Abstract
Most reported isolates of Sarcocystis spp. derived from Brazilian opossums (Didelphis sp.) have genetic characteristics distinct from the known species of Sarcocystis, but behave similarly as Sarcocystis falcatula, as they are infective to budgerigars. In previous studies, these Brazilian isolates, classified as Sarcocystis falcatula-like, were originated from South and Southeast regions of Brazil. In the current work, we aimed to culture and to perform multilocus sequence analysis of Sarcocystis sp. derived from a Brazilian opossum (D. aurita/D. marsupialis) that inhabited the city of Salvador, Bahia, in the Northeast of Brazil. The parasite was isolated in Vero cells, referred here as Sarco-BA1, and propagated in avian cells (DF-1). Molecular analysis of Sarco-BA1 revealed that the nucleotide sequence of the internal transcribed spacer 1 (ITS1) of the rDNA was identical to all isolates (n = 19) of Sarcocystis spp. reported in two studies from South and Southeast regions of the country. Two budgerigars were inoculated with 10 and 1000 sporocysts of Sarco-BA1, respectively, and developed acute sarcocystosis, showing that the parasite behaves like S. falcatula. It was interesting to observe that Sarco-BA1 had almost identical ITS1 and SAG sequences to all 16 isolates of S. falcatula-like recently described in Magellanic penguins (Spheniscus magellanicus) rescued on the coast of Espírito Santo state, Brazil. Our results suggest that Sarco-BA1 and S. falcatula-like may represent a single species of Sarcocystis. Propagation of the parasite in a permanent avian cell line significantly improved the yield of merozoites in cell culture. To our knowledge, this is the first molecular study and in vitro isolation of S. falcatula-like derived from Northeastern Brazil. Studies are under way to determine the infectivity of Sarco-BA1 to other animal species, as well as to investigate serological cross-reactivity among Sarco-BA1, S. neurona and related species.
Keywords: Didelphis sp., Sporocyst, ITS1, Surface antigen, DNA
Graphical abstract
Highlights
-
•
Sarcocystis falcatula-like was isolated for the first time in Northeastern Brazil.
-
•
Its genetic pattern was similar to isolates from South and Southeastern Brazil.
-
•
A permanent avian cell line was successfully used to propagate the parasite.
1. Introduction
Sarcocystis spp. are cyst-forming coccidia that are able to infect a large spectrum of hosts, including mammals, birds and reptiles (Dubey et al., 2015). Opossums (Didelphis spp.), which inhabit exclusively the Americas, were identified as definitive hosts of at least four known species of Sarcocystis; S. falcatula (Box et al., 1984), S. neurona (Fenger et al., 1995) and S. speeri (Dubey and Lindsay, 1999) were originally reported in North America, whereas S. lindsayi (Dubey et al., 2001c) was described in South America. S. neurona and S. falcatula are well known for causing encephalomyelitis in equids (Reed et al., 2016) and sarcocystosis in birds (Box et al., 1984; Smith et al., 1990), respectively. S. lindsayi is also infective to birds, but was proposed as a new species mostly due to differences observed in the internal transcribed spacer 1 (ITS1) locus of the rDNA (Dubey et al., 2001c). S. speeri resembles S. neurona, because under experimental conditions both were infective to immunodeficient mice and did not induce infection in birds (Dubey and Lindsay, 1999). S. speeri was proposed a new species based on morphological and antigenic differences from S. neurona (Dubey and Lindsay, 1999). Natural intermediate hosts for S. lindsayi and S. speeri are unknown.
In earlier reports on Sarcocystis spp. derived from Didelphis spp. in South and Southeastern Brazil, some intriguing results have been observed. Firstly, S. neurona, that had been reported in Brazil infecting the South American opossum D. albiventris (Dubey et al., 2001a), has never been genetically confirmed in the country. Secondly, most isolates of Sarcocystis spp. derived from opossums reported in Brazil are S. falcatula-like organisms, as they are infective to birds (Acosta et al., 2018; Cesar et al., 2018; Dubey et al., 2001b; Gallo et al., 2018; Gondim et al., 2017; Konradt et al., 2017; Stabenow et al., 2008; Valadas et al., 2016). In studies employing different genetic markers, in particular ITS1, almost all examined isolates differ from the known species of Sarcocystis that use opossum as definitive hosts (Acosta et al., 2018; Cesar et al., 2018; Gondim et al., 2017; Konradt et al., 2017; Valadas et al., 2016). In the entire North America there is a single species of opossum (Didelphis virginiana), whereas in Brazil, four species of Didelphis are observed (D. albiventris, D. aurita, D. marsupialis and D. imperfecta) (Cerqueira, 1985). The occurrence of four Didelphis species residing in Brazil, associated with the great fauna diversity in the country, probably account for the genetic differences observed among Sarcocystis spp. derived from opossums in Brazilian studies (Acosta et al., 2018; Cesar et al., 2018; Gondim et al., 2017; Konradt et al., 2017; Monteiro et al., 2013; Valadas et al., 2016).
Unfortunately, until the completion of the current study, there were no available opossum Sarcocystis spp. isolates derived from other parts of the country, such as North, Northeast and Central regions, whose states possess country-like dimensions (https://www.britannica.com/place/Brazil). Genetic comparisons of isolates from geographically distant regions would provide more robustness to characterize the isolates that may represent new species of the parasite. The aim of the present work was to characterize by genetic and biological tools an isolate of Sarcocystis sp. obtained from an opossum in Salvador city, located in Northeastern Brazil.
2. Material and methods
2.1. Opossum and source of sporocysts
An adult female black-eared opossum (Didelphis aurita/Didelphis marsupialis) was rescued from the campus at Federal University of Bahia (UFBA) in Ondina neighborhood and referred to the Clinic for Rehabilitation of Wildlife at UFBA. D. marsupialis and D. aurita are morphologically similar species, so additional tools are probably necessary to accurately identify each of these opossums’ species (Cerqueira, 1985). In the current study, the examined opossum was referred as D. aurita/D. marsupialis. The place where the animal was rescued is located in the urban area of Salvador, about 600 m from the Atlantic coast. The opossum was severely injured and died ten days after presentation to the clinic. Opossums of the genus Didelphis are commonly observed on UFBA campus (personal observation, Gondim, L.F.P.).
The small intestine of the animal was longitudinally sectioned and its entire internal surface was scraped by using a microscopic glass slide, according to previously described methods (Dubey et al., 2015). The intestinal content (10 ml) was placed in a 500 ml Becker and homogenized on a magnetic stirrer for 10 min with 20 ml of a commercial sodium hypochlorite solution (2.5% of active chlorine) and 70 ml of distilled water. The mixture was filtered in gauze, placed in 50 ml tubes and centrifuged for 10 min (1000 g at 4 °C). Two additional rounds of centrifugations (1000 g at 4 °C for 10 min) with PBS (pH 7.2) were performed to remove sodium hypoclorite from the solution. A small drop of the sediment was examined using light and fluorescence microscope and revealed great numbers of coccidian sporocysts/oocysts. Ten volumes of a sucrose flotation solution were gradually added to the sediment under continuous homogenization. Then, the solution was centrifuged at 1200 g at 4 °C for 10 min and 50% of the upper portion of the tube was collected. Sucrose was removed from the suspension by adding ten volumes of distilled water followed by centrifugation. The obtained sediment was mixed with a commercial 100x concentrated antibiotic/antimycotic solution (10,000 units/mL of penicillin, 10,000 μg/mL of streptomycin, and 25 μg/mL of amphotericin B) (Gibco-Invitrogen, Carlsbad, CA, USA) and sored at 4 °C for posterior use. The number of parasites was determined using a hemocytometer and corresponded to 3.9 × 107 sporocysts.
2.2. In vitro isolation of Sarcocystis sp. and propagation of the parasite in avian cells
Sporocysts of Sarcocystis sp. from opossum, which had been stored for 30 days, were adjusted to the number of 1 × 106. The sporocysts were mixed with 2.5% of sodium hypochlorite solution in a 15 ml sterile tube and homogenized for 30 min. The solution was washed in RPMI medium for three times by centrifugation and resuspended to 700 μl with RPMI medium supplemented with 1% antibiotic/antymicotic (100 units/mL of penicillin, 100 μg/mL of streptomycin and 0.25 μg/mL of amphotericin B) and 5% of bovine calf serum. A 1.5 ml tube containing a volume of 0.4 ml of glass beads (400–600 μm in diameter) was simultaneously treated with 2.5% of sodium hypochlorite solution for 30 min and washed three times with RPMI medium. The 700 μl of sporocyst solution was added to the 1.5 ml tube with glass beads. The mixture was vortexed at maximum speed for 2 min and the final solution was aspirated from the tube. The beads were washed with RPMI to retrieve parasites which were still in the beads. A drop of the lysed sporocysts’ solution was observed under the microscope to examine for released sporozoites. This solution was passed through a sterile 5 μm-pore size filter and the filtered sporozoites were saved for cell culture.
One milliliter of RPMI containing free sporozoites was cultured in Vero cells on two 25 mm2 cell culture flasks. The cells were maintained with RPMI-1650+L-glutamin (Invitrogen/Gibco®, Carlsbad, USA), supplemented with 1% antibiotic-antimycotic: 100 units/mL of penicillin, 100 μg/mL of streptomycin and 0.25 μg/mL of amphotericin B (Gibco®, Carlsbad, USA) and 5% of inactivate bovine serum (Invitrogen/Gibco®, Auckland, NZ), at 37 °C in a humidified incubator containing 5% CO2. The flasks were examined daily for parasite growth.
A permanent chicken cell line (UMNSAH/DF-1) (Foster and Foster, 1997) was employed to propagate the parasite. The culture conditions for DF-1 cells were identical to those employed for Vero cells.
2.3. DNA extraction and amplification
DNA was extracted from opossum's sporocysts using a commercial fecal DNA extraction kit (ZR Fecal DNA MiniPrep™, USA) according to the manufacturer's instructions. Seven genetic loci were PCR amplified from the obtained DNA.
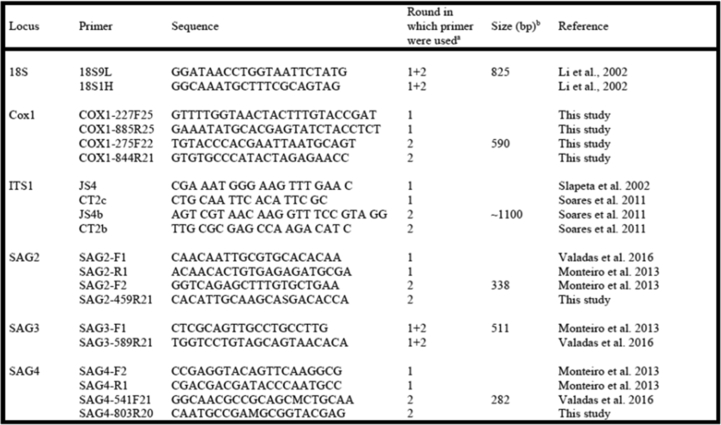
Conventional PCR were conducted in order to amplify genetic sequences of S. neurona/S. falcatula using the primers JNB25 and JD396 (Tanhauser et al., 1999), and nested PCR was used to amplify Sarcocystis spp. sequences of 18S small subunit rRNA region, gene coding for cytochrome c oxidase subunit I, internal transcribed spacer 1, SAG2, SAG3 and SAG4. Nested PCR directed to 18S (nPCR-18S) was performed using primers 18S9L and 18S1H (Li et al., 2002). DNA amplification of Sarcocystis spp. cytochrome c oxidase subunit I (nPCR-COX1) was performed using primers designed in this study, whereas complete ITS1 was nested PCR amplified using primers directed to 18S and 5.8S flanking ITS1 (nPCR-ITS1), as described elsewhere (Soares et al., 2011; Acosta et al., 2018). Finally, genetic sequences of SAG2, SAG3 and SAG4 were nested PCR amplified using the primers as designed in Monteiro et al., (2013), Valadas et al., (2016) and in the present study. Primers used to amplify genetic sequences of Sarcocystis spp. using nested PCR are available as a supplementary file (S.1).
2.3.1. Conventional PCR
Conventional PCR for S. neurona/S. falcatula using the primers JNB25 and JD396 (Tanhauser et al., 1999)was performed with the following conditions: 0.5 μl of each primer (50 pmol of each), 12.5 μl of PCR commercial mix (master Mix, Promega, Madison, WI), 10.5 μl of ultrapure water and 1.0 μl of sample DNA. The PCR was carried out as follows: an initial cycle of 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 62 °C for 1 min, 72 °C for 45 s, and final extension at 72 °C for 6 min. The PCR products were run in GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) thermal cycler. Ultra-pure water was used as a negative control. DNA from S. neurona cultured merozoites was employed as positive control. The expected amplicon for S. neurona/S. falcatula PCR was 334 base pairs. The amplicons were analyzed by electrophoresis in a 1.5% agarose gel and a buffered Syber gold was used as a DNA stain. A 100 bp DNA ladder was used in each gel.
2.3.2. Nested PCR
Amplification of ITS1 was conducted with primers that anneal to coding regions of the ribosomal units 18S and 5.8S (Slapeta et al., 2002; Soares et al., 2011), as previously described (Soares et al., 2011). Amplifications of SAG2, SAG3 and SAG4 were carried out using primers for multilocus differentiation of S. falcatula-like organisms (Monteiro et al., 2013; Valadas et al., 2016). The sequences of primers used in these PCRs are are available as a supplementary file.
The first rounds of amplifications (external primers) for all nested PCRs were conducted by adding 4 μL of extracted DNA, 2.5 μL of 10x PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0), 1 μL of MgCL2 (50 mM), 0.5 μL of mixed dNTPs (10 mM), 0.35 μL of each primer (10 pmol/uL), 0.2 μL of Taq DNA Polimerase Platinum-Invitrogen (5 U/μL), and 16.1 μL of ultrapure autoclaved water to a volume of 25 μL per reaction. The PCR conditions consisted of an initial cycle of denaturation at 94 °C for 3 min, followed by 40 cycles (94 °C for 30 s, 56 °C for 30 s, 72 °C for 50 s) and a final extension cycle at 72 °C for 5 min.
For the second rounds of amplifications (internal primers), 2 μL of template derived from the first reactions were added to 2.5 μL of 10x PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0), 1 μL of MgCL2 (50 mM), 0.5 μL of mixed dNTPs (1.25 mM), 1 μL of each primer (10 pmol/uL), 0.2 μL of Taq DNA Polimerase Platinum-Invitrogen (5 U/μI) and 16.8 μL of ultrapure autoclaved water to a volume of 25 μL per reaction. The PCR conditions were the same used in the first amplifications.
2.4. Detection and sequencing of amplified PCR products
The PCR products were analyzed by electrophoresis in 2% agarose gels. Aliquots of 10 μL of each amplified product were mixed with 5 μL of a loading dye (30% of glycerol and 0.25 of bromophenol blue) and deposit in the gel immersed in Tris-borate-EDTA buffer (0.045M Tris-borate; 1 mM EDTA). A multiple 100 bp DNA ladder (GeneRulerTM 100 pb DNA Ladder, Thermo Scientific, Carlsbad, California, USA) was added in each gel. After electrophoresis, the gel was stained with ethidium bromide (0.5 μg/mL) for 60 min and visualized under an ultraviolet transiluminator. The targeted DNA bands were excised from the gel and purified using a commercial purification kit (Illustra GFXTM PCR DNA and Gel Band Purification, Amersham Biosciencies) according to the manufacturer's instructions. The purified samples were sequenced using the Kit ABI PRISM Big Dye TerminatorTM (Applied Biosystem) following manufacturer protocol.
2.5. Sequence editing and sequence analysis
The assessing of the quality of nucleotide sequences, the contig assembly and sequence edition were done using the program Phred-Phrad in Codon Code Aligner, version 4.2.1. The final nucleotide sequences were analyzed using Blast tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). SAG sequences were aligned using the program Crustal W in BioEdit Sequence Alignment Editor with homologues sequences available in GeneBank. By using MEGA 7, Dendrograms were inferred using UPGMA method, computing evolutionary distance using p-distance method.
Nucleotide sequence data reported in this paper are available in the GenBank™ database under the following accession numbers: 18S (MK803217), COX1 (temporary number: 2215653), ITS1 (MK803362) SAG2 (temporary number: 2215645), SAG3 (temporary number: 2215650) and SAG4 (temporary number: 2215651).
2.6. Bioassay in budgerigars
Two female, five months old Melopsittacus undulatus were acquired from a local breeder and maintained for one week in the animal facility of the Veterinary Hospital at Federal University of Bahia. The two animals were orally inoculated by gavage with 10 and 103 Sarcocystis sp. sporocysts, respectively, which had been stored for 70 days in antibiotic/antimycotic solution. The animals were caged individually and observed daily for clinical signs. Tissues from inoculated animals, after euthanasia, were employed for isolation in cell culture according to methods reported in a previous study (Gondim et al., 2017).
The use of animals in the study was approved by the University Local Committee in compliance with Ethical Principles in Animal Research adopted by the Brazilian College of Animal Experimentation (License number:44/2018).
3. Results
3.1. Sporocysts and direct isolation of Sarcocystis sp. in cell culture
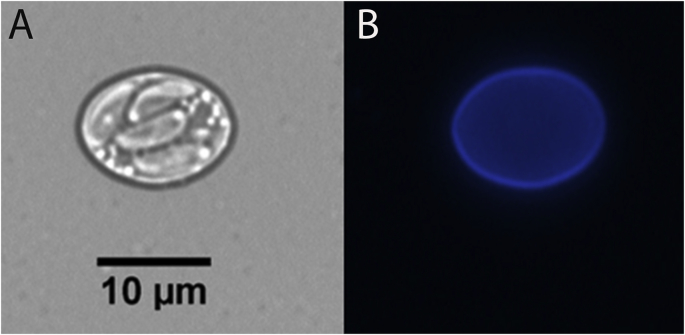
Opossum's sporocysts which had been stored in antibiotic/antimycotic solution for 30 days, were used for direct isolation of the parasite in cell culture. Sporocysts were 11.07–12.0 μm x 7.51–8.59 μm (n = 23) in size. Sporozoites were visualized inside most sporocysts and autofluorescence of the sporocyst wall was observed after excitation with ultraviolet (Fig. 1). Sporozoites were released from sporocysts using mechanical lysis by vortexing with glass beads and developed into schizonts/merozoites in Vero cells between six and eight days after inoculation of parasites' sporozoites on cell monolayers. Due to the slow growth and low number of parasites, the flasks were maintained without replication to new flasks for at least 40 days. Parasite schizonts in different degrees of maturation were visualized in the host cells. New monolayers of Vero cells were prepared and scraped cells from the original flasks were used for inoculations into these uninfected cells. The newly isolated strain is referred here and throughout the manuscript as Sarco-BA1. Aliquots of Sarco-BA1 containing merozoites and schizonts of the parasite inside DF-1 cells were frozen in liquid nitrogen.
Fig. 1.
Sporocyst of Sarcocystis falcatula-like. Four sporozoites are visualized inside the sporocyst by light microscopy (A). Autofluorescence of the sporocyst wall is observed after excitation with ultraviolet on a fluorescence microscope (B).
3.2. Propagation of Sarco-BA1 in avian cells
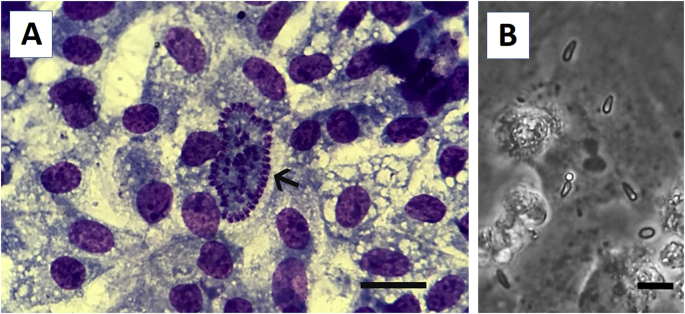
Merozoites were initially isolated in Vero cells, however, the growth of merozoites was slow. Sixty days after inoculation of merozoites in Vero cell monolayer, the multiplication of the parasites decreased, and between 90 and 120 days of culture in Vero cells, free merozoites were rarely seen in the culture flasks. In order to improve the propagation of the parasite, merozoites were cultured in avian cells (DF-1). A fragment from a 25 mm2 flask contained schizonts was cut and stained by May-Grüenwald-Giemsa (Fig. 2A). Interestingly, growth of parasites was dramatically improved in DF-1 cells and free merozoites were easily observed after 60 days of culture (Fig. 2B). Vero cells were no longer used to culture Sarco-BA1 and the parasite has been cultured in DF-1 cells.
Fig. 2.
(A) A mature schyzont of Sarcocystis falcatula-like (Sarco-BA1 strain) in a permanent chicken cell line (UMNSAH/DF-1). May-Grüenwald-Giemsa stain. Bar = 20 μm. (B) Extracellular merozoites of Sarco-BA1 on a monolayer of UMNSAH/DF-1 cells. Bar = 10 μm.
3.3. Bioassay in budgerigars
Two budgerigars were orally inoculated with parasites’ sporocysts. The animal inoculated with 103 sporocysts developed dyspneia and lethargy on the 21st day after inoculation (DAI). The animal was immediately euthanised and lung tissues were aseptically removed for parasite isolation. Merozoites were seen in the inoculated flask, however, on the sixth day after tissue inoculation into the flask, bacterial contamination was seen and the flask was discarded. The second budgerigar that was inoculated with 10 sporocysts, became dyspneic on the 34th DAI and was euthanised immediately after showing clinical signs. Lung tissue was processed for in vitro isolation of the parasite, but no merozoites were observed during 40 days of culture.
3.4. Molecular analysis
The conventional PCR using the primers JNB25 and JD396 (Tanhauser et al., 1999) generated a product of approximately 330 bp, which is compatible with S. neurona/S. falcatula, and also with Sarcocystis spp. shed by Didelphis spp. in Brazil (Gondim et al., 2017).
Sequencing of the entire PCR-18S nested products of Sarco-BA1, which corresponds to 825 bp (primers included), disclosed a 100% identity and 100% coverage to S. falcatula detected in a confirmed acute, fatal Sarcocystis infection in the Rainbow Lorikeets (Trichoglossus moluccanus) (Verma et al., 2018) (GenBank accession number: MH626537, from nucleotide position 12–794). Only a G-A substitution differs Sarco-BA1 from S. speeri KT207459 at nucleotide position 30 (taking KT207459 as reference). Only 2 single nucleotide polymorphism and no gap differ S. falcatula MH626537 from S. speeri KT207459, among 1618 paired nucleotides, represented by G-A and C-A substitutions at nucleotide positions 30 and 1563, respectively (taking MH626537 as reference).
Sequencing of the entire nested PCR-COX1 products, which corresponds to 590 bp (primers included), disclosed a 100% identity and 100% coverage to the S. falcatula detected in fatal infection in the Rainbow Lorikeets (Verma et al., 2018) (GenBank accession number: MH665257) and S. speeri (KT207461), from nucleotide position 239–785 and from positions 272–818, respectively. At this locus, S. falcatula MH665257 and S. speeri KT207461 disclosed 100% identity and 100% coverage among 1002-paired nucleotides.
Multilocus analysis based on ITS1, SAG2, SAG3 and SAG4 was performed as previously described to identify Sarcocystis spp. in budgerigars that were experimentally infected with opossums’ sporocysts (Cesar et al., 2018; Gondim et al., 2017), except for the fact that the almost complete nested PCR-ITS1 amplicon was sequenced in this study. The ITS1 sequence of Sarco-BA1 was identical to all 19 sequences derived from the abovementioned studies, and to a S. falcatula isolate (59-2016-RS-BR) from a naturally infected barefaced ibis (Phimosus infuscatus) (KX265018) (Konradt et al., 2017). The 1013 sequenced nucleotides from nested PCR-ITS1 products disclosed 99.9% identity and 100% coverage to S. falcatula detected in Rainbow Lorikeets (Verma et al., 2018) (GenBank accession number MH626538, from nucleotide position 68–1081). These two sequences differ from each other in only one gap, at nucleotide position 80 (taking MH626538 as reference).
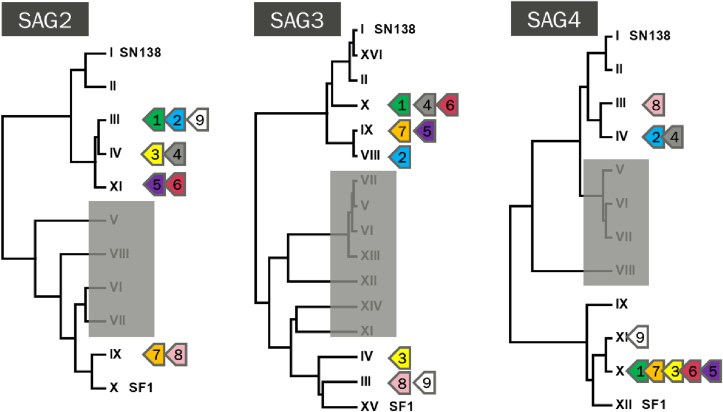
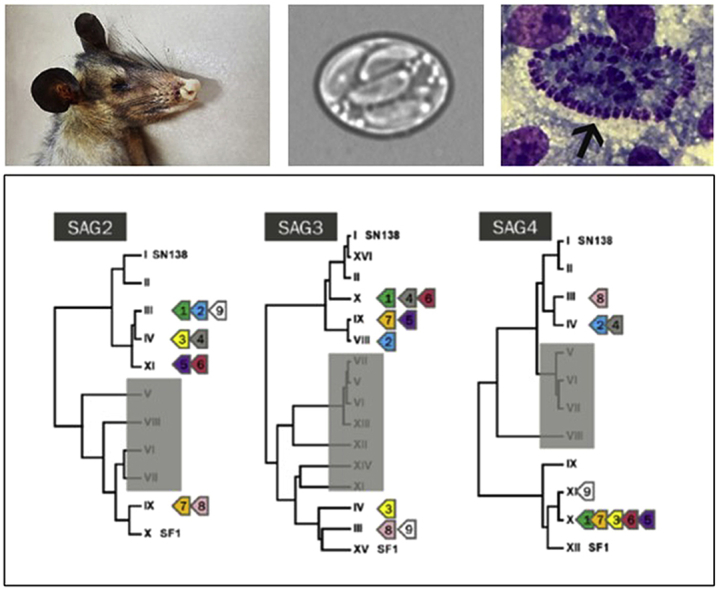
Genetic sequences of SAG2, SAG3 and SAG4 were nested PCR amplified and the total length of each nested PCR fragment obtained were 338, 511 and 282 bp, respectively. SAG genotype derived from Sarco-BA1 was related, but distinct, after comparison with 19 isolates from previous studies (Gondim et al., 2017; Cesar et al., 2018). SAG sequences (SAG2, SAG3 and SAG4) of Sarco-BA1 were 100% identical to sequences of homologous alleles of S. falcatula-like identified in sporocysts of Brazilian opossums, classified by Monteiro et al. (2013) as types III (JN185358), III (JN185386) and XI (JN185400), for SAG2, SAG3 and SAG4, respectively. At all these loci, Sarco-BA1 disclosed 100% identity to Sarcocystis spp. detected in carcasses of Magellanic penguins (Spheniscus magellanicus) rescued on the coast of Brazil (Acosta et al., 2018). Coverage of SAG3 and SAG4 to homologous from Sarcocystis spp. from Magellanic penguins were less than 100% because the SAG3 and SAG4 fragments derived from Sarco-BA1 are larger than those sequenced from penguins. Although identical at SAG2, SAG3 and SAG4, Sarco-BA1 and Sarcocystis spp. from Magellanic penguins slightly differed at ITS1. Alignment of both sequences disclosed an insertion of eight nucleotides in ITS1 from penguins (MG493471). SAG genotypes from the current isolate and from those reported in previous studies are shown in Fig. 3.
Fig. 3.
Dendograms on SAG2, SAG3 and SAG4 genotypes from Sarcocystis spp. that use Brazilian Didelphis spp. as definitive hosts, as proposed by Monteiro et al. (2013) and Valadas et al. (2016). Arrows 1 to 7 correspond to Sarcocystis spp. genotypes derived from budgerigars that were experimentally infected with Didelphis spp. sporocysts (Cesar et al., 2018; Gondim et al., 2017). Arrow 8 identifies a genotype of S. falcatula isolated from a naturally infected bare-faced ibis (Phimosus infuscatus) (Konradt et al., 2017). Arrow 9 represents the current genotype (Sarco-BA1) and those observed in Magellanic penguins (Acosta et al., 2018). SN138 (Lindsay et al., 2004) and SF1 (Marsh et al., 1997) are reference strains of Sarcocystis neurona and Sarcocystis falcatula, respectively. Alleles in shaded boxes correspond to genotypes identified in opossums-derived sporocysts that have not been associated with S. falcatula so far.
4. Discussion
In the present work, S. falcatula-like (Sarco-BA1) was in vitro isolated from sporocysts obtained from a Brazilian opossum (D. aurita/D. marsupialis) which inhabited the urban area of Salvador, Bahia. Opossums of the species D. aurita and D. marsupialis are morphologically similar (Cerqueira, 1985), so additional tools are probably required to accurately identify each of these species. The opossum, whose sporocysts were derived, was referred here as D. aurita/D. marsupialis. Finding sporocysts in intestinal scraping of the animal was not expected, because in a previous study, intestinal scrapings of 39 Didelphis spp. that resided in Salvador did not contain parasites’ sporocysts (Gondim et al., 2017); these authors reported that opossums inhabiting the urban area of the city had easy access to food from humans and minor chances of acquiring infection from intermediate hosts.
DNA extracted from sporocysts was amplified using primers directed to S. neurona/S. falcatula (Tanhauser et al., 1999) that are also known to amplify DNA from other Sarcocystis species shed by opossums (Gondim et al., 2017). Nested PCR for ITS1, followed by nucleotide sequencing, revealed that Sarco-BA1 differed from the known species of Sarcocystis shed by Didelphis sp. (S. neurona, S. falcatula, S. speeri and S. lindsayi), but had identical ITS1 sequences to 19 isolates derived from two studies (Cesar et al., 2018; Gondim et al., 2017). Interestingly, Sarco-BA1 possessed identical SAG (SAG2, SAG3 and SAG4) and almost identical ITS1 sequences to Sarcocystis spp. detected in 16 Magellanic penguins (Spheniscus magellanicus) rescued on the coast of Espírito Santo state, Brazil (Acosta et al., 2018). Bahia, where Sarco-BA1 was derived, neighbors Espírito Santo, therefore, the geographical proximity of these states possibly explains the identical SAG sequences observed for Sarco-BA1 and Sarcocystis spp. detected in the penguins.
Due to the genetic similarity among Sarco-BA1 and previously isolated Brazilian strains of Sarcocystis spp. from opossums (Cesar et al., 2018; Gondim et al., 2017), bioassay of the parasite was attempted in budgerigars, which were susceptible to infections in earlier studies on S. falcatula-like organisms (Cesar et al., 2018; Gondim et al., 2017). The two birds were inoculated with small numbers of parasites (10 and 103 sporocysts) and developed sarcocystosis on the 21st and 34th DAI, confirming that Sarco-BA1 was highly virulent for this avian species. In vitro isolation of merozoites from budgerigars' tissues was attempted, however, bacterial contamination was observed in the culture flask derived from the first budgerigar, and no merozoites were obtained from the second infected bird. Obtaining Sarcocystis sp. merozoites derived from bioassay was desired to minimize isolation of mixed species, because it is known that an infected opossum, as Didelphis virginiana, may have sporocysts of more than one Sarcocystis species (Dubey, 2000). In the current study, the opossum was rescued in the urban area of Salvador city and was probably exposed to a narrow range of potential intermediate hosts. Merozoites of Sarco-BA1 isolated directly from opossum's sporocysts are probably originated from a single species.
In vitro isolation of Sarco-BA1 was initially conducted using Vero cells. However, multiplication of merozoites in this cell line was poor and led to minimal number of free parasites in the culture. Due to the ability of Sarco-BA1 to actively propagate in tissues from an avian species (Mellopsitacus undulatus), the authors attempted to culture the parasite in a permanent avian cell line (DF-1) derived from chicken. The multiplication of Sarco-BA1 was dramatically improved in DF-1 cells. In contrast to Vero cell monolayers, where free merozoites (outside the cells) were rarely seen after 60 days of cultivation, the growth of the parasite in DF-1 cells led to great numbers of free merozoites, as well as intracellular parasites. Therefore, Vero cells have been substituted for DF-1 to culture Sarco-BA1.
To our knowledge, this is the first molecular study and in vitro isolation of S. falcatula-like derived from Northeastern Brazil. Previous molecular analysis of S. falcatula-like isolates in Brazil employed parasites derived from opossums in regions located more than 1,600 km far from Salvador (Cesar et al., 2018; Gondim et al., 2017; Monteiro et al., 2013; Siqueira et al., 2010; Valadas et al., 2016). Finding S. falcatula-like in the Northeast of Brazil, whose genetic and biological characteristics match with isolates detected in at least four different Brazilian states, reinforces the premise that these isolates probably represent a new species shed by opossums, besides S. neurona, S. falcatula, S. speeri and S. lindsayi. Studies are under way to determine infectivity of Sarco-BA1 to other animal species, as well as to investigate serological cross-reactivity among Sarco-BA1, S. neurona and related species.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank to CAPES (Coordination for Higher Education Staff Development) for providing fellowships to Aline S. Tavares and Waléria B. Silva. This work was supported by the Brazilian Research Council (CNPq), process number 308795/2015-6. Luís F.P. Gondim, Rodrigo M. Soares and Horwald A.B. Llano are recipients of fellowships by CNPq.
We acknowledge Dr. Gereon Schares (Friedrich-Loffler-Institut, Germany) for donating the UMNSAH/DF-1 cells.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2019.08.008.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
References
- Acosta I.C.L., Soares R.M., Mayorga L., Alves B.F., Soares H.S., Gennari S.M. Occurrence of tissue cyst forming coccidia in Magellanic penguins (Spheniscus magellanicus) rescued on the coast of Brazil. PLoS One. 2018;13 doi: 10.1371/journal.pone.0209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box E.D., Meier J.L., Smith J.H. Description of Sarcocystis falcatula stiles, 1893, a parasite of birds and opossums. J. Protozool. 1984;31:521–524. doi: 10.1111/j.1550-7408.1984.tb05495.x. [DOI] [PubMed] [Google Scholar]
- Cerqueira R. The distribution of Didelphis in South America (Polyprotodontia, Didelphidae) J. Biogeogr. 1985;12:135–145. [Google Scholar]
- Cesar M.O., Matushima E.R., Zwarg T., de Oliveira A.S., Sanches T.C., Joppert A.M., Keid L.B., Oliveira T., Ferreira H.L., Llano H.A.B., Konradt G., Bianchi M.V., Gregori F., Gondim L.F.P., Soares R.M. Multilocus characterization of Sarcocystis falcatula-related organisms isolated in Brazil supports genetic admixture of high diverse SAG alleles among the isolates. Exp. Parasitol. 2018;188:42–49. doi: 10.1016/j.exppara.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Dubey J., Calero-Bernal R., Rosenthal B., Speer C., Fayer R. CRC Press; 2015. Sarcocystosis of Animals and Humans. [Google Scholar]
- Dubey J.P. Prevalence of sarcocystis species sporocysts in wild-caught opossums (Didelphis virginiana) J. Parasitol. 2000;86:705–710. doi: 10.1645/0022-3395(2000)086[0705:POSSSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dubey J.P., Lindsay D.S. Sarcocystis speeri N. sp. (Protozoa: Sarcocystidae) from the opossum (Didelphis virginiana) J. Parasitol. 1999;85:903–909. [PubMed] [Google Scholar]
- Dubey J.P., Lindsay D.S., Kerber C.E., Kasai N., Pena H.F., Gennari S.M., Kwok O.C., Shen S.K., Rosenthal B.M. First isolation of Sarcocystis neurona from the South American opossum, Didelphis albiventris, from Brazil. Vet. Parasitol. 2001;95:295–304. doi: 10.1016/s0304-4017(00)00395-2. [DOI] [PubMed] [Google Scholar]
- Dubey J.P., Lindsay D.S., Rosenthal B.M., Kerber C.E., Kasai N., Pena H.F., Kwok O.C., Shen S.K., Gennari S.M. Isolates of Sarcocystis falcatula-like organisms from South American opossums Didelphis marsupialis and Didelphis albiventris from Sao Paulo, Brazil. J. Parasitol. 2001;87:1449–1453. doi: 10.1645/0022-3395(2001)087[1449:IOSFLO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dubey J.P., Rosenthal B.M., Speer C.A. Sarcocystis lindsayi n. sp. (Protozoa: Sarcocystidae) from the South American opossum, Didelphis albiventris from Brazil. J. Eukaryot. Microbiol. 2001;48:595–603. doi: 10.1111/j.1550-7408.2001.tb00196.x. [DOI] [PubMed] [Google Scholar]
- Fenger C.K., Granstrom D.E., Langemeier J.L., Stamper S., Donahue J.M., Patterson J.S., Gajadhar A.A., Marteniuk J.V., Xiaomin Z., Dubey J.P. Identification of opossums (Didelphis virginiana) as the putative definitive host of Sarcocystis neurona. J. Parasitol. 1995;81:916–919. [PubMed] [Google Scholar]
- Foster, D.N., Foster, L.K., 1997. Immortalized cell lines for virus growth. US Patent5,672,485 dated Sep 30.
- Gallo S.S.M., Lindsay D.S., Ederli N.B., Matteoli F.P., Venancio T.M., de Oliveira F.C.R. Identification of opossums Didelphis aurita (Wied-Neuweid, 1826) as a definitive host of Sarcocystis falcatula-like sporocysts. Parasitol. Res. 2018;117:213–223. doi: 10.1007/s00436-017-5695-4. [DOI] [PubMed] [Google Scholar]
- Gondim L.S.Q., Jesus R.F., Ribeiro-Andrade M., Silva J.C.R., Siqueira D.B., Marvulo M.F.V., Aléssio F.M., Mauffrey J.-F., Julião F.S., Savani E.S.M.M., Soares R.M., Gondim L.F.P. Sarcocystis neurona and Neospora caninum in Brazilian opossums (Didelphis spp.): molecular investigation and in vitro isolation of Sarcocystis spp. Vet. Parasitol. 2017;243:192–198. doi: 10.1016/j.vetpar.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Konradt G., Bianchi M.V., Leite-Filho R.V., da Silva B.Z., Soares R.M., Pavarini S.P., Driemeier D. Necrotizing meningoencephalitis caused by Sarcocystis falcatula in bare-faced ibis (Phimosus infuscatus) Parasitol. Res. 2017;116:809–812. doi: 10.1007/s00436-016-5341-6. [DOI] [PubMed] [Google Scholar]
- Li Q.Q., Yang Z.Q., Zuo Y.X., Attwood S.W., Chen X.W., Zhang Y.P. A PCR-based RFLP analysis of Sarcocystis cruzi (Protozoa: Sarcocystidae) in Yunnan Province, PR China, reveals the water buffalo (Bubalus bubalis) as a natural intermediate host. J. Parasitol. 2002;88:1259–1261. doi: 10.1645/0022-3395(2002)088[1259:APBRAO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lindsay D.S., Mitchell S.M., Vianna M.C., Dubey J.P. Sarcocystis neurona (Protozoa: Apicomplexa): description of oocysts, sporocysts, sporozoites, excystation, and early development. J. Parasitol. 2004;90:461–465. doi: 10.1645/GE-230R. [DOI] [PubMed] [Google Scholar]
- Marsh A.E., Barr B.C., Tell L., Koski M., Greiner E., Dame J., Conrad P.A. In vitro cultivation and experimental inoculation of Sarcocystis falcatula and Sarcocystis neurona merozoites into budgerigars (Melopsittacus undulatus) J. Parasitol. 1997;83:1189–1192. [PubMed] [Google Scholar]
- Monteiro R.M., Keid L.B., Richtzenhain L.J., Valadas S.Y., Muller G., Soares R.M. Extensively variable surface antigens of Sarcocystis spp. infecting Brazilian marsupials in the genus Didelphis occur in myriad allelic combinations, suggesting sexual recombination has aided their diversification. Vet. Parasitol. 2013;196:64–70. doi: 10.1016/j.vetpar.2013.01.019. [DOI] [PubMed] [Google Scholar]
- Reed S.M., Furr M., Howe D.K., Johnson A.L., MacKay R.J., Morrow J.K., Pusterla N., Witonsky S. Equine Protozoal myeloencephalitis: an updated consensus statement with a focus on parasite biology, diagnosis, treatment, and prevention. J. Vet. Intern. Med. 2016;30:491–502. doi: 10.1111/jvim.13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira D.B., Aessio F.M., Mota R.A., Marvulo M.F., Mauffrey J.F., Monteiro S.R., Farias R.C., Cunha R.C., Oliveira R.L., Souza T.C., Medeiros E.S., Silva J.C. Staphylococcus aureus mastitis in a white-eared opossum (Didelphis albiventris) in the Atlantic Forest of northeast Brazil. J. Zoo Wildl. Med. 2010;41:526–529. doi: 10.1638/2009-0079.1. [DOI] [PubMed] [Google Scholar]
- Slapeta J.R., Koudela B., Votypka J., Modry D., Horejs R., Lukes J. Coprodiagnosis of Hammondia heydorni in dogs by PCR based amplification of ITS 1 rRNA: differentiation from morphologically indistinguishable oocysts of Neospora caninum. Vet. J. 2002;163:147–154. doi: 10.1053/tvjl.2001.0599. [DOI] [PubMed] [Google Scholar]
- Smith J.H., Neill P.J., Dillard E.A., 3rd, Box E.D. Pathology of experimental Sarcocystis falcatula infections of canaries (Serinus canarius) and pigeons (Columba livia) J. Parasitol. 1990;76:59–68. [PubMed] [Google Scholar]
- Soares R.M., Lopes E.G., Keid L.B., Sercundes M.K., Martins J., Richtzenhain L.J. Identification of Hammondia heydorni oocysts by a heminested-PCR (hnPCR-AP10) based on the H. heydorni RAPD fragment AP10. Vet. Parasitol. 2011;175:168–172. doi: 10.1016/j.vetpar.2010.09.022. [DOI] [PubMed] [Google Scholar]
- Stabenow C.S., De Oliveira F.C., Albuquerque G.R., Lopes C.W. Sarcocystis lindsayi-like (Apicomplexa: Sarcocystinae) of the opossum (Didelphis aurita) from Southeastern Brazil. Rev. Bras. Parasitol. Vet. 2008;17(Suppl. 1):342–344. [PubMed] [Google Scholar]
- Tanhauser S.M., Yowell C.A., Cutler T.J., Greiner E.C., MacKay R.J., Dame J.B. Multiple DNA markers differentiate Sarcocystis neurona and Sarcocystis falcatula. J. Parasitol. 1999;85:221–228. [PubMed] [Google Scholar]
- Valadas S.Y., da Silva J.I., Lopes E.G., Keid L.B., Zwarg T., de Oliveira A.S., Sanches T.C., Joppert A.M., Pena H.F., Oliveira T.M., Ferreira H.L., Soares R.M. Diversity of Sarcocystis spp. shed by opossums in Brazil inferred with phylogenetic analysis of DNA coding ITS1, cytochrome B, and surface antigens. Exp. Parasitol. 2016;164:71–78. doi: 10.1016/j.exppara.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Verma S.K., Trupkiewicz J.G., Georoff T., Dubey J.P. Molecularly confirmed acute, fatal Sarcocystis falcatula infection in the Rainbow Lorikeets (Trichoglossus moluccanus) at the philadelphia zoo. J. Parasitol. 2018;104:710–712. doi: 10.1645/18-78. [DOI] [PubMed] [Google Scholar]