Abstract
Persistent inflammation promotes development and progression of heart failure (HF). TWEAK (TNF-Related WEAK Inducer Of Apoptosis), a NF-κB- and/or AP-1-responsive proinflammatory cytokine that signals via TWEAK receptor (TWEAKR), is expressed at high levels in human and preclinical models of HF. Since the adapter molecule TRAF3IP2 (TRAF3 Interacting Protein 2) is an upstream regulator of various proinflammatory pathways, including those activated by NF-κB and AP-1, we hypothesized that targeting TRAF3IP2 inhibits TWEAK induced proinflammatory and pro-fibrotic responses in vitro and in vivo. Consistent with the hypothesis, forced expression of TRAF3IP2 upregulated TWEAK and its receptor expression in cultured adult mouse cardiac fibroblasts (CF). Further, exogenous TWEAK upregulated TRAF3IP2 expression in a time- and dose-dependent manner, suggesting a positive-feedback regulation of TRAF3IP2 and TWEAK. TWEAK also promoted TRAF3IP2 nuclear translocation. Confirming its critical role in TWEAK signaling, silencing TRAF3IP2 inhibited TWEAK autoregulation, TWEAKR upregulation, p38 MAPK, NF-κB and AP-1 activation, inflammatory cytokine expression, MMP and TIMP1 activation, collagen expression and secretion, and importantly, proliferation and migration. Recapitulating these in vitro results, continuous infusion of TWEAK for 7 days increased systolic blood pressure (SBP), upregulated TRAF3IP2 expression, activated p38 MAPK, NF-κB and AP-1, induced the expression of multiple proinflammatory and pro-fibrotic mediators, and interstitial fibrosis in hearts of wild type mice. These proinflammatory and pro-fibrotic changes occurred in conjunction with myocardial hypertrophy and contractile dysfunction. Importantly, genetic ablation of TRAF3IP2 inhibited these TWEAK-induced adverse cardiac changes independent of increases in SBP, indicating that TRAF3IP2 plays a causal role, and thus a therapeutic target, in chronic inflammatory and fibro-proliferative diseases.
Keywords: Fn14, CIKS, Act1, adverse cardiac remodeling, migration
1. Introduction
TWEAK (TNF-Related WEAK Inducer Of Apoptosis), a secreted ligand in the TNF superfamily of ligands (TNF Superfamily member12 or TNFSF12), is a multifunctional protein that exerts proinflammatory, pro-mitogenic, pro-migratory and pro-apoptotic effects, depending on the cellular context and the disease process investigated [1–3]. It signals mainly via TWEAK receptor (TWEAKR; also known as Fn14, Fibroblast Growth Factor-Inducible Immediate-Early Response Protein 14) [4]. Both TWEAK and TWEAKR are expressed at low basal levels in various organs, including the heart.
In the heart, cardiac fibroblasts (CF) express both TWEAK and TWEAKR [1–3,5,6]. Though genetic ablation of TWEAK or its receptor failed to result in developmental defects, metabolic alterations, or death [7,8], their sustained expression has been shown to amplify inflammatory responses in the heart, resulting in adverse myocardial remodeling and HF development. For example, subjects with ST-elevation myocardial infarction, a predisposing factor for HF development, showed increased circulating levels of TWEAK, with as high a level as 7500 pg/ml in some individuals compared to ~524 pg/ml in normal controls [9], possibly contributing to adverse myocardial remodeling in those subjects. In fact, transgene-mediated sustained elevation in systemic soluble TWEAK (a 1000-fold increase compared to non-transgenic littermate controls) or adenoviral-mediated overexpression of full-length TWEAK has been shown to result in dilated cardiomyopathy, fibrosis, contractile dysfunction, HF development and early mortality [10]. In those transgenic animal models, TWEAK expression was under the regulation of liver-specific apolipoprotein E enhancer/human α-antitrypsin promoter, indicating that sustained increases in systemic TWEAK levels are detrimental to the heart, irrespective of origin [10]. Surprisingly, though elevated soluble or membranous TWEAK promoted dilated cardiomyopathy, no cardiomyocyte death was detected in those models. Importantly, genetic ablation of TWEAKR blocked those TWEAK-induced adverse cardiac changes [10], suggesting that targeting TWEAK or its receptor has the potential to inhibit persistent inflammation and HF development.
The intracellular domain of TWEAKR contains an amino acid motif (-PIEET-) that binds various members of the TRAF (TNF receptor-associated factors) family of signaling adapters, resulting in the activation of multiple downstream signaling pathways [11,12]. For example, association between TWEAKR and TRAF2 promotes recruitment of cellular inhibitors of apoptosis and TNFR death domain proteins, resulting in cell survival or death [13,14]. TWEAK induces TRAF1 expression in an NF-κB-dependent manner [15], and the interaction between TRAF1 and TRAF2 has been shown to facilitate a crosstalk between TWEAKR and the receptors for TNF, resulting in amplification of inflammatory signaling. In a myoblast cell line, the interaction between TWEAKR and TRAF6 has been shown to inhibit anabolic pathways, but results in the activation of the ubiquitin-proteasome and the NF-κB systems, culminating in atrophy [16]. These reports indicate that the association between TWEAKR and various TRAFs results in the activation of multiple proinflammatory pathways, majority of which lead to NF-κB activation. In addition, interactions between TWEAKR and TRAFs have also been shown to activate JNK, AP-1 and p38 MAPK [17,18], whose sustained activation is well known to affect adversely both cardiac structure and function through sustained inflammation [19,20].
In addition to activating NF-κB and AP-1, TWEAK and TWEAKR are also NF-κB and/or AP-1-responsive genes [21,22]. Thus, upon induction, TWEAK/TWEAKR signaling perpetuates inflammation by the sustained induction of their own and that of various other inflammatory mediators, through a NF-κB- and/or AP-1-dependent mechanism. Of note, sustained activation of NF-κB and AP-1 has been shown to contribute causally to myocardial injury, hypertrophy, fibrosis, contractile dysfunction and HF development [21–23]. We and others have previously reported that the cytoplasmic adapter molecule TRAF3 Interacting Protein 2 (TRAF3IP2, also known as CIKS [Connection To IKK And SAPK/JNK] or Act1 [NF-Kappa-B Activator 1]) is an upstream regulator of both NF-κB and AP-1, and plays a causal role in various autoimmune and inflammatory diseases [24–26].
Structurally, TRAF3IP2 is a 555 amino acid adapter molecule that contains two TRAF-interacting motifs, a helix-loop-helix (HLH) domain to activate IKK, a U-box that plays a role in ubiquitination of TRAF6 at Lys63 resulting in its activation, and a STIR (similar expression to fibroblast growth factor genes and interleukin-17 receptors [SEFIR] + Toll-IL-1 receptor [TIR]) domain that interacts with other SEFIR-domain containing proteins (e.g., IL-17 receptors) [24–26]. Though TRAF3IP2 contains two TRAF-interacting motifs (-EESE-), these sites appear to be mutually redundant since signaling was impaired only when both TRAF-interacting motifs are mutated. Lys63-mediated polyubiquitination of TRAF6 activates NF-κB via mechanisms similar to those established for IL-18, IL-1, Toll, and CD40 receptors; i.e., via recruitment of TAK1 (TGF-beta activated kinase 1) and the ubiquitinbinding proteins TAB1/2 (TGF-β Activated Kinase 1 and 2), resulting in IKK (IκB kinase) activation [27,28]. We recently demonstrated that TRAF3IP2 physically associates with IKKγ/NEMO and JNK, and silencing TRAF3IP2 inhibits IKKγ/JNK association and JNK activation, indicating that TRAF3IP2 may act as a scaffold in IKKγ and JNK physical association, and AP-1 activation [26].
Similar to the pathological role played by the enhanced expression of TWEAK and its receptor, cardiomyocyte-specific transgenic overexpression of TRAF3IP2 results in sustained activation of the IKK/NF-κB and JNK/AP-1-dependent inflammatory signaling and the spontaneous development myocardial hypertrophy, fibrosis and contractile dysfunction [29]. Since TWEAK/TWEAKR signaling activates NF-κB and AP-1 via TRAF-dependent mechanisms, and as TRAF3IP2 is an upstream regulator of both transcription factors, we hypothesized that targeting TRAF3IP2 will not only inhibit TWEAK/TWEAKR-induced NF-κB and AP-1 activation, but also TWEAK autoregulation and TWEAKR upregulation, resulting in reduced proinflammatory and profibrotic responses both in vitro and in vivo.
2. Materials and methods
2.1. Materials
The materials used in the present study are described in Supplementary methods.
2.2. In vitro studies
2.2.1. Animals and cardiac fibroblast (CF) isolation and treatment
This investigation conforms to the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health (DRR/National Institutes of Health, 1996). All protocols were approved by the Institutional Animal Care and Use Committees at the University of Texas Health Science Center, San Antonio, TX and the Tulane University, New Orleans, LA. CF were isolated from wild type male C57BL/6 mice weighing ~25–30 g and aged 3 to 4 months as discussed in detail in Supplementary methods. Freshly isolated and up to third passage CF were used in all experiments. At 50–70% confluency, the cells were made quiescent by incubating in a medium containing 0.5% BSA (serum-free) for 48 h. These growth-arrested and synchronized CF were treated with recombinant mouse TWEAK as described in Supplementary methods.
2.2.2. Lentiviral and adenoviral infection
Lentiviral short hairpin RNA (shRNA) against TRAF3IP2, TWEAKR, p65 subunit of NF-κB, and c-Jun subunit of AP-1 were purchased from Sigma-Aldrich (St. Louis, MO, USA), and are described in Supplementary methods. Lentiviral shRNA against green fluorescent protein (GFP) served as a non-targeting shRNA, and was purchased from Santa Cruz Biotechnology, Inc. (Supplementary Table 1). Adenoviral vectors expressing TRAF3IP2, enhanced GFP (eGFP), or silencing siRNA against NOX4 or GFP are also described in Supplementary Table 1. Infection with lentiviral and adenoviral vectors is detailed in Supplementary methods.
2.2.3. In vitro Detection of hydrogen peroxide by Amplex Red assay
Quiescent CF treated with TWEAK (10 ng/ml) for 1 h were analyzed for H2O2 production using a commercially available kit (Amplex® Red Hydrogen Peroxide/Peroxidase assay kit, Thermo Fisher Scientific). The protocol is described in Supplementary methods. Studies were also performed after TWEAKR or NOX4 knockdown prior to TWEAK addition for 1 h.
2.2.4. TWEAK and TWEAKR promoter-reporter activity
A 3 kb fragment of the 5′-flanking region of the murine TWEAK gene was amplified using mouse genomic DNA as a template by the following primers containing the restriction sites KpnI and XhoI; Forward: 5′-GCCGGTACCAGTCTCTCAGTTCATGGTCACTTTGTAC-3′ and Reverse: 5;-GCCCTCGAGGGGGGCGGGGGGCTGTGCCTGCCTCACC-3′. The amplified 3 kb fragment was cloned into pGL4.10[luc2] vector upstream of the luciferase gene (pGL4.10-TWEAK-3 kb) and sequence verified (Supplementary Fig. 1). The 3 kb fragment of the 5′ flanking region of murine TWEAKR in pGL3-Basic vector (pGL3-TWEAKR-3 kb) is previously described [30]. pGL4.10 and pGL3-Basic served as vector controls. CF were transduced with lentiviral shRNA against p65, c-Jun or TRAF3IP2 (moi0.5). Twenty four hours later, the transduced cells were transfected (2 μg) with pGL4.10-TWEAK-3 kb or pGL3-TWEAKR-3 kb or respective empty vectors along with 100 ng of the control Renilla luciferase vector pRL-TK using the Neon transfection system (MPK-5000, Invitrogen) with the following parameters: pulse voltage: 1400 V, pulse width: 20 ms, pulse number: 2, and the tip type: 10 μl. Cell viability was 89%, and the transfection efficiency was 71%. Twenty four-hours later, cells were incubated with TWEAK (10 ng/ml) for 12 h, harvested, and analyzed with the dual luciferase reporter assay. Data were normalized by dividing firefly luciferase activity with that of the corresponding Renilla luciferase. The activity was expressed as fold change after considering the data obtained with respective empty vectors alone as 1. All plasmid preparations were endotoxin-free.
2.2.5. mRNA expression by quantitative RT-PCR (RT-qPCR)
Total RNA was isolated from frozen LV tissues using Trizol reagent (Sigma) and 0.5 μg of RNA was reverse transcribed into cDNA using a reverse transcription kit (Agilent Technologies). IL-1β, IL-6, IL-18, TNF-α, TNFR1, CT-1, CXCL1, CXCL2, CXCL5, MCP-1, ICAM1, Selectin E, MMP-13, MMP-14, TIMP-1, ANP, Collagen Iα1 and Collagen IIIα1 mRNA levels were determined by RT-qPCR using best coverage TaqMan® probes (Supplementary Table 2). GAPDH served as the endogenous invariant control gene. All data were normalized to corresponding GAPDH levels and analyzed using the 2−ΔΔCt method.
2.2.6. Immunoblotting and ELISA
Preparation of whole cell lysates, membrane extracts, immunoblotting, detection of the immuno-reactive bands by enhanced chemiluminescence (ECL Plus; GE Healthcare), and semi-quantification by densitometry were all described previously [26,29,31–33]. Secreted IL-1β, TNF-α and MCP-1 levels were quantified by ELISA using equal amounts of culture supernatants. The antibodies used in immunoblotting and the ELISA kits are listed in Supplementary Tables 3 and 4.
2.2.7. Immunofluorescence
Expression and localization of TRAF3IP2 and TWEAKR were analyzed by immunofluorescence using the antibodies described under immunoblotting/immunofluorescence in Supplementary Table 3.
2.2.8. Collagen secretion by hydroxyproline assay
Total collagen content in equal amounts of culture supernatants was analyzed as previously described [34]. In brief, culture supernatants after 24 h TWEAK treatment were digested with 6 N hydrochloric acid for 12 h at 110 °C, vacuum dried, resuspended in citrate-acetate buffer, and a colored reaction was performed by adding isopropyl alcohol, chloramine T, and Ehrlich’s reagent. The samples were then incubated at 25 °C for 18 h, and intensity of the red color was measured at 558 nm. A standard curve was generated using known concentrations of hydroxyproline. After calculating hydroxyproline content in the unknown samples, the amount of collagen was calculated by multiplying hydroxyproline content by a factor of 8.2.
2.2.9. Cell proliferation
The effects of TWEAK on CF proliferation were analyzed according to manufacturer’s protocol using the CyQUANT® assay (Invitrogen) [35]. Briefly, second-passage CF were plated at 2000 cells per well into 96-well clear bottom, black-sided plates (VWR Scientific Products, West Chester, PA), and allowed to attach overnight. After 24 h, the cells were fed with serum-free medium containing 0.5% BSA and incubated for an additional 24 h. Cells were then continuously stimulated with TWEAK (10 ng/ml) in serum-free medium for 24 h. After removing the medium, the plates were frozen at −80 °C for 2 h before assay. Plates were then thawed, stained with CyQUANT® GR dye according to manufacturer’s protocol, and assayed on a FLX800 microplate fluorescence reader (Bio-Tek Instruments, Winooski, VT) using 485/20 excitation and 528/20 emission filters, and analyzed using KC4 software (Bio-Tek). Proliferation assays were carried out 6 times. To determine the role of TWEAKR or TRAF3IP2 in TWEAK-induced CF proliferation, CF were plated in complete medium, and after 24 h, cell were infected with lentiviral shRNA against TWEAKR or TRAF3IP2 (moi0.5). One hour later, the medium with virus was changed to medium containing 0.5% BSA for an additional 24 h. The cells were then incubated with TWEAK as described above. Lentiviral shRNA against eGFP at a similar moi and duration served as a control.
2.2.10. Cell migration
CF migration was quantified as described previously using BioCoat™ Matrigel™ migration chambers and 8.0-μm pore polyethylene terephthalate membranes with a thin layer of Matrigel™ basement membrane matrix [35]. Cultured CF were trypsinized and suspended in RPMI + ITS 1×), and 1 ml containing 2.0 × 105 cells was layered on the coated insert filters. Cells were stimulated with TWEAK (10 ng/ml). The lower chamber contained medium with 10% serum. Plates were incubated at 37 °C for 12 h. The membranes were washed with PBS, and non-invading cells on the upper surface were removed using cotton swabs. CF invading into and through the Matrigel™ matrix were quantified by MTT assay. Numbers of CF migrating in response to TWEAK were normalized to those of untreated cells and expressed as fold change from untreated. Migration assays were carried out 6 times. To determine the role of TWEAKR or TRAF3IP2 in TWEAK-induced CF proliferation, CF infected with lentiviral TWEAKR or TRAF3IP2 (moi0.5 for 48) were layered on the coated insert filters and treated with TWEAK (10 ng/ml) for 12 h. Lentiviral shRNA against eGFP at a similar moi and duration served as a control.
2.2.11. Cell death detection by ELISA
After 48 h incubation in DMEM containing 0.5% BSA, CFs were treated with TWEAK (0.1–1000 ng/ml) 24 h. Cells were harvested and analyzed for mono- and oligo-nucleosomes in the cytoplasmic fraction of cell lysates by a cell death detection ELISA as detailed in Supplementary Methods. H2O2 (100 μM) serves as a positive control.
2.3. In vivo studies
2.3.1. Animals
Male wild type (WT) C57Bl/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The TRAF3IP2-knockout mice (TRAF3IP2-KO) on C57Bl/6 background were previously described [26]. All mice used in the study were male, aged 3–4 months and weighed ~25–30 g. However, in TUNEL assay, ovaries from 3 month old naïve female C57Bl/6 mice were used as a positive control to detect cells undergoing apoptosis. The TRAF3IP2-KO mice exhibited no basal phenotypic abnormalities. We had 4 groups of 5 mice each: Group 1, WT mice infused with physiological saline, Group 2: WT mice infused with recombinant TWEAK, Group 3: TRAF3IP2-KO mice infused with physiological saline, and Group 4: TRAF3IP2-KO mice infused with recombinant TWEAK. Both WT and TRAF3IP2-KO mice were administered with recombinant mouse TWEAK (10 μg in physiological saline/kg body weight/day) for 7 days using subcutaneously implanted osmotic minipumps. Control animals were implanted with sterile salinefilled pumps. The dose of TWEAK was chosen based on previous published reports, where recombinant TWEAK was administered to mice between 2 and 200 micrograms, either as a single bolus or repeatedly, and either intraperitoneally or continuously via subcutaneously implanted osmotic minipumps for up to 14 days [36,37]. At the end of the 7-day study period, systolic blood pressure (SBP) was measured by tailcuff plethysmography. Myocardial function was analyzed by 2D echocardiography.
2.3.2. Systolic blood pressure (SBP) by tail-cuff plethysmography
Prior to initiation of study, animals were trained for SBP measurement using a tail-cuff method without anesthesia (CODA Noninvasive Blood Pressure System, Kent Scientific, Torrington, CT). SBP was also measured at 1, 2, and 7 days post-TWEAK administration.
2.3.3. Echocardiography
Left ventricular (LV) function was analyzed by 2D echocardiography (Acuson 128XP/10) as described before [31]. We measured LV Posterior Wall thickness at end-systole (LVPWs), LV Posterior Wall thickness at end-diastole (LVPWd), LV Internal Dimension at end-systole (LVIDs), LV Internal Dimension at end-diastole (LVIDd), LV Volume at end-systole (LVVols) and Ejection Fraction (EF).
2.3.4. Myocardial hypertrophy
After recording body weights, mice were deeply anesthetized with ketamine:xylazine (9:1; 0.1 ml, IM). After thoracotomy, blood was drawn by heart puncture, perfused with physiological saline, and hearts removed. After recording heart weights, LV was separated, cut into three pieces, and used for histology and biochemical/molecular analysis. Myocardial hypertrophy was determined by three different methods: heart weight to body weight ratios, quantification of cardiomyocyte cross-sectional area by WGA staining of fixed tissue sections (Supplementary methods and Supplementary Table 3), and fetal gene (ANP) expression by RT-qPCR. Hypertrophic response was also analyzed by normalizing heart weights to tibia length. To measure tibia length, hind limbs were incubated in 1 M NaOH for 5 h at 37 °C with occasional agitation to digest skin, fat, and muscle tissue. After digestion, the tibia was collected, rinsed in physiological saline, dried on a paper towel, and the length measured using a digital caliper.
2.3.5. Cardiac fibrosis
Cardiac fibrosis was analyzed by three different methods: (i) Masson’s trichrome staining using 6 μM-thick sections, (ii) collagens Ia1, collagen IIIa1, and fibronectin mRNA expression by RT-qPCR, and (iii) collagens Ia1, collagen IIIa1, and fibronectin protein expression by immunoblotting, and are described in Supplementary Methods. Measurement of histological myocardial fibrosis following Masson’s trichrome staining was performed by the NIH Image J software, and presented as percent area occupied by collagens.
2.3.6. Terminal transferase-mediated dUTP-biotin nick end-labeling (TUNEL) assay
In situ detection of DNA stand breaks was analyzed by TUNEL assays using the ApopTag® Peroxidase In Situ Apoptosis Detection Kit. The assays were performed essentially as described by the manufacturer. Spleen, testis and ovary from naïve WT mice were used as positive controls.
2.4. Statistical analysis
All data are expressed as mean ± SE. Statistical significance was determined by one-way analysis of variance followed by Tukey’s posthoc test (GraphPad Prism software, San Diego, CA). Prior to performing one-way analysis of variance, normality of distribution and equality of variance were considered. In addition, all 4 groups contained equal number of animals. Differences are considered significant if the P value is less than or equal to 0.05. A power analysis was performed prior to initiating the study to ensure that type II errors are unlikely. In Fig. 6, the panel describing LVPWd in WT mice infused with TWEAK, the Grubbs’ method in GraphPad identified an outlier. Therefore, the data in that specific panel represent 4 animals. Further, though a representative immunoblot is shown in the main figures, changes in protein/phosphorylation levels from three to four independent experiments were semi-quantified by densitometry, and are shown as ratios and fold changes from untreated or respective controls at the bottom or side of respective panels.
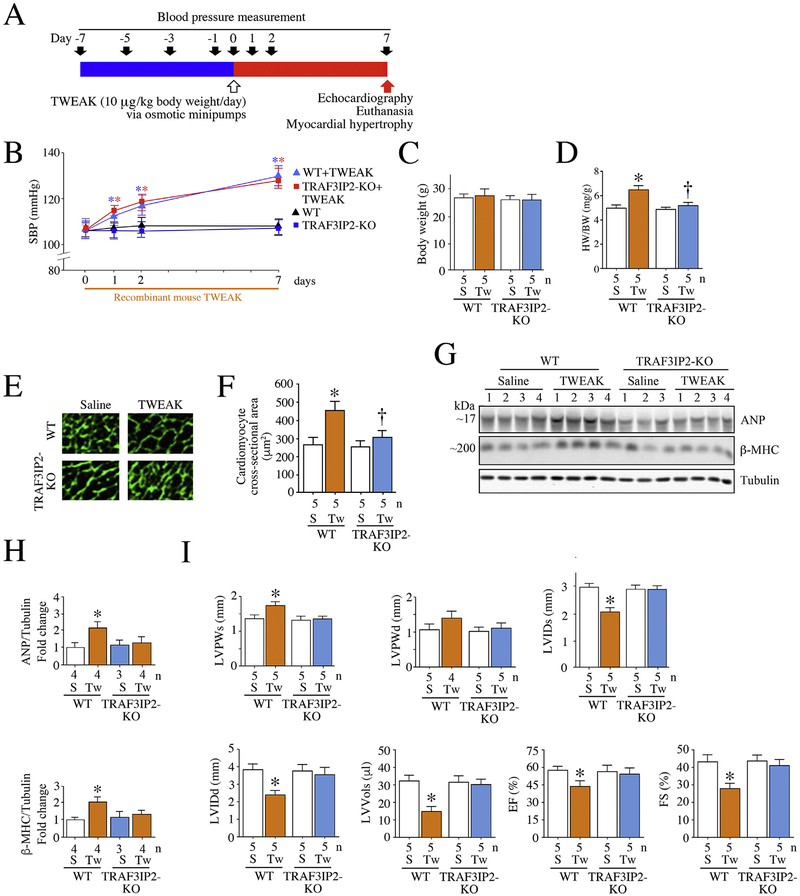
Fig. 6.
TRAF3IP2 gene deletion blunts TWEAK-induced myocardial hypertrophy and contractile dysfunction. Both wild type (WT) and homozygous TRAF3IP2 knockout mice (TRAF3IP2-KO) were continuously infused with recombinant mouse TWEAK (10 μg/kg body weight/day) for 7 days via subcutaneously implanted osmotic minipumps (A). Saline-infused animals served as controls. Systolic blood pressure was measured by tail-cuff plethysmography (B). Neither saline nor TWEAK affected body weights (C). However, TWEAK infusion resulted in myocardial hypertrophy, as evidenced by increased heart weight to body weight ratios (D), cardiomyocyte cross-sectional area (WGA staining; E and quantification, F) and ANP mRNA expression (RT-qPCR; G) in WT mice, but not in TRAF3IP2-KO mice. Further, echocardiography revealed myocardial dysfunction in TWEAK-infused WT, but not TRAF3IP2-KO mice (H), as evidenced by significant increases in LVPWs (left ventricular posterior wall thickness during systole), LVPWd (left ventricular posterior wall thickness during diastole), LVIDd (left ventricular internal diameter during diastole), LVVols (left ventricular volume in systole) and EF (percent Ejection Fraction), and reduced LVIDs. B, C, D, F–H, *P < 0.05 versus Saline, †P < 0.05 versus TWEAK (n = 5/group). HW: heart weight, BW: body weight, S: saline; Tw: TWEAK; WT: wild type; TRAF3IP2-KO: homozygous TRAF3IP2 knockout mice.
3. Results
3.1. TWEAK upregulates TRAF3IP2 expression in cultured cardiac fibroblasts (CF) in part via oxidative stress
The results in Fig. 1A show that TRAF3IP2 is expressed at low basal levels in CF, and treatment with TWEAK for 2 h upregulated its expression in a dose-dependent manner, with peak levels detected ~10 ng/ml. At higher concentrations, TWEAK did not further increase TRAF3IP2 expression, but in fact suppressed its levels. At higher levels, TWEAK reduced fibroblast viability, as evidenced by increased levels of monoand oligonucleosomal fragmented DNA in the cytoplasmic extracts, as well as active (cleaved) caspase-3 (Fig. 1B), suggesting that at higher concentrations TWEAK affects CF survival. Therefore, TWEAK was used at 10 ng/ml in all subsequent experiments. At this concentration, TWEAK upregulated TRAF3IP2 expression in a time-dependent manner with peak levels detected after 2 h treatment (Fig. 1C). Interestingly, even after 24 h, TRAF3IP2 levels remained above basal levels (Fig. 1C), suggesting persistent induction. Consistent with immunoblotting data, immunofluorescence revealed low levels of TRAF3IP2 at basal conditions, localized predominantly in cytoplasm (Fig. 1D). However, treatment with TWEAK increased TRAF3IP2 expression and showed cytoplasmic, perinuclear and nuclear localization (Fig. 1D), suggesting that TWEAK treatment promotes TRAF3IP2 nuclear localization. Immunoblotting using cytoplasmic and nuclear extracts confirmed these results (Fig. 1E). Since TWEAK signals mainly via TWEAKR, we next investigated whether silencing or neutralizing TWEAKR blunts TWEAK-induced TRAF3IP2 expression. The results in Fig. 1F show that silencing TWEAKR by lentiviral shRNA and preincubation with ITEM-4, an anti-TWEAKR neutralizing antibody, each inhibited TWEAK-induced TRAF3IP2 upregulation (knockdown of TWEAKR was confirmed by immunoblotting and is shown on the right in panel G). gp130 served as an off target, and its expression was not affected by the shRNA directed against TWEAKR (Fig. 1G). Since TRAF3IP2 is a redox-sensitive adapter molecule [33], we next investigated whether TWEAK-induced TRAF3IP2 expression is regulated by oxidative stress. Indeed, TWEAK significantly increased H2O2 generation (Fig. 1H), and silencing TWEAKR and NOX4 each attenuated its production and TRAF3IP2 induction (Fig. 1I), indicating that TWEAK upregulates TRAF3IP2 expression in CF via TWEAKR and oxidative stress (Fig. 1).
Fig. 1.
TWEAK induces ROS-dependent TRAF3IP2 expression in isolated adult mouse cardiac fibroblasts (CF). TWEAK induced TRAF3IP2 protein expression in CF in a dose-dependent manner (0.1 to 1000 ng/ml for 1 h; A), with cell death (treatment for 12 h) detected only at higher doses (B). In cell death analysis (B), H2O2 (100 μM) served as a positive control. Further, TWEAK (10 ng/ml) induced TRAF3IP2 expression in a time-dependent manner (C), with peak levels detected at ~1 h. Immunofluorescence revealed low levels of TRAF3IP2 expression in saline-treated CF and its increased expression following TWEAK treatment (10 ng/ml for 2 h), localized in cytoplasm (arrow), perinuclear (arrowhead) and nuclear (block arrow) regions (D). Immunoblotting using cytoplasmic and nuclear extracts confirmed its nuclear localization (E). Silencing TWEAKR with lentiviral shRNA (moi0.5 for 48 h) or pre-incubation with TWEAKR neutralizing antibodies (ITEM-4, 10 μg/ml for 1 h) inhibited TWEAK (10 ng/ml for 1 h)-induced TRAF3IP2 expression (F). Knockdown of TWEAKR was confirmed by immunoblotting and is shown on the right (G). gp130 served as an off-target. Lentiviral shRNA directed against eGFP as well as normal mouse IgG2b served as controls. Moreover, TWEAK (10 ng/ml for 30 min) induced oxidative stress as evidenced by increased generation of hydrogen peroxide (H2O2), an effect attenuated by TWEAKR (lentiviral TWEAKR shRNA, moi 0.5 for 48 h) or NOX4 (Ad.siNOX4, moi100 for 24 h) knockdown (H). Further, TWEAKR and NOX4 knockdown each attenuated TWEAK (10 ng/ml for 1 h) -induced TRAF3IP2 expression (I). A, C, E, F, H, I: A representative image from three independent immunoblotting experiments with similar results is shown on the left or at the top, and the summarized semi-quantification of intensity of immunoreactive bands by densitometry from all three experiments on the right or at the bottom. D, Scale bar: 100 μm. min: minutes, h: hours. B and H, n = 6. *P < at least 0.05 versus untreated, †P < at least 0.05 versus TWEAK, §P < 0.05 versus Untreated.
3.2. TRAF3IP2 mediates TWEAK-induced p38 MAPK, NF-κB and AP-1 activation in CF
Persistent inflammation, even at a low grade, promotes cardiac fibrosis [38]. The mitogen-activated protein kinase p38 and the redoxsensitive nuclear transcription factors NF-κB and AP-1 not only play a role in signal transduction of various inflammatory cytokines, but also regulate their induction. Since TRAF3IP2 is an upstream regulator of p38 MAPK, NF-κB, and AP-1 [29,32], we investigated whether TWEAK regulates their activation in CF via TRAF3IP2. Indeed, TWEAK induced p38 MAPK activation in CF in a time-dependent manner as evidenced by an increase in its Thr180/Tyr182 phosphorylated form (Fig. 2A), and this effect was markedly suppressed by TRAF3IP2 knockdown or pretreatment with the p38 MAPK-specific inhibitor SB 239063 (Fig. 2B; ~70% knockdown of TRAF3IP2 by lentiviral shRNA as shown on the right). Similarly, TWEAK induced NF-κB activation in a time-dependent manner (Fig. 2C), as evidenced by an increase in Ser536-phophorylated p65, an effect also inhibited by TRAF3IP2 knockdown (Fig. 2D) and SB 239063 (Fig. 2D). TWEAK also induced JNK activation, as evidenced by a time-dependent increase in Thr183/Tyr185-phopsorylated form (Fig. 2E, upper panel). Further, TWEAK induced Ser63 phosphorylation of c-Jun, one of the subunits of AP-1 and a substrate of JNK, indicating its activation (Fig. 2E, lower panels). Importantly, silencing TRAF3IP2 and pretreatment with the JNK inhibitor SP600125 attenuated JNK activation and c-Jun phosphorylation (Fig. 2E). These results indicate that TWEAK induces p38 MAPK, NF-κB, and JNK/AP-1 activation in CF in part via TRAF3IP2 (Fig. 2).
Fig. 2.
TWEAK induces TRAF3IP2-dependent p38 MAPK, NF-κB, and AP-1 activation in cardiac fibroblasts. TWEAK (10 ng/ml) induced time-dependent p38 MAPK activation (Thr180/Tyr182 phosphorylation) in CF (A), an effect inhibited by TRAF3IP2 knockdown (lentiviral shRNA, moi0.5 for 48 h, shown on the right) or pretreatment with the p38 MAPK-specific inhibitor SB239063 (10 μM in DMSO for 1 h; B). Similarly, TWEAK induced NF-κB activation (Ser536 phosphorylation of p65) in a time-dependent manner, an effect also inhibited by TRAF3IP2 knockdown or pre-treatment with SB239063 (D). Further, TWEAK induced time-dependent JNK (Thr183/Tyr185 phosphorylation) and AP-1 activation (Ser63 phosphorylation of c-Jun) in CF (E), an effect blunted by TRAF3IP2 knockdown (F). A-E: A representative image from three independent immunoblotting experiments with similar results is shown at the top and summarized semi-quantification of intensity of immunoreactive bands by densitometry from all three experiments in the respective bottom panels. *P < 0.05 versus Untreated, †P < 0.05 versus TWEAK, §P < 0.05 versus Untreated.
3.3. TRAF3IP2 and its downstream signaling intermediates mediate TWEAK autoregulation and TWEAKR upregulation in CF
The 5′-regulatory regions of TWEAK and TWEAKR contain binding sites for several putative transcription factors, including NF-κB for TWEAK [21] and AP-1 for TWEAKR [30]. Since TRAF3IP2 is an upstream regulator of both transcription factors [24,25], we next investigated whether TRAF3IP2 mediates TWEAK-induced TWEAK and its receptor expression. The results in Fig. 3A show that CF express low levels of TWEAK at basal conditions, and treatment with TWEAK rapidly and significantly upregulated its expression in a time dependent manner, with increased levels detected for up to 2 h. This increased expression represents, at least initially, both exogenous and endogenous TWEAK. Importantly, silencing TRAF3IP2 blunted TWEAK auto-regulation (Fig. 3A). Similarly, TWEAKR expression was detected at low basal levels, and treatment with TWEAK upregulated its expression in a time-dependent manner, and in part via TRAF3IP2 (Fig. 3B). Confirming the immunoblotting results, immunofluorescence revealed low expression of TWEAKR in CF at basal conditions, and its increased levels after 2 h treatment with TWEAK, with both membrane and cytoplasmic localization, and its reduced expression following TRAF3IP2 knockdown (Fig. 3C). Further, silencing TRAF3IP2 or its downstream signaling intermediates NF-κB and AP-1, or pharmacological inhibition of p38MAPK, markedly inhibited TWEAK autoregulation and TWEAKR upregulation (Fig. 3D; p65 and c-Jun knockdown is shown on the right in panel E). Promoter reporter assays demonstrated that TWEAK induces transcription of both TWEAK and its receptor in CF. By analyzing promoter regions 3 kb upstream of TWEAK and TWEAKR for putative transcription factor binding sites using the PROMO (Prediction of transcription factor binding sites) program [39,40], we identified nine NF-κB and five AP-1 potential binding sites in TWEAK promoter (Supplementary Fig. 1). Similarly, we identified seven NF-κB and five AP-1 potential binding sites in TWEAKR promoter. Importantly, silencing TRAF3IP2, p65 or c-Jun attenuated the promoter-reporter activities of both TWEAK and TWEAKR (Fig. 3G), suggesting that TRAF3IP2 activates their transcription possibly via a NF-κB- and AP-1-dependent pathway. Together, these results demonstrate that TRAF3IP2 plays a critical role in TWEAK/TWEAKR induction in CF. To further demonstrate its critical role, we overexpressed TRAF3IP2 by adenoviral transduction, and investigated whether overexpression of TRAF3IP2 by itself induces TWEAK/TWEAKR expression. Indeed, ectopic expression of TRAF3IP2 at various multiplicities of infection (moi) increased TRAF3IP2 expression in a dose-dependent manner, with peak levels detected at a moi of 50 (Fig. 3H). However, ectopic expression of eGFP, used as a control, failed to modulate TRAF3IP2 expression (Fig. 3H). Importantly, forced expression of TRAF3IP2, by itself, induced TWEAK and TWEAKR expression (Fig. 3I). These results demonstrate a positive feedback regulation between TRAF3IP2 and TWEAK/TWEAKR signaling (Fig. 3).
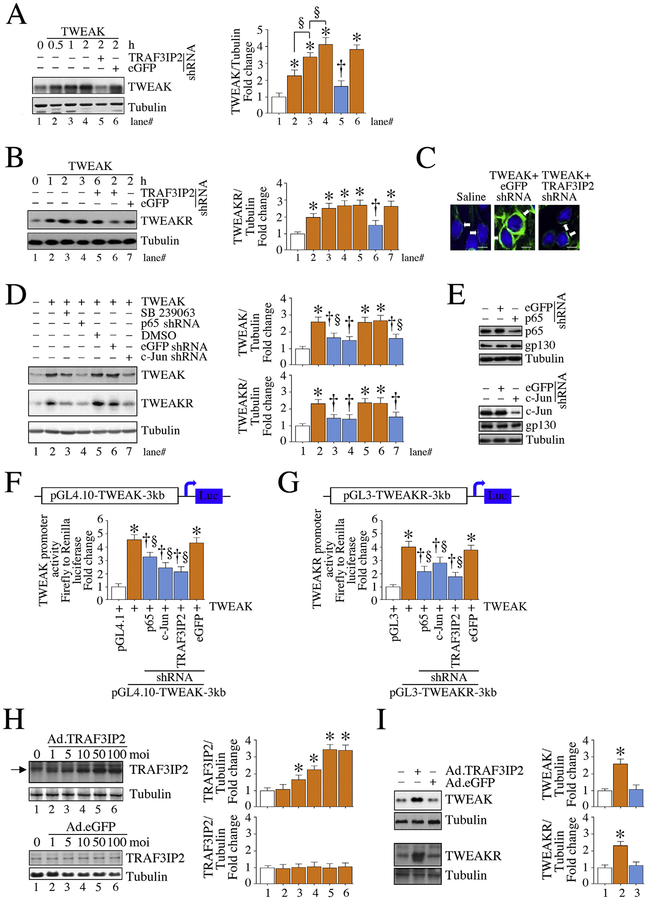
Fig. 3.
TWEAK induces its own expression and that of its receptor (TWEAKR) via TRAF3IP2 and its downstream signaling intermediates NF-κB and AP-1 in cardiac fibroblasts. TWEAK (10 ng/ml) induced TWEAK expression in a time-dependent manner in CF, and lentiviral TRAF3IP2 knockdown (moi0.5 for 48 h) inhibited this effect (A; time ‘0’ represents no exogenous addition of TWEAK). Further, TWEAK upregulated TWEAKR expression in a time-dependent manner, an effect also blunted by TRAF3IP2 knockdown (B). Immunofluorescence revealed low levels of TWEAKR expression at basal conditions, and its increased levels after TWEAK treatment (10 ng/ml for 2 h), localized predominantly in the membrane at cytoplasm, and its reduced levels following TRAF3IP2 knockdown (C; arrows denote membrane and cytoplasmic localization of TWEAKR). Moreover, pretreatment with the p38 MAPK-specific inhibitor SB239063 (10 μM in DMSO for 1 h) and lentiviral-mediated p65 (moi0.5 for 48 h) and c-Jun (moi0.5 for 48 h) knockdown each attenuated TWEAK autoregulation (2 h) and its receptor upregulation (2 h; D). Knockdown of p65 and c-Jun was confirmed by immunoblotting and is shown on the right in panel E. gp130 served as an off-target. Further, silencing TRAF3IP2, p65 and c-Jun (moi0.5 for 48 h) attenuated TWEAK-induced TWEAK (F) and TWEAKR (G) promoter-reporter activities (both TWEAK and TWEAKR reporters contain 3 kb DNA fragments upstream of transcription start site and 5’-UTR). Importantly, adenoviral transduction of TRAF3IP2, but not control GFP, increased TRAF3IP2 protein expression in a dose-dependent manner (moi1–100 for 24 h) (H), and at a moi50, induced TWEAK and its receptor expression (I). A, B, D, F, H, I: A representative image from three independent immunoblotting experiments with similar results is shown on the left and summarized semi-quantification of intensity of immunoreactive bands by densitometry from all three experiments on the right. F, G, n = 6. C, Magnification: 40×. *P < at least 0.05 versus untreated, †P < 0.05 versus TWEAK, §P < 0.05 (n = 3–6).
3.4. TRAF3IP2 regulates TWEAK-induced proinflammatory responses in CF
Several inflammatory mediators are p38 MAPK, NF-κB and/or AP-1 responsive genes. Since TWEAK induced TRAF3IP2-dependent p38 MAPK, NF-κB and AP-1 activation in CF (Fig. 2), we next investigated whether TWEAK induces proinflammatory responses in CF in part via TRAF3IP2. Indeed, TWEAK enhanced the mRNA expression of multiple inflammatory mediators, including cytokines (IL-1β, IL-6, IL-18, TNF-α), cytokine receptors (TNFR1), growth factors (CT-1), chemokines (CXCL1, CXCL2, CXCL5, MCP-1) and adhesion molecules (ICAM1, Selectin E), an effect markedly inhibited by TRAF3IP2 knockdown (Fig. 4A). Silencing of TRAF3IP2 also inhibited TWEAK-induced IL-1β, TNF-α and MCP-1 secretion (Fig. 4B). Confirming reduced mRNA expression (Fig. 4A), silencing TRAF3IP2 also inhibited ICAM1, VCAM1 and Selectin E protein levels (Fig. 4C). These results indicate that TRAF3IP2 is a critical signaling intermediate in TWEAK-induced proinflammatory responses in CF (Fig. 4).
Fig. 4.
TWEAK induces the expression of multiple inflammatory mediators in cardiac fibroblasts in part via TRAF3IP2. Silencing TRAF3IP2 by lentiviral shRNA (moi0.5 for 48 h) inhibited TWEAK (10 ng/ml for 2 h)-induced cytokine, chemokine, growth factor and adhesion molecule mRNA expression (RT-qPCR, A). Further, silencing TRAF3IP2 reduced TWEAK-induced cytokine and chemokine secretion (ELISA, B) and protein expression of adhesion molecules (Immunoblotting, C). C, A representative image from three independent immunoblot experiments with similar results is shown at the top with summarized semi-quantification of intensity of immunoreactive bands by densitometry from all three experiments in the respective bottom panels. A, B, n = 6, C, n = 3. A-C, *P < at least 0.01 versus untreated, †P < 0.05 versus TWEAK, §P < 0.05 versus Untreated.
3.5. TRAF3IP2 mediates TWEAK-induced collagen expression, MMP activation, and CF proliferation and migration
At basal conditions, a fine balance between deposition and proteolytic enzyme-mediated degradation of extracellular matrix proteins (e.g., fibrillar collagens) plays a vital role in maintaining normal myocardial structure and function. However, when ECM deposition exceeds degradation, and if the process persists, fibrosis ensues resulting in adverse cardiac remodeling, contractile dysfunction and HF development [41]. Since fibroblasts are one of the major cell types responsible for collagen synthesis, secretion, and deposition in the heart, and as COL1A1 and COL3A1 are redox-sensitive and NF-κB- and/or AP-1-responsive genes, we hypothesized that TRAF3IP2 mediates TWEAK-induced collagen expression in CF. Indeed, TWEAK upregulated collagens Iα1 and IIIα1 expression in a time-dependent manner (Fig. 5A), and in TRAF3IP2-dependent manner (Fig. 5B). Further, TWEAK promoted collagen secretion in part via TRAF3IP2 (Fig. 5C). Collagen expression and degradation are regulated by MMPs and their tissue inhibitors TIMPs. Fibrillar collagens are initially degraded by collagenases followed by gelatinases. Therefore, we analyzed MMP-13, a major collagenase in mice, and its activator MMP-14 or MT1-MMP. The data show that TWEAK induced mRNA and protein expression of both MMPs 13 and 14 in CF in a TRAF3IP2-dependent manner (Fig. 5D). Further, TWEAK also induced TIMP-1 expression in a TRAF3IP2-dependent manner (Fig. 5D). CF express the glatinases MMPs 2 and 9, and both MMPs are NF-κB and/or AP-1 responsive genes. Therefore, we next analyzed activation of MMPs 2 and 9 by immunoblotting using antibodies that detect both pro and active forms. The data in Fig. 5E and F show that TWEAK increases activation of both MMPs as evidenced by increased levels of respective active forms, and their significant inhibition by TRAF3IP2 knockdown. Since increased fibroblast proliferation and migration are two major players in cardiac fibrosis [38,41], we hypothesized that TRAF3IP2 mediates TWEAK-induced CF proliferation and migration. Indeed, TWEAK significantly increased CF proliferation (Fig. 5G) and migration (Fig. 5H), and silencing TRAF3IP2 or TWEAKR blunted its mitogenic and migratory effects. These results indicate that targeting TRAF3IP2 blunts TWEAKinduced pro-fibrotic responses in CF (Fig. 5).
Fig. 5.
TWEAK induces collagen, collagenase and gelatinase expression, and cardiac fibroblast proliferation and migration in part via TRAF3IP2. TWEAK (10 ng/ml) induced the expression of fibrillar collagens Iα1 and IIIα1 in a time-dependent manner (A), an effect blunted by TRAF3IP2 knockdown by lentiviral TRAF3IP2 shRNA (moi0.5 for 48 h) (B). Further, silencing TRAF3IP2 inhibited TWEAK-induced collagen secretion at 24 h (C; hydroxyproline assay), the collagenase MMP-13, MMP-14 and TIMP-1 mRNA (RT-qPCR) and protein expression (immunoblotting) at 2 h (D; protein expression is shown in insets at the top). Moreover, silencing TRAF3IP2 inhibited activation of the gelatinases MMPs 2 and 9 (E, F; immunoblotting using antibodies that detect both pro and active forms), as well as CF proliferation (CyQUANT® assay, G) and migration (Matrigel™ migration assay, H). A, B, D, E, A representative image from three independent immunoblot experiments with similar results is shown at the top and summarized semi-quantification of intensity of immunoreactive bands by densitometry from all three experiments in the respective right hand or bottom panels. C, G, H, n = 6. A-H, *P < at least 0.05 versus untreated, †P < 0.05 versus TWEAK, §P < 0.05 versus Untreated.
3.6. TRAF3IP2 gene ablation blunts TWEAK-induced myocardial hypertrophy and contractile dysfunction in a blood pressure-independent manner
Since silencing TRAF3IP2 blunted TWEAK-induced proinflammatory, pro-mitogenic and pro-migratory effects in cultured CF (Figs. 1–5), we next hypothesized that targeting TRAF3IP2 ameliorates TWEAK-induced cardiac fibrosis and adverse remodeling in vivo. Both WT and homozygous TRAF3IP2-KO mice were continuously infused with recombinant mouse TWEAK for 7 days using subcutaneously implanted osmotic minipumps (Fig. 6A). Tail-cuff plethysmography demonstrated a significant time-dependent increase in SBP in WT mice infused with TWEAK, but not saline used as a solvent control (Fig. 6B). Interestingly, TWEAK infusion increased SBP to a similar extent in the TRAF3IP2-KO mice (Fig. 6B). Further, no changes in body weights were detected in WT and TRAF3IP2-KO mice infused with TWEAK for 7 days (Fig. 6C). However, TWEAK infusion for 7 days resulted in myocardial hypertrophy in WT mice, as evidenced by increased heart weight to body weight ratios (Fig. 6D). Heart weights normalized to tibia length also showed a similar trend (data not shown). Notably, its pro-hypertrophic effects were blunted in the TRAF3IP2-KO mice (Fig. 6D). Increased cardiomyocyte cross-sectional area (Fig. 6E and F) and increased expression of fetal genes ANF and β-MHC (Fig. 6G) confirmed the pro-hypertrophic responses of TWEAK in the WT mice. Further, echocardiography revealed contractile dysfunction in TWEAK-infused WT, but not TRAF3IP2-KO mice, as evidenced by significant increases in LVPWs and LVPWd), and decreased LVIDs and LVIDd, LVVols and EF (Fig. 6H). Importantly, no cell death was detected in the hearts of TWEAK-infused WT or TRAF3IP2-KO mice (Supplementary Fig. 2). These results indicate that TRAF3IP2 gene ablation blunts TWEAK-induced adverse cardiac remodeling and contractile dysfunction, independent of increases in SBP (Fig. 6).
3.7. TRAF3IP2 gene ablation blunts TWEAK-induced cardiac fibrosis
Adverse cardiac remodeling is characterized by increased deposition of extracellular matrix proteins, including fibrillar collagens and fibronectin, in the heart [38,41]. The results show that while the baseline values of fibrosis (percent area occupied by collagens) are similar between WT and TRAF3IP-KO mice (data not shown), continuous infusion of TWEAK for 7 days (Fig. 7A) significantly increased interstitial fibrosis in hearts from the WT, but not TRAF3IP2-KO, mice (Fig. 7B). Further, TWEAK infusion increased mRNA (Fig. 7C) and protein expression of both fibrillar collagens I and III, as well as fibronectin, in the WT mouse heart (Fig. 7D; densitometric analysis is summarized in panel E). Importantly, TRAF3IP2 gene deletion blunted these TWEAK-induced profibrotic responses (Fig. 7B–E), indicating its critical role in TWEAK-induced cardiac fibrosis (Fig. 7).
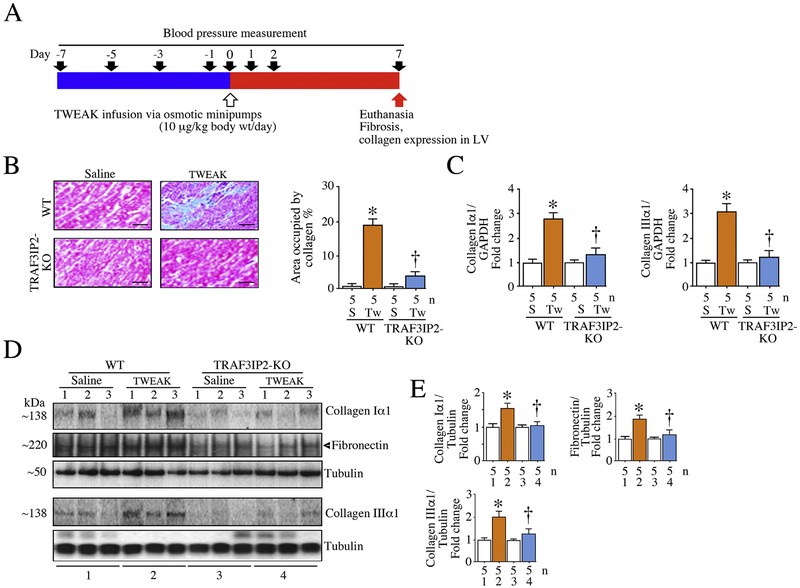
Fig. 7.
TRAF3IP2 gene deletion blunts TWEAK-induced myocardial fibrosis. Both wild type and TRAF3IP2-KO mice were continuously infused with TWEAK (10 μg/kg body weight/day) for 7 days via subcutaneously implanted osmotic minipumps (A). Cardiac fibrosis was analyzed by Masson’s trichrome (B; quantitative measurement of fibrosis is shown on the right), collagen Iα1 and collagen IIIα1 mRNA expression (RT-qPCR; C), and collagen and fibronectin protein expression (immunoblotting; D) in WT mice, and TRAF3IP2 gene ablation blunted these TWEAK-induced pro-fibrotic responses. While the immunoblots in D show three individual animals selected at random in each group, the corresponding summarized semi-quantification data in panel E includes all 5 animals/group. C, E, *P < 0.05 versus untreated, †P < 0.05 versus TWEAK (n = 5/group).
3.8. TRAF3IP2 gene ablation blunts TWEAK-induced p38 MAPK, NF-κB and AP-1 activation, and inflammatory responses in the heart
Our in vitro results showed that TWEAK stimulates ROS generation (Fig. 1), TRAF3IP2 upregulation (Fig. 1), TRAF3IP2-dependent p38 MAPK, NF-κB and AP-1 activation (Fig. 2), and TRAF3IP2-dependent proinflammatory cytokine expression in cultured CF (Fig. 4). Of note, several of these inflammatory mediators not only exert pro-hypertrophic and pro-fibrotic responses, but also induce contractile dysfunction [42]. In fact, recapitulating the in vitro data, TWEAK infusion for 7 days (Fig. 8A) increased oxidative stress in the hearts of WT mice, as evidenced by a significant increase in lipid peroxides (Fig. 8B), a surrogate marker of oxidative stress. Further, TWEAK infusion upregulated TRAF3IP2 expression (Fig. 8C), and p38 MAPK, NF-κB, and AP-1 activation in the hearts of WT mice. Importantly, TRAF3IP2 gene ablation blunted, but not abrogated, these responses (Fig. 8C). Furthermore, TRAF3IP2 deletion inhibited TWEAK-induced TWEAK and TWEAKR expression, and the induction of multiple inflammatory mediators, including IL-18, TNF-α, TNFR1, MCP-1, ICAM1 and iNOS in the heart (Fig. 8C; summarized densitometric analyses are shown at the bottom in panel D). These results demonstrate the critical role of TRAF3IP2 in TWEAK-induced myocardial inflammation (Fig. 8).
Fig. 8.
TRAF3IP2 gene deletion blunts TWEAK-induced NF-κB, AP-1 and p38 MAPK activation, TWEAK and its receptor upregulation, and induction of other inflammatory mediators in the hypertrophied heart. Both wild type and TRAF3IP2-KO mice were continuously infused with TWEAK (10 μg/kg body weight/day) for 7 days via subcutaneously implanted osmotic minipumps (A). TWEAK significantly increased oxidative stress, as evidenced by increased lipid peroxides (B). Further, TWEAK upregulated TRAF3IP2 expression (C), NF-κB (p65), AP-1 (c-Jun) and p38 MAPK activation (C), and proinflammatory cytokine (TNF-α, IL-18), chemokine (MCP-1), adhesion molecule (ICAM1) and iNOS expression (C). These TWEAK-induced pro-oxidant and proinflammatory effects were blunted by TRAF3IP2 gene deletion (C). While all the immunoblots in C include three individual animals selected at random in each group, the corresponding summarized semi-quantification data in panel D includes all 5 animals/group. B, D, *P < 0.05 versus untreated, †P < 0.05 versus TWEAK (n = 5/group).
4. Discussion
The major findings of this study are: TWEAK upregulates TRAF3IP2 expression in cultured cardiac fibroblasts. TWEAK also promotes its nuclear localization. Silencing TRAF3IP2 inhibits TWEAK-induced p38 MAPK, NF-κB and AP-1 activation, its own and that of its receptor (TWEAKR) expression, various other inflammatory mediators, collagen expression and release, and induction of MMPs that exert collagenolytic activity. Importantly, silencing TRAF3IP2 inhibits TWEAK-induced CF proliferation and migration. Further supporting its critical role, forced expression of TRAF3IP2, by itself, upregulates TWEAK and its receptor expression, suggesting a feed-forward loop between TWEAK and TRAF3IP2 in CF. Recapitulating the in vitro results, continuous infusion of TWEAK for 7 days increases SBP, upregulates TRAF3IP2 expression, activates p38 MAPK, NF-κB and AP-1, enhances the expression of various cytokines, chemokines, and adhesion molecules, and increases collagen expression and interstitial fibrosis in hearts from wild type mice. These proinflammatory and pro-fibrotic changes occur in conjunction with myocardial hypertrophy and contractile dysfunction. Importantly, TRAF3IP2 gene ablation ameliorates these adverse cardiac changes, independent of increases in SBP. These results indicate that TRAF3IP2 plays a causal role, and thus a potential therapeutic target, in inflammatory cardiac diseases (Fig. 9).
Fig. 9.
Schema showing a critical role for TRAF3IP2 in TWEAK-induced adverse cardiac remodeling, contractile dysfunction, and heart failure development. TWEAK induced its own expression and that of TWEAKR by inducing TRAF3IP2-dependent p38 MAPK, NF-κB and AP-1 activation, pro-inflammatory and pro-fibrotic cytokin, chemokine, adhesion molecule, and matrix metalloproteinase (MMP) expression, and fibroblast migration and proliferation, resulting in adverse cardiac remodeling, contractile dysfunction, and possible development of heart failure. The membrane localized TWEAK (mTWEAK) is processed by the serine protease furin, resulting in the release of soluble TWEAK (sTWEAK) that forms a homotrimer and binds the TWEAKR to initiate the downstream inflammatory signaling. Notably, TRAF3IP2 deletion blunts these deleterious effects of TWEAK. Arrowheads denote interventions used in delineating the possible signaling pathways activated by TWEAK. Broken arrows: TWEAK has previously been shown to activate MMPs.
TWEAK and its receptor are expressed at low basal levels and play a role in normal development and immune regulation. However, their increased expression for brief periods following injury or infection contributes to tissue repair and resolution of inflammation [12]. However, when expressed at high levels and for prolonged periods, this ligand/receptor pair plays a deleterious role in various models of inflammation, including myocardial inflammation, injury and HF development. In many inflammatory diseases, including that of the heart, the signal transduction pathways activated by TWEAK/TWEAKR culminate in NF-κB and AP-1 activation, involving one of the TRAF family members [11]. For example, the interaction between TWEAKR and TRAF6 has been shown to activate NF-κB via a canonical or a noncanonical pathway involving the IKK signalosome that contains the catalytic subunits IKKα and IKKβ, and the regulatory subunit IKKγ or NEMO. Interactions between TWEAKR and TRAF6 also activate AP-1, in part via the MAP kinase JNK. In turn, NF-κB and AP-1, either alone or in cooperation, can transcriptionally upregulate the expression of various genes involved in inflammation, proliferation, survival, or death [11].
Persistent activation of MAP kinases plays a pathological role in cardiac diseases [43]. Our data show that TWEAK induces p38 MAPK activation in part via TRAF3IP2, and targeting its activation by a pharmacological inhibitor attenuates NF-κB activation, suggesting a crosstalk between p38 MAPK and NF-κB in amplifying TWEAK-induced inflammatory signaling in CF. Previously, we reported that targeting p38 MAPK also inhibited IL-17-induced AP-1 activation in human CF, suggesting that targeting p38 MAPK can inhibit TWEAK-induced activation of both NF-κB and AP-1. Though we have not investigated the underlying molecular mechanisms involved in TWEAK-induced TRAF3IP2-dependent p38 MAPK activation, it is plausible that oxidative stress and interactions between TWEAKR and members of the TRAF family might have contributed to its activation. For example, TRAF3IP2 might have induced p38 MAPK activation via TRAF6/TAK1/ROS or TRAF6/MKK (Mitogen-activated Protein Kinase Kinase)3/6. In fact, our data show that TWEAK induces NOX4-dependent ROS generation in CF. More importantly, targeting TRAF3IP2 suppresses p38 MAPK-, NF-κB- and/or AP-1-responsive inflammatory gene expression, including IL-1β, IL-6, IL-18, TNF-α, CT-1, CXCL1, CXCL2, CXCL5, MCP-1, ICAM1, and Selectin A.
Previously, TWEAK was shown to stimulate collagen synthesis and proliferation of rat CF [6]. Here we demonstrated that TWEAK upregulates collagen expression and secretion by mouse CF in a TRAF3IP2- dependent manner. Interestingly, though TWEAK enhanced collagen expression, it also promoted induction and activation of MMPs with collagenolytic activity. For example, TWEAK upregulated the expression of MMP-13, the predominant collagenase in mice. It also upregulated the expression of MMP-14, a membrane type 1 MMP (MT1-MMP), that plays a role in MMP-13 activation by processing pro-MMP-13 to a fully active form [44]. Interestingly, TWEAK also induced TIMP-1 expression. It also induced activation of the gelatinases MMP-2 and MMP-9 that are known to further degrade denatured collagens, indicating that TWEAK promotes both synthesis and degradation of collagens, with a net positive effect on synthesis, resulting ultimately in increased fibrosis. In fact, TWEAK infusion for 7 days induced interstitial fibrosis in WT mouse heart, and its significant inhibition in TRAF3IP2-KO mice. Of note, MMPs 2, 9, 13 and 14, and TIMP-1 are all NF-κB and/or AP-1 responsive genes. Targeting TRAF3IP2 also inhibited TWEAK-induced CF migration and proliferation, further supporting a causal role for TRAF3IP2 in TWEAK-induced proinflammatory and pro-fibrotic responses in CF.
Interestingly, some of the proinflammatory mediators induced by TRAF3IP2 also regulate TRAF3IP2 expression. For example, IL-1, TNF-α and LPS have all been shown to upregulate TRAF3IP2 expression in human lung epithelial cells [45]. We have previously demonstrated that IL-18, which signals via the cytoplasmic adapter molecule MyD88, promotes CF migration and differentiation in part via TRAF3IP2 [46], indicating a potential positive feedback loop between inflammatory mediators and TRAF3IP2 in perpetuating inflammation. It also suggests that TRAF3IP2 could play a role in the signal transduction of cytokines other than IL-17 either directly or through TIR-signaling intermediates. Though our data show a critical role of TRAF3IP2 in TWEAK/TWEAKR signaling, it is not known whether a physical interaction between TRAF3IP2 and TWEAKR is necessary to activate downstream proinflammatory signaling, such as those reported for TRAF3IP2 and IL-17R [24,25,47]. Interestingly, both TRAF3IP2 and TWEAKR contain TRAF-interacting domains. For example, similar to TWEAKR binding to various TRAFs via a –PIEET-motif at the C-terminus cytoplasmic tail [11,12], TRAF3IP2 is known to bind various TRAFs via two TRAF-interacting motifs [48]. Therefore, it is plausible that TRAF3IP2 might physically associate with TWEAKR by forming complexes involving TRAF proteins, considering that TRAF proteins oligomerize through their RING domain interaction. Our future studies will determine this possibility.
Originally, TRAF3IP2 has been shown to localize in cytoplasm [24,25]. However, in 2012, we reported its nuclear localization in Angiotensin II-treated isolated cardiomyocytes [26]. Here, as reported in cardiomyocytes, TWEAK promoted TRAF3IP2 nuclear localization in CF. Though we have not investigated the significance of its nuclear localization, a recent report also demonstrated TRAF3IP2 nuclear localization in IL-17-treated airway epithelial cells [49]. Those authors further demonstrated that the translocated TRAF3IP2 transcriptionally upregulated beta-defensin B4 (DEFB4) [49], an IL-17-responsive gene, induced locally by inflammation, by binding a large portion of its promoter region encompassing several NF-κB binding sites. However, mutating those NF-κB-binding sites failed to modulate DEF4B transcription [49], suggesting that though TRAF3IP2 is an upstream regulator of NF-κB, its effects on transcription of some of the IL-17-responsive genes are not NF-κB dependent. Together, these reports demonstrate that TRAF3IP2 not only resides in cytoplasm and functions as an E3-ubiquitin ligase [47], it translocates to the nucleus to transcriptionally upregulate some IL-17-responsive genes via mechanisms not fully understood. Since members of the IKK complex shuttle between cytoplasm and nucleus, and as TRAF3IP2 is known to bind all members of the IKK signalosome [24], we hypothesize that its physical interaction with IKKs is one of the mechanisms responsible for its nuclear translocation. Our future studies will investigate this hypothesis. In addition, we will also determine whether nuclear TRAF3IP2 transcriptionally upregulates genes involved in inflammation and fibrosis in CF treated with TWEAK and other pro-fibrotic mediators, including Angiotensin II [26].
Recapitulating the in vitro results, our in vivo results show that infusion of TWEAK for 7 days increases SBP, contractile dysfunction, TRAF3IP2 expression, p38 MAPK, NF-κB, and AP-1 activation, cytokine and collagen expression, and interstitial fibrosis in a WT mouse heart. Importantly, genetic ablation of TRAF3IP2 inhibited these deleterious effects, without affecting increases in SBP. Of note, we previously reported that TRAF3IP2 deletion blunts angiotensin II- and aldosteroneinduced adverse myocardial remodeling without modulating increases in SBP [26,50]. In those models, lack of TRAF3IP2 blunted oxidative stress and expression of various inflammatory markers, indicating that the effects of lack of TRAF3IP2 on hypertrophy and fibrosis are blood pressure independent, and likely due to suppression of oxidative stress and inflammation. For example, low doses of spironolactone, a mineralocorticoid receptor antagonist, was previously reported to improve diastolic dysfunction without lowering blood pressure in transgenic (mRen2)27 [TG(mRen2)27] rats that are severely hypertensive and express high levels of tissue angiotensin II and plasma aldosterone [51], indicating a disconnect between therapeutic improvements in cardiac function and structure and blood pressure. Thus, it is not entirely surprising that TRAF3IP2 deletion blunted adverse remodeling in the absence of improvement in blood pressure in TWEAK-infused mice, and that this improvement in remodeling is likely due to reductions in oxidative stress and inflammation. These results also indicate that TRAF3IP2 is a critical player in TWEAK-induced adverse cardiac remodeling and contractile dysfunction, and thus a potential therapeutic target.
TWEAK has been targeted in various autoimmune and inflammatory diseases like rheumatoid arthritis (RA) and lupus nephritis. In a phase I clinical trial using RA subjects (ClinicalTrials.gov identifier: NCT00771329), Wisniacki, et al. reported that a single dose of a TWEAK-blocking monoclonal antibody (BIIB023) suppressed serum levels of soluble TWEAK for several days. In those subjects BIIB023 showed a favorable safety and tolerability profile [52]. However, in a phase II clinical trial (ClinicalTrials.gov Identifier: NCT01499355), the same antibody failed to show improvements in subjects with lupus nephritis, and therefore had to be discontinued. In an ongoing independent phase I clinical trial (ClinicalTrials.gov Identifier: NCT02628795), it will be determined whether targeting TWEAK signaling will restore muscle and mobility in subjects with end-stage osteoarthritis, a chronic inflammatory disease. These clinical trials suggest that targeting TWEAK/TWEAKR or their signaling is beneficial in slowing the progression of some autoimmune and inflammatory diseases. Since TRAF3IP2 is a critical player in various autoimmune and inflammatory diseases, and TWEAK-induced inflammatory signaling and adverse cardiac remodeling are markedly inhibited by TRAF3IP2 deletion, we hypothesize that TRAF3IP2 serves as a potential novel therapeutic target in inflammatory cardiac diseases.
A limitation of the study is that though TRAF3IP2 deletion significantly reduced TWEAK-mediated inflammatory response and fibrosis, it did not totally reverse TWEAK’ effects on these critical phenomena, indicating TRAF3IP2-independent effects of TWEAK on adverse cardiac remodeling and contractile dysfunction. In fact, using bioinformatics, Bhattacharjee, et al. reported activation and interaction of diverse signaling pathways in various cell types treated with TWEAK [11], a majority of which involve interaction with one or more members of the TRAF family, ultimately resulting in the activation of NF-κB and AP-1. Interestingly, TRAF3IP2 also interacts with these TRAFs in a cell type and context-dependent manner, resulting in activation of NF-κB and AP-1, and activation of downstream inflammatory and pro-fibrotic pathways. Because of interactions with members of TRAF family and activation of NF-κB and AP-1, we strongly believe in a critical role for TRAF3IP2 in TWEAK signaling, and its potential as a therapeutic target in TWEAK-associated pathologies. Another limitation of the study is that no diastolic parameters were analyzed in TWEAK-infused animals as fibrosis is known to be associated with diastolic dysfunction.
Supplementary Material
Acknowledgements
BC is a recipient of the Department of Veterans Affairs Research Career Scientist award (#IK6BX004016–01), and is supported by the U.S. Department of Veterans Affairs, Office of Research and Development Biomedical Laboratory Research and Development (ORD-BLRD) Service Award VA-I01-BX002255. Work in SM’s laboratory is supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI119131. TY is supported by the American Heart Association Scientist Development Grant (15SDG25240022) and a University of Missouri Research Council grant. The contents of this report do not represent the views of the Department of Veterans Affairs or the United States government.
Abbreviations:
- Act1
NF-kappa B Activator 1
- ANP
atrial natriuretic peptide
- AP-1
activator protein-1
- CCL
C-C Motif Chemokine Ligand
- CD
Cluster of Differentiation
- CF
cardiac fibroblasts
- CIKS
Connection to IKK and SAPK/JNK
- ColIα1
collagen Iα1
- ColIIIα1
collagen IIIα1
- CT1
Cardiotrophin 1
- ColIα1
Collagen Type I Alpha 1 chain
- CTGF
connective tissue growth factor
- CXCL
C-X-C Motif Chemokine Ligand
- DMEM
Dulbecco’s Modified Eagle Medium
- DMSO
dimethyl sulfoxide
- E3
ubiquitin-protein isopeptide ligase
- ECM
extracellular matrix
- EF
ejection fraction
- eGFP
enhanced green fluorescent protein
- ELISA
Enzyme Linked Immunosorbent Assay
- Fn14
Fibroblast Growth Factor-Inducible Immediate-Early Response Protein 14
- GAPDH
Glyceraldehyde 3-Phosphate Dehydrogenase
- siGFP
silencer RNA against Green Fluorescent Protein
- gp130
glycoprotein 130
- HLH
Helix-Loop-Helix
- ICAM1
Intercellular Adhesion Molecule 1
- IκB
inhibitory kappa B
- IKK
IκB kinase
- IL
interleukin
- IM
intramuscular
- iNOS
inducible nitric oxide synthase
- ITS
Insulin-Transferrin-Sodium selenite
- JNK
c-JunN-terminal kinase
- KO
knockout
- LV
left ventricle/Left Ventricular
- LVPWs
Left ventricular posterior wall end systole
- LVPWd
Left ventricular posterior wall end diastole
- LVIDs
Left ventricular internal diameter end diastole
- LVIDd
Left ventricular internal diameter end diastole
- LVVols
left ventricular end systole volume
- MAPK
Mitogen-Activated Protein Kinase
- MCP-1
Monocyte Chemoattractant Protein 1
- MKK
Mitogen-activated Protein Kinase Kinase
- MMP
Matrix Metalloproteinase
- moi
multiplicity of infection
- MT1-MMP
Membrane type I metalloprotease
- MTT
(3-(4,5-dimethylthiazolyl-2)-25-diphenyltetrazolium bromide)
- NADPH
nicotinamide adenine dinucleotide phosphate reduced
- NEMO
NF-κB Essential Modulator
- NF-κB
nuclear factor κB
- NOX
NADPH oxidase
- siNOX4
silencer RNA against NOX4
- OD
optical density
- qPCR
quantitative PCR
- RA
Rheumatoid Arthritis
- ROS
Reactive Oxygen Species
- SAPK
Stress-Activated Protein Kinase
- SBP
Systolic Blood Pressure
- SEFIR
Similar Expression to Fibroblast Growth Factor and Interleukin-17 Receptors
- shRNA
short hairpin RNA
- STIR
SEFIR+TIR
- TAK1
Transforming Growth Factor-Beta-Activated Kinase 1
- TIMP-1
Tissue Inhibitor of Metalloproteinase 1
- TIR
Toll-IL-1 Receptor
- TNFRSF12A
Tumor Necrosis Factor Receptor Superfamily Member 12A
- TNFSF12
TNF Superfamily Member 12
- TRAF3IP2
TRAF3 Interacting Protein 2
- TRAF
TNFR-Associated Factor
- TUNEL
Terminal transferase-mediated dUTP-biotin nick end-labeling
- TWEAK
TNF-Related Weak Inducer Of Apoptosis
- TWEAKR
TWEAK receptor
- VCAM1
Vascular Cell Adhesion Molecule 1
- WGA
Wheat Germ Agglutinin
- WT
wild type
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yjmcc.2018.07.003.
References
- [1].Novoyatleva T, Sajjad A, Engel FB, TWEAK-Fn14 Cytokine-Receptor Axis: A New Player of Myocardial Remodeling and Cardiac Failure, Front. Immunol 5 (2014) 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Blanco-Colio LM, TWEAK/Fn14 Axis: A Promising Target for the Treatment of Cardiovascular Diseases, Front. Immunol 5 (2014) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ren MY, Sui SJ, The role of TWEAK/Fn14 in cardiac remodeling, Mol. Biol. Rep 39 (2012) 9971–9977. [DOI] [PubMed] [Google Scholar]
- [4].Meighan-Mantha RL, Hsu DK, Guo Y, Brown SA, Feng SL, Peifley KA, et al. , The mitogen-inducible Fn14 gene encodes a type I transmembrane protein that modulates fibroblast adhesion and migration, J. Biol. Chem 274 (1999) 33166–33176. [DOI] [PubMed] [Google Scholar]
- [5].Chorianopoulos E, Heger T, Lutz M, Frank D, Bea F, Katus HA, et al. , FGF-inducible 14-kDa protein (Fn14) is regulated via the RhoA/ROCK kinase pathway in cardiomyocytes and mediates nuclear factor-kappaB activation by TWEAK, Basic Res. Cardiol 105 (2010) 301–313. [DOI] [PubMed] [Google Scholar]
- [6].Chen HN, Wang DJ, Ren MY, Wang QL, Sui SJ, TWEAK/Fn14 promotes the proliferation and collagen synthesis of rat cardiac fibroblasts via the NF-small ka, CyrillicB pathway, Mol. Biol. Rep 39 (2012) 8231–8241. [DOI] [PubMed] [Google Scholar]
- [7].Maecker H, Varfolomeev E, Kischkel F, Lawrence D, Leblanc H, Lee W, et al. , TWEAK attenuates the transition from innate to adaptive immunity, Cell 123 (2005) 931–944. [DOI] [PubMed] [Google Scholar]
- [8].Jakubowski A, Ambrose C, Parr M, Lincecum JM, Wang MZ, Zheng TS, et al. , TWEAK induces liver progenitor cell proliferation, J. Clin. Invest 115 (2005) 2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chorianopoulos E, Jarr K, Steen H, Giannitsis E, Frey N, Katus HA, Soluble TWEAK is markedly upregulated in patients with ST-elevation myocardial infarction and related to an adverse short-term outcome, Atherosclerosis 211 (2010) 322–326. [DOI] [PubMed] [Google Scholar]
- [10].Jain M, Jakubowski A, Cui L, Shi J, Su L, Bauer M, et al. , A novel role for tumor necrosis factor-like weak inducer of apoptosis (TWEAK) in the development of cardiac dysfunction and failure, Circulation 119 (2009) 2058–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bhattacharjee M, Raju R, Radhakrishnan A, Nanjappa V, Muthusamy B, Singh K, et al. , A Bioinformatics Resource for TWEAK-Fn14 Signaling Pathway, J. Signal. Transduct 2012 (2012) 376470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Burkly LC, Regulation of Tissue Responses: The TWEAK/Fn14 Pathway and Other TNF/TNFR Superfamily Members That Activate Non-Canonical NFkappaB Signaling, Front. Immunol 6 (2015) 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Salzmann S, Lang I, Rosenthal A, Schafer V, Weisenberger D, Carmona Arana JA, et al. , TWEAK inhibits TRAF2-mediated CD40 signaling by destabilization of CD40 signaling complexes, J. Immunol 191 (2013) 2308–2318. [DOI] [PubMed] [Google Scholar]
- [14].Ikner A, Ashkenazi A, TWEAK induces apoptosis through a death-signaling complex comprising receptor-interacting protein 1 (RIP1), Fas-associated death domain (FADD), and caspase-8, J. Biol. Chem 286 (2011) 21546–21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Carmona Arana JA, Seher A, Neumann M, Lang I, Siegmund D, Wajant H, TNF Receptor-Associated Factor 1 is a Major Target of Soluble TWEAK, Front. Immunol 5 (2014) 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dogra C, Changotra H, Wedhas N, Qin X, Wergedal JE, Kumar A, TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine, FASEB J. 21 (2007) 1857–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Martinez-Miguel P, Medrano-Andres D, Griera-Merino M, Ortiz A, RodriguezPuyol M, Rodriguez-Puyol D, et al. , Tweak up-regulates endothelin-1 system in mouse and human endothelial cells, Cardiovasc. Res 113 (2017) 207–221. [DOI] [PubMed] [Google Scholar]
- [18].Li H, Mittal A, Paul PK, Kumar M, Srivastava DS, Tyagi SC, et al. , Tumor necrosis factor-related weak inducer of apoptosis augments matrix metalloproteinase 9 (MMP-9) production in skeletal muscle through the activation of nuclear factor-kappaB-inducing kinase and p38 mitogen-activated protein kinase: a potential role of MMP-9 in myopathy, J. Biol. Chem 284 (2009) 4439–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hamid T, Guo SZ, Kingery JR, Xiang X, Dawn B, Prabhu SD, Cardiomyocyte NF-kappaB p65 promotes adverse remodelling, apoptosis, and endoplasmic reticulum stress in heart failure, Cardiovasc. Res 89 (2011) 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ricci R, Eriksson U, Oudit GY, Eferl R, Akhmedov A, Sumara I, et al. , Distinct functions of junD in cardiac hypertrophy and heart failure, Genes Dev. 19 (2005) 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Baxter FO, Came PJ, Abell K, Kedjouar B, Huth M, Rajewsky K, et al. , IKKbeta/2 induces TWEAK and apoptosis in mammary epithelial cells, Development 133 (2006) 3485–3494. [DOI] [PubMed] [Google Scholar]
- [22].Kaiser RA, Liang Q, Bueno O, Huang Y, Lackey T, Klevitsky R, et al. , Genetic inhibition or activation of JNK1/2 protects the myocardium from ischemia-reperfusion-induced cell death in vivo, J. Biol. Chem 280 (2005) 32602–32608. [DOI] [PubMed] [Google Scholar]
- [23].Milano G, Morel S, Bonny C, Samaja M, von Segesser LK, Nicod P, et al. , A peptide inhibitor of c-Jun NH2-terminal kinase reduces myocardial ischemia-reperfusion injury and infarct size in vivo, Am. J. Physiol. Heart Circ. Physiol 292 (2007) H1828–H1835. [DOI] [PubMed] [Google Scholar]
- [24].Leonardi A, Chariot A, Claudio E, Cunningham K, Siebenlist U, CIKS, a connection to Ikappa B kinase and stress-activated protein kinase, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li X, Commane M, Nie H, Hua X, Chatterjee-Kishore M, Wald D, et al. , Act1, an NF-kappa B-activating protein, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 10489–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Valente AJ, Clark RA, Siddesha JM, Siebenlist U, Chandrasekar B, CIKS (Act1 or TRAF3IP2) mediates Angiotensin-II-induced Interleukin-18 expression, and Nox2-dependent cardiomyocyte hypertrophy, J. Mol. Cell. Cardiol 53 (2012) 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ha HL, Wang H, Pisitkun P, Kim JC, Tassi I, Tang W, et al. , IL-17 drives psoriatic inflammation via distinct, target cell-specific mechanisms, Proc. Natl. Acad. Sci. U. S. A 111 (2014) E3422–E3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, et al. , The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease, Nat. Immunol 8 (2007) 247–256. [DOI] [PubMed] [Google Scholar]
- [29].Yariswamy M, Yoshida T, Valente AJ, Kandikattu HK, Sakamuri SS, Siddesha JM, et al. , Cardiac-restricted Overexpression of TRAF3 Interacting Protein 2 (TRAF3IP2) Results in Spontaneous Development of Myocardial Hypertrophy, Fibrosis, and Dysfunction, J. Biol. Chem 291 (2016) 19425–19436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tajrishi MM, Shin J, Hetman M, Kumar A, DNA methyltransferase 3a and mitogen-activated protein kinase signaling regulate the expression of fibroblast growth factor-inducible 14 (Fn14) during denervation-induced skeletal muscle atrophy, J. Biol. Chem 289 (2014) 19985–19999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Prabhu SD, Wang G, Luo J, Gu Y, Ping P, Chandrasekar B, Beta-adrenergic receptor blockade modulates Bcl-X(S) expression and reduces apoptosis in failing myocardium, J. Mol. Cell. Cardiol 35 (2003) 483–493. [DOI] [PubMed] [Google Scholar]
- [32].Erikson JM, Valente AJ, Mummidi S, Kandikattu HK, Demarco VG, Bender SB, et al. , Targeting TRAF3IP2 by Genetic and Interventional Approaches Inhibits Ischemia/Reperfusion-induced Myocardial Injury and Adverse Remodeling, J. Biol. Chem 292 (2017) 2345–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Valente AJ, Yoshida T, Clark RA, Delafontaine P, Siebenlist U, Chandrasekar B, Advanced oxidation protein products induce cardiomyocyte death via Nox2/Rac1/superoxide-dependent TRAF3IP2/JNK signaling, Free Radic. Biol. Med 60 (2013) 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Venkatachalam K, Mummidi S, Cortez DM, Prabhu SD, Valente AJ, Chandrasekar B, Resveratrol inhibits high glucose-induced PI3K/Akt/ERK-dependent interleukin-17 expression in primary mouse cardiac fibroblasts, Am. J. Physiol. Heart Circ. Physiol 294 (2008) H2078–H2087. [DOI] [PubMed] [Google Scholar]
- [35].Valente AJ, Sakamuri SS, Siddesha JM, Yoshida T, Gardner JD, Prabhu R, et al. , TRAF3IP2 mediates interleukin-18-induced cardiac fibroblast migration and differentiation, Cell. Signal 25 (2013) 2176–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Akahori H, Karmali V, Polavarapu R, Lyle AN, Weiss D, Shin E, et al. , CD163 interacts with TWEAK to regulate tissue regeneration after ischaemic injury, Nat. Commun 6 (2015) 7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shi J, Jiang B, Qiu Y, Guan J, Jain M, Cao X, et al. , PGC1alpha plays a critical role in TWEAK-induced cardiac dysfunction, PLoS One 8 (2013) e54054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Suthahar N, Meijers WC, Sillje HHW, de Boer RA, From Inflammation to Fibrosis-Molecular and Cellular Mechanisms of Myocardial Tissue Remodelling and Perspectives on Differential Treatment Opportunities, Curr Heart Fail Rep. 14 (2017) 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM, PROMO: detection of known transcription regulatory elements using species-tailored searches, Bioinformatics 18 (2002) 333–334. [DOI] [PubMed] [Google Scholar]
- [40].Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. , Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN, Nucleic Acids Res. 31 (2003) 3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kong P, Christia P, Frangogiannis NG, The pathogenesis of cardiac fibrosis, Cell. Mol. Life Sci 71 (2014) 549–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mann DL, Innate immunity and the failing heart: the cytokine hypothesis revisited, Circ. Res 116 (2015) 1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Petrich BG, Wang Y, Stress-activated MAP kinases in cardiac remodeling and heart failure; new insights from transgenic studies, Trends Cardiovasc Med. 14 (2004) 50–55. [DOI] [PubMed] [Google Scholar]
- [44].Knauper V, Will H, Lopez-Otin C, Smith B, Atkinson SJ, Stanton H, et al. , Cellular mechanisms for human procollagenase-3 (MMP-13) activation. Evidence that MT1-MMP (MMP-14) and gelatinase a (MMP-2) are able to generate active enzyme, J. Biol. Chem 271 (1996) 17124–17131. [DOI] [PubMed] [Google Scholar]
- [45].Zhao Z, Qian Y, Wald D, Xia YF, Geng JG, Li X, IFN regulatory factor-1 is required for the up-regulation of the CD40-NF-kappa B activator 1 axis during airway inflammation, J. Immunol 170 (2003) 5674–5680. [DOI] [PubMed] [Google Scholar]
- [46].Valente AJ, Yoshida T, Izadpanah R, Delafontaine P, Siebenlist U, Chandrasekar B, Interleukin-18 enhances IL-18R/Nox1 binding, and mediates TRAF3IP2-dependent smooth muscle cell migration. Inhibition by simvastatin, Cell. Signal 25 (2013) 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu C, Qian W, Qian Y, Giltiay NV, Lu Y, Swaidani S, et al. , Act1, a U-box E3 ubiquitin ligase for IL-17 signaling, Sci. Signal 2 (2009) ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Amatya N, Garg AV, Gaffen SL, IL-17 Signaling: The Yin and the Yang, Trends Immunol. 38 (2017) 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Velichko S, Zhou X, Zhu L, Anderson JD, Wu R, Chen Y, A Novel Nuclear Function for the Interleukin-17 Signaling Adaptor Protein Act1, PLoS One 11 (2016) e0163323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sakamuri SS, Valente AJ, Siddesha JM, Delafontaine P, Siebenlist U, Gardner JD, et al. , TRAF3IP2 mediates aldosterone/salt-induced cardiac hypertrophy and fibrosis, Mol. Cell. Endocrinol 429 (2016) 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Habibi J, Demarco VG, Ma L, Pulakat L, Rainey WE, Whaley-Connell AT, et al. , Mineralocorticoid receptor blockade improves diastolic function independent of blood pressure reduction in a transgenic model of RAAS overexpression, Am. J. Physiol. Heart Circ. Physiol 300 (2011) H1484–H1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wisniacki N, Amaravadi L, Galluppi GR, Zheng TS, Zhang R, Kong J, et al. , Safety, tolerability, pharmacokinetics, and pharmacodynamics of anti-TWEAK monoclonal antibody in patients with rheumatoid arthritis, Clin. Ther 35 (2013) 1137–1149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.