Dear Editor,
Congenital bilateral absence of the vas deferens (CBAVD), a complete or partial defect of the Wolffian duct derivatives, is found in >25% of men with obstructive azoospermia (OA), but the underlying pathological mechanism remains poorly understood. Previous research has shown that the most common disease associated with CBAVD is cystic fibrosis (CF), the predominant manifestations of which include progressive lung disease, pancreatic dysfunction, elevated sweat chloride electrolyte, meconium ileus, and male infertility.1 CF is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which encodes a cyclic adenosine monophosphate (cAMP)-activated chloride channel protein that plays a vital function in maintaining chloride and bicarbonate homeostasis.2 Therefore, it is generally presumed that the absence of vas deferens is caused by a progressive atrophy related to abnormal electrolyte balance and fluid transport in the male excurrent ducts.3 Approximately 80% of CBAVD cases are caused by CFTR mutations, and mutations of the adhesion G protein-coupled receptor G2 (ADGRG2) gene have recently been found in CBAVD patients;4 however, for the remaining patients, the origin of CBAVD is unknown. Indeed, mutations in several specific gene loci have been confirmed to lead to male infertility, while recent studies have also suggested that most infertility is not caused by simple genetic variations.5 To effectively determine the cause of CBAVD, other pathological mechanisms, such as changes in gene transcriptional levels or epigenetic regulations, should also be considered in CBAVD patients without gene mutations.
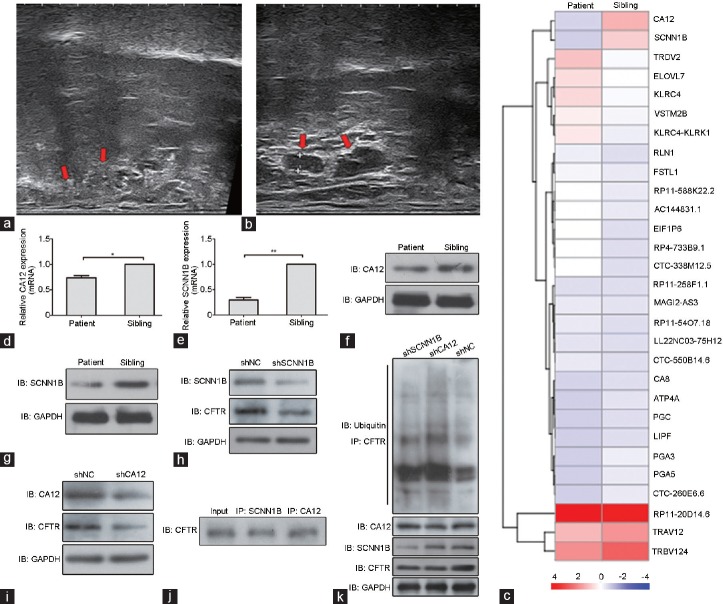
Here, we report the case of an infertile man with CBAVD in the context of assisted reproductive procedures; interestingly, his monozygotic twin brother did not present this phenotype. Several initial examinations were performed on the patient as well as his brother, including physical examination, rectal ultrasound examination, and basal hormonal examination. Consequently, aside from the absence of bilateral vas deferens and an elevated sweat chloride concentration (213.50 mmol l−1) detected only in the patient (Figure 1a and 1b), the other examination indexes for the two brothers were normal (Table 1). The study was approved by the Ethical Review Board of West China Second University Hospital, Sichuan University, Chengdu, China. Informed consent was obtained from two brothers in our study.
Figure 1.
Analysis of the CBAVD phenotype in the patient. (a) The patient showing a complete absence of bilateral vas deferens (red arrow). (b) The sibling of the patient showing normal development of bilateral vas deferens (red arrow). (c) The heat map of RNA expression in the two brothers. SCNN1B and CA12 RNA levels were decreased in the patient compared to his sibling. The patient showed the lower expression of (d) CA12 and (e) SCNN1B according to his sibling by RT-PCR. The results are presented as the mean ± s.d. (n = 3). *P < 0.05 and **P < 0.01 according to Student's t-test. The reduced protein amount of (f) CA12 and (g) SCNN1B was observed in the patient compared to his sibling by western blot. The western blot results represented that the downregulation of CFTR was observed when (h) SCNN1B and (i) CA12 were knocked down in the 293T and A549 cells, respectively. (j) The interactions were observed between CFTR and SCNN1B, or CA12 in 293T and A549 cells, respectively. (k) The increased ubiquitination degradation of CFTR was observed when SCNN1B and CA12 were downregulated in 293T and A549 cells, respectively. CBAVD: congenital bilateral absence of the vas deferens; SCNN1B: sodium channel epithelial 1 beta subunit; CA12: carbonic anhydrase 12; CFTR: cystic fibrosis transmembrane conductance regulator; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; NC: negative control; IB: immunoblotting; RT-PCR: real-time polymerase chain reaction; IP: immunoprecipitation; s.d.: standard deviation; TRDV2: T-cell receptor delta variable 2; ELOVL7: ELOVL fatty acid elongase 7; KLRC4: killer cell lectin-like receptor C4; VSTM2B: V-set and transmembrane domain containing 2B; KLRC4-KLRK1: KLRC4-KLRK1 readthrough; RLN1: relaxin 1; FSTL1: follistatin-like 1; EIF1P6: eukaryotic translation initiation factor 1 pseudogene 6; MAGI2-AS3: MAGI2 antisense RNA 3; TRBV124: T-cell receptor beta variable 12-4; TRAV12: T-cell receptor alpha variable 1-2; PGA5: pepsinogen 5, Group I (pepsinogen A); PGA3: pepsinogen 3, Group I (pepsinogen A); LIPF: lipase F, gastric type; PGC: progastricsin; ATP4A: ATPase, H+/K+ exchanging, gastric, alpha polypeptide; CA8: carbonic anhydrase 8.
Table 1.
Reproductive system-related index analysis of the two brothers
| Case | Semen analysis | Testis size (ml) | Epididymal size (ml) | Reproductive hormones | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Volume (ml) | Sperm count (106 per ml) | Left | Right | Left | Right | E2 (pg ml−1) | T (ng ml−1) | LH (IU l−1) | FSH (IU l−1) | PRL (ng ml−1) | |
| Patient | 0.7 | Azoospermia | 17.1 | 22.8 | 1.4 | 1.1 | 48.0 | 6.92 | 6.0 | 6.9 | 6.4 |
| Sibling | 1 | 18 | 15.8 | 16.8 | 0.06 | 0.06 | 38.1 | 7.20 | 3.4 | 6.4 | 6.8 |
E2: estradiol; T: testosterone; LH: luteinizing hormone; FSH: follicle-stimulating hormone; PRL: prolactin
Then, we performed whole-genome sequencing to search for candidate genetic variations responsible for the patient's phenotype (detailed method can be seen in whole genome sequencing of Supplementary Information (56.4KB, pdf) ). As a result, seven gene mutations, namely, agrin (AGRN), centrosomal protein 104 (CEP104), androglobin (ADGB), neutrophil cytosolic factor 1 (NCF1), polyhomeotic homolog 1 (PHC1), ubiquitin B (UBB), and ribosomal protein S15 (RPS15), were detected only in the patient. Unfortunately, all of them were single nucleotide polymorphisms (SNPs) and synonymous mutations, and all were predicted to be benign by bioinformatic tools including SIFT (http://provean.jcvi.org/seq_submit.php), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml), and M-CAP (http://bejerano.stanford.edu/MCAP/). Thus, we speculated that the CBAVD in the patient was caused by transcriptome or posttranslational regulation rather than genemutations. Epigenetic changes can affect gene expression or function independent of changes in DNA sequence. DNA methylation is a crucial epigenetic modification and has been found associated with multiple diseases including cancer, neurodegenerative and cardiovascular disease, and autoimmune syndromes, such as lupus and rheumatoid arthritis (RA). Furthermore, the altered DNA methylation levels of CFTR were found in several CF patients.6 In this study, we further dissected the whole DNA methylation patterns of the two brothers through chip-based 850K whole DNA methylome analysis (Shanghai OE Biotech Inc., Shanghai, China; detailed method can be seen in whole DNA methylome analysis of Supplementary Information (56.4KB, pdf) ). We observed increased levels of DNA methylation on several genes in the patient compared with his sibling; however, after bioinformatics filtering analysis, none of these genes were related to CBAVD (Supplementary Figure 1 (264.2KB, tif) ).
Finally, we considered whether there was variation in the transcription levels of CBAVD-related genes between the two brothers. Using high-throughput RNA sequencing (Novogene Bioinformatics Technology Co., Ltd., Beijing, China), a total of 162 downregulated genes were found in the patient compared to his brother (Figure 1c; detailed method can be seen in RNA sequencing of Supplementary Information (56.4KB, pdf) ). Eventually, through bioinformatics analysis and a comprehensive review of the literature, we focused on two genes, sodium channel epithelial 1 beta subunit (SCNN1B) and carbonic anhydrase 12 (CA12), which were included in the multigene panel for CF molecular genetic testing. SCNN1Bis a subunit of epithelial sodium channel (ENaC), which regulates Na+ transport by interacting with CFTR.7 It is reported that the loss of functional interaction between CFTR and ENaC can result in a Na+ transport defect in CF patients.8 In addition, SCNN1B mutations have been found in nonclassical CF patients without CFTR mutations.9 Taken together, these findings emphasize the potential role of SCNN1B in CF, the disease most relevant for CBAVD. CA12 is a type I membrane protein gene-encoding carbonic anhydrase XII, and mutations in this gene may result in hyperchlorhidrosis, which could be the reason that the patient with decreased CA12 expression had excessive chloride secretion in his sweat compared to his sibling.10 Furthermore, patients with loss-of-function variants in CA12 potentially resulting in decreased expression of CA12 have developed CF syndrome.11 Thus, it is clear that low expression of SCNN1B and CA12 together might be the cause of CBAVD for this patient. In addition, we further confirmed the reduced expression of the two genes in the patient by real-time polymerase chain reaction (RT-PCR; Figure 1d and 1e) and western blot (Figure 1f and 1g). To further elucidate the mechanism of how decreased SCNN1B and CA12 expression could lead to CBAVD, we knocked down SCNN1B and CA12 in human embryonic kidney cells 293T and adenocarcinomic human alveolar basal epithelial cells A549, respectively. The western blot results showed decreased expression of CFTR in the knocked-down cells compared with the control cells (Figure 1h and 1i). Furthermore, co-immunoprecipitation showed that CFTR could bind to SCNN1B and CA12 in 293T and A549 cells, respectively (Figure 1j). In addition, we found that the ubiquitin-mediated degradation of CFTR was increased when SCNN1B and CA12 were knocked down (Figure 1k). Together, these data suggest that the downregulation of SCNN1B and CA12 might be related to the occurrence of CBAVD through regulation of CFTR expression.
In summary, our work suggests a new pathophysiological mechanism in which the compound action of SCNN1B and CA12 may play a vital role in CBACD through regulating the expression of CFTR via the ubiquitination pathway. These findings may have potential significance in explaining CBAVD in patients without CFTR mutations and provide new insight into the identification of the pathogenic genes underlying this condition. Further epidemiological and functional research on the association of SCNN1B and CA12 with CBAVD may provide more evidence on the contribution of these two genes in CBAVD.
AUTHOR CONTRIBUTIONS
YS and XHJ conceived and designed the experiments; FPL and HXY participated in the acquisition of patients’ data; JGJ and DMC performed the ultrasound examination; YS, XLL, and WRZ performed the experiments; FYH and QW analyzed the data; YS wrote the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
The heat map of DNA methylation levels in wholegenome for the two brothers. No DNA methylation difference was observed in genes associated with CBAVD between the two brothers.
ACKNOWLEDGMENTS
We thank the two brothers voluntarily participated in this study, and we thank all collaborators of West China Hospital, West China Second University Hospital, Sichuan University, and Southwest Medical University, for their clinic support in this research.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Dequeker E, Cuppens H, Dodge J, Estivill X, Goossens M, et al. Recommendations for quality improvement in genetic testing for cystic fibrosis. European concerted action on cystic fibrosis. Eur J Hum Genet. 2000;8(Suppl 2):S2–24. doi: 10.1038/sj.ejhg.5200487. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Chen Z, Ni Y, Li Z. CFTR mutations in men with congenital bilateral absence of the vas deferens (CBAVD): a systemic review and meta-analysis. Hum Reprod. 2012;27:25–35. doi: 10.1093/humrep/der377. [DOI] [PubMed] [Google Scholar]
- 3.Gaillard DA, Carre-Pigeon F, Lallemand A. Normal vas deferens in fetuses with cystic fibrosis. J Urol. 1997;158:1549–52. [PubMed] [Google Scholar]
- 4.Patat O, Pagin A, Siegfried A, Mitchell V, Chassaing N, et al. Truncating mutations in the Adhesion G protein-coupled receptor G2 gene ADGRG2 cause an X-Linked congenital bilateral absence of vas deferens. Am J Hum Genet. 2016;99:437–42. doi: 10.1016/j.ajhg.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aston KI, Carrell DT. Genome-wide study of single-nucleotide polymorphisms associated with azoospermia and severe oligozoospermia. J Androl. 2009;30:711–25. doi: 10.2164/jandrol.109.007971. [DOI] [PubMed] [Google Scholar]
- 6.Magalhães M, Rivals I, Claustres M, Varilh J, Thomasset M, et al. DNA methylation at modifier genes of lung disease severity is altered in cystic fibrosis. Clin Epigenetics. 2017;9:19. doi: 10.1186/s13148-016-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy MM, Light MJ, Quinton PM. Activation of the epithelial Na+ channel (ENaC) requires CFTR Cl- channel function. Nature. 1999;402:301–4. doi: 10.1038/46297. [DOI] [PubMed] [Google Scholar]
- 8.Anguiano A, Oates RD, Amos JA, Dean M, Gerrard B, et al. Congenital bilateral absence of the vas deferens. A primarily genital form of cystic fibrosis. JAMA. 1992;267:1794–7. [PubMed] [Google Scholar]
- 9.Sheridan MB, Fong P, Groman JD, Conrad C, Flume P, et al. Mutations in the beta-subunit of the epithelial Na+ channel in patients with a cystic fibrosis-like syndrome. Hum Mol Genet. 2005;14:3493–8. doi: 10.1093/hmg/ddi374. [DOI] [PubMed] [Google Scholar]
- 10.Feldshtein M, Elkrinawi S, Yerushalmi B, Marcus B, Vullo D, et al. Hyperchlorhidrosis caused by homozygous mutation in CA12, encoding carbonic anhydrase XII. Am J Hum Genet. 2010;87:713–20. doi: 10.1016/j.ajhg.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M, Vecchio-Pagan B, Sharma N, Waheed A, Li X, et al. Loss of carbonic anhydrase XII function in individuals with elevated sweat chloride concentration and pulmonary airway disease. Hum Mol Genet. 2016;25:1923–33. doi: 10.1093/hmg/ddw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The heat map of DNA methylation levels in wholegenome for the two brothers. No DNA methylation difference was observed in genes associated with CBAVD between the two brothers.