Abstract
Chronic hepatitis B virus (HBV) infection is a global public health challenge on the same scale as tuberculosis, HIV, and malaria. The International Coalition to Eliminate HBV (ICE-HBV) is a coalition of experts dedicated to accelerating the discovery of a cure for chronic hepatitis B. Following extensive consultation with more than 50 scientists from across the globe, as well as key stakeholders including people affected by HBV, we have identified gaps in our current knowledge and new strategies and tools that are required to achieve HBV cure. We believe that research must focus on the discovery of interventional strategies that will permanently reduce the number of productively infected cells or permanently silence the covalently closed circular DNA in those cells, and that will stimulate HBV-specific host immune responses which mimic spontaneous resolution of HBV infection. There is also a pressing need for the establishment of repositories of standardised HBV reagents and protocols that can be accessed by all HBV researchers throughout the world. The HBV cure research agenda outlined in this position paper will contribute markedly to the goal of eliminating HBV infection worldwide.
Introduction
Hepatitis B virus (HBV) is a major global public health threat with over 257 million people worldwide chronically infected; over 887 000 deaths are caused by the virus every year.1 Chronic hepatitis B causes almost 40% of cases of hepatocellular carcinoma, which is the second leading cause of cancer-related mortality worldwide.2 The current prophylactic vaccine has no effect on established chronic infection. Available treatments suppress viral replication but they are not curative, largely due to the persistence of the viral covalently closed circular DNA (cccDNA) transcriptional template in infected hepatocytes and the failure of chronically infected patients to mount an immune response that is sufficiently robust, functional, and sustained to clear the infection.3 Thus, in most cases, treatment must continue for life. However, even successfully virally suppressed patients may still develop liver cancer, especially if their livers are cirrhotic.4
Despite the huge human and economic toll of chronic hepatitis B,2 HBV research remains largely underfunded,5 to the point of recently being compared to a neglected tropical disease.6 Given recent scientific progress and the momentum created by hepatitis C virus (HCV) cure discoveries, together with the immense public health impact of chronic hepatitis B, we believe that governments, foundations, industry, and academic research institutions throughout the world can and should join forces and work together to accelerate HBV cure research.
The International Coalition to Eliminate HBV (ICE-HBV) is an international, independent, research-based forum created in 2016 to accelerate HBV cure research. ICE-HBV aims to coordinate, promote, and foster collaborative partnerships working towards an HBV cure and the alleviation of suffering for all individuals living with chronic hepatitis B.7 Following an inclusive nomination process, ICE-HBV formed international multidisciplinary scientific working groups consisting of leaders in HBV virology, immunology, technology, and clinical research, who have collaborated to identify current strengths in the HBV field that can be built upon, as well as knowledge gaps that must be addressed to achieve cure. The strategy presented here is the result of a 2 year-long process, involving over 50 scientists from five continents with input from key HBV research stakeholders from 21 countries. Through this process, we propose a strategic and collaborative HBV research agenda to advance the discovery of a safe, affordable, scalable, and effective chronic hepatitis B cure, accessible to all within the foreseeable future.
The HBV lifecycle
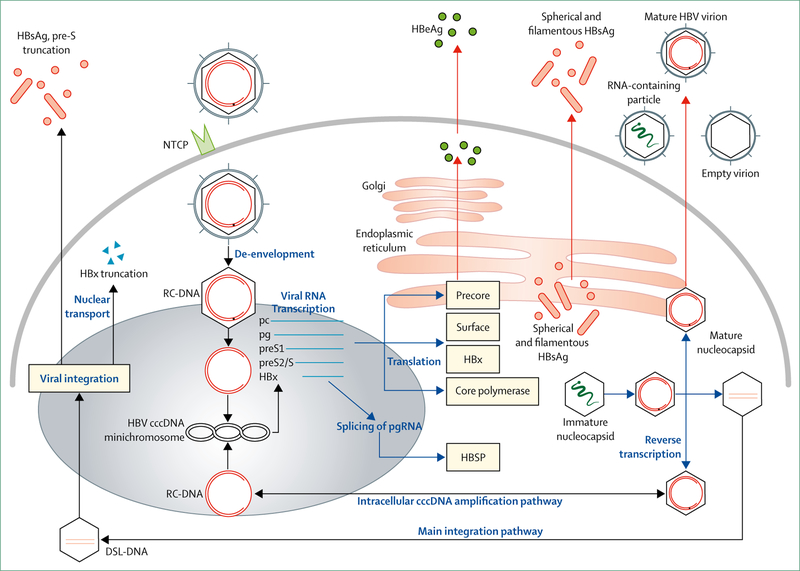
The HBV replication cycle is outlined in figure 1. There are still many gaps in our understanding of the HBV replication cycle that are major barriers to HBV cure research, particularly concerning the biogenesis, homoeostasis, and turnover of the cccDNA reservoir. However, recent technological advances, such as the discovery of the NTCP HBV entry receptor,8 have led to the development of additional cell culture models that, together with other available systems (primary human hepatocytes, HepaRG cells, transient transfection systems, stably transformed cell lines, hepatocyte-like cells derived from induced pluripotent stem cells) and the use of humanised mice, permit interrogation of the complete HBV replication cycle. These developments make it feasible to systematically study the mechanisms that control the biogenesis, homoeostasis, and decay of the cccDNA transcriptional template, and to use that information to identify vulnerabililites in cccDNA machinery that could in turn be exploited to eradicate HBV from infected cells.
Figure 1: The hepatitis B virus replication cycle.
Following viral entry via binding of the preS1 domain of the large envelope protein to the sodium-taurocholate co-transporting polypeptide (NTCP) receptor8 on the hepatocyte plasma membrane, the viral and cellular membranes fuse to release the nucleocapsid, which is transported into the cell nucleus.The nucleocapsid then disassembles to release its cargo of partially double-stranded genomic viral DNA. The host DNA repair machinery9 converts the incoming DNA to covalently closed circular DNA (cccDNA) molecules that are packaged into chromatin-like minichromosomes by histone and non-histone proteins,10,11 whereupon it acts as the HBV transcriptional template and long-lived reservoir in the hepatocyte nucleus.10 The 3∙2 kb HBV genome is organised into four overlapping but frame-shifted open-reading frames encoding the core protein (HBcAg, nucleocapsid); polymerase (P); the precore protein, which is processed to secreted hepatitis B e antigen (HBeAg);12 hepatitis B surface antigens (HBsAg, consisting of the large, medium, and small envelope proteins); and the X protein (HBx). A number of virus-encoded promoter and enhancer elements promote transcription of five major mRNAs, namely pregenomic (pgRNA, also the core mRNA), precore mRNA, preS1 and preS2/S mRNA, and HBx mRNA, which are translated into the aforementioned seven viral proteins. The pgRNA is encapsidated with P and reverse transcribed into DNA. Following viral DNA synthesis mature nucleocapsids are either packaged with envelope proteins to be exported as infectious virions, or recycled back to the nucleus for cccDNA replenishment. DSL-DNA=double-stranded linear DNA. RC-DNA=relaxed circular DNA.
Chronic hepatitis B and resolution of infection
In the absence of vaccination, over 90% of individuals infected in infancy will progress from acute hepatitis to lifelong chronic infection. By contrast, over 90% of people infected in adulthood will resolve the infection, due to robust immune responses that eliminate infected cells and produce neutralising antibodies that provide lifelong protection. Once chronic infection is established, patients are exposed to significant risk of liver disease including chronic hepatitis, cirrhosis, and hepatocellular carcinoma. The natural history of chronic HBV infection usually consists of up to five stages,13 which differ in the level of viral replication, viral antigen expression, and inflammatory activity in the liver.
Although only approximately 1% of chronically infected individuals per year mount an effective immune response to control chronic hepatitis B infection, spontaneous resolution of acute and chronic hepatitis B proves that immune control of HBV infection is possible. However, the clinical resolution of a chronic infection or of acute hepatitis does not mean complete eradication of the virus, as small numbers of cccDNA-positive infected hepatocytes can persist in these individuals14–16 and may be the source of viral reactivation during immune suppression.17 Thus, an unknown number of the two billion people alive today who have resolved previous HBV infection maintain a viral cccDNA reservoir in their liver, as demonstrated by the emergence of chronic hepatitis B infection in people who were infected and initially recovered from HBV infection but in whom the virus reactivated when exposed to therapies inducing immune suppression for other medical conditions.17 We believe that improving our understanding of the biogenesis and homoeostasis of HBV cccDNA is urgently required if we are to develop strategies to eliminate it from infected cells, or control its expression.
Immunise, test, and treat
Before chronic hepatitis B can be cured, we need to know who is infected and their stage of disease. Only 10% of the estimated number of individuals living with HBV infection in 2016 knew of their status, and of those diagnosed, only 5% of those who were eligible for treatment actually received treatment.18 WHO aims to eliminate viral hepatitis as a major public health threat by 2030,19 and while increasing preventive vaccine coverage is the main cornerstone of this global effort to achieve elimination, WHO also proposes an increase of the proportion of eligible individuals treated for chronic hepatitis B to 80%. This will require a concomitant increase in global HBV testing worldwide. Priority should be given to development of rapid point-of-care tests for use in countries of low and middle income, where HBV is highly endemic. Since there are already effective treatments that have a marked effect on HBV replication and liver disease, increased efforts must be made to improve access globally. Large-scale accessible diagnosis and treatment facilities in countries of low and middle income should be established to reduce mortality and ensure HBV cure preparedness,20 while also investing in HBV cure strategies, which could allow important cost-savings for health systems in the long run.21
Current treatments are limited to nucleos(t)ide analogues, which are direct-acting antivirals that block DNA synthesis,22 and interferon-α. Interferon-α produces its antiviral effects by several mechanisms, including direct suppression of RNA and protein production in infected cells23 and inhibition of HBV DNA replication by blocking nudeocapsid assembly,24 in addition to immunomodulatory activity, such as the activation of NK cells and T cells that can target HBV-infected cells.25–27 Thus, in a small subset of patients, interferon-α treatment can result in cure. By contrast, nucleos(t)ide analogues only inhibit HBV genome replication; they have minimal effects on the immune response or the cccDNA minichromosome reservoir, and are not curative.
Monitoring and prevention of cancer
Current antiviral therapies reduce but do not eliminate the risk of liver cancer. As curative therapies are developed, it will be important to monitor patients for progression to liver cancer, even if they have been cured of chronic HBV infection. This is particularly important for patients with cirrhosis, who are generally thought to be most at risk for development of liver cancer; however, certain HBV subgenotypes, such as African A1 and Alaskan Fib are strongly associated with liver cancer in the absence of underlying cirrhosis.28 While studies have identified HBV-related factors associated with increased likelihood of progressing to liver cancer (high viral load, HBeAg positivity, genotype C, and failure to clear HBsAg before the age of 5029,30), there are currently no predictive biomarkers that accurately predict the onset of cancer in the setting of HBV infection. Even though the cure of infection is likely to reduce the likelihood of developing liver cancer, the direct impact of integrated HBV DNA and the viral proteins (eg, HBsAg and HBx) expressed from integrants on cellular gene expression and hepatocyte homoeostasis will need to be carefully considered. Accordingly, new markers that predict the likelihood of progression of chronic HBV infection, with or without cirrhosis, to liver cancer should be developed.
Definitions of HBV cure
Discussions about HBV cure are complicated by the need to consider curing both the infection and the associated liver disease (panel 1). Previous discussions of HBV cure have focused mainly on curing the infection, either by eliminating the virus (ie, complete cure), or by controlling the virus or inducing host immune responses to eliminate the infection and prevent viral spread (ie, functional cure).7–31,32 A recent joint American Association for the Study of Liver Diseases-European Association for the Study of the Liver workshop on HBV treatment endpoints further defined complete cure as a complete sterilising cure with undetectable HBsAg in serum and eradication of HBV DNA including intrahepatic cccDNA and integrated HBV DNA.31 Functional cure was defined as sustained, undetectable HBsAg and HBV DNA in serum with or without seroconversion to hepatitis B surface antibody (anti-HBs), with the persistence of low amounts of intrahepatic cccDNA and HBV DNA intergration.31 A third definition was also proposed (partial cure), defined as detectable HBsAg but persistently undetectable HBV DNA in serum after completion of a finite course of treatment.31 Since it is currently not possible to eradicate the HBV cccDNA minichromosome reservoir or integrated HBV DNA from every infected cell, functional cure is more likely to be achievable, at least initially. Indeed, a complete sterilising cure is yet to be observed naturally in any individuals living with chronic HBV, nor in individuals who have recovered from transient acute HBV infection, and seems unlikely to be achievable therapeutically, at least in the short term.
It is questionable whether current therapies could replicate spontaneous recovery from acute hepatitis B. Studies in chimpanzees showed that in resolution of acute HBV infection, almost all infected hepatocytes are eliminated by the CD8+ T-cell response,33 although HBV is also cleared by non-cytopathic mechanisms.34 A very small proportion of infected cells remain, with infection of new cells prevented by anti-HBs.35 Such resolution and control of HBV infection has occurred naturally in millions of individuals with acute infection. Although this is rarely achieved by currently available treatments in individuals with chronic hepatitis B, it does occur and indicates that HBV need not be totally eradicated to achieve lifelong viral control by a sustained cellular and humoral immune response. With this background, we believe that the ideal aim of a curative therapeutic regimen for chronic HBV infection should be to induce resolution similar to that observed in acute or chronic hepatitis B resolvers, as long as this can be done in a safe manner.
The clinical goal of chronic hepatitis B remission is the resolution of residual liver injury including disappearance of inflammation, prevention of fibrosis progression, reversal of fibrosis and cirrhosis, prevention of liver failure, and reduction of the risk of developing hepatocellular carcinoma,31 following completion of a finite course of treatment. Great progress has been made in this regard, with the latest generation nucleos(t)ide analogues demonstrating reversal of fibrosis and reduced likelihood of progression to hepatocellular carcinoma in many, but not all, patients.36,37 However, treatment is long term and a high priority of curative treatments is that treatment be finite, while still meeting the goals of disease remission. Remission in early chronic hepatitis B, where no fibrosis is yet present, is analogous to a person who clears acute HBV infection, but retains HBV cccDNA in some of their hepatocytes—there is no evidence of ongoing liver disease, but the viral reservoir is still present, albeit at very low levels tightly constrained by the humoral and cellular immune responses. Although the remission of liver disease in chronic hepatitis B does not preclude potential reactivation of disease, it largely meets the societal goal of cure, namely the prevention of adverse outcomes, restoration of health, prolongation of life expectancy after cessation of therapy, elimination of transmission risks, and removal of stigma associated with having a chronic communicable disease,38 all of which should be the guiding principles in the ultimate adoption of a definition.
As a result of academic and industrial efforts to identify viral targets for drug discovery, a number of drugs are being evaluated in preclinical models or are in the first phases of clinical development (figure 2).40 These include drugs interfering with viral entry, drugs or other methods targeting cccDNA or viral RNA, novel polymerase or ribonuclease H inhibitors, core protein allosteric modulators and capsid assembly modulators, drugs interfering with HBx function, drugs targeting viral and HBsAg egress, or agents affecting the viral proteome (eg, small interfering RNA and RNA destabilisers). Some of these compounds appear promising, with marked reductions in HBV viral load or circulating HBsAg, or both, and the emergence of anti-HBs, but to date none seem to be capable of curing HBV infection on their own.7,32 None of these approaches directly target the HBV cccDNA reservoir, but it will be important to evaluate these approaches in combination, to determine their effectiveness in the kinetics of cccDNA clearance or in their capacity to improve the restoration of antiviral immune responses. Earlier attempts to boost innate immunity and adaptive immune responses did not lead to improved cure rates, as defined by HBsAg loss. It is therefore critical to design drug combinations with the aim that they are curative. To this end, following extensive discussion among the ICE-HBV working groups, we have prioritised the research necessary to achieve HBV cure and the tools that will be required to achieve this goal. These are described in the context of key concepts that should be addressed to eliminate HBV from infected cells and induce effective immune control.
Figure 2: Current and future HBV virological and immunological targets that will be necessary for treatment and cure of chronic hepatitis B.
Adapted from reference 39. CpAMs/CAMs=core protein allosteric modulators/capsid assembly modulators. CAR =chimeric antigen receptor. cccDNA=covalently closed circular DNA. IL=interleukin. IFN-α=interferon-α. KC=Kupffer cell. NK=natural killer cell. NKT=natural killer T cell. pDC=plasmocytoid dendritic cell. RC DNA=relaxed circular DNA.
We believe that research efforts should focus on reducing the number of cccDNA molecules per cell and the number of cccDNA-containing cells, and on inducing HBV-specific immune responses to achieve what the host is capable of achieving during the resolution of infection, namely profound reduction of the number of infected cells and induction of HBV-specific immune responses that prevent viral spread from those that remain infected. Such approaches need to be done in a manner that is safe, durable, and affordable. Increased efforts will be needed to improve our understanding of the biogenesis, homoeostasis, transcriptional regulation, and decay of the viral cccDNA reservoir,9 together with a deeper understanding of factors required to induce host immune responses that control or eliminate HBV infection in individuals with chronic hepatitis B. However, it is also important that research continues to gain knowledge about many other aspects of the HBV replication cycle, including viral entry and uncoating, mechanisms regulating cccDNA activity, gene expression including mRNA translation and splicing, and packaging of pgRNA and its reverse transcription forming the main replication complexes, subviral particle formation, and viral egress.
To achieve our goal of HBV cure, ICE-HBV proposes two strategies, namely curing of HBV infection without killing infected cells and inducing immune control to safely eliminate infected cells. Each of these approaches will need to be underpinned by coordinated clinical studies to advance HBV cure. Detailed descriptions of each of these strategies, together with the clinical approaches necessary to advance HBV cure, are presented below.
Strategy 1: curing HBV infection without killing infected cells
More than 50 years have elapsed since the discovery of HBsAg.41 The discovery of the HBV entry receptor NTCP in 20128 revolutionised our ability to study in vitro the HBV replication cycle from infection to egress, and the development of new animal models that permit studies of immunopathogenesis42–44 and preclinical antiviral evaluation. However, these models still have considerable limitations, and important gaps in our understanding of the HBV replication cycle and the host immune response must be addressed to expand the exploitable vulnerabilities in the replication cycle that can be targeted therapeutically to cure the infection.
Targeting the cccDNA minichromosome reservoir
The HBV cccDNA minichrommosome reservoir in the nucleus of infected hepatocytes is the single most important barrier to curing chronic HBV infection. We suggest that research directed towards elimination of cccDNA or permanently silencing cccDNA transcription should be prioritised.
cccDNA elimination will be the most direct and efficient strategy to cure chronic HBV infection. Current antiviral drugs do not eliminate cccDNA, even after prolonged treatment either in patients or experimental cell culture or animal models. The observation that cytokine administration, including interferon-α, may induce APOBEC-dependent deamination of viral DNA45–47 has led to considerable discussion of whether this could lead to a substantial decrease of cccDNA in infected hepatocytes.48–50 Approaches that directly target cccDNA (eg, CRISPR/Cas950 or other gene editing methodologies51) have shown promising results in laboratory studies, but issues including hepatocellular delivery, off-target effects, and the likelihood that they will also cleave chromosomally integrated HBV DNA and thus trigger unpredictable consequences of chromosomal DNA recombination will need to be carefully considered. Because of these potential dangers and difficulties, approaches that target viral or host proteins that are critical for cccDNA formation, stability, and expression (eg, HBx) must be further explored. An intense and coordinated effort will be required to discover the fundamental biochemical, cellular, and physiological basis of cccDNA production, maintenance, and turnover, about which very little is known. Now that it is possible to study the complete HBV replication cycle, we believe these studies are imperative because cccDNA elimination is the only strategy that has the potential to definitively produce a safe, durable cure of HBV infection.
Since cccDNA is not completely eradicated during spontaneous or treatment-induced resolution of acute or chronic infection,52 it is unlikely that total eradication of cccDNA will be achievable pharmacologically. Transcriptional silencing of the residual pool of cccDNA remaining after most of the cccDNA is eliminated or antibody-mediated prevention of virus spread from the remaining infected cells (or both) will probably also be necessary. While modification of epigenetic control of cccDNA by general epigenetic modifiers would probably be too risky in terms of potential adverse events on cell homoeostasis, other strategies targeting viral factors involved in the regulation of cccDNA transcriptional activity should be pursued. Transcriptional control of cccDNA expression may be achieved by targeting the HBx protein, which is critical for cccDNA expression or stability.53–56 Greater insight into the role of HBx, HBc,57 and other viral proteins in the control of cccDNA homoeostasis is thus mandatory. The Smc5/6 complex is a host restriction factor that, in the absence of HBx protein, represses cccDNA expression.58–60 It has recently emerged that HBx promotes cccDNA transcription by hijacking cellular DDB1-containing E3 ubiquitin ligase to target the Smc5/6 complex for degradation.58 Therefore, therapeutic approaches targeting the HBx-DDB1 binding interaction are also worthy of investigation. Post-transcriptional control of cccDNA or integrated HBV DNA expression can be achieved using RNA interference; this is currently being pursued, with promising results.61,62 The overlapping nature of the HBV genome suggests that RNA interference should impede expression of all HBV transcripts, and it has recently been shown that truncated forms of transcripts derived from integrated viral DNA can also be targeted.61 There is also a pressing need for the development of in-situ, single-cell, singlemolecule, live-cell assays, to visualise the localisation and trafficking of cccDNA,63,64 and cccDNA-dependent surrogate reporter cell systems for high throughput screening of cccDNA inhibitors.65–67
New models are required
Although it is now possible to interrogate the complete HBV replication cycle in cell culture, the current models still have some limitations. For example, most studies to date have been limited to HBV genotype D, the only genotype for which stably transformed cell lines are readily available to produce HBV at sufficient multiplicity of infection for downstream experiments. Additionally, the transformed nature of hepatoma cells means that they do not fully reflect primary liver cells. Primary human hepatocytes—the natural target cells for HBV infection in vivo—are available and support the complete HBV replication cycle, and are presumably more physiologically relevant to hepatocytes in the liver. However, their use has limitations: they require polyethylene glycol, dimethylsulfoxide, and high amounts of input virus for infection; they are expensive; they have a limited polarisation and life-span in vitro; and batch-to-batch variations affect infection efficiency and responses to infection.68
In-vitro models that would eliminate the need for polyethylene glycol and provide consistency and uniformity of infection from lab to lab over time are needed to accelerate the discovery process and to confirm the reproducibility of promising new data. Systems that have the chance of satisfying these criteria include human hepatocyte-like cells differentiated in vitro from embryonic stem cells or induced pluripotent stem cells69–71 and three-dimensional microfluidic liver cultures, which permit analysis of the complete HBV replication cycle.72 However, all available HBV infection systems only support limited viral spreading in the cultures. Culture systems that support robust amplification of HBV are needed.
Furthermore, none of the current animal models for HBV infection is optimal,68 especially in the context of HBV cure research. Despite the existence of immune-deficient humanised mouse models permitting studies of HBV infection and spreading in vivo, as well as to evaluate the efficacy of direct antiviral compounds in HBV-infected primary human hepatocytes, immunocompetent mouse models that are susceptible to HBV infection are only now starting to become available. Humanised mouse models will also enable studies on long-term effects of drugs on cccDNA maintenance and loss in non-dividing versus dividing hepatocytes.73 Further development of double humanised mouse models (liver and immune system) will permit studies of immune-mediated clearance.42,43,74 Breaching the current intracellular block to HBV infection and cccDNA formation in mouse hepatocytes75 would generate a small animal model, enabling analysis of the immune response to HBV that may facilitate immune-mediated cccDNA elimination and control, similar to clearance of acute HBV infection. Thus, we believe that the further development of humanised mouse models amenable to HBV infection should receive high priority because of their great potential to enable studies of mechanisms regulating cccDNA metabolism and clearance.
With the loss of the chimpanzee model of HBV infection, the only primate model available to the HBV research community is the tree shrew (Tupaia belangeri chinensis).76,77 Research with this model, however, is severely limited by the absence of a deep portfolio of Tupaia-based reagents for virological and immunological analysis. However, the discovery that HBV uses NTCP as its receptor raised the possibility of creating a macaque model for HBV infection by expressing human NTCP on macaque hepatocytes after infection with recombinant adeno-associated virus vectors, leading to HBV infection for at least 6 weeks.78 If improved, this model has the potential to not only better understand the immunobiology and pathogenesis of HBV infection and test immunomodulatory approaches to cure chronic HBV infection, but also to study the therapeutic potential of HBV-targeted drugs, including those that target cccDNA. A macaque model would benefit immediately from the wealth of information and reagents that have been developed during the use of macaques to study simian immunodeficiency virus immunobiology and pathogenesis.79 We believe that the establishment of a human NTCP transgenic macaque model should be of high priority for the HBV cure research community and for international governmental research funding agencies interested in or committed to curing chronic HBV infection.
Since all current experimental models have limitations, and differences between HBV and HBV-related viruses have emerged, it is likely that observations previously made with HBV-related viruses require confirmation using HBV and human hepatocytes.80 The research priorities and tools required for HBV elimination are shown in panel 2.
Strategy 2: inducing immune control to safely eliminate HBV-infected cells
A complex interplay of innate and adaptive immune responses is essential for viral clearance and a failure of these responses can result in liver pathogenesis. CD8+ T cells are the main effector cells that eliminate the virus by cytolytic and non-cytolytic effector functions.33,34 However, sufficient CD4 T-cell activity and the production of neutralising anti-HBV envelope antibodies are required for protective immunity.85 A major effort has been made to compare the efficient integrated response resulting in HBV clearance of acute infection to the dysregulated response observed in patients with chronic hepatitis B. This knowledge has led to the development of the first generation of immune-based therapies and a second generation of therapeutic vaccines entering phase 1 and 2 clinical trials.
Current immunotherapies for chronic HBV infection
Key points of immune regulation that can be targeted for therapy have been identified in studies done in animal models and HBV-infected patients. Injection of ligands targeting Toll-like receptors (TLRs) or RIG-I leads to suppression of viral replication in HBV mouse models.86,87 This observation has led to the development of drugs targeting TLR-7,88 TLR-8, and RIG-I89 for use in chronically HBV-infected patients. Numerous experiments have revealed that blocking inhibitory receptors can enhance expansion and function of HBV-specific T cells, both in vitro90–94 and in vivo.95,96 This has motivated the use of anti-PD-1/PD-L1 antibodies in phase 1 studies in chronically HBV-infected patients. Furthermore, interleukin-12 appears critical to restoring T-cell expansion and function in chronic infection.97 This has led to interleukin-12, or adjuvants inducing it, being incorporated into the new generation of therapeutic vaccines for chronic HBV.98
That immunological observations have translated to new experimental therapies is encouraging. Now, a critical step is to ensure proper immunological evaluation of these strategies to improve their efficacy for future generations of immunotherapy. Additionally, continued investigation into the dysregulation of antiviral responses in HBV infection is key to expand the repertoire of immune-based therapies. Further integration of data is key to effective T-cell-based therapies, defining networks of T-cell exhaustion mechanisms and the extent to which T-cell functionality can be restored. Since anti-HBs is considered the clinical indicator of HBV resolution, we need a better understanding of the role of B cells and neutralising antibody response in the natural history of chronic hepatitis B and how they can be effectively enhanced. Mounting evidence for organ-specific immunity necessitates analysis of the liver environment.99–101 Furthermore, accurate quantification of the number of infected hepatocytes in patients with chronic hepatitis B is important to avoid immune-related toxicity (panel 3). We highlight areas of research that are central to addressing these knowledge gaps to improve the application of immune-based therapies.
Methodology to improve ex-vivo analysis of HBV-specific immunity
Compared with chronic infection with HCV, the frequency of virus-specific peripheral blood T cells in patients with chronic hepatitis B is 10–100 fold lower,102 and often undetectable using standard assays directly ex vivo. Better and more sensitive assays or novel strategies for ex-vivo measurement of HBV-specific immunity in peripheral blood and liver are needed—eg, to correlate HBV-specific immunity with stage of disease, responses to immunotherapy, and antiviral drugs. They are also needed to define biomarkers that best identify patients who can safely stop antiviral therapy and reflect intrahepatic immunity in the peripheral blood.
Similarly, novel approaches are needed to study the comprehensive phenotypic and functional characteristics of the intrahepatic T-cell responses using limited number of cells from individual biopsies. These data need to be compared with responses in the blood and then applied as immune-based biomarkers that predict or reflect viral control and inflammation. This comparison needs to be done in chronically HBV-infected patients at different stages of disease. Additionally, assays that allow the analysis of T cells of different epitope specificity need to be further developed to overcome limitations imposed by pentamer/tetramer staining that only partially reflect the overall antiviral immune response.
Mechanisms of HBV-mediated T-cell exhaustion and recovery
HBV persistence is associated with CD8+ T-cell exhaustion and (to a lesser extent) with viral escape. Persistent exposure to high antigen loads and antigen presentation by hepatocytes play a key role in CD8+ T-cell exhaustion.95,103 The expression of inhibitory receptors (predominantly PD-1),90,95,96 depletion of amino acids (eg, arginine and tryptophan),104 and mitochondrial alterations105 contribute to T-cell dysfunction (figure 3). Additionally, lack of CD4+ T-cell helper activity,85,106 immunosuppressive cytokines,107 regulatory T (Treg) cells,108 granulocytic myeloid-derived suppressor cells,109 and NK cells110 also contribute to CD8+ T-cell failure.25,111However, the relative contribution of these different mechanisms to T-cell exhaustion are not well characterised and should be actively pursued. Also, whether functional T-cell restoration is possible, and if so, how much restoration is required for HBV control, is not fully understood. Understanding these aspects will refine our targets to restore T-cell functionality and improve immunotherapy.
Figure 3: Mechanisms of CD8+ T-cell failure and potential targets for curative immunotherapeutic approaches in the setting of chronic hepatitis B.
HBsAg=hepatitis B surface antigen. IL=interleukin. MDSC=myeloid-derived suppressor cells. NK=natural killer. TCR=T-cell receptor. TGF=transforming growth factor.
Comprehensive analysis of the immune responses of patients with active and inactive chronic HBV infection (with and without antiviral therapy), patients with resolved HBV infection, chronically infected patients with HBsAg loss, and vaccinated healthy individuals will help define the level of immune function or reconstitution needed for viral control. Recent evidence suggests we need a better understanding of metabolic immune regulation, how this relates to overt, detectable inhibitory receptor expression in immune cells, and how this is linked to a broader profile of immune dysfunction. To evaluate the efficacy of checkpoint blockade, the possibility that differential baseline expression of inhibitory molecules and their ligands differ in chronically infected patients versus resolvers should be assessed. A better understanding of the relative contribution of inhibitory receptors (eg, PD-1, CTLA-4, Tim-3) at different stages of disease is required to determine the patient populations that are most likely to benefit from checkpoint blockade therapy.
B cells in the natural history of disease
Anti-HBV antibodies are used as biomarkers for resolution of HBV infection but the immunological impact of B cells and antibodies on the natural history and resolution of chronic HBV infection, or on seroconversion in chronic HBV infection, have been under-investigated.112 B cells can produce the immune-suppressive cytokine interleukin-10 but their depletion can also result in clinical reactivation in both patients with resolved infection and those with chronic hepatitis B.107,113,114 Therefore, fundamental questions related to B-cell biology, antibody specificity, and neutralising responses need to be addressed so that B cells can be incorporated more effectively into new immunotherapeutic approaches.
Assays and reagents need to be developed and optimised for the detection and quantification of HBV-specific B cells. ELISPOT assays are needed for accurate and reproducible quantification of B cells producing HBV-specific antibodies for research and clinical trial monitoring. Flow cytometry-based reagents are needed to investigate the phenotypic, functional, and genetic profile of HBV-specific B cells. Deeper analysis of antibody specificity, isotype, and functional responses in different stages of HBV infection should be a key area of future research.
Accurately measuring the frequency of infected hepatocytes
The primary safety concern with immunotherapy in chronic HBV infection is the induction of fulminant hepatitis. Therefore, it is important to know the actual number of infected hepatocytes, the number of hepatocytes expressing viral antigens, and the number of hepatocytes that can be destroyed in an individual patient without causing hepatic deficiency. HBV integration occurs early in cell culture infection but it is unclear how cytolytic versus non-cytolytic mechanisms will target infected hepatocytes (with replicating HBV) versus those with integration.61,115 It is imperative to understand the frequency of infected and integrated hepatocytes and how hepatocytes are cleared of HBV or HBsAg to minimise the risk of hepatic decompensation.
In the short term, better methods to quantify both infected and integrated hepatocytes in biopsies or through serum biomarkers are needed. An in-vivo diagnostic capable of visualising the infected liver on medical imaging instruments would be an ideal longterm goal. A better understanding of cytolytic clearance of infected cells versus non-cytolytic inhibition of viral replication in surviving hepatocytes is needed in humans. Different effector functions may be used by T cells or innate effectors (eg, NK, γδ, MAIT cells) depending on the quantity or phenotype of newly infected, persistently infected, or integrated hepatocytes. The development of less invasive technologies based on fine needle aspiration instead of liver biopsies will help to understand these interactions, since they will be important to maximise antiviral efficacy and minimise potential toxicity.
Standardisation of immune monitoring in clinical trials
Standardisation of immune monitoring during clinical trials is a monumental but necessary task. To improve consistency of immune monitoring between different sites, we propose the development of centrally curated and widely available HBV peptide libraries and tetramers. Coupled with these would be validated protocols for ELISPOT and flow cytometry staining. Such approaches would help promote consistent T-cell monitoring.
Sampling during clinical trials for immunological analysis will be critical to improve and advance immune-based therapies. Blood and serum will remain most accessible. Adoption of fine needle aspiration should be considered where possible to allow for intrahepatic sampling and single cell analyses.116 Timing is key, as innate immune modulators will have rapid responses whereas T-cell and B-cell responses will require months of follow-up. Comparison of positive responses to the drug or vaccine in patients with chronic HBV should not be gauged against baseline responses alone. The magnitude of responses in patients with chronic hepatitis B should be compared to phase 1 data in healthy individuals or immune responses detected in patients with resolved HBV infection whenever possible. This will provide a clearer assessment of immunogenicity in patients with chronic hepatitis B. Immune monitoring in early stage drug development should focus on a broader assessment of immunity—eg, high frequency populations, immune profiling, ELISPOT with large peptide pools—rather than attempts at fine specificity. Even negative data is useful and will promote progress and innovation.
Future of immunotherapy
Just as we are now seeing an expansion of drug targets specific to the virus, expansion of our knowledge in key areas related to immune failure and reconstitution will expand the breadth of potential immune-based therapies. Identifying the inflection points of immune exhaustion for restoration of T-cell and B-cell immunity will improve specificity and reduce toxicity. Checkpoint inhibitors that are currently tested in phase 1 trials for their potential to restore HBV-specific CD8+ T-cell function offer a promising perspective for HBV immunotherapy. However, important questions need to be addressed in future trials about the extent to which T cells can be restored, the dose of anti-PD-1 antibodies, the selection of patients likely to respond based on biomarkers or disease stage, and the risk of side-effects due to tissue damage by restored T cells. In this context, it will also be important to determine the efficacy of combination therapies—eg, checkpoint inhibitors with different antiviral therapies or therapeutic vaccinations—as has been shown to be effective in preclinical animal models.117 For therapies targeting innate immunity, separating antiviral efficacy from inflammation with pattern recognition receptor agonists could improve their therapeutic index. In addition to refining immunotherapy approaches, pushing forward with strategies to engineer antiviral immunity hold promise. T-cell receptor and chimeric antigen receptor gene therapies circumvent the need for restoration.43,118–122 However, whether long-term engraftment of genetically engineered T cells specific for HBV is needed to maintain control of viral spread from infected cells that escape elimination, similar to patients with resolved infection, has yet to be addressed. Each of these immunotherapeutic strategies should be considered and investigated in the context of combination therapy. Rationally pairing antiviral and immunotherapeutic drugs based on complementary mechisms of action will hopefully ensure potent antiviral effects with elimination of infected hepatocytes and improve HBV cure rates.
Coordinated clinical studies to advance HBV cure
While the pharmaceutical industry will develop novel drugs and evaluate them in clinical trials, collaborations with clinical scientists outside of industry will be instrumental to the success of drug development, by facilitating clinical studies, characterising correlates of cure, refining treatment endpoints, and identifying the best patients for clinical trials according to the mode of action of the tested drugs.
The timing for initiation of curative regimens will also require careful consideration. There is emerging evidence that the HBV disease time-clock commences ticking earlier than previously appreciated. This is demonstrated by the finding of integrated HBV DNA in infected hepatocytes within a few days of viral infection in vitro123 and in patients during the so-called immune tolerant stage of HBV natural history.115 There is also increasing evidence that HBV DNA integrations are associated with liver cancer,124 and it remains to be determined whether early initiation of treatment would reduce the likelihood of progression to hepatocellular carcinoma by preventing integration, inflammation, and cell turnover, and in turn clonal expansion of hepatocytes harbouring integrated sequences.
The identification and characterisation of the virological, immunological, and other host correlates or determinants of HBV cure would be of utmost importance to guide the clinical development of future curative regimens for patient selection, monitoring of treatment with novel biomarkers, and design of clinical trials according to patient characteristics and the modes of action of the tested treatments. A repository of serum samples from patients who have resolved their HBV infection, including patients with HBsAg loss and those exhibiting immune control (anti-HBs), with appropriate matched controls prior to resolution, should be established and made accessible to researchers across the globe. This would generate considerable statistical power, permitting investigation of serum biomarkers associated with HBV resolution, such as HBsAg levels, HBcrAg, and circulating HBV RNA. For some samples, matching liver biopsy material and peripheral blood mononuclear cells should be obtained, enabling direct comparison of cccDNA (from liver samples) and immune responses (from blood samples) with HBV serum markers in patients with chronic hepatitis B who lost HBsAg either spontaneously or with currently available treatments.
There is also an urgent need for centralised repositories of HBV-related materials that are readily accessible to HBV researchers globally, similar to resources available to colleagues in the HIV field. Critical to this will be quality assurance of the samples, and the availability of matching clinical data. This repository could be accessed by basic researchers, clinicians, biotechnology, and pharmaceutical companies, to facilitate studies and development of new drugs. A representative list of reagents required needs to be developed by the HBV community and some of the reagents are shown in panel 4.
Conclusions
The time for development and implementation of a safe, affordable, widely available cure for chronic hepatitis B for the 257 million people who are affected by HBV globally is now. Panel 5 shows the key points that must be urgently prioritised to achieve this goal.
HBV research funding has for too long been woefully inadequate; this must be addressed. New research programmes targeting the cccDNA reservoir and stimulating the host immune response in a safe and durable manner must be a high priority for researchers, foundations, and governments. We support the recent R01 call from the US National Institutes of Health for projects on HBV or HIV/HBV immunology, virology, and therapeutics.127 ICE-HBV seeks to achieve its goals by fostering collaborative partnerships with researchers (both within the HBV field and outside), clinicians, the pharmaceutical industry, and a range of stakeholders, including communities affected by chronic hepatitis B; we invite these groups to join forces with ICE-HBV in a global effort to discover, develop, test, and implement HBV cure strategies, to help ensure that the WHO goal of HBV elimination as a public health threat by 2030 is achieved.
Supplementary Material
Panel 1: Definitions of curative HBV treatment outcomes.
Cure of infection—complete sterilising cure with undetectable HBsAg in serum and eradication of HBV DNA including intrahepatic cccDNA and integrated HBV DNA31
Functional cure of HBV infection—sustained, undetectable HBsAg and HBV DNA in serum with or without seroconversion to hepatitis B surface antibody (anti-HBs), with the persistence of low levels of intrahepatic cccDNA31
Remission of liver disease—resolution of residual liver injury including reversal of fibrosis, prevention of fibrosis progression, remission of disease, and reducing risk of hepatocellular carcinoma
Panel 2: Research priorities and tools for HBV elimination.
Priorities
Develop standardised methods to quantify cccDNA and study mechanisms of cccDNA homoeostasis and processes affecting its biogenesis, homoeostasis, structure, transcriptional control, and decay
Define mechanisms determining establishment of HBV infection: characterise all steps from cell entry to cccDNA minichromosome formation and maintenance
Improve methods for the study of cccDNA processing and virus-host interactions to reveal new targets for therapeutic approaches to clear cccDNA by applying state-of-the-art “omics” approaches to increase understanding of HBV-host interactions at a genome-wide level
Develop and validate new serum markers (eg, core related antigens [HBcrAgs81], HBV RNA82–84) as reliable bio markers of cccDNA activity in the liver. Once markers are identified and characterised, ensure they are standardised
Develop methods to specifically degrade HBV cccDNA
Develop methods to prevent transcription of cccDNA and integrated HBV DNA
Continue to develop methods to inhibit other key steps of the viral replication cycle that could be included in combination strategies to cure the infection
Tool kit
Develop efficient and convenient in-vitro functional cccDNA systems—eg, human hepatocyte-like cells differentiated in vitro from embryonic stem cells or induced pluripotent stem cells;primary human hepatocytes and human hepatocyte-like cells transplanted into the liver of immune deficient mice for expansion after controlled removal of endogenous mouse hepatocytes;68 robust, specific, and reliable cccDNA reporter cell culture systems
Develop convenient in-vivo model systems, particularly immunocompetent non-human primate and mouse models susceptible to HBV infection—eg, non-human primate HBV infection animal models (eg, h-NTCP macaque79); humanised mouse models permissive for HBV for immunological studies; mouse models fully permissive to HBV infection in murine hepatocytes
Panel 3: Research priorities and tools for HBV immune control.
Priorities
Conduct clinical studies with existing immune interventions
Determine the relative contribution of different components of the immune system to viral clearance versus viral persistence, immunopathology, and treatment response among neonates, children, adolescents, and adults
Identify the mechanisms of T-cell exhaustion and the extent to which T-cell restoration is reversible, durable, and needed for viral control
Determine the role of B cells in the natural history of disease and how they can be effectively monitored for research and clinical trials
Investigate the impact of the liver microenvironment on the composition and function of innate and adaptive cells and identification of biomarkers in the blood that best reflect the intrahepatic immune response
Determine the number of infected hepatocytes in each category of patients and the degree of immune-mediated destruction that is required for clearance but can still be tolerated before hepatic decompensation occurs
Tool kit
Develop overlapping peptide libraries to measure virus-specific T-cell responses covering the five dominant HBV genotypes
Develop HLA-tetramer libraries of validated HBV epitopes
Develop validated flow cytometry and cytokine panels available to investigators for immune-profiling
Develop validated ELISPOT protocols for HBV-specific T and B cells
Develop validated protocols for stimulation and functional monitoring of innate and adaptive immune cells
Panel 4: Reagent and specimen repositories required to advance HBV cure research.
Viral DNA, RNA, and protein standards
Monoclonal antibodies against HBV proteins
Replication competent HBV clones of various genotypes and subgenotypes
Stably transfected cell lines of various genotypes and subgenotypes
Cell lines susceptible to HBV replication and cell to cell spread
Mouse and primate models susceptible to HBV infection
Chemical libraries for studies in experimental models
Collection of serum and liver biopsy samples from cohort study for research purposes
Peptide libraries for clinical immunology studies
Panel 5: Immediate and future actions required to achieve HBV cure—the ICE-HBV strategy.
INCREASE funding for individual and collaborative cure-related research projects by governmental and private funding agencies and philanthropic benefactors. Consideration should be given to establishing international research consortia, similar to the Martin Delaney Collaboration for HIV research managed by the National Institutes of Health in the USA. HBV cure research investment strategies should be prioritised in national HBV plans globally. We also believe the WHO hepatitis elimination strategy19 should be funded in full, with particular focus on delivery of birth dose vaccines and substantially increased investments in HBV research. We support recent calls from the Hepatitis B Foundation for increased HBV cure research funding125,126
CONCENTRATE on the discovery of interventional strategies that will permanently reduce the number of productively infected cells or permanently silence the cccDNA in those cells and also stimulate HBV-specific T cells and the production of neutralising antibodies that will prevent viral spread to uninfected cells, mimicking spontaneous resolution of acute HBV infection
ESTABLISH repositories of standardised HBV reagents and protocols, facilitate access to all researchers across the world, and support the development of new animal models of HBV infection
Search strategy and selection criteria.
ICE-HBV has facilitated scientific discussions within and between working groups on virology, immunology, and innovative tools with input from clinical advisors, to identify gaps in HBV cure research and perform the research needed to address these. Together these groups form the ICE-HBV scientific working group, which met in person in April, 2017, September, 2017, and April, 2018, and worked online via Zoom and email to draft this position paper. They produced a gap analysis of HBV cure research that served as a basis for developing the scientific roadmap for HBV cure. ICE-HBV has gathered key stakeholders from around the world to provide input on the scientific roadmap. Two consultations were organised in person in Sao Paulo and Melbourne. 152 stakeholders from 21 countries and five continents were consulted.
Acknowledgments
This work was supported by the Australian Academy of Science, France Recherche (ANRS), and the Peter Doherty Institute for Infection and Immunity. This work was supported in part by the intramural research programme of NIDDK, NIH (general grant to Barbara Rehermann, a member of the ICE-HBV Working Groups).
Declaration of interests
PAR reports grants from Gilead Sciences, outside the submitted work. FVC reports personal fees from Gilead Sciences, Avalia Immunotherapies, and Vir Biotechnology, outside the submitted work. MD reports grants from Roche, personal fees from Gilead Sciences, research contracts from Novira Therapeutics, Humabs BioMed, and Hepatera, outside the submitted work. AJG reports grants from Gilead Sciences and Janssen Pharmaceuticals and personal fees from SpringBank Pharmaceuticals, Aicuris, Arbutus, and Vir Biotechnology, outside the submitted work. HG reports grants from Arbutus Biopharma, Spring Bank Pharmaceuticals, and Aligos Therapeutics and personal fees from Assembly Biosciences and Aligos Therapeutics, outside the submitted work. JH reports personal fees from Arbutus, Gilead, and Sanofi and grants from Gilead and Roche, outside the submitted work. HLAJ reports grants from AbbVie, Bristol-Myers Squibb, Gilead Sciences, Janssen, Medimmune, Merck, and Roche and personal fees from AbbVie, Benitec, Bristol-Myers Squibb, Gilead Sciences, Janssen, Medimmune, Merck, Roche, Arbutus, and Vir Biotechnology, outside the submitted work. PL reports personal fees from BMS, Roche, Gilead, GlaxoSmithKline, MSD, and Janssen, outside the submitted work. ML reports grants and non-financial support from Bristol-Myers Squibb, grants from Contravir, personal fees from Galapagos and Arbutus, personal fees and has acted on speakers’ bureau from Gilead and Janssen, and acted on speakers’ bureau for Roche and MSD, outside the submitted work. S-GL reports grants and personal fees from Gilead Sciences, Merck Sharpe and Dohme, Roche, and Abbott Diagnostics, and personal fees from Abbvie, Springbank, outside the submitted work. MCP reports personal fees from Janssen, outside the submitted work. FZ reports grants from Roche, Janssen, and Evotec and personal fees from Arbutus, Gilead, Myr Pharma, Transgene, and Vir Biotechnology, outside the submitted work. JMB, AK, WL, TJL, FL, JET, and RT declared no competing interests.
References
- 1.WHO. Global hepatitis report, 2017. https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/ (accessed April 4, 2019).
- 2.Stanaway JD, Flaxman AD, Naghavi M, et al. The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet 2016; 388:1081–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol 1995; 13: 29–60. [DOI] [PubMed] [Google Scholar]
- 4.Grossi G, Vigano M, Loglio A, Lampertico P. Hepatitis B virus long-term impact of antiviral therapy nucleot(s)ide analogues (NUCs). Liver Int 2017; 37 (suppl 1): 45–51. [DOI] [PubMed] [Google Scholar]
- 5.NIH. Estimates of funding for various research, condition, and disease categories (RCDC), 2017. https://report.nih.gov/categorical_spending.aspx (accessed April 8, 2019).
- 6.O’Hara GA, McNaughton AL, Maponga T, et al. Hepatitis B virus infection as a neglected tropical disease. PLoS Negl Trop Dis 2017; 11: e0005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Revill P, Testoni B, Locarnini S, Zoulim F. Global strategies are required to cure and eliminate HBV infection. Nat Rev Gastroenterol Hepatol 2016; 13: 239–48. [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Zhong G,Xu G, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012; 1: e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassal M HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 2015; 64:1972–84. [DOI] [PubMed] [Google Scholar]
- 10.Newbold JE, Xin H, Tencza M, et al. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol 1995; 69: 3350–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bock CT, Schwinn S, Locarnini S, et al. Structural organization of the hepatitis B virus minichromosome. J Mol Biol 2001; 307:183–96. [DOI] [PubMed] [Google Scholar]
- 12.Messageot F, Salhi S, Eon P, Rossignol JM. Proteolytic processing of the hepatitis B virus e antigen precursor. Cleavage at two furin consensus sequences. J Biol Chem 2003; 278: 891–95. [DOI] [PubMed] [Google Scholar]
- 13.EASL EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 2017; 67: 370–98. [DOI] [PubMed] [Google Scholar]
- 14.Michalak TI, Pasquinelli C, Guilhot S, Chisari FV. Hepatitis B virus persistence after recovery from acute viral hepatitis. J Clin Invest 1994; 93: 230–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehermann B, Ferrari C, Pasquinelli C, Chisari FV The hepatitis B virus persists for decades after patients’ recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat Med 1996; 2:1104–08. [DOI] [PubMed] [Google Scholar]
- 16.Werle-Lapostolle B, Bowden S, Locarnini S, et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 2004; 126:1750–58. [DOI] [PubMed] [Google Scholar]
- 17.Seetharam A, Perrillo R, Gish R. Immunosuppression in patients with chronic hepatitis B. Curr Hepatol Rep 2014; 13: 235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol 2018; 3: 383–403. [DOI] [PubMed] [Google Scholar]
- 19.WHO. Global health sector strategy on viral hepatitis 2016–2021 Towards ending viral hepatitis. Geneva: World Health Organization, 2016. [Google Scholar]
- 20.Lazarus JV, Block T, Brechot C, et al. The hepatitis B epidemic and the urgent need for cure preparedness. Nat Rev Gastroenterol Hepatol 2018; 15: 517–18. [DOI] [PubMed] [Google Scholar]
- 21.Nayagam S, Thursz M, Sicuri E, et al. Requirements for global elimination of hepatitis B: a modelling study. Lancet Infect Dis 2016; 16:1399–408. [DOI] [PubMed] [Google Scholar]
- 22.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 2009; 137:1593–608. [DOI] [PubMed] [Google Scholar]
- 23.Belloni L, Allweiss L, Guerrieri F, et al. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome.J Clin Invest 2012; 122: 529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci USA 2005; 102: 9913–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill US, Peppa D, Micco L, et al. Interferon alpha induces sustained changes in NK cell responsiveness to hepatitis B viral load suppression in vivo. PLoS Pathog 2016; 12: e1005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maini MK, Gehring AJ. The role of innate immunity in the immunopathology and treatment of HBV infection. J Hepatol 2016; 64(suppl 1): S60–70. [DOI] [PubMed] [Google Scholar]
- 27.Maini MK, Peppa D. NK cells: a double-edged sword in chronic hepatitis B virus infection. Front Immunol 2013; 4: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genotypes Kramvis A. and genetic variability of hepatitis B virus. Intervirology 2014; 57:141–50. [DOI] [PubMed] [Google Scholar]
- 29.Chen CJ, Iloeje UH, Yang HI. Long-term outcomes in hepatitis B: the REVEAL-HBV study. Clin Liver Dis 2007; 11: 797–816. [DOI] [PubMed] [Google Scholar]
- 30.Yuen MF, Wong DK, Fung J, et al. HBsAg seroclearance in chronic hepatitis B in Asian patients: replicative level and risk of hepatocellular carcinoma. Gastroenterology 2008; 135:1192–99. [DOI] [PubMed] [Google Scholar]
- 31.Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis B cure: from discovery to regulatory approval. Hepatology 2017; 66:1296–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeisel MB, Lucifora J, Mason WS, et al. Towards an HBV cure: state-of-the-art and unresolved questions—report of the ANRS workshop on HBV cure. Gut 2015; 64:1314–26. [DOI] [PubMed] [Google Scholar]
- 33.Thimme R, Wieland S, Steiger C, et al. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol 2003; 77: 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV Viral clearance without destruction of infected cells during acute HBV infection. Science 1999; 284: 825–29. [DOI] [PubMed] [Google Scholar]
- 35.Rehermann B Immune responses in hepatitis B virus infection. Semin Liver Dis 2003; 23: 21–38. [DOI] [PubMed] [Google Scholar]
- 36.Hosaka T, Suzuki F, Kobayashi M, et al. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology 2013; 58: 98–107. [DOI] [PubMed] [Google Scholar]
- 37.Papatheodoridis GV, Idilman R, Dalekos GN, et al. The risk of hepatocellular carcinoma decreases after the first 5 years of entecavir or tenofovir in Caucasians with chronic hepatitis B. Hepatology 2017; 66:1444–53. [DOI] [PubMed] [Google Scholar]
- 38.Sugarman J, Revill P, Zoulim F, et al. Ethics and hepatitis B cure research. Gut 2017; 66: 389–92. [DOI] [PubMed] [Google Scholar]
- 39.Levrero M, Testoni B, Zoulim F. HBV cure: why, how, when? Curr Opin Virol 2016; 18:135–43. [DOI] [PubMed] [Google Scholar]
- 40.Hepatitis B Foundation. Drug watch. http://www.hepb.org/treatment-and-management/drug-watch (accessed Oct 1, 2018). [Google Scholar]
- 41.Blumberg BS, Alter HJ, Visnich S. A “new” antigen in leukemia sera. JAMA 1965; 191: 541–46. [DOI] [PubMed] [Google Scholar]
- 42.Strick-Marchand H, Dusseaux M, Darche S, et al. A novel mouse model for stable engraftment of a human immune system and human hepatocytes. PLoS One 2015; 10: e0119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kah J, Koh S, Volz T, et al. Lymphocytes transiently expressing virus-specific T cell receptors reduce hepatitis B virus infection. J Clin Invest 2017; 127: 3177–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klumpp K, Shimada T, Allweiss L, et al. Efficacy of NVR 3–778, alone and in combination with pegylated interferon, vs entecavir in uPA/SCID mice with humanized livers and HBV infection. Gastroenterology 2018; 154: 652–62. [DOI] [PubMed] [Google Scholar]
- 45.Lucifora J, Xia Y, Reisinger F, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014; 343:1221–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M, Li J, Yue L, et al. Establishment of cre-mediated HBV recombinant cccDNA (rcccDNA) cell line for cccDNA biology and antiviral screening assays. Antiviral Res 2018; 152: 45–52. [DOI] [PubMed] [Google Scholar]
- 47.Xia Y, Stadler D, Lucifora J, et al. Interferon-gamma and tumor necrosis factor-alpha produced by T cells reduce the HBV persistence form, cccDNA, without cytolysis. Gastroenterology 2016; 150:194–205. [DOI] [PubMed] [Google Scholar]
- 48.Chisari FV, Mason WS, Seeger C. Comment on “Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA”. Science 2014; 344:1237. [DOI] [PubMed] [Google Scholar]
- 49.Xia Y, Lucifora J, Reisinger F, Heikenwalder M, Protzer U. Response to Comment on “Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA”. Science 2014; 344:1237. [DOI] [PubMed] [Google Scholar]
- 50.Seeger C, Sohn JA. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol Ther 2016; 24:1258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bloom K, Ely A, Arbuthnot P. A T7 endonuclease I assay to detect talen-mediated targeted mutation of HBV cccDNA. Methods Mol Biol 2017; 1540: 85–95. [DOI] [PubMed] [Google Scholar]
- 52.Yang HC, Kao JH. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerg Microbes infect 2014; 3: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belloni L, Pollicino T, De Nicola F, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA 2009; 106:19975–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol 2009; 51: 581–92. [DOI] [PubMed] [Google Scholar]
- 55.Lucifora J, Arzberger S, Durantel D, et al. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol 2011; 55: 996–1003. [DOI] [PubMed] [Google Scholar]
- 56.Pollicino T, Belloni L, Raffa G, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 2006; 130: 823–37. [DOI] [PubMed] [Google Scholar]
- 57.Guo YH, Li YN, Zhao JR, Zhang J, Yan Z. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics 2011; 6: 720–26. [DOI] [PubMed] [Google Scholar]
- 58.Decorsiere A, Mueller H, van Breugel PC, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016; 531: 386–89. [DOI] [PubMed] [Google Scholar]
- 59.Livingston CM, Ramakrishnan D, Strubin M, Fletcher SP, Beran RK. Identifying and characterizing interplay between hepatitis B virus X protein and Smc5/6. Viruses 2017; 9: E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy CM, Xu Y, Li F, et al. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep 2016; 16: 2846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wooddell CI, Yuen MF, Chan HL, et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017; 9: doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maepa MB, Ely A, Grayson W, Arbuthnot P. Sustained inhibition of HBV replication in vivo after systemic injection of AAVs encoding artificial antiviral primary microRNAs. Mol Ther Nucleic Acids 2017; 7:190–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Lu W, Zheng Y, et al. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest 2016; 126:1079–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M, Sohn JA, Seeger C. Distribution of hepatitis B virus nuclear DNA. J Virol 2018; 92: e01391–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai D, Wang X, Yan R, et al. Establishment of an inducible HBV stable cell line that expresses cccDNA-dependent epitope-tagged HBeAg for screening of cccDNA modulators. Antiviral Res 2016; 132: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo JT, Guo H. Metabolism and function of hepatitis B virus cccDNA: implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res 2015; 122: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li F, Cheng L, Murphy CM, et al. Minicircle HBV cccDNA with a Gaussia luciferase reporter for investigating HBV cccDNA biology and developing cccDNA-targeting drugs. Sci Rep 2016; 6: 36483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas E, Liang TJ. Experimental models of hepatitis B and C- new insights and progress. Nat Rev Gastroenterol Hepatol 2016; 13: 362–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xia Y, Carpentier A, Cheng X, et al. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J Hepatol 2017; 66: 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaneko S, Kakinuma S, Asahina Y, et al. Human induced pluripotent stem cell-derived hepatic cell lines as a new model for host interaction with hepatitis B virus. Sci Rep 2016; 6: 29358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakurai F, Mitani S, Yamamoto T, et al. Human induced-pluripotent stem cell-derived hepatocyte-like cells as an in vitro model of human hepatitis B virus infection. Sci Rep 2017; 7: 45698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ortega-Prieto AM, Skelton JK, Wai SN, et al. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat Commun 2018; 9: 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allweiss L, Volz T, Giersch K, et al. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut 2018; 67: 542–52. [DOI] [PubMed] [Google Scholar]
- 74.Dusseaux M, Masse-Ranson G, Darche S, et al. Viral load affects the immune response to HBV in mice with humanized immune system and liver. Gastroenterology 2017; 153:1647–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui X, Guo JT, Hu J. Hepatitis B virus covalently closed circular DNA formation in immortalized mouse hepatocytes associated with nucleocapsid destabilization. J Virol 2015; 89: 9021–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glebe D, Aliakbari M, Krass P, Knoop EV, Valerius KP, Gerlich WH. Pre-s1 antigen-dependent infection of Tupaia hepatocyte cultures with human hepatitis B virus. J Virol 2003; 77: 9511–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walter E, Keist R, Niederost B, Pult I, Blum HE. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology 1996; 24:1–5. [DOI] [PubMed] [Google Scholar]
- 78.Burwitz BJ, Wettengel JM, Muck-Hausl MA, et al. Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat Commun 2017; 8: 2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clements JE, Gama L, Graham DR, Mankowski JL, Zink MC. A simian immunodeficiency virus macaque model of highly active antiretroviral treatment: viral latency in the periphery and the central nervous system. Curr Opin HIV AIDS 2011; 6: 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Allweiss L, Dandri M. The role of cccDNA in HBV maintenance. Viruses 2017; 9: E156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu J, Liu K. Complete and incomplete hepatitis B virus particles: formation, function and application. Viruses 2017; 9: doi:10:3390/V9030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giersch K, Allweiss L, Volz T, Dandri M, Lutgehetmann M. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol 2017;66:460–62. [DOI] [PubMed] [Google Scholar]
- 83.van Bommel F, Bartens A, Mysickova A, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 2015; 61: 66–76. [DOI] [PubMed] [Google Scholar]
- 84.Wang J, Shen T, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016; 65: 700–10. [DOI] [PubMed] [Google Scholar]
- 85.Asabe S, Wieland SF, Chattopadhyay PK, et al. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J Virol 2009; 83: 9652–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ebert G, Poeck H, Lucifora J, et al. 5’ triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology 2011; 141: 696–706. [DOI] [PubMed] [Google Scholar]
- 87.Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol 2005; 79:7269–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Janssen HLA, Brunetto MR, Kim YJ, et al. Safety, efficacy and pharmacodynamics of vesatolimod (GS-9620) in virally suppressed patients with chronic hepatitis B. J Hepatol 2018; 68: 431–40. [DOI] [PubMed] [Google Scholar]
- 89.Suresh M, Korolowicz KE, Balarezo M, et al. Antiviral efficacy and host immune response induction during sequential treatment with SB 9200 followed by entecavir in woodchucks. PLoS One 2017; 12: e0169631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bengsch B, Martin B, Thimme R. Restoration of HBV-specific CD8+ T cell function by PD-1 blockade in inactive carrier patients is linked to T cell differentiation. J Hepatol 2014; 61:1212–19. [DOI] [PubMed] [Google Scholar]
- 91.Boni C, Fisicaro P, Valdatta C, et al. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J Virol 2007; 81: 4215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fisicaro P, Valdatta C, Massari M, et al. Antiviral intrahepatic T-cell responses can be restored by blocking programmed death-1 pathway in chronic hepatitis B. Gastroenterology 2010; 138: 682–93. [DOI] [PubMed] [Google Scholar]
- 93.Nebbia G, Peppa D, Schurich A, et al. Upregulation of the Tim-3/galectin-9 pathway of T cell exhaustion in chronic hepatitis B virus infection. PLoS One 2012; 7: e47648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schurich A, Khanna P, Lopes AR, et al. Role of the coinhibitory receptor cytotoxic T lymphocyte antigen-4 on apoptosis-prone CD8 T cells in persistent hepatitis B virus infection. Hepatology 2011; 53:1494–503. [DOI] [PubMed] [Google Scholar]
- 95.Isogawa M, Chung J, Murata Y, Kakimi K, Chisari FV. CD40 activation rescues antiviral CD8(+) T cells from PD-1-mediated exhaustion. PLoS Pathog 2013; 9: e1003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maier H, Isogawa M, Freeman GJ, Chisari FV. PD-1:PD-L1 interactions contribute to the functional suppression of virus-specific CD8+ T lymphocytes in the liver. J Immunol 2007; 178: 2714–20. [DOI] [PubMed] [Google Scholar]
- 97.Schurich A, Pallett LJ, Lubowiecki M, et al. The third signal cytokine IL-12 rescues the anti-viral function of exhausted HBV-specific CD8 T cells. PLoS Pathog 2013; 9: e1003208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gehring AJ. New treatments to reach functional cure: rationale and challenges for emerging immune-based therapies. Best Pract Res Clin Gastroenterol 2017; 31: 337–45. [DOI] [PubMed] [Google Scholar]
- 99.Pallett LJ, Davies J, Colbeck EJ, et al. IL-2(high) tissue-resident T cells in the human liver: sentinels for hepatotropic infection. J Exp Med 2017; 214:1567–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stegmann KA, Robertson F, Hansi N, et al. CXCR6 marks a novel subset of T-bet(lo) Eomesjhi) natural killer cells residing in human liver. Sci Rep 2016; 6: 26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boeijen LL, Hoogeveen RC, Boonstra A, Lauer GM. Hepatitis B virus infection and the immune response: the big questions. Best Pract Res Clin Gastroenterol 2017; 31: 265–72. [DOI] [PubMed] [Google Scholar]
- 102.Rehermann B, Chang KM, McHutchinson J, et al. Differential cytotoxic T-lymphocyte responsiveness to the hepatitis B and C viruses in chronically infected patients. J Virol 1996; 70:7092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Knolle PA, Thimme R. Hepatic immune regulation and its involvement in viral hepatitis infection. Gastroenterology 2014; 146:1193–207 [DOI] [PubMed] [Google Scholar]
- 104.Das A, Hoare M, Davies N, et al. Functional skewing of the global CD8 T cell population in chronic hepatitis B virus infection. J Exp Med 2008; 205: 2111–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fisicaro P, Barili V, Montanini B, et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat Med 2017; 23: 327–36. [DOI] [PubMed] [Google Scholar]
- 106.Urbani S, Boni C, Amadei B, et al. Acute phase HBV-specific T cell responses associated with HBV persistence after HBV/HCV coinfection. Hepatology 2005; 41: 826–31. [DOI] [PubMed] [Google Scholar]
- 107.Das A, Ellis G, Pallant C, et al. IL-10-producing regulatory B cells in the pathogenesis of chronic hepatitis B virus infection. J Immunol 2012; 189: 3925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu D, Fu J, Jin L, et al. Circulating and liver resident CD4+CD25+ regulatory T cells actively influence the antiviral immune response and disease progression in patients with hepatitis B. J Immunol 2006;177:739–47 [DOI] [PubMed] [Google Scholar]
- 109.Pallett LJ, Gill US, Quaglia A, et al. Metabolic regulation of hepatitis B immunopathology by myeloid-derived suppressor cells. Nat Med 2015; 21: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peppa D, Gill US, Reynolds G, et al. Up-regulation of a death receptor renders antiviral T cells susceptible to NK cell-mediated deletion. J Exp Med 2013; 210: 99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bertoletti A, Ferrari C. Adaptive immunity in HBV infection. J Hepatol 2016; 64(1 suppl): S71–83. [DOI] [PubMed] [Google Scholar]
- 112.Vanwolleghem T, Hou J, van Oord G, et al. Re-evaluation of hepatitis B virus clinical phases by systems biology identifies unappreciated roles for the innate immune response and B cells. Hepatology 2015; 62: 87–100. [DOI] [PubMed] [Google Scholar]
- 113.Paul S, Dickstein A, Saxena A, et al. Role of surface antibody in hepatitis B reactivation in patients with resolved infection and hematologic malignancy: a meta-analysis. Hepatology 2017; 66: 379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perrillo RP, Gish R, Falck-Ytter YT. American Gastroenterological Association Institute technical review on prevention and treatment of hepatitis B virus reactivation during immunosuppressive drug therapy. Gastroenterology 2015; 148: 221–44. [DOI] [PubMed] [Google Scholar]
- 115.Mason WS, Gill US, Litwin S, et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 2016; 151: 986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tjwa ET, Zoutendijk R, van Oord GW, et al. Similar frequencies, phenotype and activation status of intrahepatic NK cells in chronic HBV patients after long-term treatment with tenofovir disoproxil fumarate (TDF). Antiviral Res 2016; 132: 70–75. [DOI] [PubMed] [Google Scholar]
- 117.Liu J, Zhang E, Ma Z, et al. Enhancing virus-specific immunity in vivo by combining therapeutic vaccination and PD-L1 blockade in chronic hepadnaviral infection. PLoS Pathog 2014; 10: e1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bohne F, Chmielewski M, Ebert G, et al. T cells redirected against hepatitis B virus surface proteins eliminate infected hepatocytes. Gastroenterology 2008; 134: 239–47 [DOI] [PubMed] [Google Scholar]
- 119.Gehring AJ, Xue SA, Ho ZZ, et al. Engineering virus-specific T cells that target HBV infected hepatocytes and hepatocellular carcinoma cell lines. J Hepatol 2011; 55:103–10. [DOI] [PubMed] [Google Scholar]
- 120.Koh S, Shimasaki N, Suwanarusk R, et al. A practical approach to immunotherapy of hepatocellular carcinoma using T cells redirected against hepatitis B virus. Mol Ther Nucleic Acids 2013; 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krebs K, Bottinger N, Huang LR, et al. T cells expressing a chimeric antigen receptor that binds hepatitis B virus envelope proteins control virus replication in mice. Gastroenterology 2013; 145: 456–65. [DOI] [PubMed] [Google Scholar]
- 122.Qasim W, Brunetto M, Gehring AJ, et al. Immunotherapy of HCC metastases with autologous T cell receptor redirected T cells, targeting HBsAg in a liver transplant patient. J Hepatol 2015; 62: 486–91. [DOI] [PubMed] [Google Scholar]
- 123.Tu T, Budzinska MA, Vondran FWR, Shackel NA, Urban S. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via NTCP-dependent uptake of enveloped virus particles. J Virol 2018; 92: pii: e02007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao LH, Liu X, Yan HX, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun 2016; 7:12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alter H, Block T, Brown N, et al. A research agenda for curing chronic hepatitis B virus infection. Hepatology 2018; 67:1127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Block TM, Alter H, Brown N, et al. Research priorities for the discovery of a cure for chronic hepatitis B: report of a workshop. Antiviral Res 2018; 150: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.NIH. Research to advance HBV cure: HIV/HBV co-infection and HBV mono-infection (R01 clinical trial not allowed). https://grants.nih.gov/grants/guide/pa-files/PAS-19-097.html?utm_campaign=+35600459&utm_content=&utm_medium=email&utm_source=govdelivery&utm_term=) (accessed Dec 1, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.