Abstract
Objective:
To identify the effect of curcumin on tumor suppression and the possible molecular pathways involved.
Methods:
The expression of long noncoding RNA neighbor of BRCA1 lncRNA 2 (NBR2) was quantified using reverse transcription-polymerase chain reaction on cultured colorectal cancer cells. Next, we used Western blot to measure the activation of adenosine monophosphate-activated protein kinase and mechanistic target of rapamycin kinase (mTOR) signaling molecules. Both cell proliferation and viability were measured via MTT assay, and the cell ratio and S phase were detected by BrdU assay. Colorectal cancer cells were pretreated with curcumin or transfected with shNBR2 or adenosine monophosphate-activated protein kinase inhibitor Compound C to examine the molecular pathway involved.
Results:
Current data showed that glucose deficiency increased the expression of NBR2 in colorectal cancer cells, and NBR2 knockdown affected the progression of colorectal cancer cells under glucose starvation conditions. When NBR2 was silenced in the treated colorectal cancer cells, the proliferation, the clone formation, and the percentage of S-phase cells suppressed by glucose deprivation were compromised. Furthermore, NBR2 knockdown could suppress glucose deprivation-induced adenosine monophosphate-activated protein kinase activation plus mTOR inactivation. Similarly, when colorectal cancer cells were treated with curcumin, the expression of NBR2 was significantly increased. NBR2 knockdown reversed curcumin-suppressed proliferation, clone formation, and the percentage of S-phase colorectal cancer cells. Furthermore, NBR2 knockdown abolished curcumin-induced activation of adenosine monophosphate-activated protein kinase and inactivation of the mTOR signaling pathway.
Conclusion:
This study revealed a novel mechanism by which long noncoding RNA NBR2 mediates curcumin suppression of colorectal cancer proliferation by activating adenosine monophosphate-activated protein kinase and inactivating the mTOR signaling pathway.
Keywords: curcumin, colorectal cancer, NBR2, AMPK
Introduction
Colorectal cancer (CRC) has become the third most prevalent malignant tumor worldwide and one of the leading causes of cancer-induced mortality.1 Despite recent progress in early-stage screening and the development of more effective treatments for CRC, the prognosis of advanced stage CRC is still unfavorable.2 Therefore, predictive biomarkers are clearly and urgently needed to detect CRC and to identify the most effective treatments for specific patients with CRC.
Long noncoding RNAs (lncRNAs) are a group of endogenous RNA molecules that are more than 200 nucleotides in length but lack an open reading frame.3 A previous study revealed that lncRNAs impact a variety of important biological processes.4 Liang et al indicated that lncRNA H19 promotes the process of epithelial to mesenchymal transition as 1 microRNA (miRNA) “sponge” in CRC.5 In addition, another lncRNA molecule, DANCR, has been indicated to be an independent predictor of unfavorable prognosis and shortened survival in CRC. Furthermore, lncRNA GAS5 facilitates cell proliferation and works as an indicator for poor prognosis in patients with CRC.6
NBR2 is a noncoding sequence that resides adjacent to the well-known tumor suppressor gene BRCA1. Its biological function has remained unknown for many years since its initial discovery.7 A previous study found that NBR2 interacts with adenosine monophosphate-activated protein kinase (AMPK) and promotes the kinase activity of AMPK, and NBR2 deficiency dampens glucose starvation-induced AMPK activation and AMPK-mediated downstream biological processes.8
Adenosine monophosphate-activated protein kinase is one intracellular energy sensor. Structural analysis reveals that it has a heterotrimeric form consisting of 1 catalytic subunit (α) plus 2 regulatory subunits (β and γ).9 Once activated, AMPK can phosphorylate various downstream effector substrates, such as those that serve to shut down anabolic processes and facilitate adenosine triphosphate-generating catabolic metabolism, thus helping to restore energy homeostasis after stress challenge.10 Although we have plenty of evidence showing that AMPK regulates downstream targets and their biological implications, the mechanisms of AMPK activation itself remain unclear. A previous study demonstrated the direct interaction between NBR2 and the kinase domain of the AMPK complex, and an in vitro kinase assay also suggested that NBR2 potentiated AMPK kinase activity. Moreover, the interaction between NBR2 and AMPK is highly correlated with its capacity to promote the kinase activity of AMPK, suggesting that the regulation of AMPK activation by NBR2 seems to be mediated via its interaction with AMPK.8
Curcumin is identified in the rhizomes of Curcuma longa and has been shown to have multiple biological functions.11 Thus far, most of the effect of curcumin is focused on cancer treatment,11 as the single or combined use of curcumin has been approved for the intervention of human cancers, including CRC.12 However, the molecular mechanism that curcumin plays in colorectal cancer is only sparsely understood. We thus performed an in vitro assay using curcumin to treat cultured CRC cells and to analyze the molecular mechanism involved. Our results supported the working model that curcumin suppressed tumor growth by inducing the expression of lncRNA NBR2, which could activate AMPK and inactivate the mechanistic target of rapamycin kinase (mTOR) signaling pathway for cell death.
Methods
Cell Culture
A total of 4 human CRC cell lines, including HCT116, HCT8, SW480, and SW620, were purchased from the Type Culture Collection, Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (Gibco, Waltham, MA, USA). Cells were cultured in a humidified incubator with 5% CO2, and the temperature was 37°C. The concentration of curcumin used in the cell treatment was 10 μM based on a previous study.13 In glucose starvation assays, cells were treated with different concentrations of glucose (0, 1, or 25 mM) within DMEM containing 10% fetal bovine serum.
Cell Transfection
LncRNA NBR2 shRNA was purchased from GenePharma (Shanghai, China). HCT116 and SW480 CRC cells were seeded at a density of 1.8 × 105 cells per well in a 12-well plate. Transient transfection was conducted using a Lipofectamine 2000 kit (Invitrogen, Carlsbad, California, United States) following the instructions of the manufacturer.
MTT Assay
HCT116 and SW480 CRC cells were treated with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (10% dilution in DMEM) at 37°C until the appearance of visible color. The cell proliferation rate was determined at 24, 48, and 72 hours posttransfection.
Colony Formation Assay
The colony formation assay was performed by seeding 200 cells into each well of the 6-well plate. After 14 days of incubation, cells were treated with 0.05% Crystal violet for 1 hour at room temperature. Under light field microscopy, the number of colonies in each well was enumerated. All groups were tested in triplicate.
BrdU Assay
HCT116 and SW480 CRC cells were seeded at 1.8 × 105 cells/well in DMEM containing 10 μM BrdU. After 3 hours incubation, cells were fixed for 15 minutes using 4% paraformaldehyde and were washed with phosphate-buffered saline (PBS) buffer followed by 0.5 mL Triton X-100. After further rinsing, the cells were treated with DNase (Tiangen, China) for 20 minutes at room temperature. Primary antibody against BrdU (Abcam, Cambridge, MA, USA) was then added following PBS washing for 12 hours incubation at 4°C. On the next day, cells were washed with PBS and incubated in secondary antibody for 60 minutes at room temperature. BrdU-positive cells were detected by flow cytometry.
RNA Isolation and Reverse Transcription-Polymerase Chain Reaction
Total cellular RNA was extracted using a Trizol kit (Takara, Kusatsu, Shiga, Japan) according to the manufacturer’s instructions. Cellular messenger RNA levels were quantified using SYBR PrimeScript RT-PCR Kit (Takara). The expression of NBR2 was measured using specific primers (Forward, 5′-GAGGTCTCCAGTTTCGGTAA-3′; Reverse, 5′-GAACCAAGGTGAAGGACCAA-3′). For the analysis of miRNA, we used stem-loop primers for reverse transcription and reverse transcription-polymerase chain reaction for miRNAs. These primers were purchased from RiboBio (Guangzhou, China). The relative expression of miRNAs was normalized to the internal control of the U6 RNA molecule (Forward, 5′-CGCTTCGGCAGCACATATAC-3′; Reverse, 5′-ATGGAACGCTTCACGAATTT-3′).
Western Blotting
Primary antibodies used in Western blotting, including anti-GAPDH (#5174), anti-p-AMPK (#4186), anti-AMPK (#2532), anti-p-ACC (#11818), anti-ACC (#3662), anti-p-S6 K (#2708), anti-S6 K (#9202), anti-p-S6 (#9209), and anti-S6 (#2317), were all purchased from Cell Signaling Technology (Danvers, MA, USA). In brief, cells were first lysed in Radio Immunoprecipitation Assay (RIPA) buffer. Proteins were quantified using the Bicinchoninic Acid (BCA) approach and were loaded onto 1× sodium dodecyl sulfate polyacrylamide gel electrophoresis for separation. Proteins were then transferred onto polyvinyl difluoride membranes (Millipore), which were incubated sequentially with primary and secondary antibodies (1:2000 dilutions for anti-GAPDH and 1:1000 for other antibodies).
Data Analysis
All statistical analyses were performed using the SPSS software package (version 17.0, IBM, Armonk, NY, USA). All data are shown as the mean ± standard error. Student t test was used for between-group comparisons, and 1-way analysis of variance was adopted for comparisons among multiple groups. P values <.05 were considered statistically significant.
Results
Energy Starvation Could Increase the Expression of NBR2
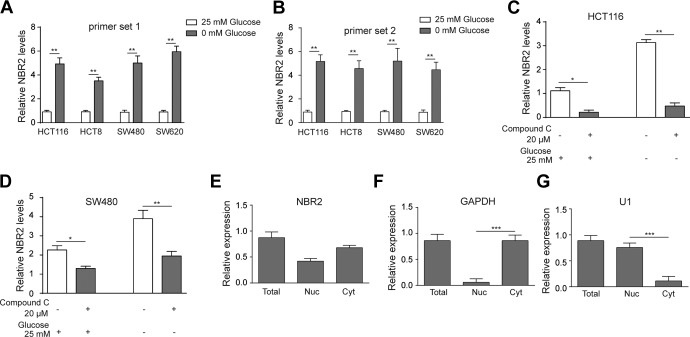
Previous knowledge has revealed the activation of the AMPK pathway by starvation of cells. To verify the connection between glucose starvation-induced AMPK activation and NBR2 in CRC cells, we tested whether glucose-free medium would induce the change of NBR2 in CRC cell lines. The results revealed that glucose deficiency induced the increased expression of NBR2 in HCT116, HCT8, SW480, and SW620 cells compared to 25 mM glucose-treated cells (Figure 1A and 1B, P < .01). Furthermore, we used the AMPK inhibitor Compound C to test the expression of NBR2 in HCT116 and SW480 cells. The results showed that Compound C could suppress the expression of NBR2 in either HCT116 or SW480 cells with or without glucose (Figure 1C and D, P < .05 or P < .01, respectively). Further substantiation came from the observation that NBR2 was expressed in both nuclear and cytosolic fractions (Figure 1E). These results showed that glucose deprivation-induced NBR2 upregulation in CRC cells, at least partly via an AMPK-dependent pathway.
Figure 1.
Glucose starvation-induced NBR2 upregulation in cultured colorectal cancer (CRC) cells. A and B, Reverse transcription-PCR showed the expression change of NBR2 in HCT116, HCT8, SW480, and SW620 cells treated with 0 or 25 mM glucose for 24 hours using 2 primer sets. C and D, Reverse transcription-PCR showed the expression of NBR2 in HCT116 and SW480 cells treated with 0 or 25 mM glucose plus Compound C treatment for 24 hours. E to G, Total, nuclear (Nul), and cytosolic (Cyt) expression of NBR2 plus normalization genes GAPDH1 and U1. Three independent experiments were performed, and the results are presented as the mean ± standard deviation. *P < .05, **P < .01. RT-PCR indicates reverse transcription-polymerase chain reaction.
NBR2 Knockdown Suppressed the Glucose Deprivation-Induced AMPK/mTOR Signaling Pathway
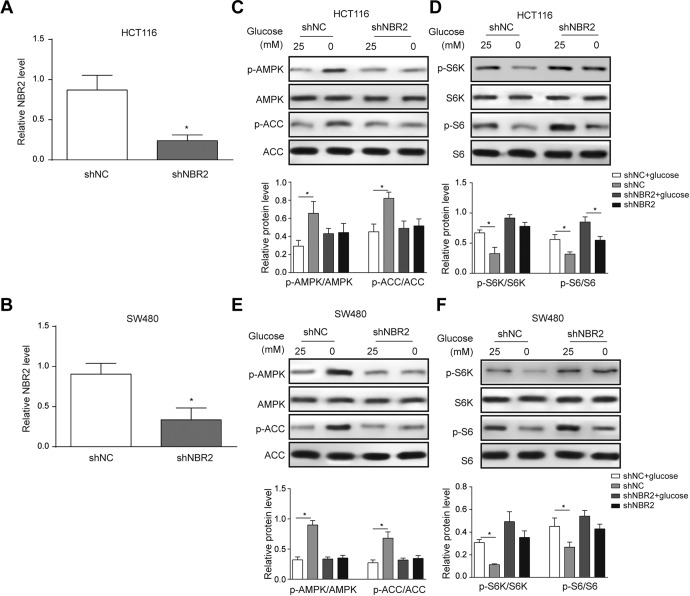
To investigate the downstream signaling pathway of NBR2, we suppressed NBR2 expression in both HCT116 and SW480 cells. First, the transfection of shNBR2 effectively suppressed NBR2 transcript levels in both HCT116 (Figure 2A, P < .05 compared to shNC group) and SW480 cells (Figure 2B). Next, we found that shNBR2 transfection effectively abolished the glucose deprivation-induced upregulation of phosphorylated AMPK (Figure 2C and E, P < .05 compared to the shNC + glucose deprivation group). Since AMPK activation is known to suppress mTOR phosphorylation, we thus examined indicators of the mTOR signaling pathway, S6K and S6. The results showed that in the shNBR2 transfection group, phosphorylation of both S6K and S6 was activated in both HCT116 (Figure 2C and D) and SW480 cells (Figure 2E and F). Therefore, NBR2 could activate the AMPK pathway to suppress mTOR activation, and NBR2 knockdown reversed this process to activate mTOR signaling.
Figure 2.
Knockdown NBR2 in HCT116 and SW480 cells and the detected expression of glucose starvation-induced changes in protein phosphorylation in AMPK/mTOR signaling. A and B, Reverse transcription-PCR showed the level of NBR2 in both HCT116 and SW480 cells after transfection with shNBR2. C-F, Western blotting protein bands and quantitative analysis showed changes in the phosphorylation levels of AMPK, ACC, S6K, and S6 proteins in both HCT116 and SW480 cells treated with 0 or 25 mM glucose for 24 hours. Three independent experiments were performed, and the results are presented as the mean ± standard deviation. *P < .05. AMPK indicates adenosine monophosphate-activated protein kinase; RT-PCR, reverse transcription-polymerase chain reaction.
NBR2 Knockdown Affects the Progression of CRC Cells Under Glucose Starvation Conditions
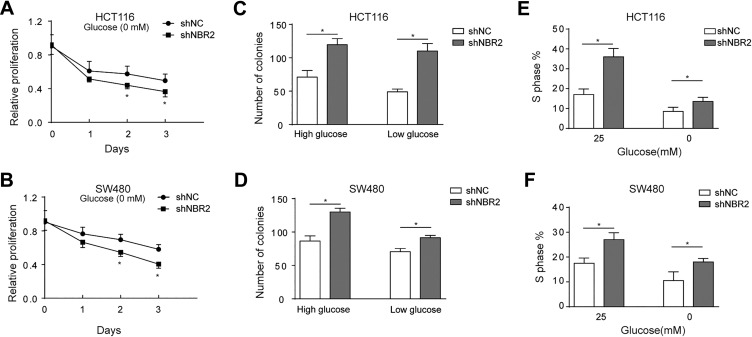
Then, we detected whether NBR2 affects the progression of CRC cells under glucose starvation conditions. First, MTT assay results revealed that shNBR2 suppresses the proliferation of either HCT116 or SW480 cells when compared to the control group under glucose deprivation conditions (Figure 3A and B, P <.05). Second, we measured the effect of NBR2 on the colony formation potency of HCT116 or SW480 cells, and the results showed that shNBR2 increases the clone formation both at high-glucose or low-glucose conditions in HCT116 and SW480 cells (Figure 3C and D, P < .05). Third, a BrdU assay was performed to detect the role of NBR2 in the regulation of the S-phase proportion. We found that shNBR2 notably increased the percentage of S-phase cells at high-glucose or low-glucose conditions in HCT116 and SW480 cells (Figure 3E and F, P < .05).
Figure 3.
NBR2 knockdown affects the progression of colorectal cancer (CRC) cells under glucose starvation conditions. A and B, MTT assay results revealed the proliferation of HCT116 and SW480 cells after transfection with shNBR2 and treatment with 0 or 25 mM glucose for 24 hours. C and D, Clonal formation assay showed the proliferation of HCT116 and SW480 cells under high-glucose or low-glucose conditions with shNBR2 transfection and treated with 1 or 25 mM glucose for 24 hours. E and F, BrdU assay showed the percentage of S-phase cells under high-glucose or low-glucose conditions in HCT116 and SW480 cells treated with 0 or 25 mM glucose for 24 hours. Three independent experiments were performed, and the results are presented as the mean ± standard deviation. *P < .05.
NBR2 Knockdown Abolished the Inhibitory Effect of Curcumin in CRC Cells
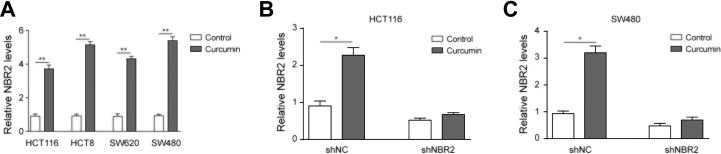
Due to the known AMPK activation by curcumin, we further detected the role that curcumin played in HCT116 and SW480 cells, as curcumin significantly upregulated NBR2 in HCT116, HCT8, SW480, and SW620 cells (Figure 4A, P < .01 compared to control cells). With shNBR2 transfection into HCT116 or SW480 cells, the expression of NBR2 was notably suppressed regardless of pretreatment with curcumin (Figure 4B and C, P < .05). Therefore, curcumin exerted these effects to upregulate NBR2 in CRC cells.
Figure 4.
Curcumin elevated the level of NBR2 in CRC cells. A, RT-PCR showed the expression level of NBR2 in HCT116, HCT8, SW480, and SW620 cells after treatment with 0 or 10 μM curcumin for 24 hours. B, C, Reverse transcription-PCR showed NBR2 levels in curcumin-treated cells after shNBR2 transfection treated with 0 or 10 μM curcumin for 24 hours. Three independent experiments were performed, and the results are presented as the mean ± standard deviation. *P < .05, **P < .01. CRC indicates colorectal cancer; RT-PCR indicates reverse transcription-polymerase chain reaction.
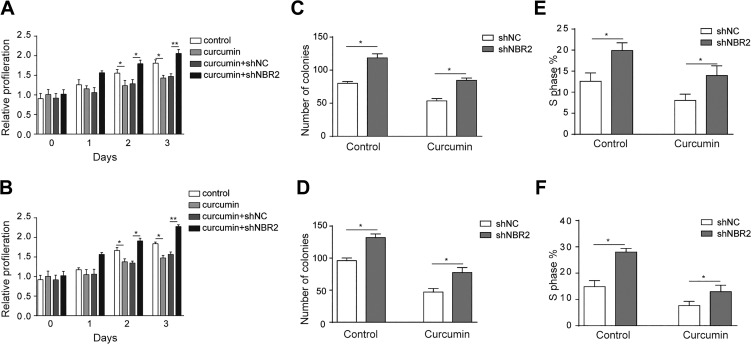
We further examined whether the suppression role of curcumin was dependent on NBR2. MTT results revealed a curcumin-induced suppression of proliferation of HCT116 and SW480 cells (Figure 5A and B, P < .05); however, when NBR2 was knocked down, this inhibitory effect was compromised (Figure 5A and B, P < .05, P < .01). Moreover, shNBR2 could elevate the suppressed colony formation by curcumin in HCT116 and SW480 cells (Figure 5C and D). Consistent results were found in cell cycle analysis, as shNBR2 increased the percentage of cells in the S-phase attenuated by curcumin treatment in HCT116 and SW480 cells (Figure 5E and F, P < .05).
Figure 5.
Inhibition effect of curcumin in CRC is mediated by NBR2. A and B, MTT assay revealed the proliferation of HCT116 and SW480 cells after treatment with 0 or 10 μM curcumin for 24 hours. C and D, Clone formation in curcumin-treated HCT116 and SW480 cells after shNBR2 transfection and treated with 0 or 10 μM curcumin for 24 hours. E and F, BrdU assay showed the percentage of S-phase cells induced by curcumin in HCT116 and SW480 cells after shNBR2 transfection and treated with 0 or 10 μM curcumin for 24 hours. Three independent experiments were performed, and the results are presented as the mean ± standard deviation. *P < 0.05, **P < .01. CRC indicates colorectal cancer.
NBR2 Knockdown Eliminated the Effect of Curcumin on AMPK/mTOR Signaling
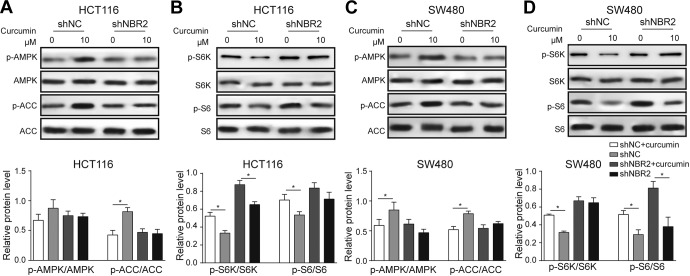
Finally, we investigated p-AMPK, p-ACC, p-S6K, and p-S6 in HCT116 and SW480 cells treated with curcumin. Similar to the glucose starvation treatment, curcumin significantly decreased the phosphorylation of p-AMPK and p-ACC (Figure 6A and C) and increased the level of p-S6K against total S6K (Figure 6B and D). With NBR2 knockdown in the curcumin-treated cells, the suppression of p-AMPK and p-ACC induced by curcumin was notably abolished, while the levels of p-S6K and p-S6 were unchanged upon curcumin treatment (Figure 6A-D). These results demonstrated that NBR2 is mediated by curcumin regulating AMPK/mTOR signaling.
Figure 6.
NBR2 knockdown compromised curcumin-induced changes in protein phosphorylation in AMPK/mTOR signaling. A and B, Western blotting bands and quantitative analysis showed phosphorylation of AMPK, ACC, S6K, and S6 proteins in curcumin-treated HCT116 cells with shNBR2 transfection and treated with 0 or 10 μM curcumin for 24 hours. C and D, Phosphorylation of AMPK, ACC, S6K, and S6 proteins in curcumin-treated SW480 cells with shNBR2 transfection and treated with 0 or 10 μM curcumin for 24 hours. Three independent experiments were performed, and the results are presented as the mean ± standard deviation. AMPK indicates adenosine monophosphate-activated protein kinase.
Effects of Curcumin on AMPK/mTOR Signaling Under Glucose Deprivation
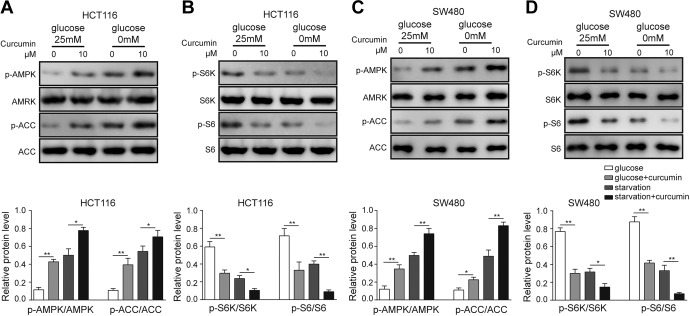
To further elucidate the mechanism of curcumin, we treated cells under glucose-deprivation conditions. Under glucose-supplemented conditions, curcumin treatment significantly activated p-AMPK and p-ACC (Figure 7A and C) and suppressed the p-S6K/p-S6 pathway (Figure 7B and D). Furthermore, glucose-deficient conditions facilitated the curcumin-activated AMPK-ACC pathway and inactivated the S6K-S6 pathway (Figure 7). These results demonstrated that curcumin and glucose deficiency have a synergetic effect in regulating the AMPK/mTOR pathway under glucose-deficient conditions.
Figure 7.
Regulation of the AMPK/mTOR pathway by curcumin under glucose-replenishment and glucose-deficient conditions. A, Western blotting and quantitative analysis of p-AMPK and p-ACC in HCT116 cells by curcumin treatment. B, Western blotting for the relative expression of the p-S6K/p-S6 pathway in HCT116 cells. C, Western blotting and quantitative analysis of p-AMPK and p-ACC in SW480 cells by curcumin treatment. D, Western blotting for the relative expression of the p-S6K/p-S6 pathway in SW480 cells. Three independent experiments were performed, and the results are presented as the mean ± standard deviation. *P < .05, **P < .01. AMPK indicates adenosine monophosphate-activated protein kinase.
Discussion
In this study, we showed that NBR2 expression was strongly induced by glucose deprivation in cultured CRC cells. Activated NBR2 may further activate the AMPK signaling pathway and thus suppress the proliferation of tumor cells. These results are consistent with previous reports showing the tumor suppressor function of NBR2.8 Both RNA sequencing and analysis of noncoding RNA profiles have identified NBR2 as a candidate lncRNA with a response to glucose starvation.14 In the present study, we found that glucose deficiency induces the expression of NBR2 in CRC cells via the energy stress sensor AMPK. This NBR2 upregulation sequentially inactivated the mTOR signaling pathway and activated AMPK, thus forming a positive feedback loop to further potentiate AMPK under energy stress. These cellular events further led to cell proliferation suppression, whereas NBR2 knockdown abolished this process.
NBR2 has been demonstrated as one tumor suppression factor via transregulation with the BRCA1 promoter using a breast cancer cell model.15 Our study is the first to report NBR2 upregulation induced by glucose deprivation in CRC cells in vitro. This is consistent with the consequent suppression of cell proliferation, which received further evidence that NBR2 knockdown could protect CRC cells from glucose deprivation-induced cell death. We further studied possible downstream signaling pathways of NBR2. As previous studies have shown that NBR2 could interfere with the AMPK signaling pathway to mediate cancer cell sensitivity,7 we thus examined the phosphorylated activation level of AMPK. The results showed that glucose deprivation could effectively induce AMPK signaling phosphorylation and that shNBR2 transfection blocked these effects. A further downstream signaling of NBR2 we identified in this study was mTOR, which is known to be inhibited by AMPK phosphorylation.16,17 It has been reported that several lncRNA molecules could mediate tumor growth by regulating mTOR and more specifically the mTOR complex 1 (mTORC1) system.18 In this study, we found that glucose deprivation prominently suppressed phosphorylation of S6K or S6, which are major effector proteins of mTORC1, and NBR2 blockade effectively abolished the selective suppression of mTORC1. In summary, NBR2 mediates glucose deprivation and reduces CRC tumor cell growth via sequential activation of AMPK, followed by mTOR inactivation. It is interesting that NBR2 knockdown induced increased cells in the S-phase under starvation conditions. Adenosine monophosphate-activated protein kinase signaling acts as the “checkpoint” for stressed cells, as inhibition of AMPK leads to mTOR activation and cell proliferation.19 Therefore, NBR2 knockdown tends to have similar effects. As a traditional Indian medicine, curcumin has a long history of usage in medical practice. Recently, curcumin has been proven to suppress the progression of various cancers, including tumors in the pancreas, lung, brain, ovary tissues, and breast gland.20 A previous study identified curcumin in suppressing CRC. For example, curcumin was found to target cancer stem cells of CRC.21,22 Other studies supported a possible mechanism of curcumin in elevating chemotherapy sensitivity of CRC cells.23 , 24 More importantly, curcumin was found to mediate the expression of microRNA molecules in CRC cells for further downstream effects.25,26 The involvement of AMPK and the mTOR signaling pathway in curcumin-mediated functions has also been reported.27,28 Our current data showed that curcumin also has an inhibitory effect on CRC. Moreover, curcumin significantly increased NBR2 expression in CRC cells; when NBR2 was suppressed, the inhibitory effect of curcumin on CRC was eliminated. These results indicated that curcumin regulates CRC progression by regulating the level of NBR2 and then activating AMPK signaling. In addition, curcumin affects downstream mTOR signaling in CRC cells, and curcumin regulated this signaling pathway through NBR2. Together, these results revealed that curcumin regulated the activated AMPK pathway via NBR2 upregulation and further suppressed mTOR to inhibit CRC cell proliferation.
In conclusion, this study revealed the inhibitory effect of curcumin on the progression of CRC via modulating the NBR2/AMPK/mTOR pathway. Further studies are thus required to illustrate the in vivo effects of curcumin in tumor models. Nevertheless, these data provide further evidence of curcumin in tumor suppression and may provide new insights for CRC therapy.
Abbreviations
- lncRNA
long noncoding RNAs
- CRC
colorectal cancer
- AMPK
adenosine monophosphate-activated protein kinase
- DMEM
Dulbecco Modified Eagle Medium
- miRNA
micro RNA
- PBS
phosphate-buffered saline
Footnotes
Authors’ Note: Our study did not require an ethical board approval because it did not contain human or animal trials.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The project was supported by the Ningbo Natural Science Foundation (Grant Nos: 2018277842, 2018262634, 2017A610145, 2017A610158, 2016A610135) and the Zhejiang Medical and Health Bureau (2017KY593, 2017KY594).
ORCID iD: Binbin Xu  https://orcid.org/0000-0002-3240-8970
https://orcid.org/0000-0002-3240-8970
References
- 1. Thomas J, Ohtsuka M, Pichler M, Ling H. MicroRNAs: clinical relevance in colorectal cancer. Int J Mol Sci. 2015;16(12):28063–28076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149(5):1204–1225.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. [DOI] [PubMed] [Google Scholar]
- 4. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015:6(26):22513–22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):253. [DOI] [PubMed] [Google Scholar]
- 7. Liu X, Gan B. lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle. 2016;15(24):3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu X, Xiao ZD, Han L, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18(4):431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13(9):1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Devassy JG, Nwachukwu ID, Jones PJ. Curcumin and cancer: barriers to obtaining a health claim. Nutr Rev. 2015;73(3):155–165. [DOI] [PubMed] [Google Scholar]
- 12. He ZY, Shi CB, Wen H, Li FL, Wang BL, Wang J. Upregulation of p53 expression in patients with colorectal cancer by administration of curcumin. Cancer Invest. 2011;29(3):208–213. [DOI] [PubMed] [Google Scholar]
- 13. Tong W, Wang Q, Sun D, Suo J. Curcumin suppresses colon cancer cell invasion via AMPK-induced inhibition of NF-kappaB, uPA activator and MMP9. Oncol Lett. 2016;12(5):4139–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao ZD, Liu X, Zhuang L, Gan B.NBR2: a former junk gene emerges as a key player in tumor suppression. Mol Cell Oncol. 2016;3(4):e1187322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suen TC, Tang MS, Goss PE. Model of transcriptional regulation of the BRCA1-NBR2 bi-directional transcriptional unit. Biochim Biophys Acta. 2005;1728(3):126–134. [DOI] [PubMed] [Google Scholar]
- 16. Faubert B, Vincent EE, Poffenberger MC, Jones RG. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett. 2015;356(2 Pt A):165–170. [DOI] [PubMed] [Google Scholar]
- 17. Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 2012;52:381–400. [DOI] [PubMed] [Google Scholar]
- 18. Xiao ZD, Zhuang L, Gan B. Long non-coding RNAs in cancer metabolism. Bioessays. 2016;38(10):991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu X, Zhu Y. Curcumin inhibits human non-small cell lung cancer xenografts by targeting STAT3 pathway. Am J Transl Res. 2017;9(8):3633–3641. [PMC free article] [PubMed] [Google Scholar]
- 21. Zang S, Liu T, Shi J, Qiao L. Curcumin: a promising agent targeting cancer stem cells. Anticancer Agents Med Chem. 2014;14(6):787–792. [DOI] [PubMed] [Google Scholar]
- 22. Ramasamy TS, Ayob AZ, Myint HH, Thiagarajah S, Amini F. Targeting colorectal cancer stem cells using curcumin and curcumin analogues: insights into the mechanism of the therapeutic efficacy. Cancer Cell Int. 2015;15:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Toden S. Curcumin mediates chemosensitization to 5-fluorouracil through miRNA-induced suppression of epithelial-to-mesenchymal transition in chemoresistant colorectal cancer. Carcinogenesis. 2015;36(3):355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. James MI, Iwuji C, Irving G, et al. Curcumin inhibits cancer stem cell phenotypes in ex vivo models of colorectal liver metastases, and is clinically safe and tolerable in combination with FOLFOX chemotherapy. Cancer Lett. 2015;364(2):135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toden S, Okugawa Y, Buhrmann C, et al. Novel evidence for curcumin and boswellic acid-induced chemoprevention through regulation of miR-34a and miR-27a in colorectal cancer. Cancer Prev Res (Phila). 2015;8(5):431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mudduluru G, George-William JN, Muppala S, et al. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci Rep. 2011;31(3):185–197. [DOI] [PubMed] [Google Scholar]
- 27. Chen L, Li WF, Wang HX, et al. Curcumin cytotoxicity is enhanced by PTEN disruption in colorectal cancer cells. World J Gastroenterol. 2013;19(40):6814–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee YK, Park SY, Kim YM, Park OJ. Regulatory effect of the AMPK-COX-2 signaling pathway in curcumin-induced apoptosis in HT-29 colon cancer cells. Ann N Y Acad Sci. 2009;1171:489–494. [DOI] [PubMed] [Google Scholar]