Abstract
CD44 is involved in malignant processes including cell motility, tumor growth and angiogenesis. To explore the potential role of CD44 as a prognostic biomarker in low grade gliomas (LGG), the mRNA expression levels of CD44 in tissues from 12 patients with glioma were evaluated by microarray analysis. The mRNA level of CD44 in LGG and glioblastoma multiforme (GBM) were analyzed using datasets downloaded from the publicly available Oncomine database. Reverse transcription-quantitative PCR and western blotting were used to further analyze the CD44 expression level in a set of 53 patients. Kaplan-Meier analysis was performed to identify the prognostic roles of CD44 mRNA in LGG and GBM, with data obtained from the OncoLnc and Gene Expression Profiling Interactive Analysis databases and clinical follow-ups. The present results revealed that CD44 mRNA expression levels were elevated in LGG and GBM compared with normal brain tissues. Furthermore, increased CD44 expression was associated with poor survival rates in LGG. The present study suggested that CD44 may act as an independent prognostic factor for LGG, and may be a potential therapeutic target for gliomas.
Keywords: CD44, glioma, prognosis, bioinformatics analysis
Introduction
Gliomas are the most common malignant primary brain tumors, and are categorized, according to the 2016 World Health Organization (WHO) classification, as low-grade (grade I–II) and high-grade (grade III–IV) (1). Low-grade gliomas (LGGs) are infiltrative neoplasms that most frequently arise in the cerebral hemispheres of adults, and include astrocytomas, oligodendrogliomas and oligoastrocytomas (1). LGGs generally occur in young adults between 35 and 44 years of age (2). Despite a better prognosis compared with HGG, complete neurosurgical resection of LGGs is challenging due to their invasive nature, increased vascularity and lack of a well-defined tumor capsule. Furthermore, certain cases LGG may rapidly progress to WHO grade IV glioma, becoming a glioblastoma multiforme (GBM), whereas others may remain stable for a long time (3). The survival time of patients with LGG ranges from 1 to 15 years (4).
Although traditional histopathological methods are regarded as the gold standard for LGG classification, they do not adequately predict clinical outcome. Consequently, an increasing number of studies have shown that biomarkers may improve the clinical decision-making process (5). Hypermethylation of the O6-methylguanine-DNA methyltransferase promoter can effectively predict the responsiveness to temozolomide (6). Mutations in isocitrate dehydrogenase (NADP+) 1 (IDH1) and IDH2 may result in an improved prognosis for patients with LGG. Mutations in IDH and the heterozygous deletion at the chromosomal position 1p/19q may result in improved responses to radiotherapy and chemotherapy, and patients with these mutations have longer survival times than patients without these mutations (7–10).
The present study investigated novel prognostic and predictive biomarkers for LGG using an mRNA PCR array to screen for genes with altered expression in LGG. The prognostic value of newly identified biomarkers was subsequently investigated using the OncoLnc and Gene Expression Profiling Interactive Analysis (GEPIA) databases, and biomarker mRNA level was analyzed using the Oncomine database.
Materials and methods
Patient specimens
The study included 53 glioma tissues and formalin-fixed paraffin-embedded tumor tissues. Glioma tissues were fixed overnight at 4°C in 10% neutral-buffered formalin, dehydrated by soaking in 70, 85, 95 and 100% ethanol for 45 min each time at room temperature, washed in xylene twice for 15 min each at room temperature, soaked twice in paraffin for 1 h each at 56°C, embedded in paraffin and subsequently stored at 4°C until use. Six patients with LGG and six patients with GBM were recruited from the Neurology Institute of Lanzhou University Second Hospital (Lanzhou, China) between January 2013 and December 2017. Of the patients recruited in the present study, 31 were male and 22 were female, with a mean age of 48 years (range, 20–72 years). A total of five normal brain tissue samples were obtained from patients without glioma who underwent surgery for other reasons, including cerebral trauma. The glioma and normal tissue specimens were snap-frozen in liquid nitrogen following surgery, and subsequently stored at −80°C until use. Histological diagnosis was based on the criteria stated by the WHO. According to the 2016 World Health Organization Classification of Tumors of the Central Nervous System (1), gliomas were divided into four categories: i) WHO I (pilocytic astrocytoma); ii) WHO II (diffuse astrocytoma, oligoastrocytoma and oligodendroglioma); iii) WHO III (anaplastic astrocytoma, anaplastic oligodendroglioma and anaplastic oligoastrocytoma); and iv) WHO IV (glioblastoma). Patients undergoing surgery who had not been previously treated with radiotherapy or chemotherapy were included in the present study. Approval for the study was obtained from The Medical Ethics Committee of The Affiliated Second Hospital of Lanzhou University. Written informed consent was obtained from all recruited patients.
Human mRNA PCR array
After dewaxing, total RNA was extracted from the tumor samples of 12 randomly selected patients using TRIzol® reagent (Thermo Fisher Scientific, Inc.). First strand cDNA was synthesized using the mRNA First-Strand cDNA Synthesis kit (cat. no. AS-MR-004; Arraystar, Inc.) according to the manufacturer's protocol. The cDNA products were amplified using TB SYBR green Premix Ex Taq II (Takara Biotechnology Co., Ltd.). The Human mRNA PCR Array (cat. no. AS-MR-0033, Arraystar, Inc.) was used with the ABI PRISM7900 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling condition of the qPCR were: Initial denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 15 sec, and annealing and extension at 60°C for 30 sec. Each 364-well miRStar Human Cancer Focus miRNA and Target mRNA PCR Array (Arraystar, Inc.) contains 177 genes associated with human cancer, six wells for housekeeping genes, a genomic DNA contamination control, three replicates of reverse transcription (RT) controls and three replicates of positive PCR controls. The raw data were processed by performing the following analyses: Background detection, robust multi-array average global background correlation, quantile normalization, median adjustment and log2-transformation with miRNA QC tool version 1.1 (Affymetrix; Thermo Fisher Scientific, Inc.).
Oncomine database analysis
The expression level of CD44 in LGG, GBM and matched normal tissues were retrieved from the Oncomine database (http://www.oncomine.org). The expression analysis of CD44 in the Oncomine database was based on a previous study (11). A total of 157 brain and central nervous system tumors and 23 normal brain samples were analyzed on an Affymetrix U133 Plus 2.0 platform (Affymetrix; Thermo Fisher Scientific, Inc.). Normal tissue samples were provided in the Oncomine database. The-fold-change of mRNA expression in LGG and GBM samples, compared with their matched normal tissues, was determined using the following parameters: i) P<1×10−3; and ii) fold-change ≥2-fold.
Kaplan-Meier plotter analysis
The prognostic value of CD44 was analyzed using OncoLnc (www.oncolnc.org/search_results/?q=CD44) and GEPIA (gepia.cancer-pku.cn). Patients were divided into high-level and low-level groups based on the median value of CD44. The overall survival (OS) rates of the patients in the high-level and low-level CD44 groups were evaluated using Kaplan-Meier analysis (gepia.cancer-pku.cn/detail.php?gene=&clicktag=survival).
RT-quantitative PCR (RT-q) PCR
Total RNA was extracted from normal brain and glioma tissues using Trizol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA (2 µg) was reverse transcribed using the PrimeScript RT reagent kit with genomic DNA Eraser (Takara Biotechnology Co., Ltd.). Reverse transcription was performed as follows: 37°C for 15 min and 85°C for 5 sec. The cDNA products were amplified using TB SYBR green Premix Ex Taq II (Takara Biotechnology Co., Ltd.) on a Bio-Rad CFX96 real-time PCR system (Bio-Rad Laboratories, Inc.). The qPCR procedure was performed as follows: Pre-denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 15 sec, and annealing and extension at 60°C for 30 sec. The relative mRNA expression data were analyzed using the 2−ΔΔCq method (10), and expression values were normalized to the internal control GAPDH. The following primers were used: CD44 forward, 5′-CCAACTCCATCTGTGCAG-3′ and reverse, 5′-AACCTCCTGAAGTGCTGC-3′; GAPDH, forward 5′-GGACCTGACCTGCCGTCTAG-3′ and reverse, 5′-TAGCCCAGGAGGATGCCCTTGAG-3′.
Western blotting
The glioma tissues were homogenized and lysed with cell lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.) containing 1% PMSF. The lysates were centrifuged at 12,000 × g for 5 min at 4°C. The protein concentration was determined using a bicinchoninic acid assay. The proteins (30 µg total protein/lane) were separated using SDS-PAGE on a 12% gel. Proteins were transferred to PVDF membranes (EMD Millipore), blocked with 5% fat-free milk for 25°C for 1 h and incubated with primary antibodies against CD44 (cat. no. D190741; 1:750; Sangon Biotech Co., Ltd.) and GAPDH (cat. no. TA-08; 1:1,000; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) at 4°C overnight. The membranes were washed with PBS and 0.1% Tween, and incubated with goat anti-mouse secondary antibody (cat. no. abs20001; 1:2,000; Absin Bioscience Inc.) at 25°C for 1 h. Protein bands were visualized using the enhanced chemiluminescence (PerkinElmer, Inc.) method and a Bio-Rad gel imaging system (Bio-Rad Laboratories, Inc.), and densitometry analysis was performed using ImageJ version 1.8.0 software (National Institutes of Health).
Data analysis
The median mRNA expression levels of CD44 mRNA was used as the cut-off point, and the patients were divided into a high-level and a low-level group. The χ2 test was used to analyze the association between CD44 mRNA expression and clinicopathological characteristics. The OS rates of LGG in the high- and low-level groups were evaluated using Kaplan-Meier analysis. Furthermore, univariate and multivariate Cox proportional hazard regression analysis was performed to evaluate the prognostic value of multiple variables, including CD44 mRNA expression, sex, age and Karnofsky Performance Scale (KPS) (12).
Statistical analysis
All analyses were performed using SPSS software (version 17; SPSS, Inc.). The data are presented as the mean ± SEM. Data were analyzed using one-way ANOVA followed by the least significant difference post hoc test or an unpaired Student's t-test. Survival curves were estimated using the Kaplan-Meier method with the log-rank test. Univariate and multivariate Cox regression analysis was employed to estimate the risk factor of gliomas. P<0.05 was considered to indicate a statistically significant difference. The analyses were performed using GraphPad Prism software (version 5; GraphPad Software, Inc.).
Results
Gene expression analysis
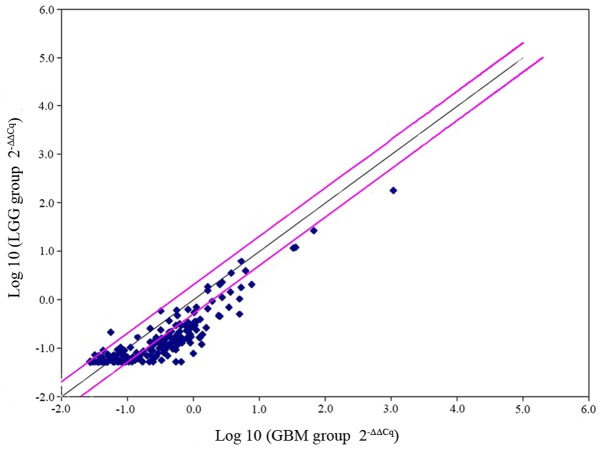
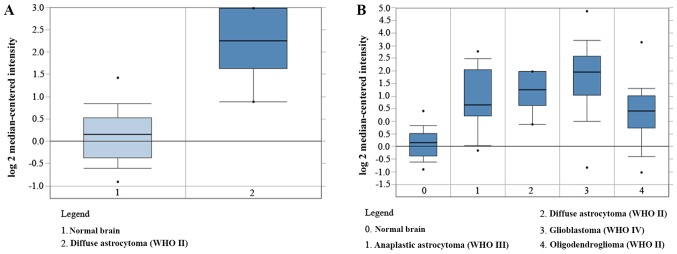
The mRNA profile was analyzed for LGG (n=6) and GBM (n=6) using the miRStar Human Cancer Focus mRNA PCR Array. The median expression levels of each mRNA were calculated in both groups, and the differences between them were determined using a t-test. P<0.05 was considered to indicate a significant difference and a-fold change >2 was used as the cut-off value. The results indicated that of the 177 genes, 44 difference genes were upregulated. CD44 was identified to exhibit a 3-fold increase in GBM (P=0.02; Fig. 1). The results were further validated using Oncomine analysis, which demonstrated that the CD44 mRNA level was significantly higher in GBM compared with LGG and normal brain tissue (Fig. 2A and B).
Figure 1.
Screening of 177 cancer-associated genes by miRStar Human Cancer Focus miRNA and Target mRNA PCR array. The results were analyzed using a PCR array (n=6 samples per group). A total of 44 genes were identified with >2-fold change between LGG and GBM. LGG, low-grade glioma; GBM, glioblastoma multiforme.
Figure 2.
CD44 mRNA expression level was evaluated using Oncomine analysis. (A) CD44 mRNA expression in low-grade glioma. (B) CD44 mRNA expression in different grades of gliomas. Normal brain tissues, WHO II (including diffuse astrocytoma and oligodendroglioma), WHO III (anaplastic astrocytoma) and WHO IV (glioblastoma). WHO, World Health Organization.
CD44 expression in glioma tissues
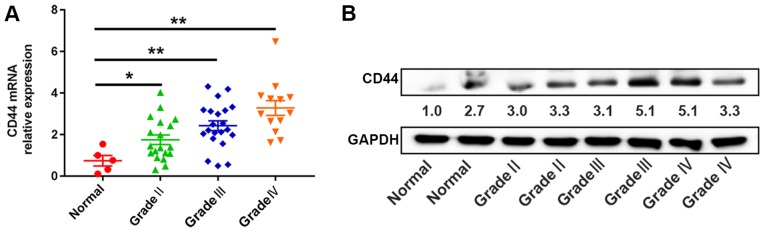
To validate the mRNA PCR array and Oncomine database findings, the expression level of CD44 was determined in the glioma tissue samples (n=53) and normal brain tissues (n=5) using RT-qPCR and western blotting. The CD44 expression level in glioma tissues was significantly increased compared with normal brain tissues (P<0.05; Fig. 3A and B). The data were consistent with the results of the mRNA PCR array and Oncomine database analysis, in which patients were divided into high-level (n=26) and low-level groups (n=27), with the median expression level of CD44 mRNA as the cut-off value. The expression level of CD44 mRNA was associated with KPS (P<0.01) and WHO grade (P<0.01; Table I). However, it was not significantly associated with other clinicopathological parameters, including age and sex.
Figure 3.
Expression levels of CD44 in glioma tissues. (A) Expression patterns of CD44 in different grade glioma tissues were analyzed by reverse-transcription quantitative PCR. (B) The protein expression levels of CD44 in patients with different grade glioma tissues were analyzed by western blotting. The numbers under the blot represents the protein expression levels of CD44 in glioma tissues of different grades relative to the respective GAPDH loading controls. *P<0.05, **P<0.01.
Table I.
Association of the expression level of CD44 mRNA with clinicopathological factors of low-grade glioma and glioblastoma multiforme.
| CD44 mRNA expression | ||||
|---|---|---|---|---|
| Clinicopathological feature | Patients, n | Low, n | High, n | P-value |
| Age | 0.126 | |||
| >50 | 24 | 15 | 9 | |
| ≤50 | 29 | 12 | 17 | |
| Sex | 0.659 | |||
| Male | 31 | 15 | 16 | |
| Female | 22 | 12 | 10 | |
| World health organization grade | <0.01 | |||
| II | 19 | 14 | 5 | |
| III–IV | 34 | 13 | 21 | |
| Karnofsky performance status | <0.01 | |||
| >80 | 19 | 15 | 4 | |
| ≤80 | 34 | 12 | 22 | |
Survival analysis
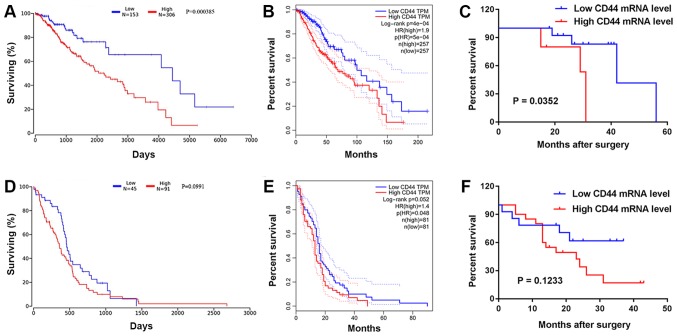
Prognostic value analysis of CD44 mRNA was performed in patients with LGG and GBM using data from the OncoLnc and GEPIA databases. Kaplan-Meier analysis revealed that a high expression level of CD44 in LGG was a predictor of short OS and poor patient outcome, as assessed by OncoLnc (P<0.01) and GEPIA (P<0.01) (Fig. 4A and B). The level of CD44 mRNA did not significantly influence patient outcome, as assessed by OncoLnc (P=0.099) and GEPIA (P=0.052) in GBM (Fig. 4D and E). In addition, Kaplan-Meier analysis and the log-rank test were used to evaluate the prognostic value of CD44 mRNA expression in patients with LGG and GBM according to clinical follow-up data. The present results suggested that patients with LGG with a high expression level of CD44 had a significantly shorter OS than the low expression group (P=0.035; Fig. 4C). There was no significant difference in patients with GBM (P=0.123; Fig. 4F). Furthermore, Cox regression analysis revealed that the expression level of CD44 was significantly associated with the OS of LGG patients (hazard ratio, 3.7012; 95% CI, 0.927–11.215; P=0.032; Table II).
Figure 4.
Kaplan-Meier curves for OS in patients with gliomas. (A) Prognostic significance of CD44 was analyzed in LGG using the OncoLnc databases. (B) Prognostic significance of CD44 was analyzed in LGG using the GEPIA databases. (C) Analysis of survival curves in patients with LGGs by Kaplan-Meier survival analysis. (D) The prognostic significance of CD44 was analyzed in GBM by OncoLnc databases. (E) The prognostic significance of CD44 was analyzed in GBM by GEPIA databases. (F) Analysis of survival curves in patients with GBM by Kaplan-Meier survival analysis. OS, overall survival; LGG, low-grade glioma; GEPIA, Gene Expression Profiling Interactive Analysis; GBM, glioblastoma multiforme; HR, hazard ratio.
Table II.
Univariate and multivariate analyses of the prognostic parameters of patients with low-grade glioma.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (>50 vs. ≤50) | 2.932 | 0.679–5.214 | 0.127 | 1.515 | 0.212–2.534 | 0.099 |
| Sex (male vs. female) | 0.501 | 0.038–3.011 | 0.450 | 3.715 | 0.611–8.425 | 0.679 |
| Karnofsky performance status (>80 vs. ≤80) | 0.246 | 0.001–1.091 | 0.014 | 0.417 | 0.046–1.914 | 0.034 |
| CD44 mRNA expression (low vs. high) | 3.7012 | 0.927–11.215 | 0.032 | 2.862 | 0.436–5.534 | 0.042 |
HR, hazard ratio.
Discussion
Despite recent progresses in neurosurgery, the survival rate of patients with LGG varies widely. To improve prediction accuracy for LGG, molecular markers such as ATRX chromatin remodeler, p53 and telomerase reverse transcriptase have been established (13,14). Furthermore, IDH mutation and 1p/19q codeletion were used in the 2016 WHO classification of central nervous system tumors (1). IDH-wild-type and IDH-mutants are observed in 90 and 10%, respectively, of all GBM cases (15). Present studies for the classification of glioma are focused on high-grade glioma; therefore, biomarkers relevant for the prognostic stratification of patients with LGG remain limited.
In recent years, there have been rapid developments in molecular biology techniques, including chromatin immunoprecipitation and high-throughput sequencing, as well as in the application of bioinformatics methods. Advances in these techniques have allowed the investigation of the occurrence and development of glioma at the molecular level (16–18). The present study used a PCR array, and identified CD44 expression level to be associated with the histopathological grade of gliomas. Western blotting suggested that CD44 expression was higher in patients with high-grade glioma compared with patients with LGG and controls. Similar results were obtained using an mRNA PCR array and Oncomine analysis. However, there was no significant difference between grade III and IV glioma.
CD44 primarily regulates cell-to-cell and cell-to-extracellular matrix adhesion to preserve the organizational structure of tissues and organ. Additionally, CD44 is involved in cell migration, signaling, proliferation, angiogenesis, lymphocyte function and metastasis (19). In glioma, CD44 serves as an oncogene, and can promote GBM cell migration and invasion by influencing the mitogen-activated protein kinase/EPH receptor B2, epidermal growth factor and protein kinase B signaling pathways (20–22). Xu et al (23) demonstrated that CD44 is upregulated in GBM and promoted GBM cell growth and increased resistance to reactive oxygen species- and cytotoxic agent-induced stress by inhibiting the Hippo signaling pathway. Although CD44 expression was positively correlated with malignancy in GBM, higher levels of CD44 were additionally correlated with decreased survival rates in these patients (24). However, the present study showed no statistically significant association between CD44 expression and OS in patients with GBM. The reason may be that the tumorigenicity of primary GBM differs between CD44low/CD133high and CD44high/CD133low for gene expression profiles (25). Furthermore, molecular heterogeneity among tumors affects the prognostic value of CD44 in GBM (26).
CD44 is highly upregulated in prostate, lung and pancreatic cancer, and is associated with poor prognosis (27–29). Nevertheless, the expression pattern and functional role of CD44 in LGG has not been fully elucidated. The present study identified that a high expression level of CD44 in patients with LGG were significantly associated with poor OS, and Kaplan-Meier analysis demonstrated that the expression level of CD44 may be used as a prognostic marker for patients. A recent study revealed that cells with low expression level of CD44 exhibited more glioma stem cell (GSC) traits, suggesting that CD44 is not an appropriate marker for GSCs, but CD44 can be used for predicting glioma-associated invasion and migration (30).
In conclusion, CD44 expression levels were associated with the histopathological grade of gliomas. Higher expression levels of CD44 mRNA were found in LGG and GBM tissues compared with normal brain tissues. A high expression level of CD44 mRNA was identified to be associated with a poor survival rate in LGG. The present results suggested that CD44 may be a novel prognostic biomarker that may improve OS prediction in LGG and serve as a potential therapeutic target for glioma. However, a larger sample size is required to further investigate the present results. Further experimental studies are required to elucidate the functions of CD44 in the progression of glioma.
Acknowledgements
Not applicable.
Funding
This project was supported by the Natural Science Foundation of Gansu (grant nos. 18JR3RA365 and 18JR3RA309), the Research Fund from The Project of the Health and Family Planning Commission of Gansu (grant no. GSWSKY-2014-31/2015-58), the Lanzhou Science and Technology Bureau Project (grant no. 2018-1-109), and the Doctoral Research Fund and Cuiying Science and Technology fund of Lanzhou University Second Hospital (grant nos. YNBSKYJJ2015-1-02/2015-2-11/2015-2-5 and CY2017-MS12/-MS15/CYXZ-01).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the OncoLnc (http://www.oncolnc.org/search_results/?q=CD44) and GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=&clicktag=survival) databases.
Authors' contributions
YP, GY and QD conceived the project. QD, QL, MW and JH performed the experiments. QD, QL and GY analyzed the data. QL, JH, LN and JD interpreted the data and revised the manuscript. YP, GY, QL and QD wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by The Ethical Committee of The Second Hospital of The Lanzhou University (Gansu, China). Written informed consent was provided by each patient or their guardians prior to their participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarellabranger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016;18(Suppl_5):v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Bent MJ. Practice changing mature results of RTOG study 9802: Another positive PCV trial makes adjuvant chemotherapy part of standard of care in low-grade glioma. Neuro Oncol. 2014;16:1570–1574. doi: 10.1093/neuonc/nou297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behin A, Hoangxuan K, Carpentier AF, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379:1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 5.Roszkowski K, Furtak J, Zurawski B, Szylberg T, Lewandowska M. Potential role of methylation marker in glioma supporting clinical decisions. Int J Mol Sci. 2016;17:1876. doi: 10.3390/ijms17111876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan G, Niu L, Zhang Y, Wang X, Ma K, Yin H, Dai J, Zhou W, Pan Y. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J Neurooncol. 2017;133:193–201. doi: 10.1007/s11060-017-2433-9. [DOI] [PubMed] [Google Scholar]
- 7.Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, Paris S, Boisselier B, Idbaih A, Laigle-Donadey F, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75:1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 8.Felsberg J, Wolter M, Seul H, Friedensdorf B, Göppert M, Sabel MC, Reifenberger G. Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol. 2010;119:501–507. doi: 10.1007/s00401-010-0647-4. [DOI] [PubMed] [Google Scholar]
- 9.Delgado-López PD, Corrales-García EM. Survival in glioblastoma: A review on the impact of treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 10.Phan K, Ng W, Lu VM, McDonald KL, Fairhall J, Reddy R, Wilson P. Association between IDH1 and IDH2 mutations and preoperative seizures in patients with low-grade versus high-grade glioma: A systematic review and meta-analysis. World Neurosurg. 2018;111:e539–e545. doi: 10.1016/j.wneu.2017.12.112. [DOI] [PubMed] [Google Scholar]
- 11.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Chambless LB, Kistka HM, Parker SL, Hassam-Malani L, Mcgirt MJ, Thompson RC. The relative value of postoperative versus preoperative Karnofsky Performance Scale scores as a predictor of survival after surgical resection of glioblastoma multiforme. J Neurooncol. 2015;121:359–364. doi: 10.1007/s11060-014-1640-x. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Shen J, Cao J, Zhou G, Lei T, Sun Y, Gao H, Ding Y, Xu W, Zhan Z, et al. Alternative splicing of human telomerase reverse transcriptase in gliomas and its modulation mediated by CX-5461. J Exp Clin Cancer Res. 2018;37:78. doi: 10.1186/s13046-018-0749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takano S, Ishikawa E, Sakamoto N, Matsuda M, Akutsu H, Noguchi M, Kato Y, Yamamoto T, Matsumura A. Immunohistochemistry on IDH 1/2, ATRX, p53 and Ki-67 substitute molecular genetic testing and predict patient prognosis in grade III adult diffuse gliomas. Brain Tumor Pathol. 2016;33:107–116. doi: 10.1007/s10014-016-0260-x. [DOI] [PubMed] [Google Scholar]
- 15.Gulluoglu S, Tuysuz EC, Sahin M. Simultaneous miRNA and mRNA transcriptome profiling of glioblastoma samples reveals a novel set of OncomiR candidates and their target genes. Brain Res 1700. 2018:199–210. doi: 10.1016/j.brainres.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 16.Valder CR, Liu JJ, Song YH, Luo ZD. Coupling gene chip analyses and rat genetic variances in identifying potential target genes that may contribute to neuropathic allodynia development. J Neurochem. 2003;87:560–573. doi: 10.1046/j.1471-4159.2003.02016.x. [DOI] [PubMed] [Google Scholar]
- 17.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update-an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39(Database Issue):D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sedlazeck FJ, Lee H, Darby CA, Schatz MC. Piercing the dark matter: Bioinformatics of long-range sequencing and mapping. Nat Rev Genet. 2018;19:329–346. doi: 10.1038/s41576-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 19.Dalal S, Zha Q, Daniels CR, Steagall RJ, Joyner WL, Gadeau AP, Singh M, Singh K. Osteopontin stimulates apoptosis in adult cardiac myocytes via the involvement of CD44 receptors, mitochondrial death pathway, and endoplasmic reticulum stress. Am J Physiol Heart Circ Physiol. 2014;306:H1182–H1191. doi: 10.1152/ajpheart.00954.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng C, Zhang Y, Yin J, Li J, Abounader R, Zuo Z. Regulatory factor X1 is a new tumor suppressive transcription factor that acts via direct downregulation of CD44 in glioblastoma. Neuro Oncol. 2014;16:1078–1085. doi: 10.1093/neuonc/nou010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monaghan M, Mulligan KA, Gillespie H, Trimble A, Winter P, Johnston PG, McCormick D. Epidermal growth factor up-regulates CD44-dependent astrocytoma invasion in vitro. J Pathol. 2015;192:519–525. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH784>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Zhao LH, Lin QL, Wei J, Huai YL, Wang KJ, Yan HY. CD44v6 expression in patients with stage II or stage III sporadic colorectal cancer is superior to CD44 expression for predicting progression. Int J Clin Exp Pathol. 2015;8:692–701. [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the Hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70:2455–2464. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei KC, Huang CY, Chen PY, Feng LY, Wu TW, Chen SM, Tsai HC, Lu YJ, Tsang NM, Tseng CK, et al. Evaluation of the prognostic value of CD44 in glioblastoma multiforme. Anticancer Res. 2010;30:253–259. [PubMed] [Google Scholar]
- 25.Fu J, Yang QY, Sai K, Chen FR, Pang JC, Ng HK, Kwan AL, Chen ZP. TGM2 inhibition attenuates ID1 expression in CD44-high glioma-initiating cells. Neuro Oncol. 2013;15:1353–1365. doi: 10.1093/neuonc/not079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishikawa M, Inoue A, Ohnishi T, Kohno S, Ohue S, Matsumoto S, Suehiro S, Yamashita D, Ozaki S, Watanabe H, et al. Significance of Glioma Stem-Like cells in the tumor periphery that express high levels of CD44 in tumor invasion, early progression, and poor prognosis in glioblastoma. Stem Cells Int 2018. 2018:5387041. doi: 10.1155/2018/5387041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng Y, Wodzenski D, Gao D, Shiraishi T, Terada N, Li Y, Vander Griend DJ, Luo J, Kong C, Getzenberg RH, Kulkarni P. Stress-response protein RBM3 attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res. 2013;73:4123–4133. doi: 10.1158/0008-5472.CAN-12-1343. [DOI] [PubMed] [Google Scholar]
- 28.Guo JY, Hsu HS, Tyan SW, Li FY, Shew JY, Lee WH, Chen JY. Serglycin in tumor microenvironment promotes non-small cell lung cancer aggressiveness in a CD44-dependent manner. Oncogene. 2017;36:2457–2471. doi: 10.1038/onc.2016.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S, Chen C, Chang K, Karnad A, Jagirdar J, Kumar AP, Freeman JW. CD44 expression level and isoform contributes to pancreatic cancer cell plasticity, invasiveness and response to therapy. Clin Cancer Res. 2016;22:5592–5604. doi: 10.1158/1078-0432.CCR-15-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HH, Liao CC, Chow NH, Huang LL, Chuang JI, Wei KC, Shin JW. Whether CD44 is an applicable marker for glioma stem cells. Am J Transl Res. 2017;9:4785–4806. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the OncoLnc (http://www.oncolnc.org/search_results/?q=CD44) and GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=&clicktag=survival) databases.