Abstract
Prostate cancer is a common malignant tumor in elderly men. As a novel metabolic-reprogramming molecule, the role of ankyrin repeat domain 22 (ANKRD22) in the tumorigenesis and progression of prostate cancer remains unknown. In the present study, mouse monoclonal antibodies against human ANKRD22 were prepared using recombinant ANKRD22 from prokaryotic expression and validated. Subsequently, these antibodies were used to evaluate ANKRD22 levels via immunohistochemical staining in prostate cancer tissues. Finally, the association between ANKRD22 levels and prostate cancer progression was analyzed in 636 samples of prostate cancer using The Cancer Genome Atlas (TCGA) database. A total of four anti-ANKRD22 monoclonal antibodies were generated and validated, which could be effectively blocked by recombinant ANKRD22 protein. Using these antibodies for immunohistochemical staining, ANKRD22 was detected in prostate cancer cells in both the cytoplasm and nucleus. Bioinformatics analysis demonstrated that the mRNA level of ANKRD22 was inversely associated with prostate cancer stage (P<0.05) and Gleason score (P<0.01) in TCGA database. Patients with higher ANKRD22 mRNA levels exhibited longer disease-free survival following radical prostatectomy. These findings suggest that ANKRD22 may negatively regulate the progression of prostate cancer. The prepared ANKRD22 antibodies with high specificity provide a powerful tool in ANKRD22 research.
Keywords: prostate cancer, ankyrin repeat domain 22, antibodies, immunohistochemical staining
Introduction
Prostate cancer is the most frequently diagnosed type of cancer in men and the second leading cause of cancer-associated mortality in Western countries (1). In 2013, the incidence rate of prostate cancer in China was ~9.9/100,000, ranking as the sixth leading cause of malignancy in men (2,3). Due to changes in lifestyle and dietary habits, the incidence rate of prostate cancer has risen significantly in recent years (2,3). Clinically significant prostate cancer is rarely diagnosed under the age of 40 years, with a steady rise in incidence and prevalence after 50 years of age (4,5). Classical treatments for prostate cancer include surgical resection, radiotherapy, chemotherapy and endocrine therapy (6). However, the efficacy of these therapies remains unsatisfactory (6,7). Immunotherapy may be another potential treatment modality (8), which requires further research. It is necessary to identify novel prostate cancer regulators for the diagnosis and treatment of prostate cancer.
To date, the underlying molecular mechanism of prostate cancer remains to be fully elucidated. Abnormal activation of androgen receptor is currently considered as the most important mechanism of prostate cancer (7). Abnormalities in other signaling molecules, such as overexpression of transcription factors and oncogenes, and inactivation of tumor suppressors also participate in different stages of prostate cancer development (9). Metabolic reprogramming is one of the important molecular events in the tumorigenesis and progression of prostate cancer (10). In our previous studies, ankyrin repeat domain 22 (ANKRD22) was found to be a novel metabolic-reprogramming molecule, which interacts with the gatekeeper enzyme of aerobic glycolysis, phosphoinositide-dependent kinase-1 and multiple subunits of the ATP synthase complex (Pan et al, unpublished data). ANKRD22 is an ankyrin repeat protein with four copies of the ankyrin motif. It belongs to the ankyrin repeat (ANK) family and contains 191 amino acids with a molecular weight of 21,849 Daltons (11). A number of previous studies have demonstrated that abnormal ANK family protein function is associated with disease, including hemolytic anemia, arrhythmia and autism (12–14). ANK proteins also have important roles in cancer development and progression (15,16). For example, ANKRD1 induces renal cancer cell proliferation by regulation of downstream miRNA (15). By contrast, ANKRD12 may exert antitumor effects in colorectal cancer by inhibiting liver metastasis (16). ANKRD22 expression has been detected in esophageal, gastric and colorectal epithelial cells, and in ovarian epithelial cells (17). However, the exact function of ANKRD22 in prostate cancer remains unclear.
The present study aimed to analyze the organelle distribution and expression status of ANKRD22 in prostate cancer and the function of ANKRD22 in prostate cancer progression.
Materials and methods
Clinical tissue specimens
A total of 11 patients with prostate cancer (age range, 56–76 years) and 4 male patients with gastric cancer (age range, 54–68 years) were recruited between August 2017 and March 2018 at The Second Affiliated Hospital of Zhejiang University (Hangzhou, China). None of patients with prostate cancer had received hormone therapy, radiotherapy or chemotherapy prior to radical prostatectomy. Tumor and adjacent non-cancerous tissues located at least 1 cm from the cancer margin were collected during the surgery. The tissues were fixed by 10% formaldehyde (Juhua Group Corporation) at room temperature for 6 h and embedded in paraffin following dehydration. Subsequently, the paraffin-embedded tissues were sliced into 5–7 µm sections. The hematoxylin and eosin (H&E) staining slides of prostate and gastric cancer tissues were reviewed under light-microscope at ×100 magnification and diagnoses were verified by an experienced pathologist. The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Zhejiang University, and all patients provided written informed consent.
Bioinformatics analysis
The Cancer Genome Atlas (TCGA) database (https://www.cbioportal.org/study/clinicalData?id=prad_tcga) and Gene Set Enrichment Analysis (GSEA) of 636 samples was analyzed by Hope Biotech Company (Hangzhou, China). Human prostate tissue gene expression profile data sets with corresponding clinical and follow-up information were downloaded from TCGA database. The detailed GSEA protocol was obtained from the Broad Institute GSEA website (www.broad.mit.edu/gsea). The GSEA software v2.0.13 (http://software.broadinstitute.org/gsea/downloads.jsp) was run using the JAVA 7.0 platform. The dataset and phenotype label files were created and loaded into the GSEA software. ANKRD22 mRNA level was quantiled into Q0-Q3. Patients in Q2 and Q3 were considered to exhibit high ANKRD22 expression, whereas patients in Q0 and Q1 exhibited low expression of ANKRD22 mRNA.
Expression and purification of recombinant human ANKRD22 protein
The ANKRD22 open reading frame DNA fragment in ANKRD22/pCMV5-XL5 (OriGene Technologies, Inc.) was amplified by DNA polymerase (Takara Bio, Inc.). The primers used were as follows: ANKRD22 forward, 5′-AGTTCTCCATGGAGATGGGAATCCTATACTCTGAGCCCAT-3′ and reverse, 5′-ACTGAGTGGATCTTACAATGCTTTCCTTAGCATTAATTC-3′. The following thermocycling conditions were used: Initial denaturation at 95°C for 30 sec; 35 cycles of 95°C for 10 sec, 55°C for 15 sec and 72°C for 60 sec. pET-42a vectors (Novagen, Merck KGaA) were initially digested using NcoI and BamHI endonucleases, respectively, and mixed at a ratio of 9:1 ANKRD22:pET-42a with T4 ligase at 16°C overnight. The ligation product was transformed into freshly prepared DH5α E. coli competent cells (Sangon Biotech Co., Ltd.) and DNA sequencing was performed by Sanger to determine if transfection had been successful. Once ANKRD22 was successfully subcloned into pET-42a (ANKRD22 fused with glutathione S-transferase- and histidine-tags), ANKRD22/pET-42a plasmids were transformed into Rosetta DE3 competent cells (Novagen; Merck KGaA). The recombinant proteins were induced using isopropyl-β-D-thiogalactoside (Sangon Biotech Co., Ltd.) and the inclusion bodies (IB) were collected and renatured using a Protein Refolding kit (Novagen; Merck KGaA). Briefly, the cells were resuspended with IB wash buffer (Novagen; Merck KGaA) and incubated with 100 µg/ml lysozyme (Sangon Biotech Co., Ltd.) at 30°C for 15 min. Sonication (frequency, 20–25 kHz; 90 W for 10 sec and 10 cycles of 30 sec in an ice bath) was performed until the solution was not viscous. The lysate was subsequently centrifuged at 10,000 × g at 4°C for 10 min. The pellet was washed with 1X IB wash buffer twice and the supernatant was discarded. The inclusion body pellet was then added to an appropriate amount of dissolving buffer [50 ml of dissolving buffer consisting of 5 ml 10X IB Solubilization Buffer, 0.5 ml 30% N-lauroylsarcosine, 50 µl 1 M dithiothreitol (DTT) and 44.45 ml deionized water] to make a final inclusion body concentration of 10–20 mg/ml. The mixture was fully combined, incubated at room temperature for 15 min and centrifuged at 10,000 × g at 4°C for 10 min. The supernatant was collected and dialyzed using 20 mM Tris-HCl buffer (dialysis buffer; pH 8.5) with 0.01% DTT solution twice (3 h at 4°C each time), using dialysis buffer twice (3 h at 4°C each time) and using dialysis buffer with 1 mM reduced glutathione and 0.2 mM oxidized glutathione overnight at 4°C. The renatured protein was purified through a Ni2+ column (Bio-Rad Laboratories, Inc.) and frozen following dialysis with 30% glycerol at −20°C for long-term storage (1 year).
Establishment of 293T cells overexpressing ANKRD22
Recombinant ANKRD22-expressing lentivirus particles (100 µl/ml; multiplicity of infection=100; Cyagen Inc.) were used to infect 293T cells (Cell Bank of Shanghai Branch of the Chinese Academy of Sciences) which were maintained in high-glucose DMEM (Corning, Inc.) supplemented with 10% fetal bovine serum (Corning, Inc.) and 100 µg/ml gentamicin (Sangon Biotech Co., Ltd.) at 37°C in a humidified environment with 5% CO2. Following incubation for 48 h, cells were passaged and transferred to a new six-well plate and screened with 5 µg/ml puromycin for 7–10 days. After propagation, 293T cells with high expression levels of ANKRD22 were digested with 0.25% trypsin solution (Genom Biotechnology Co. Ltd.) at room temperature for 5 min and collected by centrifugation at 1,000 × g at room temperature for 5 min and washed twice with PBS. Following aspiration of the PBS, 500 µl ice-cold radioimmunoprecipitation assay lysis buffer (pH 7.5; 50 mM Tris-HCl buffer containing 150 mM NaCl, 1 mM EDTA, 0.25% deoxycholate acid and 1% NP-40 and 0.25% deoxycholate sodium) was added, and the solution was mixed by pipetting and gentle agitation. The solution was lysed on ice for 15 min, centrifuged at 13,000 × g at 4°C for 10 min, and the supernatant was aspirated for western blot analysis.
Preparation of mouse anti-human ANKRD22 monoclonal antibodies
A total of 32 female BALB/C mice (age, 6 weeks; weight, 16–18 g) were purchased from Shanghai Laboratory Animal Research Center and maintained between February 2017 and July 2017 in the standardized specific pathogen-free-grade experimental animal center of Zhejiang Chinese Medical University authorized by the government of Zhejiang province at 22±2°C with 40–70% humidity, noise <50 dB, light intensity 150–200 Lx, 12:12 h light/dark cycle, sterile ultrapure water and sterile complete formulated nutrition granulated feed. The health and behavior of each mouse was monitored daily. Recombinant ANKRD22 was used as an immunogen to immunize the mice. The mice were placed in a 10 liter 99% CO2 chamber (3 l/min) for 5–10 min and were monitored for signs of life including heartbeat, breath and muscular tension for 5 min prior to verification of death. All procedures complied with the Laboratory Animal Guidelines for Ethical Review of Animal Welfare in China and the animal research protocol was approved by the Animal Care and Welfare Committee of Zhejiang Chinese Medical University. Lysate from overexpressed ANKRD22 293T cells was used for hybridoma screening using western blot analysis. During the course of antibody production, mice immunization and hybridoma production was performed by Hangzhou HuaAn Biotechnology Co., Ltd. Following three screening cycles, four hybridoma clones were obtained: 1A8, 1F3, 2A4 and 2E3.
Western blot analysis
A total of 20 µg protein lysate per lane, as determined by BCA method (Bio-Rad Laboratories, Inc.), was separated using 12% SDS-PAGE and loaded onto a nitrocellulose membrane. The membrane with ANKRD22 protein was cut into strips, blocked with 5% skimmed milk powder at room temperature for 1 h and washed with PBS mixed with 0.05% Tween-20 (PBST) five times (3 min each time). Each membrane strip was then incubated overnight with the aforementioned mouse anti-ANKRD22 antibodies (1:500) at 4°C. The strips were then incubated with a horseradish peroxidase-labeled goat anti-mouse immunoglobulin G (IgG) (H+L) secondary antibody (1:2,000; cat. no. 115-036-003; Jackson Immunoresearch Laboratories, Inc.) for 1 h at room temperature. The protein bands were visualized using an enhanced chemiluminescence reagent (Pierce; Thermo Fisher Scientific, Inc.) for 1 min, and the signal was detected using a digital imaging system (LI-COR Biosciences).
ELISA
For determination of the specificity of the monoclonal antibodies
A competitive-ELISA was used for validation of the specificity of the monoclonal antibodies. Briefly, 10 µg/ml aforementioned recombinant ANKRD22 protein was coated onto microplate wells and incubated at 4°C overnight. A total of 5 µg/ml truncated ANKRD22 (2nd-89th) peptide (Pan et al, unpublished data) was used as a blocking protein and incubated with the aforementioned four clones of anti-ANKRD22 monoclonal antibody (1A8, 1F3, 2A4 and 2E3; 1 µg/ml) for 30 min at room temperature prior to being added to the wells. The ANKRD22 blocking peptide was replaced with 1% calf bovine serum (Hangzhou Tianhang Biotechnology Co., Ltd.) as a negative control.
For determination of ANKRD22 protein in tissue lysates
A total of 10 µg/ml aforementioned recombinant ANKRD22 protein was coated onto microplate wells and incubated at 4°C overnight. Subsequently, 60 µl lysates from four gastric cancer tissues or recombinant ANKRD22 protein (10, 50, 200, 500 and 1,000 ng/ml) were mixed with 60 µl anti-ANKRD22 monoclonal antibody (clone 1A8; 1 µg/ml) for 60 min at room temperature prior to being added to the wells. The mixed solution was added to the wells and incubated at 37°C for 60 min.
Subsequently, the microplates were incubated at 37°C for 2 h. After washing three times with PBST, 100 µl horseradish peroxidase-labeled goat anti-mouse IgG (1:2,000; cat. no. 115-036-003, Jackson Immunoresearch Laboratories, Inc.) was added to each well and incubated at 37°C for 1 h. The samples were subsequently washed three times with PBST, and 100 µl 3,3′5,5′-tetramethylbenzidine (TMB; Sigma-Aldrich; Merck KGaA) substrate solution was added to each well, incubated at room temperature for 10 min, and terminated with 1.8 M sulfuric acid (50 µl/well). Absorbance was measured at 450 nm using a microplate reader.
Reverse transcription-quantitative PCR (RT-qPCR)
A total of four fresh human gastric cancer tissues obtained by gastrectomy were used for total RNA extraction using TRIzol® reagent (Invitrogen, Thermo Fisher Scientific, Inc.). 1.0 µg extracted RNA was quantified and reverse-transcribed to cDNA by incubation at 42°C for 60 min using a PrimeScript™ RT reagent (Takara Bio, Inc.). Subsequently, ANKRD22 was quantified using qPCR with the Premix Ex Taq™ kit (Takara Bio, Inc.) on a CFX connect system (Bio-Rad Laboratories, Inc.). The primers and probes used were as follows: ANKRD22 forward, 5′-CCAGCTTGGACTTCTAGGGA-3′ and reverse, 5′-GGCAGATGGGCTCAGAGTAT-3′; probe, 5′-FAM-TCCCATGCTGGTCCTTCACAGG-TAMRA-3′; GAPDH forward, 5′-ATCATCCCTGCCTCTACTGG-3′ and reverse, 5′-GTCAGGTCCACCACTGACAC-3′; and probe, 5′-FAM-ACCTTGCCCACAGCCTTGGC-TAMRA-3′ (all from Sangon Biotech Co., Ltd.). The following thermocycling conditions were used: Initial denaturation at 95°C for 30; 40 cycles of 95°C for 5 sec and 60°C for 30 sec. GAPDH was used as an internal reference. Relative ANKRD mRNA expression was quantified using the 2−ΔΔCq method (18).
Immunohistochemical staining
Immunohistochemical staining was performed using standard methods. Paraffin-embedded tissues were sliced into 5–7 µm sections. Following hydration, citric acid buffer was used at 95°C for 10 min for antigen retrieval. The sections were blocked at room temperature with 10% normal goat serum (Fuzhou Maxim Biotechnology Co. Ltd.) and incubated with the mouse anti-ANKRD22 primary antibody (clone 1A8; 1:200) at room temperature for 1 h. Subsequently, the sections were washed 5 times in PBST and incubated for 30 min at room temperature with ready-to-use streptavidin-peroxidase-labeled goat anti-mouse IgG solution (cat. no. KIT-9710, Fuzhou Maxim Biotechnology Co. Ltd.). The sections were developed with 3,3′-diaminobenzidine (cat. no. KIT-9710, Fuzhou Maxim Biotechnology Co. Ltd.) at room temperature for 15 min, counterstained with hematoxylin for 1 min, and examined by light-microscopy at ×400 magnification.
Statistical analysis
Data were analyzed using SPSS 22.0 software (IBM Corp.). The data from TCGA database were imported into an Excel spreadsheet for normalization, and ANKRD22 mRNA expression was subsequently retrieved from the dataset. Student's t-test was used for the comparison of ANKRD22 mRNA expression between prostate cancer and normal tissues. Kaplan-Meier analysis was used to investigate the association between ANKRD22 level and survival of prostate cancer. ANOVA with the Least Significance Difference (LSD) post hoc test was used to analyze the ANKRD22 expression and pathological parameters of prostate cancer. P<0.05 was considered to indicate a statistically significant difference.
Results
Preparation of mouse anti-human ANKRD22 monoclonal antibodies
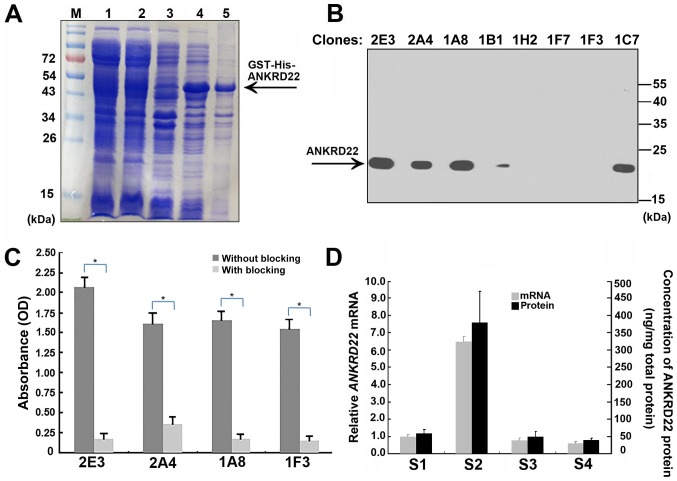
To prepare mouse anti-human ANKRD22 monoclonal antibodies, recombinant human ANKRD22 protein was produced from E. coli using the pET42a expression system. Human ANKRD22 protein exists in the form of inclusion bodies in E. coli bacteria. Following renaturing of the inclusion bodies, hydrosoluble ANKRD22 protein was then purified using Ni2+ affinity columns (Fig. 1A). BALB/c mice were immunized with the purified recombinant protein. A total of 93 candidate hybridoma cells were obtained and were further screened using lysates of 293T cells with overexpression of ANKRD22 using ELISA and western blot analysis. A total of four anti-ANKRD22 monoclonal antibodies were identified (Fig. 1B). Furthermore, all these antibodies could be almost completely blocked by the 2nd to 89th ANKRD22 peptide, indicating the high specification of anti-ANKRD22 antibodies (Fig. 1C). Finally, the association between ANKRD22 protein and ANKRD22 mRNA from gastric cancer tissues was analyzed. Protein levels determined using ELISA were in accordance with levels of ANKRD22mRNA detected using RT-qPCR, thus confirming affinity between these antibodies and human ANKRD22 protein (Fig. 1D).
Figure 1.
Preparation and identification of anti-ANKRD22 monoclonal antibodies. (A) Preparation of recombinant ANKRD22 protein in prokaryotic cells. M, Marker; lane 1 and 2, whole lysate of E. coli bacteria transformed with ANKRD22/pET-42a plasmids induced using isopropyl β-D-1-thiogalactopyranoside; lane 3, bacterial lysate supernatant; lane 4, bacterial precipitates; lane 5, partially purified recombinant ANKRD22 protein by Ni2+ affinity chromatography following renaturation. The SDS-PAGE gel was stained using Coomassie brilliant blue dye. (B) ANKRD22 protein was detected using western blot analysis with different anti-ANKRD22 monoclonal antibodies. Lysates of 293T cells with overexpression of ANKRD22 were used as test samples. A total of 4 clones of ANKRD22 monoclonal antibodies (clone no. 1A8, 1B1, 2E4 and 1F3) were successfully verified. (C) Determination of the specificity of the monoclonal antibodies was validated by competitive-ELISA. The results showed that activity of each monoclonal antibody could be blocked by ANKRD22 antigen. Each experiment was repeated 6 times. *P<0.01. (D) Analysis of the association between ANKRD22 protein and ANKRD22 mRNA. Comparison of the relative levels of ANKRD22 determined using ELISA and RT-qPCR in four gastric cancer tissue samples (S1-4). Clone 1A8 was used as the primary antibody in ELISA. GAPDH was used as the internal inference in RT-qPCR. The data were normalized to ANKRD22 mRNA of S1. ANKRD22, ankyrin repeat domain 22; GST, glutathione S-transferase; His, histidine; RT-qPCR, reverse transcription-quantitative PCR.
Expression of ANKRD22 in prostate cancer
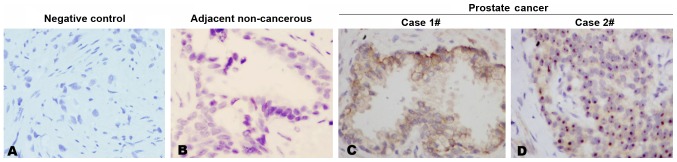
Using the generated anti-ANKRD22 monoclonal antibodies, immunohistochemical staining was performed in prostate cancer tissue. It was demonstrated that each anti-ANKRD22 monoclonal antibody exhibited a strong reaction with ANKRD22 in prostate cancer tissues and each prostate cancer tissue sample was stained positive for ANKRD22 in all 11 clinical samples. Four clones exhibited similar IHC patterns. Expression of ANKRD22 in prostate cancer was higher compared with that in adjacent non-cancerous tissue (Fig. 2A-C). ANKRD22 was located in the cytoplasm and nucleus of prostate cancer cells (Fig. 2C and D). These results indicated that ANKRD22 may be involved in the progression of prostate cancer.
Figure 2.
Detection of ANKRD22 in prostate cancer. Immunohistochemical staining with monoclonal antibody (ANKRD22 clone no. 1A8) was used to detect ANKRD22 expression in prostate cancer tissues. (A) Negative control. A section of prostate cancer tissue without primary antibodies was used as an immunohistochemical negative control. (B) Detection of ANKRD22 in adjacent non-cancerous tissue. (C and D) ANKRD22 expression in the (C) cytoplasm and (D) nucleus of prostate cancer cells. Magnification, ×400. ANKRD22, ankyrin repeat domain 22.
ANKRD22 is associated with prostate cancer aggressiveness
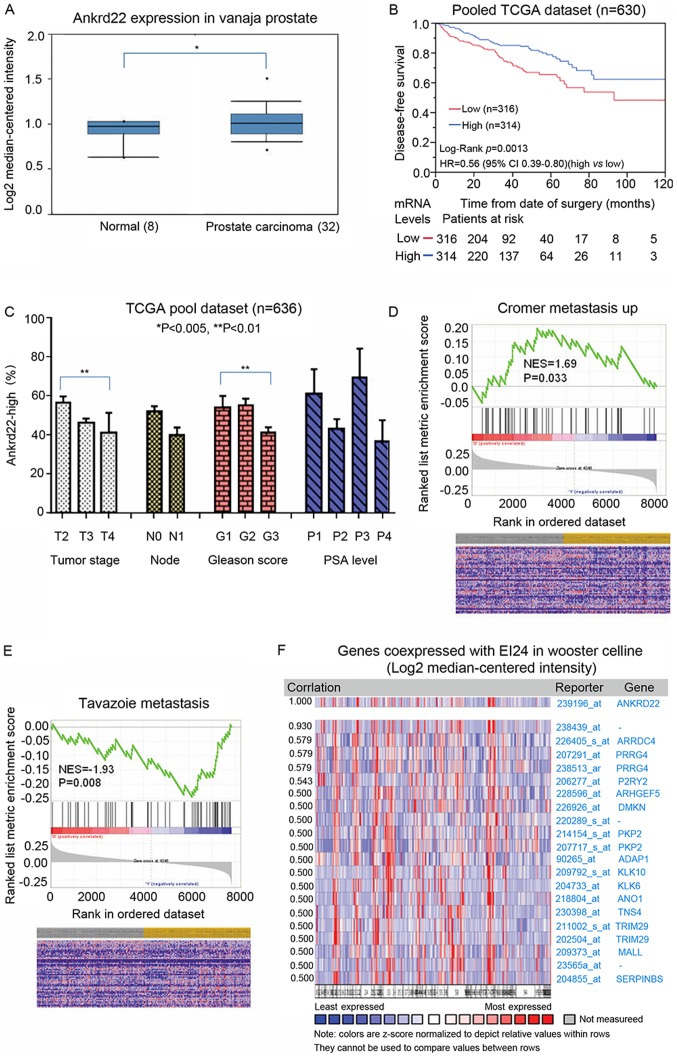
Our previous study demonstrated that ANKRD22 is a tumor suppressor in colorectal cancer (Pan et al, unpublished data). As such, the role of ANKRD22 in the aggressiveness of prostate cancer and the subsequent patient outcomes were investigated. In accordance with the immunohistochemical staining results, TCGA analysis demonstrated that ANKRD22 was higher in prostate cancer compared with that in normal prostate (Fig. 3A). Data from TCGA datasets suggested that patients with high expression of ANKRD22 exhibited significantly improved prognosis compared with those with low expression (Fig. 3B), thus suggesting that ANKRD22 is a good prognostic biomarker for prostate cancer.
Figure 3.
Analysis of the association between the expression of ANKRD22 and the prognosis of patients with prostate cancer. (A) The expression of ANKRD22 mRNA in prostate cancer tissue was higher compared with that in normal prostate tissue. ANKRD22 in normal prostate tissue and prostate cancer is available from TCGA database. (B) Analysis of the association between ANKRD22 level and survival time of patients with prostate cancer. Kaplan-Meier analysis of disease-free survival for prostate cancer with low vs. high expression levels of ANKRD22. Long-term disease-free survival time of patients with higher ANKRD22 level was longer than the lower group. (C) The association between ANKRD22 and pathological parameters of prostate cancer. Parameters of prostate cancer include staging, lymph node metastasis, Gleason score and prostate-specific antigen level. Data were obtained from TCGA database. (D and E) Gene set enrichment analysis of the association between ANKRD22 and metastasis of prostate cancer. Data from (D) Cromer and (E) Tavazoie metastasis gene sets demonstrated that ANKRD22 level was negatively associated with metastasis of prostate cancer. The grey column represents the low ANKRD22 mRNA expression group; the yellow column represents the high ANKRD22 mRNA expression group. (F) Gene expression profile analysis of co-expressed molecules with ANKRD22 from TCGA data. *P<0.05 and **P<0.01. ANKRD22, ankyrin repeat domain 22; TCGA, The Cancer Genome Atlas; NES, normalized enrichment score.
The association between ANKRD22 expression and prostate cancer staging, lymph node metastasis, Gleason score and prostate-specific antigen (PSA) level was then explored through analysis of TCGA datasets. The proportion of patients with high ANKRD22 expression in stage IV prostate cancer patients was significantly lower compared with that in patients with stage II prostate cancer, and a low Gleason score was associated with high ANKRD22 expression (G1 vs. G3; P<0.01). However, no association was observed between the proportion of ANKRD22-high patients and lymph node metastasis or PSA level (Fig. 3C). Additionally, in GSEA, high expression of ANKRD22 was negatively associated with prostate metastasis (metastatic vs. non-metastatic prostate cancer) (Fig. 3D and E) and ANKRD22 was significantly co-expressed with tumor suppressor EI24 (Fig. 3F). These results indicate that ANKRD22 is a good prognostic target for prostate cancer and may be a tumor suppressor in prostate cancer.
Discussion
To the best of our knowledge, the present study is the first to demonstrate that the level of ANKRD22 has an inverse association with prostate cancer progression. ANKRD22 may be expressed in either the cytoplasm or nuclei of prostate cancer cells, although the underlying molecular mechanism is not clear. Furthermore, it was discovered that high-grade and high-stage prostate cancer exhibited a significantly reduced ANKRD22 expression. Patients with higher ANKRD22 mRNA levels exhibited longer disease-free survival post-prostatectomy.
ANKRD22 is an ankyrin repeat protein with four copies of the ankyrin motif. Ankyrin motif is one of the most common protein-protein interaction structural motifs, located in >400 proteins (19). Each ankyrin repeat is an L-shaped structure consisting of two inverted α helices and a β hairpin, with a size of ~33 amino acid residues, and forms a molecular connection scaffold structure with high affinity (20). Unlike other protein-protein binding motifs, ankyrin repeat motifs are primarily classified based on their specific tertiary structures, which determine the proteins they interact with and therefore their specific biological functions. The ankyrin repeats are highly conserved in evolution, although they interact with a diverse range of proteins (21). This protein-protein interaction is involved in the regulation of many cellular physiological activities, such as transcriptional regulation, cytoskeleton, ion transport, signal transduction, inflammation and immunity, and tumorigenesis (22–24). This suggests, therefore, that ANK family protein members, such as ANKRD22, are involved in cancer.
ANKRD22 is expressed in normal digestive tract epithelial and tumor cells. Yin et al (11) observed high expression of ANKRD22 in non-small cell lung cancer cells, which is induced by upregulation of transcription factor E2F1. Venner et al (25) observed an increase of ANKRD22 expression in macrophages in acute renal allograft rejection, indicating that ANKRD22 is a negative regulator of the interferon pathway similar to interferon-inducible protein AIM2 (26). Both oncogenic and tumor suppressor functions have previously been reported in ANK proteins. For example, the Notch pathway receptors (Notch 1–4) are ANK family members that contain six ankyrin repeats (27). Activation of the Notch pathway increases stemness and inhibits differentiation (28). Compared with non-stem cells, ANKRD28 is upregulated in acute myeloid leukemia stem cells (29). Apoptosis-stimulation of p53 protein 2, which contains four ankyrin repeats, was previously reported to induce stemness and chemoresistance in hepatoma cells (30). By contrast, ankyrin repeat and SOCS box protein 4 is highly expressed in the early stage of placental development and mediates differentiation of embryonic stem cells into the vascular system (31). ANKRD3 is reported to be associated with differentiation of neural stem cells (32). ANKRD6 recruits casein kinase I isoform-ϵ to the β-catenin degradation complex and inhibits β-catenin/transcription factor signaling (33). In the present study, the potential role of ANKRD22 in prostate cancer as a tumor suppressor was uncovered, similar to the study by Schoenborn et al (34) where the copy number of ANKRD22 was decreased in prostate cancer. By contrast, a recent study demonstrated that ANKRD22 promoted progression of non-small cell lung cancer (11). This suggests that ANKRD22 may exhibit either pro- or antitumor function depending on the origin of the tumor.
Taken together, these results indicate that ANKRD22 may be involved in the progression of prostate cancer. The mechanisms of how ANKRD22 affects tumorigenesis of prostate cancer remains to be defined. Identifying the targets of ANKRD22 and biological functions of their interaction in prostate cancer progression remains to be further discussed.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the Natural Science of Foundation of Zhejiang Province (grant. no. Y17H050034).
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YQ and HW contributed to the conception and design of the present study. HW also provided administrative support. YQ, SY and TP performed experiments. YQ, LY and JL were responsible for the collection and assembly of data. YQ, LY and YZ performed data analysis and interpretation. All authors contributed to the writing of the manuscript and its final approval.
Ethics approval and consent to participate
The research protocol was approved by the Second Affiliated Hospital of Zhejiang University Ethics Committee, and all patients provided written informed consent. The animal research protocol was approved by the Animal Care and Welfare Committee of Zhejiang Chinese Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zhang S, Zeng H, Xia C, Zuo T, Yang Z, Zou X, He J. Cancer incidence and mortality in China 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Scher HI, Solo K, Valant J, Todd MB, Mehra M. Prevalence of prostate cancer clinical states and mortality in the United States: Estimates using a dynamic progression model. PLoS One. 2015;10:e0139440. doi: 10.1371/journal.pone.0139440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye D, Zhu Y. Epidemiology of prostate cancer in China: An overview and clinical implication. Zhonghua Wai Ke Za Zhi. 2015;53:249–252. (In Chinese) [PubMed] [Google Scholar]
- 6.Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, Tannock I, Drake CG, de Bono JS. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 7.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schepisi G, Farolfi A, Conteduca V, Martignano F, De Lisi D, Ravaglia G, Rossi L, Menna C, Bellia SR, Barone D, et al. Immunotherapy for prostate cancer: Where we are headed. Int J Mol Sci. 2017;18:E2627. doi: 10.3390/ijms18122627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrecengost R, Knudsen KE. Molecular pathogenesis and progression of prostate cancer. Semin Oncol. 2013;40:244–258. doi: 10.1053/j.seminoncol.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin J, Fu W, Dai L, Jiang Z, Liao H, Chen W, Pan L, Zhao J. ANKRD22 promotes progression of non-small cell lung cancer through transcriptional up-regulation of E2F1. Sci Rep. 2017;7:4430. doi: 10.1038/s41598-017-04818-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennink I, van Leeuwen MW, Penning LC, Piek CJ. Increased number of tissue factor protein expressing thrombocytes in canine idiopathic immune mediated hemolytic anemia. Vet Immunol Immunopathol. 2018;196:22–29. doi: 10.1016/j.vetimm.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Chen Y, Xiong X, Zhou X, Han L, Ni L, Wang W, Wang X, Zhao L, Shao D, Huang C. Peptidome analysis reveals novel serum biomarkers for children with autism spectrum disorder in China. Proteomics Clin Appl. 2018;12:e1700164. doi: 10.1002/prca.201700164. [DOI] [PubMed] [Google Scholar]
- 14.Roell W, Klein AM, Breitbach M, Becker TS, Parikh A, Lee J, Zimmermann K, Reining S, Gabris B, Ottersbach A, et al. Overexpression of Cx43 in cells of the myocardial scar: Correction of post-infarct arrhythmias through heterotypic cell-cell coupling. Sci Rep. 2018;8:7145. doi: 10.1038/s41598-018-25147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fragiadaki M, Zeidler MP. Ankyrin repeat and single KH domain 1 (ANKHD1) drives renal cancer cell proliferation via binding to and altering a subset of miRNAs. J Biol Chem. 2018;293:9570–9579. doi: 10.1074/jbc.RA117.000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai R, Li D, Shi Z, Fang X, Ge W, Zheng S. Clinical significance of Ankyrin repeat domain 12 expression in colorectal cancer. J Exp Clin Cancer Res. 2013;32:35. doi: 10.1186/1756-9966-32-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wissing ML, Kristensen SG, Andersen CY, Mikkelsen AL, Høst T, Borup R, Grøndahl ML. Identification of new ovulation-related genes in humans by comparing the transcriptome of granulosa cells before and after ovulation triggering in the same controlled ovarian stimulation cycle. Hum Reprod. 2014;29:997–1010. doi: 10.1093/humrep/deu008. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Chakrabarty B, Parekh N. Identifying tandem Ankyrin repeats in protein structures. BMC Bioinformatics. 2014;15:6599. doi: 10.1186/s12859-014-0440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller MK, Bang ML, Witt CC, Labeit D, Trombitas C, Watanabe K, Granzier H, McElhinny AS, Gregorio CC, Labeit S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003;333:951–964. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Parra RG, Espada R, Verstraete N, Ferreiro DU. Structural and energetic characterization of the ankyrinrepeat protein family. PLoS Comput Biol. 2015;11:e1004659. doi: 10.1371/journal.pcbi.1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanno H, Yamaguchi T, Goto E, Ishido S, Komada M. The Ankrd 13 family of UIM-bearing proteins regulates EGF receptor endocytosis from the plasma membrane. Mol Biol Cell. 2012;23:1343–1353. doi: 10.1091/mbc.e11-09-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wette SG, Smith HK, Lamb GD, Murphy RM. Characterization of muscle ankyrin repeat proteins in human skeletal muscle. Am J Physiol Cell Physiol. 2017;313:C327–C339. doi: 10.1152/ajpcell.00077.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan W, Sun K, Tang K, Xiao Q, Ma C, Yu C, Wei Z. Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion. J Biol Chem. 2018;293:1944–1956. doi: 10.1074/jbc.M117.815779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venner JM, Famulski KS, Badr D, Hidalgo LG, Chang J, Halloran PF. Molecular landscape of T cell-mediated rejection in human kidney transplants: Prominence of CTLA4 and PD ligands. Am J Transplant. 2014;14:2565–2576. doi: 10.1111/ajt.12946. [DOI] [PubMed] [Google Scholar]
- 26.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud Å, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubman OY, Korolev SV, Kopan R. Anchoringnotchgenetics and biochemistry; structural analysis of the ankyrin domain sheds light on existing data. Mol Cell. 2004;13:619–626. doi: 10.1016/S1097-2765(04)00120-0. [DOI] [PubMed] [Google Scholar]
- 28.Huan YW, Bengtsson RJ, MacIntyre N, Guthrie J, Finlayson H, Smith SH, Archibald AL, Ait-Ali T. Lawsonia intracellularis exploits β-catenin/Wnt and Notch signalling pathways during infection of intestinal crypt to alter cell homeostasis and promote cell proliferation. PLoS One. 2017;12:e0173782. doi: 10.1371/journal.pone.0173782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jonge HJ, Woolthuis CM, Vos AZ, Mulder A, van den Berg E, Kluin PM, van der Weide K, de Bont ES, Huls G, Vellenga E, Schuringa JJ. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia. 2011;25:1825–1833. doi: 10.1038/leu.2011.172. [DOI] [PubMed] [Google Scholar]
- 30.Xu L, Tong X, Zhang S, Yin F, Li X, Wei H, Li C, Guo Y, Zhao J. ASPP2 suppresses stem cell-like characteristics and chemoresistance by inhibiting the Src/FAK/Snail axis in hepatocellular carcinoma. Tumour Biol. 2016;37:13669–13677. doi: 10.1007/s13277-016-5246-0. [DOI] [PubMed] [Google Scholar]
- 31.Lahiani A, Zahavi E, Netzer N, Ofir R, Pinzur L, Raveh S, Arien-Zakay H, Yavin E, Lazarovici P. Human placental eXpanded (PLX) mesenchymal-like adherent stromal cells confer neuroprotection to nerve growth factor (NGF)-differentiated PC12 cells exposed to ischemia by secretion of IL-6 and VEGF. Biochim Biophys Acta. 2015;1853:422–430. doi: 10.1016/j.bbamcr.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Ju X, Cheng Y, Guo X, Wen T. Identifying Tmem59 related gene regulatory network of mouse neural stem cell from a compendium of expression profiles. BMC Syst Biol. 2011;5:152. doi: 10.1186/1752-0509-5-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hino S, Michiue T, Asashima M, Kikuchi A. Casein kinase I epsilon enhances the binding of Dvl-1 to Frat-1 and is essential for Wnt-3a-induced accumulation of beta-catenin. J Biol Chem. 2003;278:14066–14073. doi: 10.1074/jbc.M213265200. [DOI] [PubMed] [Google Scholar]
- 34.Schoenborn JR, Nelson P, Fang M. Genomic profiling defines subtypes of prostate cancer with the potential for therapeutic stratification. Clin Cancer Res. 2013;19:4058–4066. doi: 10.1158/1078-0432.CCR-12-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.