9984 |
| PCB81 | y = 7976.40x | 0.9995 | BDE99 (IS) | y = 1247.71x | 0.9979 |
| p,p'-DDE | y = 9262.12x | 0.9996 | BDE99 | y = 1261.05x | 0.9991 |
| PCB77 | y = 7984.81x | 0.9981 | BDE100 (IS) | y = 1440.93x | 0.9986 |
| o,p'-DDD | y = 14217.39x | 0.9979 | BDE100 | y = 1335.26x | 0.9981 |
| Endrin | y = 885.24x | 0.9994 | PCB206 | y = 1459.70x | 0.9990 |
| BDE28 (IS) | y = 2475.46x | 0.9984 | PCB209 | y = 3790.42x | 0.9994 |
| PCB123 | y = 7646.52x | 0.9982 | BDE153 (IS) | y = 515.91x | 0.9944 |
| PCB118 | y = 8653.99x | 0.9926 | BDE153 | y = 534.59x | 0.9975 |
| BDE28 | y = 2423.97x | 0.9979 | BDE154 (IS) | y = 368.79x | 0.9974 |
| p,p'-DDD | y = 11923.37x | 0.9967 | BDE154 | y = 347.93x | 0.9960 |
| PCB114 | y = 6956.96x | 0.9952 |
Table 5.
Method detection limit of POPs.
| Compound | MDL (pg/mL) | Compound | MDL (pg/mL) |
|---|---|---|---|
| TCMX | 0.944 | o,p'-DDT | 19.7 |
| PCB8 | 7.00 | PCB153 | 2.43 |
| α-HCH | 14.1 | PCB105 | 11.5 |
| HCB | 1.05 | p,p'-DDT | 10.0 |
| β-HCH | 2.41 | PCB138 | 9.38 |
| γ-HCH | 1.10 | PCB126 | 3.76 |
| PCB18 | 6.38 | PCB187 | 1.41 |
| δ-HCH | 3.85 | PCB167 | 9.87 |
| PCB28 | 7.69 | PCB156 | 4.78 |
| Heptachlor | 0.385 | PCB157 | 19.1 |
| PCB52 | 7.22 | PCB170 | 4.58 |
| Aldrin | 1.24 | BDE47 (IS) | 0.763 |
| PCB44 | 12.1 | BDE47 | 0.992 |
| HCEX | 4.81 | PCB169 | 14.1 |
| PCB66 | 12.9 | PCB180 | 6.44 |
| o,p'-DDE | 21.0 | PCB189 | 2.62 |
| PCB101 | 4.85 | PCB195 | 1.96 |
| PCB81 | 8.36 | BDE99 (IS) | 2.98 |
| p,p'-DDE | 30.7 | BDE99 | 3.17 |
| PCB77 | 1.86 | BDE100 (IS) | 2.92 |
| o,p'-DDD | 11.9 | BDE100 | 1.24 |
| Endrin | 22.3 | PCB206 | 4.36 |
| BDE28 (IS) | 0.923 | PCB209 | 3.03 |
| PCB123 | 14.7 | BDE153 (IS) | 25.6 |
| PCB118 | 8.86 | BDE153 | 17.7 |
| BDE28 | 3.48 | BDE154 (IS) | 4.62 |
| p,p'-DDD | 5.42 | BDE154 | 9.36 |
| PCB114 | 1.43 |
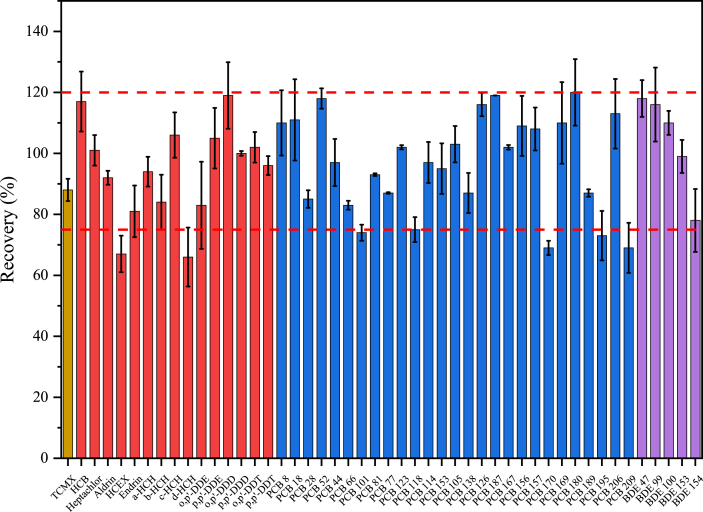
Fig. 1.
The average overall recovery of the analytes.
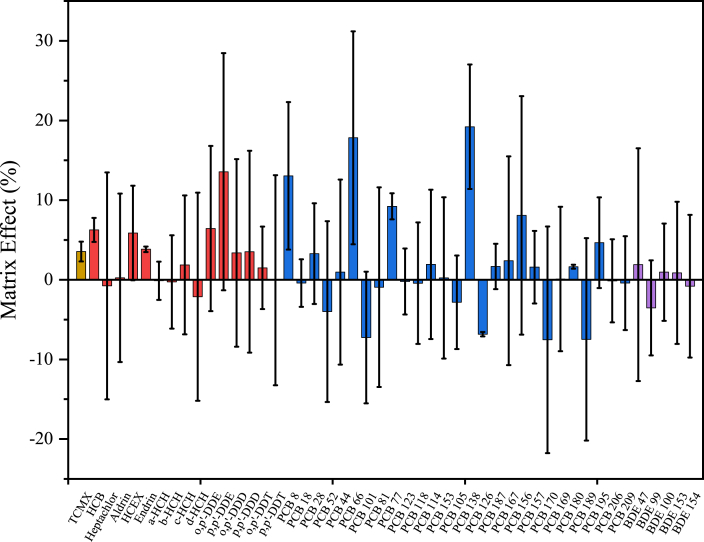
Fig. 2.
The matrix effects of the analytes.
Table 6.
The TEQ levels of DL-PCBs in POI Cases and Controls.
| DL-PCBs (pg/g lipid base) | Case |
Control |
p-Valuea | ||
|---|---|---|---|---|---|
| Median | IQR | Median | IQR | ||
| PCB 77 | 0.90 | 0.07–1.39 | 0.09 | 0.03–0.98 | <0.001 |
| PCB 81 | 2.11 | 0.34–4.21 | 0.41 | 0.35–2.75 | 0.029 |
| PCB 105 | 0.26 | 0.05–0.50 | 0.05 | 0.05–0.15 | 0.001 |
| PCB 114 | 0.01 | 0.01–0.01 | 0.01 | 0.01–0.03 | 0.051 |
| PCB 118 | 0.10 | 0.05–0.18 | 0.05 | 0.04–0.16 | 0.002 |
| PCB 123 | 0.12 | 0.07–0.24 | 0.07 | 0.06–0.19 | 0.007 |
| PCB 126 | 86.17 | 47.55–1108.55 | 58.49 | 50.24–132.37 | 0.003 |
| PCB 156 | 0.02 | 0.02–0.03 | 0.02 | 0.02–0.03 | 0.643 |
| PCB 157 | 0.08 | 0.07–0.09 | 0.08 | 0.07–0.09 | 0.204 |
| PCB 167 | 0.04 | 0.04–0.05 | 0.04 | 0.04–0.05 | 0.112 |
| PCB 169 | 51.97 | 60.01–67.30 | 62.71 | 54.03–69.16 | 0.216 |
| PCB 189 | 0.01 | 0.01–0.04 | 0.01 | 0.01–0.01 | 0.147 |
| ∑6 DL-PCBsb | 87.01 | 50.77–1116.93 | 63.52 | 53.84–135.09 | 0.005 |
| ∑12 DL-PCBsc | 151.31 | 107.48–1178.15 | 130.71 | 113.12–218.40 | 0.005 |
IQR, Interquartile range.
Mann-Whitney U test.
∑6 DL-PCBs includes PCB congeners 77, 81, 105, 118, 123, 126.
∑12 DL-PCBs includes PCB congeners 77, 81, 105, 114, 118, 123, 126, 156, 157, 167, 169, 189.
Table 7.
Association of TEQ levels with POI in Binary Logistic Regression Models.
| DL-PCBs | Unadjusted Model |
Adjusted Modela |
||
|---|---|---|---|---|
| OR (95%CIs) | p-Value | OR (95%CIs) | p-Value | |
| PCB 77 | 1.69 (1.35–2.12) | <0.001 | 1.84 (1.39–2.43) | <0.001 |
| PCB 81 | 1.40 (1.13–1.73) | 0.002 | 1.53 (1.18–1.99) | 0.001 |
| PCB 105 | 1.55 (1.24–1.93) | <0.001 | 1.88 (1.44–2.45) | <0.001 |
| PCB 118 | 1.05 (0.85–1.29) | 0.681 | 1.16 (0.90–1.50) | 0.241 |
| PCB 123 | 1.02 (0.83–1.26) | 0.854 | 1.11 (0.85–1.43) | 0.444 |
| PCB 126 | 1.52 (1.22–1.89) | <0.001 | 1.75 (1.33–2.29) | <0.001 |
| ∑6 DL-PCBsb | 1.50 (1.20–1.86) | <0.001 | 1.73 (1.32–2.26) | <0.001 |
The adjusted model includes age, BMI, parity, history of breast-feeding, age at menarche, smoking, alcohol intake, education and annual household income.
∑6 DL-PCBs includes PCB congeners 77, 81, 105, 118, 123, 126.
Table 8.
Total variance explained of principal components analysis.
| Principle Component | Initial Eigenvalues |
Extraction Sums of Squared Loadings |
Rotation Sums of Squared Loadings |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | % of Variance | Cumulative % | Total | % of Variance | Cumulative % | Total | % of Variance | Cumulative % | |
| 1 | 4.2 | 21.0 | 21.0 | 4.2 | 21.0 | 21.0 | 3.9 | 19.3 | 19.3 |
| 2 | 2.9 | 14.3 | 35.2 | 2.9 | 14.3 | 35.2 | 2.3 | 11.7 | 31.1 |
| 3 | 1.8 | 9.2 | 44.4 | 1.8 | 9.2 | 44.4 | 2.2 | 10.8 | 41.8 |
| 4 | 1.6 | 8.2 | 52.6 | 1.6 | 8.2 | 52.6 | 1.6 | 8.1 | 49.9 |
| 5 | 1.2 | 6.0 | 58.6 | 1.2 | 6.0 | 58.6 | 1.5 | 7.5 | 57.4 |
| 6 | 1.1 | 5.3 | 63.9 | 1.1 | 5.3 | 63.9 | 1.2 | 6.1 | 63.5 |
| 7 | 1.0 | 5.2 | 69.1 | 1.0 | 5.2 | 69.1 | 1.1 | 5.6 | 69.1 |
| 8 | 0.9 | 4.7 | 73.8 | ||||||
| 9 | 0.9 | 4.6 | 78.4 | ||||||
| 10 | 0.8 | 3.9 | 82.3 | ||||||
| 11 | 0.7 | 3.5 | 85.8 | ||||||
| 12 | 0.6 | 3.2 | 89.0 | ||||||
| 13 | 0.6 | 2.8 | 91.8 | ||||||
| 14 | 0.4 | 2.2 | 94.0 | ||||||
| 15 | 0.4 | 2.0 | 95.9 | ||||||
| 16 | 0.4 | 1.8 | 97.7 | ||||||
| 17 | 0.2 | 1.2 | 98.9 | ||||||
| 18 | 0.2 | 1.0 | 99.9 | ||||||
| 19 | 0.0 | 0.1 | 100.0 | ||||||
| 20 | 0.0 | 0.0 | 100.0 | ||||||
Table 9.
Principal components analyses results.
| Contaminant | PC-1 (21.0%) | PC-2 (14.3%) | PC-3 (9.2%) | PC-4 (8.2%) | PC-5 (6.0%) | PC-6 (5.3%) | PC-7 (5.2%) |
|---|---|---|---|---|---|---|---|
| PCB 8 | −0.021 | −0.036 | 0.185 | 0.070 | 0.833 | 0.120 | 0.000 |
| PCB 18 | 0.025 | 0.811 | 0.098 | 0.122 | 0.167 | −0.041 | 0.068 |
| PCB 28 | 0.184 | 0.173 | −0.103 | 0.752 | 0.035 | 0.149 | 0.122 |
| PCB 52 | 0.747 | 0.101 | −0.027 | 0.292 | −0.054 | 0.137 | 0.042 |
| PCB 77 | −0.052 | 0.685 | −0.185 | −0.287 | 0.155 | 0.016 | −0.090 |
| PCB 81 | 0.538 | 0.471 | −0.117 | −0.128 | 0.352 | −0.026 | −0.175 |
| PCB 105 | 0.170 | 0.012 | −0.096 | −0.048 | −0.092 | 0.484 | 0.408 |
| PCB 118 | 0.979 | −0.052 | −0.030 | 0.012 | −0.019 | 0.025 | 0.047 |
| PCB 123 | 0.979 | 0.004 | −0.026 | 0.046 | −0.021 | 0.076 | 0.055 |
| PCB 126 | −0.025 | 0.374 | 0.037 | −0.065 | 0.677 | −0.023 | 0.044 |
| PCB 138 | −0.012 | 0.751 | 0.355 | 0.183 | −0.108 | 0.144 | 0.032 |
| PCB 153 | 0.256 | 0.389 | 0.114 | 0.465 | −0.140 | 0.469 | 0.110 |
| PCB 187 | 0.975 | −0.058 | −0.029 | 0.002 | −0.015 | 0.024 | 0.043 |
| PCB 195 | 0.103 | 0.132 | −0.027 | 0.326 | 0.025 | −0.166 | 0.487 |
| p,p'-DDT | −0.049 | −0.256 | −0.093 | 0.677 | −0.002 | −0.163 | −0.180 |
| p,p'-DDE | −0.045 | 0.006 | 0.557 | −0.053 | 0.244 | 0.090 | 0.086 |
| β-HCH | −0.041 | 0.050 | 0.892 | −0.059 | −0.094 | −0.035 | −0.083 |
| γ-HCH | −0.027 | 0.087 | 0.881 | −0.042 | 0.102 | −0.038 | −0.057 |
| HCB | 0.037 | 0.007 | 0.052 | −0.005 | 0.204 | 0.783 | −0.223 |
| Heptachlor | −0.020 | −0.071 | 0.003 | −0.102 | 0.035 | 0.001 | 0.732 |
The bold means that the principal component has a high positive/negative loading for that contaminant.
2. Experimental design, materials and method
2.1. Optimized pretreatment
The target POPs in this study included polychlorinated biphenyls (PCBs), organochlorine pesticides (OCPs) and polybrominated diphenyl ethers (PBDEs). The pretreatment and analytical procedures were developed based on previous description with minor modification [2], [3]. A total of 0.3 mL of serum sample was spiked with 10 μL of mixture of internal standards (IS) [PCB 209, tetrachloro-m-xylene (TCMX), 13C12 isotopically labeled standards of PBDE 47, 99, 100, 153 and 154, 100 ng/mL]. Then, 0.5 mL of formic acid and 2.5 mL of ethanol were added and mixed. Ten milliliter of mixed extractant of n-hexane and dichloromethane (DCM) (1:1, v/v) was added. The mixture was ultrasonic extracted for 10 minutes and centrifuged at 2000 rpm for 10 minutes. The organic phase was transferred into a clean flat-bottomed flask. The extraction steps were repeated three times. The extracts were evaporated to about 1 mL and cleaned by a column filled with activated silica gel (6 g) and Na2SO4 (2 g). The column was eluted with 70 mL of a mixed solvent of n-hexane and DCM (1:1, v/v) before the addition of the concentrate. Then, the target compounds were eluted by another 70 mL of n-hexane and DCM (1:1, v/v). The elution was evaporated to dryness and redissolved in 50 μL of n-decane and stored in a refrigerator at 4 °C until quantification. All chemicals used above were purchased from J&K Chemical, Beijing, China.
2.2. Instrumental analysis
Gas chromatography-triple quadrupole mass spectrometry (GC-MS/MS) (Agilent 7890B GC/7000C) was used to quantitate the concentrations of POPs. The sample quantified methods were applied as described previously [2], [3]. For GC conditions, the column was DB-5ms (30 m × 0.25 mm × 0.25μm). Oven heating program was as follows: initial temperature at 80 °C hold for 1 min, and 10 °C/min to 180 °C hold for 5 min and then 20 °C/min to 220 °C (0 min) and finally 5 °C/min to 300 °C and hold for 5 min. The injector was kept at 250 °C. Carrier gas was helium (99.999% purity) at a constant flow rate of 1.0 mL/min. One microliter was splitlessly injected for each sample. The triplequad MS was operating in EI mode at 230 °C with electron ionization voltage of 70 eV and transfer line temperature at 280 °C. The multiple reaction monitoring mode was applied in the analysis process. For each analyte, two or more MRM transitions were monitored and one pair of ions with the highest peak area was chosen as the quantifier and the rest were set as qualifier. Detailed information is shown in Table 1. The quantification procedure was conducted using Agilent Masshunter Workstation Quantitative Analysis B.07.01 (Agilent Inc. Santa, Clara, CA, USA). The mass is set 0.9 or 0.1 for Agilent Workstation settings, recommended by the Agilent manual. The mass window is set at “UNIT” for both the first and second quadruple, which is 0.7 Å wide. For the retention time window, in the Agilent Masshunter, we set it at 1.0 min wide (−0.3 to +0.7) except for those with wider peaks.
2.3. Methods validation
A small-scale method validation was applied following the protocols established by the European Medicines Agency. Newborn bovine serum was used as the blank matrix. Calibration curves were analyzed in triplicates to estimate coefficients of determination (R2). Carryovers were assessed by injecting solvent blanks immediately after the analysis of the highest calibration point. Within- and between-run precision and accuracy of the methods were assessed over the course of three days using blank matrix spiked with target analytes at low (6 ng/mL of 10μL, final concentration of 0.2 ng/mL in the matrix) and high (300 ng/mL of 10μL, final concentration of 10 ng/mL in the matrix) concentrations and processed as described above. On each day, three replicates per spiking level, one blank matrix and one procedural blank were processed. All samples and blanks were spiked with IS (100 ng/mL of 10μL) prior to processing. Accuracy was calculated by subtracting the concentration measured in blank matrix from the concentration measured in low and high spiked samples. Precision and accuracy were considered satisfactory if results were <15% or <20% (for low spikes). Method detection limits (MDL) were determined using blank or low spiked blank matrix giving a signal-to-noise ratio (S/N) of 3. Recoveries of the extraction process were estimated using blank matrix spiked with native and mass labeled reference standards (at low and high concentrations) before and after extraction. Matrix effects were assessed by comparing the signal of reference standards in samples spiked after extraction with calibration standards prepared in n-decane. Background signals recorded in blank matrix samples were subtracted from analyte signals in post-extraction spikes prior to matrix effect calculation. Serum samples from three random different donors were extracted in triplicate to calculate the within-run precision using different matrices. These samples were only spiked at mid concentration.
2.4. Recovery and matrix effects
As shown in Fig. 1, the average overall recovery ranged between 78 and 113%, with relative standard deviations (RSDs) < 15% for all compounds.
Matrix effects were evaluated by comparing the signal of blank matrix spiking with native standards at low concentration (6 ng/mL of 10 μL, final concentration of 0.2 ng/mL in the matrix) or high concentration (300 ng/mL of 10 μL, final concentration of 10 ng/mL in the matrix) or IS (100 ng/mL of 10 μL) before and after extraction. In this study, corresponding IS was not available for some analytes, so matrix effects ranged from −20% to 35%, with RSDs below 15% for all compounds (Fig. 2).
2.5. Precision
For low spikes, the within- and between-run precision was lower than 20%, and for and high spikes, the precision was lower than 15% among three days for all target compounds. The inter-individual variation and the variation between the blank matrix and real human serum in precision of the method were assessed using serum samples from three random donors. The results showed the precision across different donors was acceptable (<15%) (Table 2, Table 3).
2.6. Accuracy
Low and high concentrations of target analytes were spiked into blank matrix. The nominal concentration in the guideline was defined as the sum of the background and spiked concentrations. However, as the POPs concentration in the blank matrix is lower than the MDL, the nominal concentration in this validation was set as the spiking concentration of the native standards. The accuracy for individual compounds was acceptable for all concentration levels (Bias <15%, or <20% for low spike) (Table 2, Table 3).
2.7. Calibration
Calibration curves were conducted using a mixture of native standards ranging from 0.1 ng/mL to 200 ng/mL and IS at concentration of 20 ng/mL in all calibrators. Calibration curves were computed using liner regression and were forced to pass zero. As shown in Table 4, coefficients of determination (R2) for all compounds were above 0.99.
2.8. Method detection limit (MDL)
Method detection limit (MDL) were estimated from low concentration standards giving a signal-to-noise ratio of 3 in the blank matrix. The MDL for this pretreatment process varied from 9 pg/mL to 173 pg/mL and 29 pg/mL and 575 pg/mL, respectively (Table 5).
2.9. Carry-overs
Solvent blanks (i.e. n-decane) were injected right after the highest concentration of calibration curve to assess carry-overs, which were below 20% of the MDL for all analytes. Overall, the results obtained during method validation indicate that the protocol is adapted for the analysis of targeted POPs. Thus, the method is suitable to be applied in the experiment.
2.10. Data analysis method
The TEQs were calculated by multiplying the toxic equivalence factors (TEFs) for each DL-PCB congener concentration: TEQ Σ12 DL-PCBs = PCB 77 × 0.0001 + PCB 81 × 0.0003 + PCB 105 × 0.00003 + PCB 1114 × 0.00003 + PCB 118 × 0.00003 + PCB 123 × 0.00003 + PCB 126 × 0.1 + PCB 156 × 0.00003 + PCB 157 × 0.00003 + PCB 167 × 0.00003 + PCB 169 × 0.03 + PCB 189 × 0.00003 [4]. Odds ratios (ORs) and 95% confidence intervals (CIs) for the risk of POI in association with TEQs levels were calculated by unconditional logistic regression models. The covariates included age, BMI, parity, history of breast-feeding, age at menarche, smoking, alcohol intake, education and annual household income [5], [6]. POPs concentration variables that were detected in >40% samples were subjected to principal components analysis (PCA) to produce a few number of summary PCA predictor variables. The data analysis were conducted using SPSS (version 20.0, IBM, Chicago, IL, USA).
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (2019FZJD007 and 2019QNA6008), National Natural Science Foundation of China (21876151, 21427815 and 81703236), Program for Key Subjects of Zhejiang Province in Medicine & Hygiene and Project for Zhejiang Medical Technology Program (2018KY437 and WKJ-ZJ-1621).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.104430.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Pan W., Ye X., Yin S., Ma X., Li C., Zhou J., Liu W., Liu J. Selected persistent organic pollutants associated with the risk of primary ovarian insufficiency in women. Environ. Int. 2019;129:51–58. doi: 10.1016/j.envint.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 2.Tang M., Yin S., Zhang J., Chen K., Jin M., Liu W. Prenatal exposure to polychlorinated biphenyl and umbilical cord hormones and birth outcomes in an island population. Environ. Pollut. 2018;237:581–591. doi: 10.1016/j.envpol.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 3.Xu C., Yin S., Tang M., Liu K., Yang F., Liu W. Environmental exposure to DDT and its metabolites in cord serum: distribution, enantiomeric patterns, and effects on infant birth outcomes. Sci. Total Environ. 2017;580:491–498. doi: 10.1016/j.scitotenv.2016.11.196. [DOI] [PubMed] [Google Scholar]
- 4.Van den Berg M., Birnbaum L.S., Denison M., De Vito M., Farland W., Feeley M., Fiedler H., Hakansson H., Hanberg A., Haws L., Rose M., Safe S., Schrenk D., Tohyama C., Tritscher A., Tuomisto J., Tysklind M., Walker N., Peterson R.E. The 2005 World Health Organization reevaluation of human and Mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci. : An Official Journal of the Society of Toxicology. 2006;93:223–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C., Cao M., Ma L., Ye X., Song Y., Pan W., Xu Z., Ma X., Lan Y., Chen P., Liu W., Liu J., Zhou J. Pyrethroid pesticide exposure and risk of primary ovarian insufficiency in Chinese women. Environ. Sci. Technol. 2018;52:3240–3248. doi: 10.1021/acs.est.7b06689. [DOI] [PubMed] [Google Scholar]
- 6.Ye X., Liu J. Effects of pyrethroid insecticides on hypothalamic-pituitary-gonadal axis: a reproductive health perspective. Environ. Pollut. 2019;245:590–599. doi: 10.1016/j.envpol.2018.11.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.