Abstract
The tumor microenvironment can be realistically viewed as an active battle ground between the host immune system and the growing tumor cells. This reactive space surrounding the tumor possesses several possibilities and facilitates the progression of a tumor from a neoplastic stage to that of metastasis. The contemporary approach of understanding the cancer biology from a “within the cell” perspective has been largely challenged with complex and intricate “outside the cell” events. Thus understanding the biology of the tumor microenvironment has been of scientific and clinical interest. Small non-coding microRNAs with a pleotropic and wide range of cellular gene targets can be reasonably hypothesized to regulate the events of carcinogenesis and progression. MicroRNAs have been investigated in different cancer models, and evidence of their involvement in the regulation of the tumor microenvironment has been of much interest. In particular, a major interest has been exploring the role of the tumor microenvironment in regulating the interaction of cancer cells with surrounding stromal components and the effect of such interactions on the cancer cells. Fine-tuned regulation by these microRNAs extends our contemporary understanding of these small biomolecules in epigenetic regulations. This review focuses on microRNAs that are dysregulated in ovarian carcinomas, their effect on the components of the tumor microenvironment, and the correlation of their heterogeneous expression profiles with disease severity and prognosis in patients. In addition, this paper also discusses the differential expression of exosomal microRNAs that are known to link the cancer cell with its microenvironment, facilitating the development of an improved prognostic/diagnostic marker and effective therapeutic regime.
Keywords: microRNAs, cancer-associated fibroblasts, tumor-infiltrating lymphocytes, endothelial cells, endometriosis, chemoresistance, mesenchymal cells, myeloid-derived suppressor cells
Introduction
Ovarian cancers are among the global leading cause of malignancy-associated death in women1. Each year approximately 240,000 women are diagnosed with ovarian cancer across the globe, reflecting the high prevalence and associated fatality of the disease2. The most common subtypes of these ovarian carcinomas include epithelial ovarian carcinomas (80–90%), with few cases of germ cell cancers (1–2%)3. Among the epithelial ovarian carcinomas, serous ovarian cancers (SOCs) or high-grade serous ovarian cancers (HGSOCs) result in most of the gynecological malignancies4. Research on ovarian cancer over the past few years has associated geographical variations and population-specific susceptibility factors with disease progression, and therefore needs further research and analysis4.
The most commonly adopted clinical intervention for ovarian cancers involves “mass debulking,” sometimes combined with chemotherapy and radiotherapy3. Recently the use of hormone and antibody-based treatment methodologies in patients with ovarian cancer has gained momentum5. However, these approaches of treatment still depend on the patients’ health constitution and prior medical history, and have been often associated with side-effects such as fatigue, nausea, hair loss, neuropathies, and inflammatory bowel disease, etc5. A major constraint in understanding the disease biology is the association of ovarian carcinomas with low survival rate post diagnosis, low to poor prognosis of the disease, and rapid tumor relapse (based on the repopulation hypothesis). Often cases of aggressive ovarian cancers are diagnosed among women in the age group of 50–70 years, with approximately 80% of these cases being diagnosed at the late stage of cancer4. In addition, patients with ovarian cancers are reported to have low mean 5-year survival rates; these typically range between 20% and 40%, especially for those with HGSOCs5.
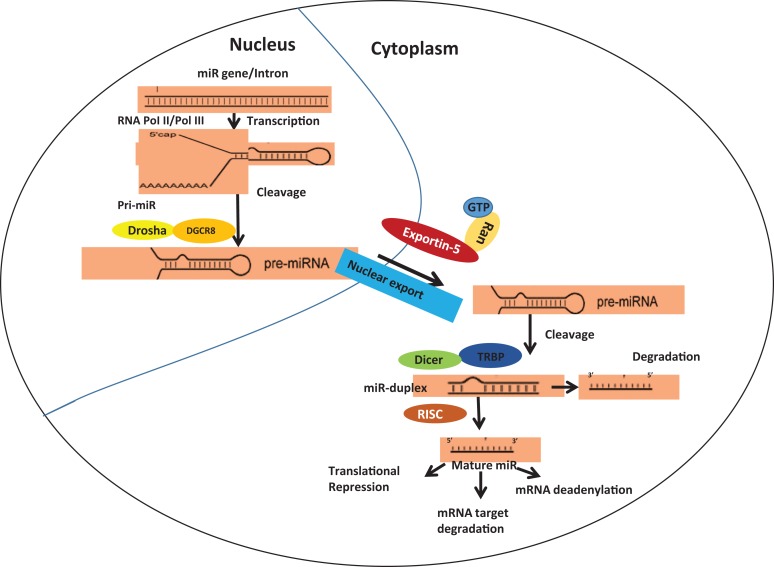
microRNAs (hereafter referred as miRs) are small non-coding ribonucleic acid (ncRNA) molecules (18–25nt long) with an ability to regulate the expression of several genes either by pre-transcriptional repression (via chromatin remodeling, which has been observed rarely) or by directing messenger RNA (mRNA) degradation of the target gene6. Such degradation of mRNA involves co- and/or post-translational modifications such as deadenylation from the 3’end, decapping of the 5’ end, mRNA cleavage, etc6. The detailed biogenesis and maturation of miRs have been discussed elsewhere (Figure 1)7–9. Several extensive studies have identified the role of miRs in immune functioning, cell growth and development, tissue remodeling, neuronal processes, cancers, and other disease models10. For any gene, the binding of miRs to the miR-binding site at the 3’untranslated region (UTR) region results in translational repression of the mRNA7. This forms one of the major axes of miR targeting. miRs are known to target several mRNAs either through complete or incomplete base pairing at the seed region7. The other aspect of the role of miRs in chromatin remodeling is debated and needs more reproducible experimental validations.
Figure 1.
Schematic representation of miR biogenesis and maturation. The biogenesis of miR involves pre-miR formation within the nucleus which subsequently gets processed and forms mature miR in the cytoplasm.
Recent advances in mathematical modeling and bioinformatics-based predictions have shown that miRs can target approximately 50% of all protein-coding genes. The abundant levels of miRs and their potential in regulating gene expression make these small molecules of significant physiological importance8. Mature miRs in the cytoplasm are known to regulate cellular processes, and abnormalities therein have been shown to cause several diseases. For example, inhibition of cell cycle check points, evasion of apoptosis-inducing factors, and resistance to cellular senescence by miRs have been widely associated with cancer onset and progression8. In addition, aberrations in the process of miR biogenesis and maturation have been shown to cause carcinogenesis11. For example, Tar-binding protein-2 (TARBP2) has been shown to incorporate a frameshift mutation resulting in a truncated protein product both in sporadic and familial carcinomas11. The lowered expression of native TARBP2 protein leads to downregulation of DICER-1-mediated maturation and processing of endogenous miRs in the cytoplasm11. Further, events involving defective or abnormal nuclear processing of pre-miRs have been correlated to cancer onset and progression11. For example, dysregulated functioning or expression of role of BCDIN3D (BCDIN3 domain-containing RNA methyltransferase) interferes with the O-methylation of 5’-monophosphate in pre-miRs leading to blockage of processing and maturation of miRs (Figure 1)11. Several other cases of errors in miR biogenesis and carcinogenesis have been discussed by others, and is beyond the scope of this review12–17. A change in the expression of several miRs has been shown in cancerous and non-cancerous cells. The differential expression of miR-10b, miR-373, and miR-520c and their role in metastasis18, expression of miR-374a, miR-200, and miR-22 and their role in epithelial and mesenchymal transitions (EMT)19, and the role of miR-126 in cancer-associated angiogenesis20 are a few such examples. In addition, approximately 186 miR genes are positioned in proximity to the fragile gene sites and other cancer-associated genes, indicating a significant role of miRs in carcinogenesis21.
In the present scenario, a need to revisit the molecular biology of ovarian carcinomas has become essential. In this review we particularly aim to decipher the factors associated with rapid disease progression and discuss the challenges faced by patients caused by the severity and aggressive metastatic ability of these cancers. The composition of the tumor microenvironment (TME) involving different modified stromal components, evasion of host immune cells, upregulation of pro-cancerous and downregulation of anti-cancerous molecular regulators, together facilitate the process of rapid transition of tumor from the neoplastic stage to that of metastatic stage (Figure 2)22. Very recently, miRs are being viewed as molecules responsible for reprogramming of the cancer microenvironment that can facilitate the growth and progression of cancer23. Therefore, critical analysis of the miRs in the cancer microenvironment holds significance, and in this paper we particularly discuss the role of miRs associated with ovarian cancers. These miRs are known for their ability to regulate the peritoneal metastasis, which forms the characteristic and confounding ability for progression of ovarian cancers23.
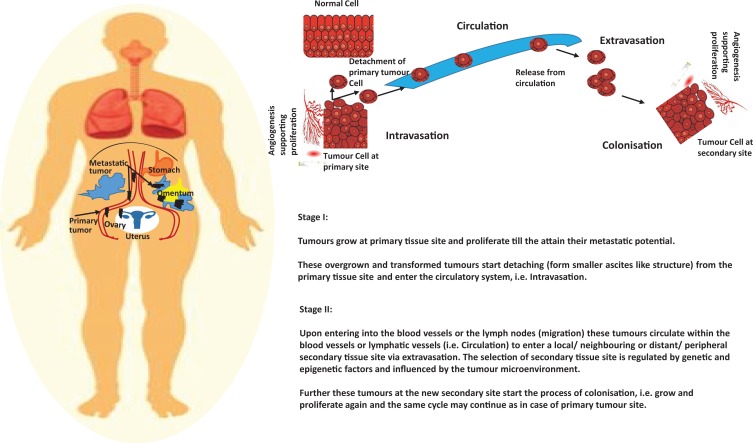
Figure 2.
Schematic representation of ovarian cancer progression and metastasis. The four stages of cancer progression and metastasis have been explained schematically which involves invasion of endothelial cells from primary tissue site, circulation, extravasation of endothelial cells at the site of secondary tumor growth, colonization, and further growth of tumor at the secondary tissue site.
Cancer Hallmarks and the Tumor Microenvironment
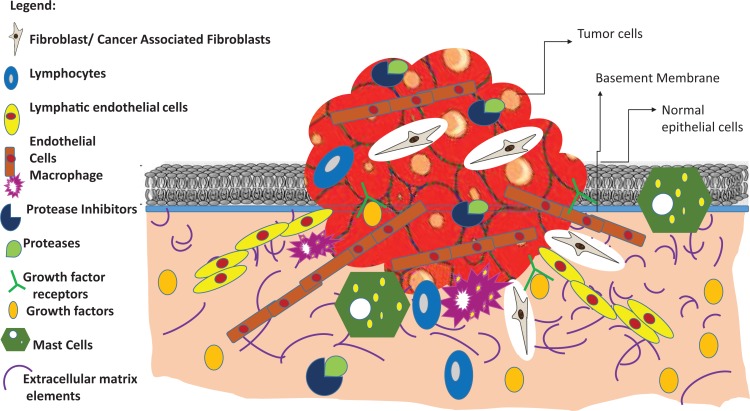
The adjoining tissues and extracellular matrix components surrounding the tumor are commonly referred to as the TME (Figure 3)24. The TME (approximately 90% of the cancer cell stroma) broadly consists of fibroblasts, immune cells, mechanically supporting cells, blood vessels, extracellular matrix, and a wide range of signaling molecules (Figure 3)25. This unique niche harbors the primary tumor that thrives, propagates, and undergoes cancerous transformations as a result of several genetic and epigenetic modifications25. One of the major roles of the components making up the microenvironment is to transform normal cells into cancer cells, which show characteristics otherwise known as “Cancer Hallmarks”26. These distinct phenotypic and molecular patterns observed specifically in cancer cells involve increased angiogenesis, abnormal vasculature of blood vessel, recruitment of endothelial cells promoted by vascular epidermal growth factors (VEGF), and suppression of apoptosis by blockage of tumor suppressor genes such as p53/p21 and retinoblastoma (pRb). In addition, some other hallmarks include blockage of cell cycle checkpoints such as cyclin-dependent kinases and cyclin D, and rapid cell proliferation and replicative immortality accelerated by sustained signaling from cell survival pathways such as PTEN/Akt, c-myc, and MAPK/ERKs. Several recent studies have also shown that genomic instability due to accumulation of mutations, and increased invasiveness leading to metastasis and evasion of cytotoxic host immune response by modulating activity of CD8 T cells, CD4 T cells, and T regulatory cells are emerging cancer hallmarks27.Therefore, this paper focuses on elucidating the role of putative and experimentally validated miRs in the attainment of the cancer cell hallmarks especially related to invasiveness and metastasis.
Figure 3.
Schematic representation of tumor microenvironment. Some of the major components of the tumor microenvironment include tumor-infiltrating lymphocytes, cancer-associated fibroblasts, macrophages, mast cells, cancer stem cells, myeloid-derived suppressor cells, proteases and protease inhibitors, growth factors and their cognate receptors etc.
Recent study of the epithelial–stromal crosstalk in ovarian cancers has indicated that miR-16-5p and miR-124-3p are differential expressed in ovarian tumor cells along with the surrounding epithelial and stromal cells28. In addition, the identification of miR-124-3p has highlighted a novel biomarker for ovarian cancers28. Emerging reports in women from the US established that there is a link between endometriosis and susceptibility for ovarian cancer. Pearce et al. have shown that a combination of environmental and genetic risk factors increase the susceptibility of women for clear-cell adenocarcinoma, low-grade serous, and ovarian endometrioid cancers29. The study carried out among women from the US has shown that approximately 73% of women with some form of self-reported endometriosis have a 4–9% lifetime risk of developing ovarian cancers29. This figure is significant considering that only 1.37% women in the US were identified as having a lifetime risk of developing ovarian cancers, with difference in severity varying among these women29.The role of differential expression of miR in endometriosis is now being associated with an increased risk for ovarian cancers.
It is evident that the crosstalk between “tumor and its niche” is critical to tumor integrity and propagation. Thus a possible bidirectional interaction may answer several confounding questions pertaining to the onset and progression of ovarian cancers. In addition, the nature of influence exerted by the TME remains largely unclear. One hypothesis is that the cancer cells influence components of the microenvironment in order to remodel both functional and structural properties. Some of these include facilitating aberrant cell growth, invasiveness, and propagation to secondary tissue locations. In addition, research is underway to explore any possibilities of cancer cell-independent regulation of the TME. Nonetheless, it is largely accepted that the constant and close interaction of the tumor and its microenvironment holds promise for development of therapeutics and evaluation of treatment outcomes for different forms of cancers. miR-based regulation of the TME is based on the early evidence of miR-302-mediated reprogramming of somatic cells into induced pluripotent stem cells30. Recent evidence of miR-9/9 and miR-124-mediated reprogramming of fibroblasts into neurons has further substantiated the role of small molecular players in cellular remodeling and carcinogenesis31. Although no current information is available about the exact mechanistic details of how these miRs influence reprogramming, further research into these aspects may be insightful for regenerative medicine. Thus the cellular and genetic plasticity conferred by these small molecule regulators remain central to oncobiology and are of high importance in the onset and progression of cancer.
miRs in the Distant and Local Tumor Microenvironment
Ovarian cancers have a characteristic property of rapid cancer progression along with formation of ascites and metastasis within the peritoneal cavity instead of the usual hematogenous metastatic spread that is seen in most other forms of cancer32. Cancer-associated fibroblasts (CAFs), endothelial cells, cancer-associated mesothelial cells, tumor-infiltrating lymphocytes (TILs), myeloid-derived suppressor cells (MDSCs), and mesenchymal stem cells (MSCs) form the TME33. In contrast, similar components found in the omentum (adipose-rich three-dimensional layered structures surrounding both the inner and outer layers of the peritoneal cavity) form the most preferred distant metastatic location of ovarian cancer cells (Figure 3)34. These consequential and direct interactions lead to a range of differentially regulated miRs comprising both the up- and downregulated miRs. However, this is in contrast to the overall profile of the miRs in cancer cells, where these small non-coding molecules are known to be downregulated with an overall tumor-suppressive phenotype35.
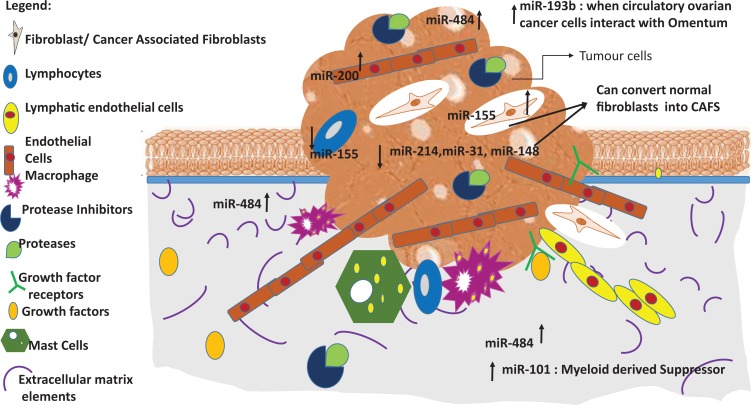
Fibroblast is one of the major stromal components that regulates a wide range of carcinogenic phenotypes such as angiogenesis, metastasis, and epithelial–mesenchymal transitions (EMTs) in the tumor cells36. Aberrant cell growth and onset of carcinogenesis result in expression of specific miRs that can regulate pro-cancerous functions of the CAFs34. These CAFs, when grown in a non-malignant environment, still exhibit their transformed characteristics, suggesting that induction and maintenance of fibroblasts into CAFs are largely irreversible and sustained epigenetic changes34. In the local ovarian TME CAFs are known to express higher levels of pro-inflammatory miR-15537 that is directly regulated by nuclear factor-kβ, i.e. NF-kβ34. The cancer cells interacting with the CAFs exhibit low levels of miR-214, miR-31, and miR-148. miR-31 is known to be downregulated in ovarian cancers35. Further, miR-31 is known to inhibit the chemokine C-C motif ligand 5 (CCL5) and promote the expression of AT-rich sequence binding protein-2 that ultimately results in increased invasiveness of endometrium and cancer growth35. miR-148 regulates the expression of Wingless-Type MMTV integration site family protein 10B (WNT10B)35. Together, miRs with differential expression in the local TME and those in the distant omentum influence the TME of ovarian cancers35. Mitra et al. have shown that a precise combinatorial choice of three different miRs (miR-214, miR-31, and miR-155) can reprogram normal fibroblasts to CAFs (Figure 4)37. miR-155 in the distant omentum is known to promote conversion of normal omental fibroblasts into CAFs35. This suggests that a cohort of miRs decide the fate of fibroblasts in the TME. Apart from the selective upregulation of certain miRs by cancer cells, fine tuning between the pro-and anti-cancerous miRs is also a key process involved in regulating the disease progression. For example, downregulation of miR-214 and miR-31 along with the upregulation of miR-155 can successfully promote CAFs and establish the miRs as regulated effector molecules35.
Figure 4.
Differential expression of miRs in tumor microenvironment. Of all the represented miRs, miR-214, miR-31, and miR-155 can act synergistically to reprogram normal fibroblasts into cancer-associated fibroblasts.
Endothelial cells in the TME promote carcinogenesis by increased angiogenesis and metastasis38. Certain miRs expressed by the endothelial cells can even inhibit several processes involved in carcinogenesis. For example, expression of miR-200 by the endothelial cells reduces migration and angiogenesis, resulting in the limited metastatic potential of ovarian carcinomas39. In addition, downregulated expression of miR-200 in ovarian cancer is known to target interleukin-8 (IL-8), resulting in a low abundance of endothelial cells40 and inhibiting the recruitment of endothelial cells to the site of the growing tumor. miR-200 has also been commonly correlated with EMT in most cancer types. An analysis using The Cancer Genome Atlas has revealed that lower levels of miR-200 and miR-484 can be related to an inferior prognosis of ovarian cancers41. miR-484 targets vascular endothelial growth factor-B (VEGFB)42 and vascular endothelial growth factor receptor-2 (VEGFR2), and thus its reduced expression has been directly linked to the progression of cancer40.
Different survival mechanisms are known to be used by cancer cells for prolonged survival of the cancer cells, especially endometriotic cells. Hypoxia is a widely correlated survival and invasion mechanism used by endometriotic cells43. Several signal transduction pathways such as STAT3 (signal transducer and activator of transcription 3)44, TGF-β45, and autophagy45 have been implicated as key mechanisms deployed by these cells for survival45. There is a decrease in the expression of miR-126 in endometriosis, endometrioid ovarian cancers, and clear-cell carcinomas. Indeed miR-126, expressed both in cancer cells and the TME, may be a potential therapeutic target for ovarian cancers.
miRs that are differentially expressed in endometriosis and endometrioid ovarian cancer cells include miRs with elevated expression such as miR-205, miR-30e-5p, miR-325, miR-492, miR-637, and miR-16-5p, and miRs with reduced expression such as let-7 f. For cases involving both clear-cell and endometrioid ovarian cancers, miRs such as miR-200a, -200c, -21, and -575 were observed to be upregulated, and miR-1, -100, -101, -105, -125a, -133a, and -137 are some that are downregulated. A detailed table summarizing these key upregulated or downregulated miRs in cases involving endometriosis and ovarian cancers can be found in Wendel et al.’s review article46.
Mesothelial cells form an integral part of the peritoneal layer of the abdominal cavity that covers peritoneum, the large and small bowel serosa, and the omentum47. As ovarian cancer cells often migrate to the peritoneal cavity and omentum, these mesothelial cells covering these preferred sites of metastasis are often driven by pro-cancerous transformations in the case of ovarian cancers. Cancer-associated mesothelial cells serve as the first point of contact for the ovarian cancer cells before they can even interact with the underlying fibroblasts48. In ovarian cancers, miR-200 is known to stimulate the mesothelial cells via TGF-β (Tumor Growth Factor-β)-mediated increase in the expression of MMP-2 and MMP-9 (Matrix Metallo-Proteinases)49. In another study, it has been reported that an ectopic expression of miR-200 in mesothelial cells inhibits the implantation and metastasis of ovarian cancer cells50. Together, these suggest a significant role of miR-200 in the onset and progression of ovarian cancers.
miR-193b is another such miR which is known to target urokinase-type plasminogen activator protein (uPA) during direct interaction of ovarian cancer cells with the mesothelial cells of the omentum. This targeted action by miR-193b downregulates the expression of uPA via the repressive methylation effect of DNA methyl transferase-151. Although several 2D in vitro assays have noted the role of miR-193b, it is interesting to understand that events such as metastasis and the spread of cancer cells can be precisely and accurately studied using a 3D organotypic model, as it resembles the interaction of ovarian cancer cells and omentum. One such study by Mitra et al., using an organotypic 3D ovarian cancer model, has identified a significant downregulation of miR-193b in progressing and metastasizing ovarian cancer cells52. Further, the authors have shown that downregulated miR-193b is critical for colonization of ovarian cancer cells followed by adhesion, invasion, migration, and colony formation of metastatic cells46.
TILs are the some of the immune cells that are recruited by cancer cells to remodel the TME. Remodeling of the TME by TILs results in evasion of recognition by the host immune system. In addition, it reduces the cytotoxicity triggered by the host immune system and interferes in the wound-healing process of the stroma surrounding the tumor cells53. Some TILs have been found to facilitate tumorigenesis52,53, and therefore any potential modifications of these TILs by miRs in the TME are of importance.
M2 macrophages, or tumor-associated macrophages (TAMs), are tumor-suppressive and anti-inflammatory immune cells that promote tumor growth and angiogenesis in addition to the evasion of immune response via specific cytokines such as IL-10, TGF-β and arginase54. In the case of TAMs associated with ovarian cancer, miR-155 is known to be downregulated, and upon exogenous restoration of miR-155 (anti-tumor responses) these TAMs promote cytotoxic T-cell functions55. miR-155 targets SOCS-1 (suppressor of cytokine signaling 1), SHIP-1 (Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 1), and C/EBPβ (CCAAT-enhancer-binding proteins), which results in transcriptional repression of cytokines related to T-helper type 1 cells49. In addition to the cytotoxic T cells (CD8+) and helper T cells (CD4+)-mediated regulation of TILs in the TME, T regulatory cells (Treg cells) are also known to regulate the expression of TILs. For example, Treg cells have been associated with dysregulated expression of miRs by TILs via Foxp356.
MDSCs form the population of tumor-suppressive immune cells of the myeloid compartment in the TME57. The direct interaction of cancer cells and MDSCs therefore must be regulated and facilitated by the reprogramming of the tumor-suppressive function of these MDSCs58. miRs have been explored for their potency in regulating the tumor-suppressive function of MDSCs. For example, expression of miR-101 is upregulated by MDSCs, which leads to increased stemness of the cancer cells59. This is achieved by miR-101-mediated targeting of co-repressor gene C-terminal binding protein 2 (CtBP2), ultimately resulting in increased expression of Sox-2 and Nanog (stemness markers) and an increase in the metastatic and/or carcinogenic potential of these cancer stem cells (CSCs)53. The rise in stemness markers indicates a higher population of cancer stem cells, which denotes a pro-tumorigenic effect (progression, metastasis, and therapeutic resistance in cancer cells) as these CSCs are better known to be cancer-initiating cells53. The characteristics identifying these CSCs have been debated over the years. The identification of these CSCs is based on markers such as CD44, CD17, CD133, CD24, and aldehyde dehydrogenase-1 A1 (ALDH1).60 Cheng et al. have shown that miR-199a targets 3’-UTR of CD44, resulting inhibition of proliferation, invasion, metastasis, and chemoresistance of ovarian CSCs61. In addition, miR-214 targets the 3’-UTR of p53 tumor suppressor gene, leading to self-renewal and expansion of ovarian CSCs62. Among the different sub-populations of CSCs, CD133+ CSCs show a downregulation of miR-200a and thereby limits the progression of cancer63. A direct correlation between the density of MDSCs/CSCs in the TME and the stage and severity of cancer can be effectively used in predicting disease prognosis and life expectancy post diagnosis57.
MSCs are stem cells actively recruited to the TME and are known for their pro-tumorigenic effects in solid tumors64. Increased recruitment of MSCs promotes angiogenesis, metastasis, and immune evasion by the cancer cells, and thus understanding the miR-based regulation of the MSCs in ovarian cancers holds significance58. Although there is no direct evidence supporting the role of miR in the regulation of MSCs, preliminary research has shown that MSCs can inhibit the expression of miR-200b and miR-200c (miR-200 family of genes) in ovarian cancer cells65. This is achieved via stimulation of TGF-β by the MSCs, which results in the reduction of miR-200b and miR-200c in MCF-7 cells. This in turn upregulates ZEB1/ZEB2 (zinc finger E box binding homeobox 1), resulting in enhanced EMT66.
miR in Chemoresistance of Ovarian Carcinomas
Cellular senescence is an important stage in the cell cycle to avoid aberrant cell growth and check carcinogenic transformations. Cells with accumulating mutations or an unstable genome usually enter senescence following stimulation from shortened telomeres and induction of oncogenic stress factors, usually associated with the expression of senescence-associated markers such as β-galactosidase67. Classically, cellular senescence has always been associated with tumor suppression, but recent studies have shown that cellular senescence can actually promote cancer growth and transformations. Weiner-Gorzel et al. have shown that miR-433 in HGSOC patients has been associated with poor progression-free survival along with lower levels of mitotic arrest deficiency protein MAD2/MAD2L1 and CDK6 (cyclin-dependent kinases 6)68. Although miR-433 was shown to target several cancer-associated genes such as GRB2, SFRP2, CREB1, and HDAC6, the study by Weiner-Gorzel et al. also established the role of miR-433 in the induction of cellular chemoresistance to paclitaxel68. The underlying reason for this miR-433-mediated chemoresistance was shown to be the inhibition of apoptosis via reduction in the levels of phosphorylated retinoblastoma protein and the induction of cellular senescence62.
miR-214 has also been shown to reduce apoptotic cell death, and thus acts as a major link for decline in chemotherapeutic sensitivity, failure of treatment regimens, and enhanced disease progression in patients with ovarian cancer69. Yang et al. have shown that miR-214 inhibits cisplatin-induced cell death in human ovarian cancers69. The major signaling comprises the PTEN/Akt pathway targeted by overexpressed miR-214 in cisplatin-induced chemotherapeutic resistance63.
miR in Hypoxic TME of Ovarian Carcinomas
In most cases the TME features limited or low oxygen availability, leading to a hypoxic TME70. The presence of a hypoxic TME can be reasoned from the extensive oxygen requirement by the rapidly proliferating tumor cells, resulting in the depletion of oxygen in the TME. This substantiates that the hypoxia-driven TME should facilitate tumor progression and metastasis71. Kim et al., using gas analyses, have shown that ovarian ascites have only 2.5% dissolved oxygen in comparison to 15–23% dissolved blood oxygen content72. Both transient and sustained hypoxic conditions have some or other pro-tumorigenic effects, and a sustained hypoxic TME promotes reorganization of cellular processes and other energy-demanding physiological functions73. It is known that HIF-1α and HIF-2α (hypoxia inducible factors) are primary regulators of hypoxic responses in TME and thus must have a role in promoting cancer metastasis and progression74. Stable complexes of HIF-1α/HIF-2α heterodimerize with HIF-1β, inducing the expression of lysyloxidase (LOX). LOX is known to result in cross-linking of collagen, which induces rapid cell migration and exfoliation of cancer cells to reach the peritoneal cavity in case of ovarian cancers75. Joshi et al. in their study have shown that miR-199a has the potency to target the 3’-UTR of HIF-1α/HIF-2α. In addition, the authors have shown that miR-199a derived from the anti-sense strand of DNM2 (dynamin 2, important for endocytosis) is downregulated in the hypoxic TME of ovarian cancers76. Thus, lower levels of DNM2 and miR-199a promote metastasis and migration of tumor cells in ovarian cancers, and thereby the levels of HIF and DNM2 are known to be reciprocally regulated70.
Exosomal miRs: Bridging the Gap between Cancer Cells and TME
Exosomal miRs consist of the miRs which are secreted in small vesicles to the TME by cancer cells, and form the link between the cancer cell’s own secretory miR and the TME77. The wide range of epigenetic changes exerted by exosomal miRs are under investigation. Undoubtedly these epigenetic changes serve as reliable basis for detection and therapeutic development in addition to our understanding of cancer biology71. These exosomal miRs secreted onto the endothelial cells in the TME facilitate angiogenesis, invasiveness, and metastasis71. In a study reported by Vaksman et al. on exosomal miRs in ovarian cancer effusion supernatants, the authors have identified miR-21, miR-23a, miR-23b, miR-29a, miR-99a, miR-125b, miR-200c, miR-320a, and miR-484.
Such a differential expression of exosomal miRs possesses statistical significance and correlation with the pathological characteristics of ovarian cancers, along with survival rates78. Further, miR-21, miR-23b, and miR-29a have been shown to be associated with poor survival. Thus exosomal miRs which are enriched in the serum, such as miR-21, miR-29a, and miR-200c, are known for their diagnostic potential for ovarian carcinomas (Figure 4)72. In addition, higher levels exosomal miR-21 and miR-23a are known to mediate chemoresistance in ovarian cancer cells via regulating the expression of PTEN (phosphatase and tensin homolog deleted on chromosome 10)72.
Contemporary Clinical Management
The present strategy for deciding the treatment of patients with ovarian cancer is based on their health status and the stage of the disease at the time of detection. The focus of several ongoing research works is to adopt and develop an optimal patient-specific treatment regime instead of a usual or routine choice of available drugs and therapeutics79. In addition to the genomic and epigenomic landscape-based studies of ovarian cancer patients, newer studies on cellular and secretory proteomics (e.g. body fluid markers in blood, serum or saliva) are being used for early diagnosis80.
Several reports on the correlation of high-risk genes such as BRAC1 and BRAC2 with the stage of ovarian cancers have proved beneficial in explaining the case of familial ovarian cancers, thereby leading to early diagnosis and improved prognostic abilities81. However, in the case of sporadic ovarian cancers, the approach largely depends on our understanding of the basic disease biology and cues from familial cases, together resulting in a limited treatment sensitivity and disease relapse.
Mathematical models have aided our contemporary treatment regimes and precise prediction of the disease82. For example, a mathematical model correlating the mutations in several high-risk genes can provide an approximate number of years a patient with ovarian cancer may survive with either or both ovaries removed83. Even the average treatment outcome for a cohort of patients with similar health conditions and disease manifestation has been possible using prognostic models. Nevertheless, such models are not precise in giving individual patient-specific information.
There are new reports in the field of ovarian carcinomas which have proved that the origin of cancerous growth at ovaries may actually be at the stage where the cancer cells migrate from the fallopian tubes (alien cancer cells of fallopian tubes). Thus treatments focusing on the mere removal of ovaries or drug-mediated apoptosis of ovarian cells are not the key solutions.84 Patients with family history of ovarian/fallopian or breast cancers with a higher possibility of mutations in BRAC genes are now being investigated for the removal of fallopian tubes first, instead of just the ovary, as the origin of these ovarian cancers has been linked to the fallopian tubes.84 Thus it is of scientific interest to investigate the miRs whose expression is shared or unique to each of the organ and is involved in ovarian carcinogenesis.
Drug-based treatment regimens, such as carboplatin (DNA-binding platinum agent) and paclitaxel (microtubule stabilizing agent), have been widely used in the past for treatment of patients with ovarian cancer85. However, several of these platinum-based drugs face the limitation of gradual cancer cell resistance to the drugs, and thus newer drug additions such as trabectedin and belotecan have shown promising results in platinum-resistant cancer cells.
Further therapies targeting the cellular processes that convert normal cells to abnormal cancerous cells are on the rise. These include Olaparib/PARP-1 inhibitors (poly-ADP ribose polymerases inhibitor that blocks cell survival signals and promotes cell death), Avastin (blocks vascular epidermal growth factors that otherwise lead to hyper-angiogenesis), Vintafolide (drugs that are used to block the folic acid receptor associated with receptor-associated ovarian cancers), Farletuzmab (monoclonal antibody against the folic acid receptors), Catumaxomab (monoclonal antibody targeting cluster of differentiation 3 and epithelial cell adhesion molecules/EpCAM), and Pazopanib (blocks angiogenesis of cancer cells and thus deprives them of the nutrient and energy supply)80. These cellular processes are known to be influenced by one or more miRNAs80. A focus on these miRs and development of gene-silencing therapies such as anti-miR or siRNAs and miR replacement therapies have proved to be instrumental in the treatment of ovarian cancers. Overall, the search for miRs that can increase the efficiency and tissue specificity of the targeted therapies holds enormous promise for the future.
Conclusion
miRs can potentially regulate cellular processes that facilitate tumorigenesis, tumor growth, and metastasis. This review discusses several miRs associated with the remodeling and/or reprogramming of the TME. Of interest have been those miRs that regulate major cellular components of the TME, such as cancer-associated fibroblasts, tumor-infiltrating cells, mesenchymal stem cells, mesothelial and endothelial cells, and cancer stem cells. In addition, the role of miRs in the hypoxic TME, with a focus on both the local and distant events of metastasis, has been discussed. Several exosomal miRs, cancer cell-associated intracellular miRs, and miRs in the TME may be of interest for the diagnosis, treatment, and prognosis of ovarian carcinomas.
The low survival rate for ovarian cancers has been mostly associated with the delayed clinical diagnosis and the lack of precise understanding of the disease biology. It seems that our present understanding of ovarian cancer has been a much simplified biological assumption where the cancer is being investigated for changes in its genome and the effect of epigenetic modifications arising from abnormal transformations. In addition, miRs, which can be effectively targeted for designing therapy for different cancers, exhibit a dichotomous and highly variable pattern of expression. Several miRs have been shown to be regulated in an opposite manner in either the same or different cancer models, thereby raising questions about the reliability of the miR data. Although variation in experimental conditions can be a major reason for these observed variations, there is also a wider acceptance of the fact that the miRs (small molecular players) can be very sensitive to cellular, extracellular, and environmental stimuli. Thus a simplified classification of miRs as either upregulated or downregulated is based on known experimentally validated results, and can be challenged at any time by opposite or dissimilar expression patterns in different experimental set-ups.
Further, such a simplistic assumption actually overshadows the complex inter and intracellular crosstalk of the cancer and its adjoining tumorous and non-tumorous tissues. In addition, the most widely used in vitro and in vivo experimental models have established the mechanisms involved in disease progression, immune evasion, and gain of metastatic ability. In vitro study of metastatic events and cancer progression is largely limited by the lack of a three-dimensional real-world approach in which the cancer cells have different ranges of cellular space to invade, migrate, and colonize. Although recent use of a 3D organotypic ex vivo omental model has been beneficial in better understanding the disease biology involved in ovarian cancers, the limited use or access to such models has further constrained the translation of the in vitro studies for clinical or therapeutic development.
Irrespective of the variability in the current studies on the expression of miRs associated with ovarian cancers, these small molecular players have been greatly appreciated for their potency to perturb or maintain several integral cellular processes. Therefore, our search to decipher and analyze the role of miRs in the progression of ovarian cancer can contribute to the development of an effective diagnostic tool, therapeutic regimen, and reduced relapse rate in ovarian cancers.
Footnotes
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Sandeep Satapathy  https://orcid.org/0000-0003-1479-1280
https://orcid.org/0000-0003-1479-1280
References
- 1. Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. [DOI] [PubMed] [Google Scholar]
- 2. du Bois A, Kristensen G, Ray-Coquard I, Reuss A, Pignata S, Colombo N, Denison U, Vergote I, Del Campo JM, Ottevanger P, Heubner M, et al. Standard first-line chemotherapy with or without nintedanib for advanced ovarian cancer (AGO-OVAR 12): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2016;17(1):78–89. [DOI] [PubMed] [Google Scholar]
- 3. Tanaka YO, Okada S, Satoh T, Matsumoto K, Oki A, Saida T, Yoshikawa H, Minami M. Differentiation of epithelial ovarian cancer subtypes by use of imaging and clinical data: a detailed analysis. Cancer Imaging. 2016;16(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang C, Bakkum-Gamez J, Mariani A, Cliby W, Goode E. Abstract B80: Tumor expression subtypes of high-grade serous ovarian cancer demonstrate associations with likelihood of complete resection: a single-center study. Clin Cancer Res. 2016;22( suppl 2):B80–B80. [Google Scholar]
- 5. Hartmann LC, Lindor NM. The role of risk-reducing surgery in hereditary breast and ovarian cancer. N Engl J Med. 2016;374(5):454–468. [DOI] [PubMed] [Google Scholar]
- 6. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. [DOI] [PubMed] [Google Scholar]
- 7. Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci. 2016;113(13):E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang S, Yan W. Current view of microRNA processing. Signal Transduct Insight. 2016;5:9–13. [Google Scholar]
- 9. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature Cell Biology. 2019;11(3):228–234. [DOI] [PubMed] [Google Scholar]
- 10. Li M, Xie H, Xiong W, Xu D, Cao K, Liu R, Luo C. [MicroRNA and metabolism regulation]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2013;38(3):318–322. [DOI] [PubMed] [Google Scholar]
- 11. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–677. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis BN, Hata A. Regulation of MicroRNA biogenesis: a miRiad of mechanisms. Cell Communicat Signal. 2009;7(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147(5):1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. [DOI] [PubMed] [Google Scholar]
- 17. Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44(3 ):424–436. [DOI] [PubMed] [Google Scholar]
- 18. Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008;10(2):203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghahhari NM, Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial–mesenchymal transition in cancer. Eur J Cancer, 2015;51(12):1638–1649. [DOI] [PubMed] [Google Scholar]
- 20. Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K, Kirita T, Kuniyasu H. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107(4):700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huppi K, Volfovsky N, Mackiewicz M, Runfola T, Jones TL, Martin SE, Stephens R, Caplen N. MicroRNAs and genomic instability. Semin Cancer Biol. 2007;17(1):65–73. Academic Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chou J, Shahi P, Werb Z. Microrna-mediated regulation of the tumor microenvironment. Cell Cycle. 2013;12(20):3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lorusso G, Rüegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem Cell Biol. 2008;130(6):1091–1103. [DOI] [PubMed] [Google Scholar]
- 25. Fukumura D, Jain RK. Tumor microenvironment abnormalities: causes, consequences, and strategies to normalize. J Cellular Biochem. 2007;101(4):937–949. [DOI] [PubMed] [Google Scholar]
- 26. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 28. Gov E, Kori M, Arga KY. Multiomics analysis of tumor microenvironment reveals gata2 and miRNA-124-3p as potential novel biomarkers in ovarian cancer. OMICS. 2017;21(10):603–615. [DOI] [PubMed] [Google Scholar]
- 29. Pearce CL, Stram DO, Ness RB, Stram DA, Roman LD, Templeman C, Lee AW, Menon U, Fasching PA, McAlpine JN, Doherty JA, et al. Population distribution of lifetime risk of ovarian cancer in the United States. Cancer Epidemiol Prevent Biomarkers. 2015;24(4):671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sandmaier SE, Telugu BPV. MicroRNA-mediated reprogramming of somatic cells into induced pluripotent stem cells. Cell Reprogram Methods Protocol. 2015;1330:29–36. [DOI] [PubMed] [Google Scholar]
- 31. Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci. 2013;110(14):5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer, 2005;5(5):355–366. [DOI] [PubMed] [Google Scholar]
- 33. Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177(3):1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17(11):1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, Visakorpi T. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59(5):671–681. [DOI] [PubMed] [Google Scholar]
- 36. Cirri P, Chiarugi P. Cancer-associated-fibroblasts and tumour cells: a diabolic liaison driving cancer progression. Cancer Metastasis Rev. 2012;31(1–2):195–208. [DOI] [PubMed] [Google Scholar]
- 37. Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, Lengyel E. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2(12):1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481(7380):190–194. [DOI] [PubMed] [Google Scholar]
- 39. Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17(12):1627–1635. [DOI] [PubMed] [Google Scholar]
- 40. Panda H, Pelakh L, Chuang TD, Luo X, Bukulmez O, Chegini N. Endometrial miR-200c is altered during transformation into cancerous states and targets the expression of ZEBs, VEGFA, FLT1, IKKβ, KLF9, and FBLN5. Reproduct Sci. 2012;19(8):786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Leva G, Croce CM. The role of microRNAs in the tumorigenesis of ovarian cancer. Front Oncol, 2013;3:153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Merhautova J, Hezova R, Poprach A, Kovarikova A, Radova L, Svoboda M, Vyzula R, Demlova R, Slaby O. miR-155 and miR-484 are associated with time to progression in metastatic renal cell carcinoma treated with sunitinib. BioMed Res Int. 2015;2015:941980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Allavena G, Carrarelli P, Del Bello B, Luisi S, Petraglia F, Maellaro E. Autophagy is upregulated in ovarian endometriosis: a possible interplay with p53 and heme oxygenase-1. Fertil. Steril. 2015;103:1244–1251.e1. doi:10.1016/j.fertnstert.201502.007. [DOI] [PubMed] [Google Scholar]
- 44. Kim BG, Yoo JY, Kim TH, Shin JH, Langenheim JF, Ferguson SD, Fazleabas AT, Young SL, Lessey BA, Jeong JW. Aberrant activation of signal transducer and activator of transcription-3 (stat3) signaling in endometriosis. Hum Reprod. 2015;30:1069–1078. doi:10.1093/humrep/dev050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin X, Dai Y, Xu W, Shi L, Jin X, Li C, Zhou F, Pan Y, Zhang Y, Lin X, Zhang S. Hypoxia promotes ectopic adhesion ability of endometrial stromal cells via tgf-beta1/smad signaling in endometriosis. Endocrinology. 2018;159(4):1630–1641. doi:10.1210/en.2017-03227. [DOI] [PubMed] [Google Scholar]
- 46. Wendel J, Wang X, Hawkins S. The endometriotic tumor microenvironment in ovarian cancer. Cancers. 2018;10(8):261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lanfrancone L, Boraschi D, Ghiara P, Falini B, Grignani F, Peri G, Mantovani A, Pelicci PG. Human peritoneal mesothelial cells produce many cytokines (granulocyte colony-stimulating factor [CSF], granulocyte-monocyte-CSF, macrophage-CSF, interleukin-1 [IL-1], and IL-6) and are activated and stimulated to grow by IL-1. Blood. 1992;80(11):2835–2842. [PubMed] [Google Scholar]
- 48. Offner FA, Obrist P, Stadlmann S, Feichtinger H, Klingler P, Herold M, Zwierzina H, Hittmair A, Mikuz G, Abendstein B, Zeimet A. IL-6 secretion by human peritoneal mesothelial and ovarian cancer cells. Cytokine. 1995;7(6):542–547. [DOI] [PubMed] [Google Scholar]
- 49. Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O’Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, Urban N, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol oncol. 2010;116(1):117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen D, Zhang Y, Wang J, Chen J, Yang C, Cai K, Wang X, Shi F, Dou J. MicroRNA-200c overexpression inhibits tumorigenicity and metastasis of CD117+ CD44+ ovarian cancer stem cells by regulating epithelial-mesenchymal transition. J Ovarian Res. 2013;6(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28(44):3937–3948. [DOI] [PubMed] [Google Scholar]
- 52. Mitra AK, Chiang CY, Tiwari P, Tomar S, Watters KM, Peter ME, Lengyel E. Microenvironment-induced downregulation of miR-193b drives ovarian cancer metastasis. Oncogene. 2015;34(48):5923–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N, Honjo T, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci. 2007;104(9):3360–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qin W, Ren Q, Liu T, Huang Y, Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587(9):1434–1439. [DOI] [PubMed] [Google Scholar]
- 56. Woo EY, Chu CS, Goletz TJ, Schlienger K, Yeh H, Coukos G, June CH. Regulatory CD4+ CD25+ T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer Res. 2001;61(12):4766–4772. [PubMed] [Google Scholar]
- 57. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182(8):4499–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Banerjee R, Mani RS, Russo N, Scanlon CS, Tsodikov A, Jing X, Cao Q, Palanisamy N, Metwally T, Inglehart RC, Tomlins S, et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 2011;30(42):4339–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13(8b):2236–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. Febs J. 2012;279(11):2047–2059. [DOI] [PubMed] [Google Scholar]
- 62. Xu CX, Xu M, Tan L, Yang H, Permuth-Wey J, Kruk PA, Wenham RM, Nicosia SV, Lancaster JM, Sellers TA, Cheng JQ. MicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/Nanog. J Biol Chem. 2012;287(42):34970–34978. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63. Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim SW, Kim YT. MicroRNA profiling of a CD133+ spheroid-forming subpopulation of the OVCAR3 human ovarian cancer cell line. BMC Med Genomics. 2012;5(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature, 2007;449(7162):557–563. [DOI] [PubMed] [Google Scholar]
- 65. Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. [DOI] [PubMed] [Google Scholar]
- 67. Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63(11):2705–2715. [PubMed] [Google Scholar]
- 68. Weiner-Gorzel K, Dempsey E, Milewska M, McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A, Sharpe D, O’Sullivan J, et al. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med. 2015;4(5):745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68(2):425–433. [DOI] [PubMed] [Google Scholar]
- 70. Kryczek I, Lange A, Mottram P, Alvarez X, Cheng P, Hogan M, Moons L, Wei S, Zou L, Machelon V, Emilie D, et al. CXCL12 and vascular endothelial growth factor synergistically induce neoangiogenesis in human ovarian cancers. Cancer Res. 2005;65(2):465–472. [PubMed] [Google Scholar]
- 71. Birner P, Schindl M, Obermair A, Breitenecker G, Oberhuber G. Expression of hypoxia-inducible factor 1α in epithelial ovarian tumors its impact on prognosis and on response to chemotherapy. Clin Cancer Res. 2001;7(6):1661–1668. [PubMed] [Google Scholar]
- 72. Kim KS, Sengupta S, Berk M, Kwak YG, Escobar PF, Belinson J, Mok SC, Xu Y. Hypoxia enhances lysophosphatidic acid responsiveness in ovarian cancer cells and lysophosphatidic acid induces ovarian tumor metastasis in vivo. Cancer Res. 2006;66(16):7983–7990. [DOI] [PubMed] [Google Scholar]
- 73. Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26(2):225–239. [DOI] [PubMed] [Google Scholar]
- 74. Wong C, Wellman TL, Lounsbury KM. VEGF and HIF-1α expression are increased in advanced stages of epithelial ovarian cancer. Gynecol Oncol. 2003;91(3):513–517. [DOI] [PubMed] [Google Scholar]
- 75. Pez F, Dayan F, Durivault J, Kaniewski B, Aimond G, Le Provost GS, Deux B, Clézardin P, Sommer P, Pouysségur J, Reynaud C. The HIF-1–inducible lysyl oxidase activates HIF-1 via the Akt pathway in a positive regulation loop and synergizes with HIF-1 in promoting tumor cell growth. Cancer Res. 2011;71(5):1647–1657. [DOI] [PubMed] [Google Scholar]
- 76. Joshi HP, Subramanian IV, Schnettler EK, Ghosh G, Rupaimoole R, Evans C, Saluja M, Jing Y, Cristina I, Roy S, Zeng Y, et al. Dynamin 2 along with microRNA-199a reciprocally regulate hypoxia-inducible factors and ovarian cancer metastasis. Proc Natl Acad Sci. 2014;111(14):5331–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. [DOI] [PubMed] [Google Scholar]
- 78. Vaksman O, Stavnes HT, Kærn J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. J Cell Mol Med. 2011;15(7):1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mandai M, Yamaguchi K, Matsumura N, Baba T, Konishi I. Ovarian cancer in endometriosis: molecular biology, pathology, and clinical management. Int J Clin Oncol. 2009;14(5):383–391. [DOI] [PubMed] [Google Scholar]
- 80. Junor EJ, Hole DJ, Gillis CR. Management of ovarian cancer: referral to a multidisciplinary team matters. Br J Cancer. 1994;70(2):363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Plaskocinska I, Shipman H, Drummond J, Thompson E, Buchanan V, Newcombe B, Hodgkin C, Barter E, Ridley P, Ng R, Miller S, et al. New paradigms for BRCA1/BRCA2 testing in women with ovarian cancer–results of the Genetic Testing in Epithelial Ovarian Cancer (GTEOC) Study. J Med Genet. 2016;53(10):655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kurman RJ, Shih IM. The dualistic model of ovarian carcinogenesis. Am J Pathol. 2016;4(186):733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhao B, Hemann MT, Lauffenburger DA. Modeling tumor clonal evolution for drug combinations design. Trends Cancer. 2016;2(3):144–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sakamoto I, Hirotsu Y, Nakagomi H, Ouchi H, Ikegami A, Teramoto K, Amemiya K, Mochizuki H, Omata M. BRCA1 and BRCA2 mutations in Japanese patients with ovarian, fallopian tube, and primary peritoneal cancer. Cancer. 2016;122(1):84–90. [DOI] [PubMed] [Google Scholar]
- 85. Kar R, Sharma C, Sen S, Jain SK, Gupta SD, Singh N. Response of primary culture of human ovarian cancer cells to chemotherapy: In vitro individualized therapy. J Cancer Res Ther. 2016;12(2):1050–1055. [DOI] [PubMed] [Google Scholar]