Abstract
N6-methyladenosine (m6A) is the most abundant eukaryote mRNA modification, modulated by regulators known as epigenetic writers, erasers and readers, which are known to serve crucial roles in mRNA metabolism. However, the role of m6A during B-cell development and B-cell tumorigenesis remains poorly understood. By analyzing the gene expression profile of 123 mantle cell lymphoma cases from the Gene Expression Omnibus database, the present study demonstrated that one-half of the m6A regulators were able to predict patient survival in mantle cell lymphoma, notably the m6A.index. The expression levels of the m6A regulators were regarded as good classifiers in mantle cell lymphoma. The m6A.index-low mantle cell lymphoma type exhibited a poor patient survival and lower mRNA levels from the total transcriptome. The m6A regulators may be associated with the cell division and the RNA metabolic pathways, which may result in poor survival of patients with mantle cell lymphoma.
Keywords: RNA, RNA modification, N6-methyladenosine, lymphoma
Introduction
N6-methyladenosine (m6A) is the most abundant eukaryote messenger RNA modification, modulated by regulators known as writers, erasers and readers (1). It is known to serve crucial roles in mRNA metabolism and is primarily involved in RNA stability, mRNA splicing and protein translation (2–9).
The proteins that are involved in m6A modifications consist of ‘writers’, ‘erasers’ and ‘readers’. The m6A writers are the m6A methyltransferase enzymes, a 70-kDa complex consisting of 3 components: Methyltransferase like (METTL) 3, METTL14 and WT1 associated protein (WTAP), which methylate the adenosine motif (A) at the N6 position (10–12). The m6A erasers comprise the m6A demethyltransferase enzymes, including alpha-ketoglutarate-dependent dioxygenase FTO (FTO) and RNA demethylase ALKBH5 (ALKBH5), which are the first and second m6A demethyltransferase enzymes (6,13–15). The m6A readers consist of effectors (m6A RNA binding protein) that decode the m6A methylation code. The YTH domain family [YTH domain-containing family protein (YTHDF) 1, YTHDF2 and YTHDF3] and ELAV-like protein 1 (ELAVL1) are known as m6A readers (2,3,16,17). The m6A modification is associated with cancer progression. The m6A demethylase ALKBH5 sustains forkhead box protein M1 expression and cell proliferation and maintains the tumorigenesis of glioblastoma stem-like cells (18). m6A RNA methylation regulates the tumorigenesis of glioblastoma and the self-renewal of glioblastoma stem cells (19).
METTL3 is an m6A writer associated with the formation of undifferentiated myeloid cells in acute myeloid leukemia (AML), as well as with chemo- and radio-resistance of pancreatic cancer cells (20,21). In addition, METTL3 serves an important role in the growth, survival and invasion of human lung cancer cells (22). Protein virilizer homolog (KIAA1429) was defined as a writer of m6A in 2014 (23). KIAA1429 is a unique type of m6A writer: i) Mammalian KIAA1429 (202 kDa) is the largest known component within the m6A methyltransferase complex; ii) among all the components examined, the depletion of KIAA1429 resulted in the largest decrease in m6A levels [KIAA1429 depletion led to a 60% decrease in m6A levels, while METTL3, METTL14, and WTAP depletion resulted in 30, 40, and 50% decreases in m6A levels, respectively (12)]; and iii) biochemical studies have indicated that KIAA1429 recruits METTL3/METTL14/WTAP, the catalytic core components (24–26). Therefore, KIAA1429 may serve as a scaffold molecule of the methyltransferase complex, and serve a unique role that is different from those of the catalytic core components METTL3, METTL14 and WTAP.
FTO is a member of the m6A eraser family of proteins and has been demonstrated to promote AML and lung squamous cell carcinoma (LUSC) progression (27,28).
The expression levels of the m6A protein readers, including YTHDF1 and YTHDF2, were identified to be markedly associated with malignancy and poor prognosis of hepatocellular carcinoma (29,30). YTHDF2 and YTHDF3 were defined as readers of m6A in 2012 (16). YTHDF2 and YTHDF3 regulate messenger RNA stability (2,31). YTHDF1 was defined as a reader of m6A in 2014. YTHDF1 facilitates messenger RNA translation initiation, while ELAVL1 inhibits protein translation (3,32). It appears that YTHDF2 and YTHDF3 are more likely to regulate mRNA stability, while YTHDF1 and ELAVL1 are more likely to regulate mRNA translation.
Mantle cell lymphoma is a type of non-Hodgkin B cell lymphoma with a median age of diagnosis at 60 years (33,34). Mantle cell lymphoma has an aggressive phenotype and a rapid rate of progression, with a short median survival of 5–7 years (35). The investigation of the molecular mechanisms that contribute to the aggressive phenotype of mantle cell lymphoma may provide novel treatment strategies. Recently, previous studies demonstrated that m6A mRNA methylation controls T cell homeostasis and modulates the characteristics of hematopoietic stem and progenitor cells (36,37). However, the role of m6A during B-cell development and B-cell tumorigenesis remains poorly understood (38). The present study demonstrated that the imbalanced expression levels of m6A regulators may be used for the prediction of poor survival in patients with mantle cell lymphoma.
Materials and methods
Data source
The Affymetrix Human Genome U133 Plus 2.0 array of 123 mantle cell lymphoma samples was retrieved from the NCBI Gene Expression Omnibus (GEO) database (GSE93291 dataset) (39,40). The detailed patient demographic data and disease characteristics for this dataset were published previously (39). The Affymetrix Human Genome U133 Plus 2.0 Array that contained 64 mantle cell lymphoma samples was retrieved from the NCBI GEO database (GSE21452) (41). GSE21452 was the first phase of the GSE93291 dataset.
Gene expression analysis
The probeset measures of all the arrays were calculated by robust multiarray averaging. The relative RNA expression values were log-transformed (log2). The data were analyzed with an unpaired Student's t-test and are presented as the mean ± standard error of the mean (SEM). P<0.05 was considered to indicate a statistically significant difference. Only genes with a fold change (log2) >1 or <-1 were defined as differentially expressed genes.
Definition of m6A.index for survival prediction
A comprehensive m6A.index was defined to predict the survival in patients with mantle cell lymphoma. The m6A.index was calculated using a previously described method (1), as follows:
where m6A.indexj represents the index of m6A of the jth sample for survival prediction. Fj represents the product of favorable gene expression of the jth sample. A total of 7 out of 10 m6A genes exhibited a hazard ratio <1 and were defined as ‘favorable genes’, which were favorable for the survival of mantle cell lymphoma. Hj represents the product of ‘harmful gene’ expression of the jth sample. A total of 3 out of 10 m6A genes (YTHDF1, KIAA1429 and ELAVL1) exhibited a hazard ratio >1 and were defined as ‘harmful genes’, which were harmful for the survival of mantle cell lymphoma.
The median of the m6A.index value from a cohort of mantle cell lymphoma, for example 123 patients with mantle cell lymphoma, was defined as the cut-off value for the m6A.index-low and m6A.index-high groups consisting of 62 and 61 samples, respectively. The correlation of the m6A.index with the gene expression levels of the marker of proliferation Ki-67 (Ki-67) was assessed using the Spearman's correlation test and the pairwise colored scatterplot was drawn based on a Kernel Density Estimation using the LSD package (version 4; cran.r-project.org/web/packages/LSD/index.html) in R.
Gene Ontology (GO) analysis
The Database for Annotation, Visualization and Integrated Discovery tool with default parameters was used for GO analysis (42). All enriched GO terms identified in the present study were manually prepared so that only selected, non-redundant GO terms in the ‘Biological Process’ category were identified.
Statistical analysis
The R software v3.1.3 (ggplot2 package) was used for the statistical analysis. Kaplan-Meier curves were used to plot survival curves of YTHDF3, METTL14, ALKBH5, ELAVL1 and KIAA1429 genes. For the YTHDF3, METTL14, ALKBH5 and ELAVL1 genes, the median of the gene expression value from a cohort of mantle cell lymphoma (123 patients with mantle cell lymphoma) was defined as the cut off value for the low and high expression groups. For KIAA1429 genes, the maximally selected rank statistics algorithm (survminer package) was used to define the low and high expression groups. Survival analysis of those genes was performed using the log-rank test. The heatmap depicted the cosine correlation similarity between 10 m6A regulators. Unpaired Student's t-tests were used for the statistical analysis of quantitative variables. The data are expressed as the mean ± SEM in scatter plots. P<0.05 was considered to indicate a statistically significant difference.
Results
Specific m6A regulators predict patient survival in mantle cell lymphoma
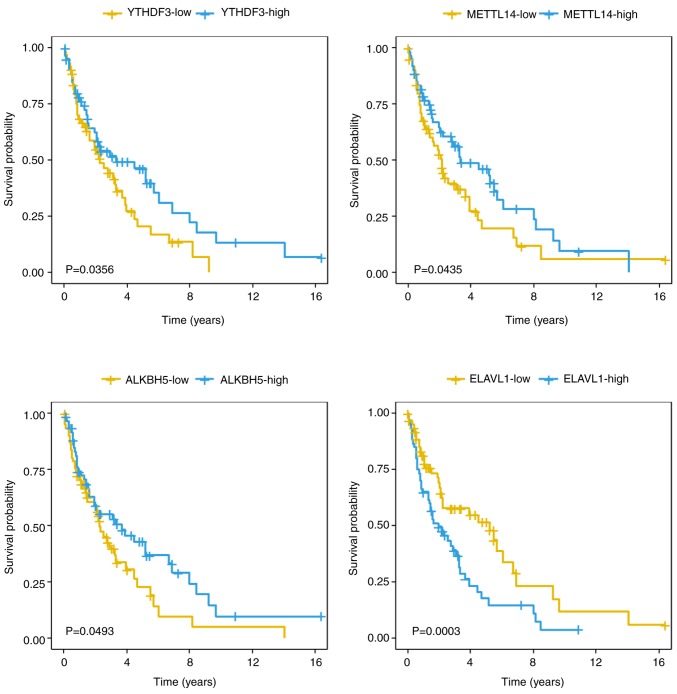
To investigate the association between the m6A regulators [METTL3, METTL14, WTAP and KIAA1429 (writers); FTO and ALKBH5 (erasers); and YTHDF1, YTHDF3, YTHDF2 and ELAVL1 (readers)] and the survival of the patients with mantle cell lymphoma, the expression profiles of 123 mantle cell lymphoma samples from the GSE21452 dataset were analyzed. A total of 5 out of 10 m6A regulators revealed expression levels that were significantly associated with the survival of patients with mantle cell lymphoma (P<0.05, log-rank test). The 10 m6A regulators were classified according to the hazard ratio values. A total of 7 out of 10 m6A genes had a hazard ratio value <1 and were defined as ‘favorable genes’. YTHDF3 exhibited the highest statistical significance of all the ‘favorable genes’ with a hazard ratio of 0.51 [95% confidence interval (CI), 0.27–0.96]. A total of 3 out of 10 m6A genes (YTHDF1, KIAA1429 and ELAVL1) exhibited a hazard ratio value of >1 and were defined as ‘harmful genes’ (Table I; Fig. 1). ELAVL1 exhibited the highest significance of all the ‘harmful genes’ with a hazard ratio of 2.46 (95% CI, 1.35–4.48). Kaplan-Meier curves were conducted to assess the overall survival of the 123 patients with mantle cell lymphoma in association with 5 m6A regulators. The comparisons were performed using a log-rank test (Fig. 2). In addition, the 123 patients with mantle cell lymphoma were divided into KIAA1429-high (n=24) and KIAA1429-low (n=99) groups using maximally selected rank statistics algorithm (survminer package), and the survival curves of the two groups were compared using a log-rank test (Fig. S1; P<0.0001). The m6A regulators of both ‘favorable’ and ‘harmful’ genes were deemed significant predictors for mantle cell lymphoma patient survival.
Table I.
Survival analysis of 10 m6A regulators.
| Gene | Hazard ratio | 95% confidence interval | P-value | m6A role |
|---|---|---|---|---|
| ELAVL1 | 2.46 | 1.35–4.48 | 0.0030 | Reader |
| KIAA1429 | 2.09 | 1.21–3.62 | 0.0086 | Writer |
| YTHDF3 | 0.51 | 0.27–0.96 | 0.0356 | Reader |
| METTL14 | 0.68 | 0.47–0.99 | 0.0435 | Writer |
| ALKBH5 | 0.70 | 0.49–1.00 | 0.0493 | Eraser |
| WTAP | 0.81 | 0.65–1.02 | 0.0683 | Writer |
| METTL3 | 0.59 | 0.32–1.07 | 0.0840 | Writer |
| FTO | 0.61 | 0.31–1.20 | 0.1519 | Eraser |
| YTHDF2 | 0.77 | 0.51–1.16 | 0.2113 | Reader |
| YTHDF1 | 1.29 | 0.51–3.30 | 0.5881 | Reader |
ELAVL1, ELAV-like protein 1; KIAA1429, Protein virilizer homolog; YTHDF, YTH domain-containing family protein; METTL, methyltransferase like; ALKBH5, RNA demethylase ALKBH5; WTAP, WT1 associated protein; FTO, alpha-ketoglutarate-dependent dioxygenase FTO.
Figure 1.
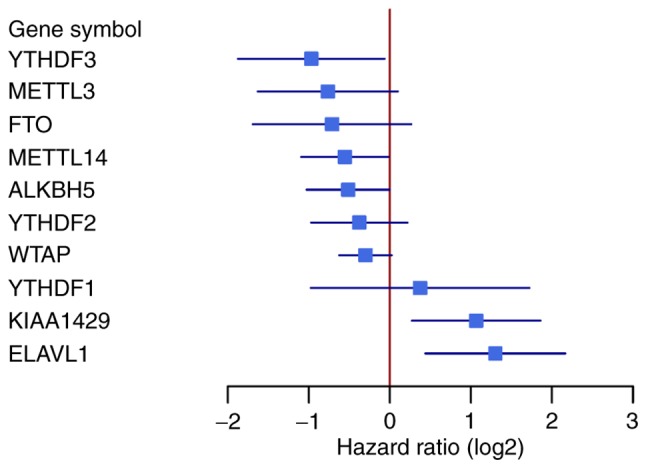
Forest plots of 10 N6-methyladenosine regulators associated with survival. The black lines indicate lower and upper 95% confidence of the hazard ratios.
Figure 2.
Kaplan-Meier curves measuring associations between overall survival and 4 N6-methyladenosine regulators in 123 patients with mantle cell lymphoma. YTHDF3 (P=0.0356), METTL14 (P=0.0435), ALKBH5 (P=0.0493) and ELAVL1 (P=0.003). The log-rank test was used to compare the Kaplan-Meier curves. YTHDF3, YTH domain-containing family protein 3; METTL14, methyltransferase like 14; ALKBH5, RNA demethylase ALKBH5; ELAVL1, ELAV-like protein 1.
Expression patterns of m6A regulators predict poor or favorable patient survival in patients with mantle cell lymphoma
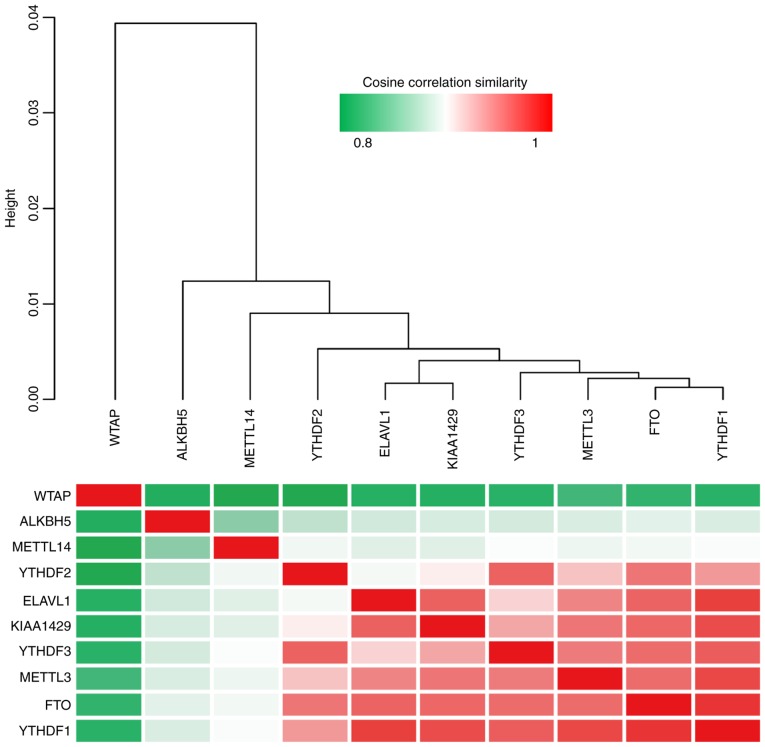
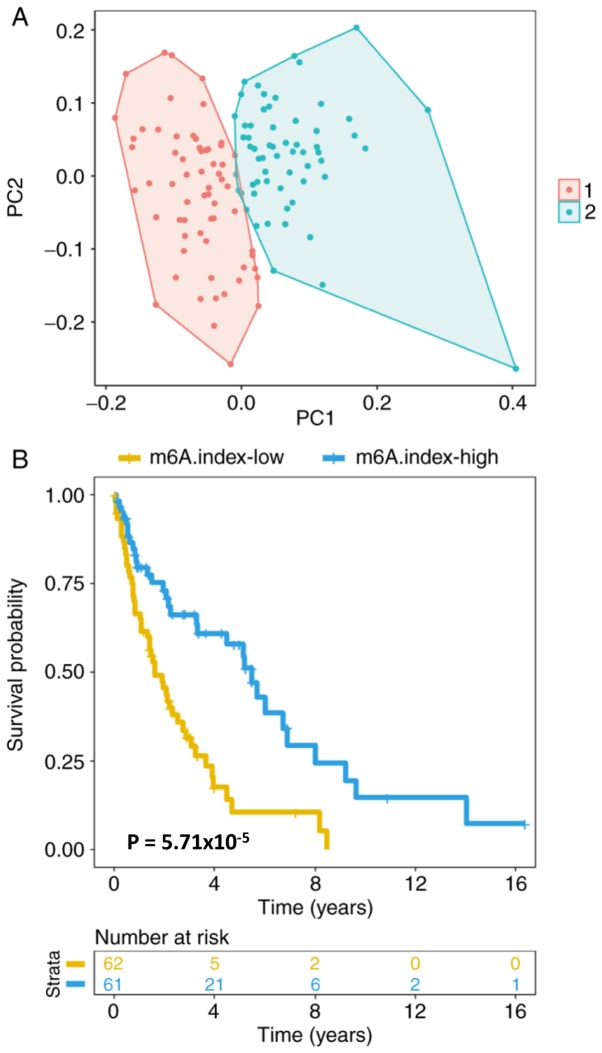
An unsupervised clustering of the expression levels of the 10 m6A regulators was conducted in 123 patients with mantle cell lymphoma, and the cosine correlation similarity was presented as a heatmap (Fig. 3). Notably, it was identified that the 10 m6A regulators were classified into two groups; WTAP, ALKBH5 and METTL14 in one group, and the other 7 genes in a second group. ELAVL1 and KIAA1429, two of the most significantly ‘harmful’ genes, were clustered together in one group. The other 3 most significantly ‘favorable’ genes, YTHDF3, METTL3 and FTO, were also clustered together in one group. The clustering of ‘harmful’ and ‘favorable’ genes together in single groups suggests that the genes that are the most closely associated with survival cluster together. Furthermore, it was observed that the mantle cell lymphoma cancer type could be classified into two groups by fuzzy clustering (Fig. 4A). The ratio of the expression levels of ‘harmful genes’ to ‘favorable genes’ was estimated, which was termed as the m6A.index. The m6A.index was highly associated with the survival of patients with mantle cell lymphoma (Fig. 4B; P<0.05). The hazard ratio of the m6A.index was 0.39 (95% CI, 0.24–0.65). The m6A.index reflected the imbalanced expression between ‘harmful genes’ and ‘favorable genes’ of the m6A regulators. The m6A.index-low group was associated with a poor patient survival in mantle cell lymphoma, while the m6A.index-high group was associated with a favorable patient survival in mantle cell lymphoma.
Figure 3.
Unsupervised clustering of the expression levels of 10 N6-methyladenosine regulators in 123 patients with mantle cell lymphoma. The cosine correlation similarity was depicted by the heatmap. ELAVL1, ELAV-like protein 1; KIAA1429, Protein virilizer homolog; YTHDF, YTH domain-containing family protein; METTL, methyltransferase like; ALKBH5, RNA demethylase ALKBH5; WTAP, WT1 associated protein; FTO, alpha-ketoglutarate-dependent dioxygenase FTO.
Figure 4.
Groups of 10 m6A regulators were used as a classifier in the 123 patients with mantle cell lymphoma. (A) The fuzzy clustering of 123 patients with mantle cell lymphoma as determined by the expression levels of the 10 m6A regulators. PC1 and PC2 were the first and second components, respectively. Each point indicated a mantle cell lymphoma sample. Colors 1 (red) and 2 (green) denote two clusters. (B) Kaplan-Meier curves for assessment of the association between overall survival of 123 patients mantle cell lymphoma with m6A.index (P<0.001). The log-rank test was used to compare the Kaplan-Meier curves. m6A, N6-methyladenosine.
The correlation of the m6A.index with the gene expression levels of marker of proliferation Ki-67 (Ki-67) in 123 mantle cell lymphoma samples was analyzed (Fig. S2; Cor =−0.52; P<0.001; Spearman's correlation test). The m6A.index exhibited a negative correlation with Ki-67 and DNA polymerase genes (DNA polymerase alpha 1, catalytic subunit, DNA polymerase alpha 2, accessory subunit, DNA polymerase delta 1, catalytic subunit, DNA polymerase delta 2, accessory subunit and DNA polymerase theta). Although, the mutational status of tumor protein 53 (TP53), ATM serine/threonine kinase (ATM) and MYC proto-oncogene, BHLH transcription factor (MYC) could not be assessed using the data from the present study, the survival analysis of 123 patients with mantle cell lymphoma was analyzed in association with Ki-67, ATM, MYC and TP53 gene expression (Fig. S2). Increased expression levels of MKI67 and MYC corresponded with poorer survival in patients with mantle cell lymphoma (Fig. S3; P=2.9×10−11 and P=4.5×10−4, respectively). The high expression levels of ATM and TP53 demonstrated the trend for predicting favorable survival in patients with mantle cell lymphoma (Fig. S3; P=7.5×10−2 and P=1.1×10−1, respectively). ATM and Ki-67 were differentially expressed between the m6A.index-low and m6A.index-high groups (Fig. S3; P<0.05), while TP53 and MYC were not differentially expressed (P>0.05). Furthermore, the correlations between the m6A.index with the expression of 17 proliferation-associated genes in 123 mantle cell lymphoma samples were calculated. A total of 16 of 17 proliferation-associated genes were highly correlated with the m6A.index (Fig. S4).
Association between the upregulation of gene expression and the m6A.index-high group in mantle cell lymphoma
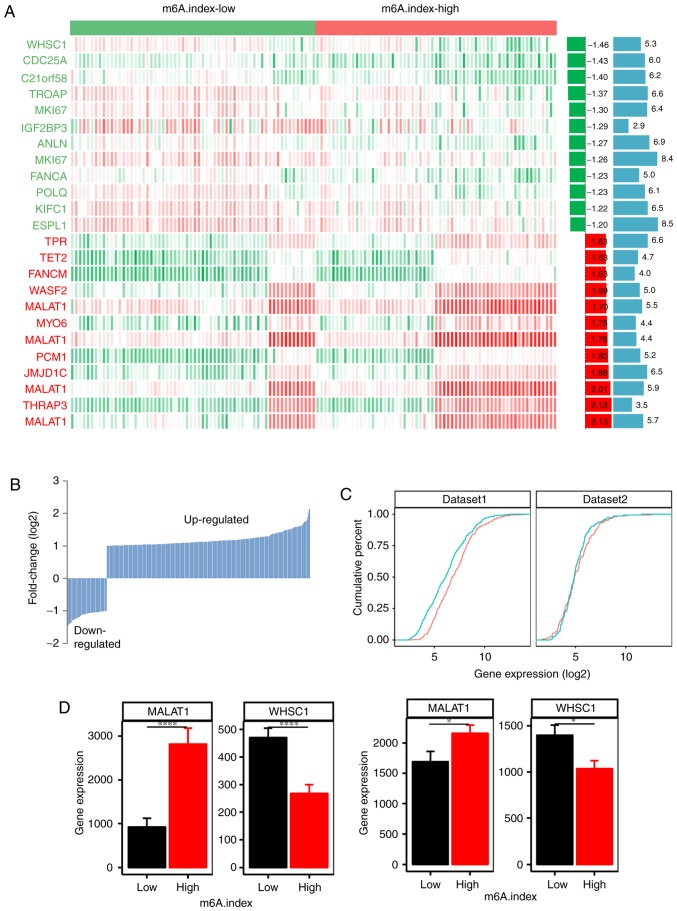
The m6A.index-high and the m6A.index-low groups were identified to be two different classes of mantle cell lymphoma. Therefore, the expression profiles of the m6A.index-high and the m6A.index-low groups in mantle cell lymphoma were compared (Fig. 5A). A total of 280 upregulated genes and 54 downregulated genes were identified between the m6A.index-high and the m6A.index-low groups in mantle cell lymphoma (Fig. 5B; P<0.05). The m6A.index-high mantle cell lymphoma group exhibited an increased number of upregulated genes compared with the m6A.index-low group, which suggested that the m6A.index-high mantle cell lymphoma type had a different RNA metabolism process from the m6A.index-low mantle cell lymphoma type. The cumulative distribution of the expression of RNA molecules corresponding to different genes with regard to the m6A.index-high and m6A.index-low mantle cell lymphoma types additionally demonstrated that the m6A.index-high mantle cell lymphoma type exhibited high RNA levels compared with the total transcript profile in the GSE93291 dataset (Fig. 5C; P<0.001). This result was also validated in the secondary GSE21452 dataset (n=64).
Figure 5.
Different expression of the genes in the m6A.index-high and the m6A.index-low groups of patients with mantle cell lymphoma. (A) The heatmap indicates the different expression of the genes in the m6A.index-high and m6A.index-low groups of patients with mantle cell lymphoma. Red, high expression; green, low expression; white, moderate expression. Only the top 12 upregulated and downregulated genes were noted. The two bar plots in the left of the heatmap refer to the fold-change (log2, left; green and red) difference and the P-value (-log10, right; blue), respectively. (B) The total number of the upregulated (280 genes) and the downregulated genes (54 genes) between the m6A.index-high and m6A.index-low mantle cell lymphoma types. (C) Cumulative distribution of RNA levels (log2-fold) of the differentially expressed genes between the m6A.index-high (red) and the m6A.index-low (green) mantle cell lymphoma groups. The left plot represents the GSE93291 dataset (n=123), and the right plot represents the GSE21452 dataset (n=64) (D) The different expression levels of the MALAT1 and the WHSC1 genes between the m6A.index-high and the m6A.index-low mantle cell lymphoma groups. The left plot corresponds to the GSE93291 dataset (n=123), and the right plot corresponds to the GSE21452 dataset (n= 64). A two sided unpaired Student's t-test was used. *P<0.05 and ****P<0.0001. m6A, N6-methyladenosine.
Cell division and RNA metabolism pathways are significantly enriched pathways of m6A in mantle cell lymphoma
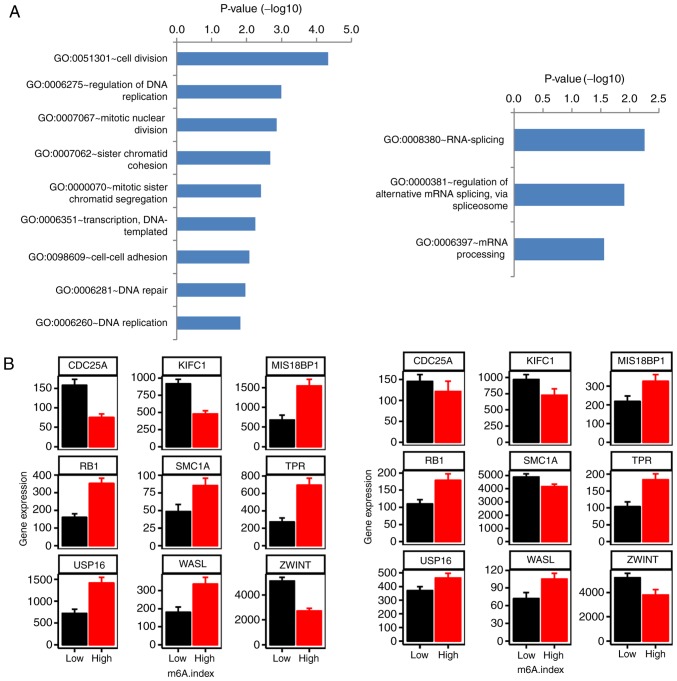
A characteristic difference between the m6A.index-high and m6A.index-low mantle cell lymphoma types was evident. WHSC1, which encodes for a histone-lysine N-methyltransferase, was the top downregulated gene in the m6A.index-high group compared with the m6A.index-low group (Fig. 5D; P=4.81×10−6). Metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) was the top upregulated gene in the m6A.index-high group compared with the m6A.index-low group (Fig. 5D; P=1.98×10−6). The differential expression of WHSC1 and MALAT1 was also validated in the GSE21452 dataset (n=64). A pathway analysis of the differentially expressed genes between the m6A.index-high and the m6A.index-low mantle cell lymphoma types was conducted. The cell division and RNA metabolism pathways were demonstrated to be the most significantly enriched pathways based on the differential expression of specific genes (Fig. 6A). The RB transcriptional corepressor 1, cell division cycle 25A and kinesin family member C1 genes were included in the cell division pathways group, and were demonstrated to be differentially expressed, out of a total of 9 genes (Fig. 6B). Therefore, the m6A regulators may regulate the cell division pathways, contributing to poor patient survival in mantle cell lymphoma.
Figure 6.
Significantly enriched pathways of differentially expressed genes between the m6A.index-high and the m6A.index-low groups of patients with mantle cell lymphoma. (A) The bar plot indicates the significantly enriched cell division (left) and the RNA metabolism (right) pathways. (B) The differentially expressed genes identified between the m6A.index-high and the m6A.index-low mantle cell lymphoma groups in the cell division pathway. The left plot corresponds to the GSE93291 dataset (n=123), while the right plot corresponds to the GSE21452 dataset (n=64). A two-sided unpaired Student's t-test was used. P<0.05 for all histograms. m6A, N6-methyladenosine.
Discussion
Recent studies have indicated that m6A mRNA methylation controls T-cell homeostasis and modulates hematopoietic stem and progenitor cell differentiation (36,37). However, the role of m6A during B-cell development and B-cell tumorigenesis remains poorly understood (38). The present study demonstrated that the imbalanced expression of m6A regulators may be used for the prediction of patient survival in mantle cell lymphoma. A previous study indicated that m6A regulator participates in the innate immunity via the RNA helicase probable ATP-dependent RNA helicase DDX46. Therefore, the m6A gene and the m6A regulators are involved in B-cell lymphoma development, T-cell homeostasis and innate immunity.
Mantle cell lymphoma is an aggressive type of B cell lymphoma with a short median patient survival time. The identification of novel biomarkers for the prediction of patient survival in mantle cell lymphoma is considered a challenging task (43). The mantle cell lymphoma international prognostic index (MIPI) score is currently the most common prognostic model for mantle cell lymphoma in clinical practice (44). MIPI includes the age, Eastern cooperative oncology group performance status, leukocyte count and lactate dehydrogenase activity (45). However, these models lack a component that incorporates gene expression analysis. The present study demonstrated that the expression levels of 5 m6A regulators were significantly associated with the survival of patients with mantle cell lymphoma. In addition, a comprehensive m6A.index was constructed to predict the survival of mantle cell lymphoma. The m6A.index was better compared with each individual m6A regulator for survival prediction with a hazard ratio of 0.39 (95% CI, 0.24–0.65).
It was surprising that half (5 of 10) of the m6A regulators were able to predict patient survival in mantle cell lymphoma (P<0.05). It appeared that the m6A regulators were commonly associated with mantle cell lymphoma patient survival. In addition, the m6A.index was a better survival predictor compared with the single m6A regulator, which suggested that the imbalanced expression of m6A regulators may predict poor patient survival in mantle cell lymphoma. Furthermore, the data from the present study provided evidence to support a marked association between m6A expression and mantle cell lymphoma incidence: i) The expression of m6A regulators was a good classifier in mantle cell lymphoma; ii) the samples of mantle cell lymphoma were divided into two groups according to the m6A.index (m6A.index-high and m6A.index-low groups), with the m6A.index-high group exhibiting high RNA levels compared with the total transcript profile; and iii) the differentially expressed genes were associated with the cell division and RNA metabolism pathways in the m6A.index-high group that may result in poor patient survival.
The m6A.index was correlated with enhanced proliferation. Ki-67 is clinically important for risk stratification and clinical management of mantle cell lymphoma (45–47). The m6A.index demonstrated a highly negative correlation with gene expressions of Ki-67 and DNA polymerases. A total of 17 proliferation-associated genes were defined as a proliferation ‘signature’ and associated with overall survival in mantle cell lymphoma (39). The present study further correlated the m6A.index with the expression of 17 proliferation-associated genes in 123 mantle cell lymphoma samples; 16 of these 17 proliferation-associated genes were highly correlated with m6A.index, which suggested that the m6A.index was associated with proliferation in mantle cell lymphoma.
Mammalian KIAA1429 is the largest known component within the m6A methyltransferase complex and serves as a scaffold of the methyltransferase complex, while METTL3/METTL14/WTAP serve as the catalytic core components (24). In the results from the present study, the majority of the m6A writer molecules, including METTL3, METTL14 and WTAP, were classified as ‘favorable genes’, while the remaining m6A writer (KIAA1429) was categorized into the group of ‘harmful genes’. Therefore, within the group of 4 m6A writers, KIAA1429 appeared to be the most significant in terms of its biological function and clinical implications.
In summary, the results from the present study demonstrated that the expression levels of m6A regulators were associated with the survival of patients with mantle cell lymphoma and may serve as potential biomarkers for prognosis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by National Natural Science Foundation of China (grant no. 81800195), interdisciplinary medicine Seed Fund of Peking University (grant no. BMU2018MB004), Beijing Natural Science Foundation (grant nos., 7132183 and 7182178) and China Health Promotion Foundation (grant no. CHPF-zlkysx-001).
Availability of data and materials
The data included in the present study have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE93291 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93291) and GSE21452 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21452). The datasets used in the present study are available from the corresponding author upon reasonable requests.
Authors' contributions
HJ and XZ conceived the project. WZ and XH analyzed the data. WZ, XH, JH, PY, CL, JW, RA, JZ, MP, KH, XK, XZ and HJ contributed towards the interpretation of the data. All authors wrote and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA Demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, Roignant JY. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–247. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 10.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 11.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q, Cheng W, Wang J, Feng Y, Chai J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464:1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 16.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 17.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al. m6A Demethylase ALKBH5 maintains Tumorigenicity of Glioblastoma Stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. m6A RNA methylation regulates the Self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, Doki Y, Mori M, Ishii H, Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621–629. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 22.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215:157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, Liu Y, Ye L, Li Y, Zhang X. MicroRNA-145 modulates N6-methyladenosine levels by targeting the 3′-untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292:3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, Xu W, Zhong L, Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 31.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Kong L, Guo S, Bu M, Guo Q, Xiong Y, Zhu N, Qiu C, Yan X, Chen Q, et al. hnRNPs and ELAVL1 cooperate with uORFs to inhibit protein translation. Nucleic Acids Res. 2016 Oct 26; doi: 10.1093/nar/gkw991. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: Perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–762. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- 34.Leux C, Maynadie M, Troussard X, Cabrera Q, Herry A, Le Guyader-Peyrou S, Le Gouill S, Monnereau A. Mantle cell lymphoma epidemiology: A population-based study in France. Ann Hematol. 2014;93:1327–1333. doi: 10.1007/s00277-014-2049-5. [DOI] [PubMed] [Google Scholar]
- 35.Vose JM. Mantle cell lymphoma: 2012 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2012;87:604–609. doi: 10.1002/ajh.23176. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 37.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwok CT, Marshall AD, Rasko JE, Wong JJ. Erratum to: Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:49. doi: 10.1186/s13045-017-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott DW, Abrisqueta P, Wright GW, Slack GW, Mottok A, Villa D, Jares P, Rauert-Wunderlich H, Royo C, Clot G, et al. New molecular assay for the proliferation signature in mantle cell lymphoma applicable to formalin-fixed paraffin-embedded biopsies. J Clin Oncol. 2017;35:1668–1677. doi: 10.1200/JCO.2016.70.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartmann EM, Campo E, Wright G, Lenz G, Salaverria I, Jares P, Xiao W, Braziel RM, Rimsza LM, Chan WC, et al. Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling. Blood. 2010;116:953–961. doi: 10.1182/blood-2010-01-263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Hoster E, Dreyling M, Klapper W, Gisselbrecht C, van Hoof A, Kluin-Nelemans HC, Pfreundschuh M, Reiser M, Metzner B, Einsele H, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111:558–565. doi: 10.1182/blood-2007-06-095331. [DOI] [PubMed] [Google Scholar]
- 44.Eskelund CW, Dahl C, Hansen JW, Westman M, Kolstad A, Pedersen LB, Montano-Almendras CP, Husby S, Freiburghaus C, Ek S, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood. 2017;130:1903–1910. doi: 10.1182/blood-2017-04-779736. [DOI] [PubMed] [Google Scholar]
- 45.Vose JM. Mantle cell lymphoma: 2017 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2017;92:806–813. doi: 10.1002/ajh.24797. [DOI] [PubMed] [Google Scholar]
- 46.Ye H, Desai A, Zeng D, Nomie K, Romaguera J, Ahmed M, Wang ML. Smoldering mantle cell lymphoma. J Exp Clin Cancer Res. 2017;36:185. doi: 10.1186/s13046-017-0652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoster E, Rosenwald A, Berger F, Bernd HW, Hartmann S, Loddenkemper C, Barth TF, Brousse N, Pileri S, Rymkiewicz G, et al. Prognostic value of Ki-67 index, cytology, and growth pattern in mantle-cell lymphoma: Results from randomized trials of the European mantle cell lymphoma network. J Clin Oncol. 2016;34:1386–1394. doi: 10.1200/JCO.2015.63.8387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data included in the present study have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE93291 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93291) and GSE21452 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21452). The datasets used in the present study are available from the corresponding author upon reasonable requests.