Abstract
Impairment of microRNA (miRNA) biogenesis may be involved in clear cell renal cell carcinoma (ccRCC). The objective of the present study was to investigate the mRNA levels of important miRNA machinery components, DICER1, DROSHA, DiGeroge syndrome critical region gene 8 (DGCR8), and Argonaute 2 (AGO2), and their correlations with clinicopathological characteristics of ccRCC using mRNA expression data from The Cancer Genome Atlas kidney clear cell carcinoma (TCGA KIRC) cohort and a Korean ccRCC cohort. mRNA levels of DICER1, DROSHA, and DGCR8 were significantly decreased in both cohorts. However, AGO2 was significantly downregulated only in the Korean ccRCC cohort. Additionally, positive correlations were observed between the altered mRNA levels of DICER1 and DROSHA as well as DROSHA and DGCR8 in both cohorts. In the TCGA KIRC cohort, alterations in the mRNA levels of DICER1 were significantly correlated with histological grade. Furthermore, the altered mRNA levels of DGCR8 showed significant associations with sex and histologic grades. However, in the Korean ccRCC cohort, no factors were significantly associated with any clinicopathological parameters, including sex, age, T stage, Fuhrman grade/The International Society of Urological Pathology grade, lymphovascular invasion, and peri-renal fat invasion. Taken together, these findings indicate that DICER1, DROSHA, DGCR8 and AGO2 are significantly dysregulated in ccRCC, suggesting that they are important in the pathophysiology of this malignancy.
Keywords: miRNA biogenesis, clear cell renal cell carcinoma, the cancer genome atlas
Introduction
Renal cell carcinoma (RCC), which originates from renal parenchyma, is the most common malignant kidney tumor in adults (87% of all renal malignancies) (1). An estimated 65,340 new cases of kidney and renal pelvis cancers will be diagnosed in the US in 2018 (42,680 in men and 22,660 in women), of which 14,970 will be fatal (2). Among the World Health Organization histological RCC subtypes, clear cell RCC (ccRCC) is the most common subtype (80–90%) (3). Because of the high aggressiveness of ccRCC, approximately 30% of patients show metastasis at the time of diagnosis, and the prognosis is typically very poor (4). Thus, studies of the underlying molecular characteristics of ccRCC are necessary to develop new biomarkers and therapeutic strategies.
MicroRNAs (miRNAs), which are short non-coding RNA molecules, regulate gene expression by cleaving transcribed target mRNAs and/or repressing protein translation. miRNA biogenesis is a well-organized process, involving ‘miRNA machinery’ (5) and is regulated by various miRNA biogenesis components including the RNase III endonuclease DROSHA (RNASEN), a double-stranded RNA binding protein known as DiGeroge syndrome critical region gene 8 (DGCR8), the RNase III ribonuclease DICER, and a component of the RNA-induced silencing complex known as Argonaute 2 (AGO2) (6). Accumulating evidence reveals that dysregulated miRNA biogenesis components have pathophysiological importance in various human cancers (7–11). Furthermore, Dicer has been reported to be down-regulated in and to suppress the malignant phenotype of ccRCC (12,13). Interestingly, significant associations were observed between haplotypes/diplotypes of the DROSHA SNPs (rs6877842 and rs10719) and ccRCC recurrence (14). However, there is still very little published study on the mRNA levels of other miRNA biogenesis components, such as DROSHA, DGCR8, and AGO2, and their clinicopathological associations in ccRCC.
In the present study, we investigated the mRNA levels of four important miRNA biogenesis components using mRNA expression data from The Cancer Genome Atlas (TCGA) KIRC cohorts and a Korean ccRCC dataset and evaluated the clinical relevance of the altered expression in both cohorts.
Materials and methods
Gene expression databases and cluster analysis
The gene expression RNA-Seq dataset (level 3) and clinical data for TCGA Kidney Clear Cell Carcinoma (TCGA KIRC) cohort were downloaded from the UCSC Xena database (xena.ucsc.edu, dataset ID is TCGA.KIRC.sampleMap/HiSeqV2). Nearly all data (496 patients) in TCGA KIRC cohort were described in detail previously (15). The four miRNA component genes present in TCGA KIRC cohort were DICER1, DROSHA (RNASEN), DGCR8, and AGO2 (EIF2C2). Comprehensive genetic information was available for 537 patients with ccRCC in TCGA KIRC cohort. Among the total data, we selected 72 paired data sets with mRNA level data for analysis of miRNA biogenesis-related genes (6) and the four miRNA biogenesis components in ccRCC tissues and adjacent non-neoplastic surround tissues (ST) from patients with ccRCC. Clinicopathological parameters of TCGA KIRC was downloaded from the UCSC Xena database (xena.ucsc.edu, dataset ID is TCGA.KIRC.sampleMap/KIRC_clinicalMatrix). This study meets the publication guidelines provided by TCGA. Cluster analysis was performed using Cluster 3.0 to classify the samples into statistically similar groups, and the resulting heatmaps were visualized in TreeView 1.6 (www.eisenlab.org/eisen).
Patients and tissues
Fifty-three patients diagnosed with ccRCC were included in this retrospective study. ccRCC tissues and adjacent non-neoplastic tissues were obtained from patients undergoing surgery in Dongsan Medical Center (Daegu, Korea) between April 2008 and November 2013. The clinicopathological characteristics and oncological outcome data were retrospectively collected from the Colorectal Cancer Database in the Department of Colorectal Cancer Surgery, Dongsan Medical Center (Daegu, Korea) and the Pathological Diagnosis Database in the Department of Pathology, Dongsan Medical Center (Daegu, Korea), respectively. Enrolled patients with ccRCC were classified according to the American Joint Committee on Cancer Tumor-Node-Metastasis (TNM) staging criteria (16). After surgery, tissue samples were immediately frozen in liquid nitrogen and stored at −196°C until RNA isolation. Tissue samples were provided by the Keimyung Human Bio-resource Bank, Korea.
RNA isolation and reverse transcription-quantitative (RT-q) PCR
Total cellular RNA was extracted from tissues using TRIzol reagent (Molecular Research Center, Inc., Cincinnati, OH, USA). RNAs were quantified using a NanoDrop 1000 (Thermo Scientific, Waltham, MA, USA). Each cDNA was synthesized from 1 µg of total RNA using Moloney murine leukemia virus reverse transcriptase (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol. Using the specific primer pairs shown in Table I and SYBR Green Premix (Toyobo, Japan), qPCR was performed on a LightCycler® 480 real-time PCR system (Roche Diagnostics GmbH, Mannheim, Germany). The following thermocycling conditions were maintained: Pre-incubation, 5 min at 95°C; amplification, 45 cycles with 10 sec at 95°C, 10 sec at 55°C, and 15 sec at 72°C; melting curve, gradually from 65°C to 95°C; cooling, 10 min at 37°C. β-Actin was used as a housekeeping gene for normalization, and a no-template sample was used as a negative control. qPCR data were analyzed using the 2−∆∆Cq method (17) in Microsoft Excel (Microsoft Corporation, Redmond, WA., USA). Three replicates were evaluated and showed similar results.
Table I.
Primer sequences of microRNA machinery components used in quantitative PCR.
| Primer | Sequence (5′-3′) |
|---|---|
| DICER1 | |
| Forward | TTAACCTTTTGGTGTTTGATGAGTGT |
| Reverse | AGGACATGATGGACAATT |
| DROSHA | |
| Forward | CTGTCGATGCACCAGATT |
| Reverse | TGCATAACTCAACTGTGCAGG |
| DGCR8 | |
| Forward | CAAGCAGGAGACATCGGACAAG |
| Reverse | CACAATGGACATCTTGGGCTTC |
| AGO2 | |
| Forward | TCATGGTCAAAGATGAGATGACAGA |
| Reverse | TTTATTCCTGCCCCCGTAGA |
| β-actin | |
| Forward | CAGCCATGTACGTTGCTATCCAGG |
| Reverse | AGGTCCAGACGCAGGATGGCATG |
DGCR8, DiGeroge syndrome critical region gene 8; AGO2, Argonaute 2.
Statistical analysis
Statistical analysis was performed with SPSS software (version 22.0; IBM SPSS, Armonk, NY, USA). Differences between groups were statistically analyzed by using paired t-test. Co-expression of mRNA levels of various miRNA biogenesis components in TCGA KIRC cohort was searched using cBioPortal (cbioportal.org). The association between inter-individual mRNA levels of miRNA biogenesis components in Korean ccRCC was assessed using the Pearson's correlation coefficient analysis for continuous variables. Clinicopathological associations with the mRNA levels of various miRNA biogenesis components in both TCGA KIRC cohort and Korean ccRCC were analyzed using the linear-by-linear association model, Pearson's Chi-square test, and Fisher's exact test for categorical variables. The mean value was used as a cut-off value (low and high) for categorical variables. P<0.05 was considered to indicate a statistically significant difference.
Results
Altered expression levels of miRNA biogenesis-related genes in TCGA KIRC cohorts
To investigate whether the miRNA biogenesis-related genes (6) are dysregulated in ccRCC tissues, we re-analyzed the sorted mRNA expression values from raw data of TCGA KIRC cohort using paired t-test. Hierarchical clustering showed that various miRNA biogenesis-related genes were dysregulated in carcinomatous tissues compared with ST of patients with ccRCC (Fig. 1). To evaluate the significance of altered mRNA levels between ccRCC and ST, paired t-test was performed. Hierarchical clustering revealed that various miRNA biogenesis-related genes were significantly dysregulated in ccRCC tissues compared with ST of the 72 patients with ccRCC (P<0.05; Fig. 2A). Among the various miRNA biogenesis-related genes, AGO1 was significantly upregulated in ccRCC tissues of TCGA KIRC cohort, while DICER1, DROSHA, DGCR8, trinucleotide repeat containing 6B (TNRC6B), trinucleotide repeat containing 6C (TNRC6C), and AGO3 were significantly downregulated (paired t-test, P<0.05; Fig. 2B).
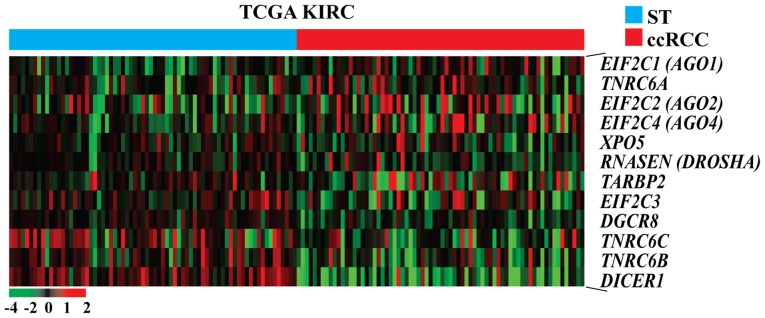
Figure 1.
Heat map presenting the mRNA expression of the miRNA biogenesis-related genes in the TCGA KIRC cohort. The data are presented in matrix format in which each row represents an individual gene and each column represents a single tissue. Each cell in the matrix represents the expression level of a gene feature in an individual tissue. Expression levels have been standardized (centered and scaled) within columns for visualization. The red and green colors in cells reflect relative high and low expression levels, respectively, as indicated in the scale bar. Images were obtained by re-analysis of the raw data of the TCGA KIRC cohort. ccRCC tissues were ordered from STs to ccRCC tissues according to the standardized expression level of each gene as indicated. TCGA KIRC, The Cancer Genome Atlas kidney clear cell carcinoma; ccRCC, clear cell renal cell carcinoma; ST, adjacent non-neoplastic surround tissues.
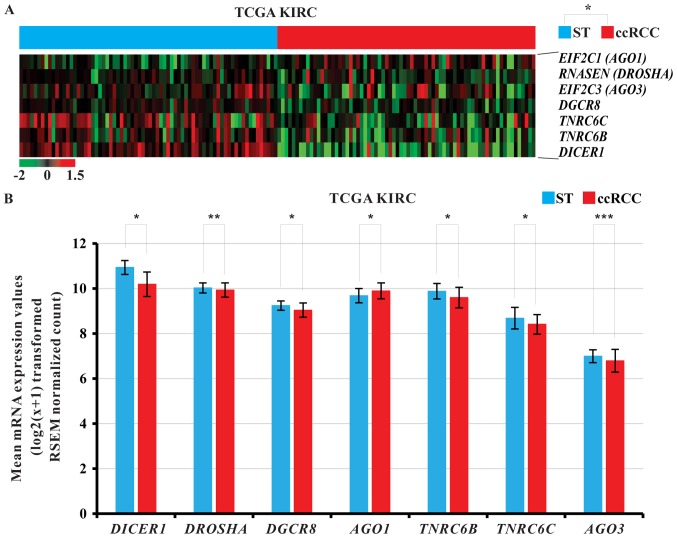
Figure 2.
(A) Heat map presenting significantly differential mRNA expression of the miRNA biogenesis-related genes in ccRCC tissues compared with the ST of TCGA KIRC cohort. The data are presented in matrix format in which each row represents an individual gene and each column represents a single tissue. Each cell in the matrix represents the expression level of a gene feature in an individual tissue. Expression levels have been standardized (centered and scaled) within columns for visualization. The red and green colors in cells reflect relative high and low expression levels, respectively, as indicated in the scale bar. Images were obtained by re-analysis of the raw data of the TCGA KIRC cohort. ccRCC tissues were ordered from STs to ccRCC tissues according to the standardized expression level of each gene as indicated. *P<0.05 (ST vs. ccRCC). (B) Relative mRNA levels of various miRNA biogenesis-related genes in ccRCC tissues and their corresponding ST. *P<0.001, **P=0.029 and ***P=0.002, as indicated. TCGA KIRC, The Cancer Genome Atlas kidney clear cell carcinoma; ccRCC, clear cell renal cell carcinoma; ST, adjacent non-neoplastic surround tissues.
Altered mRNA levels of four important miRNA biogenesis components, DICER1, DROSHA, DGCR8, and AGO2, in the Korean ccRCC cohort
To investigate whether DICER1, DROSHA, DGCR8, and/or AGO2 were dysregulated in the Korean ccRCC cohort, we measured the mRNA levels of the four miRNA biogenesis components by qPCR in 53-paired ccRCC and ST specimens from the Korean cohort. After excluding unqualified qPCR results, we analyzed the data. The mRNA levels of all four components were significantly downregulated in ccRCC tissues compared with the corresponding ST (paired t-test, DICER1: P<0.001; DROSHA: P<0.001; DGCR8: P<0.001; AGO2: P<0.001; Fig. 3).
Figure 3.
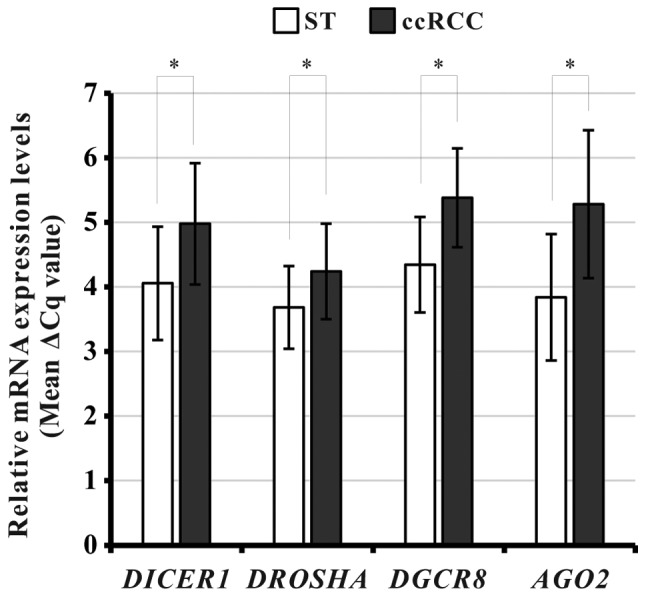
Relative mRNA levels of various microRNA biogenesis-related genes in ccRCC tissues and their corresponding ST of the Korean ccRCC cohort. *P<0.001, as indicated. ccRCC, clear cell renal cell carcinoma; ST, adjacent non-neoplastic surround tissues; DGCR8, DiGeroge syndrome critical region gene 8; AGO2, Argonaute 2.
Correlations between inter-individual mRNA levels of miRNA biogenesis components in the two cohorts
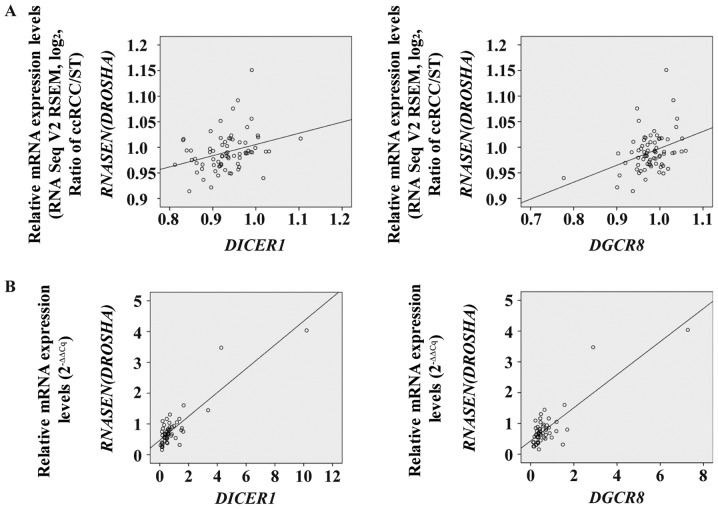
To evaluate the associations between the mRNA levels of each miRNA biogenesis component in ccRCC specimens, Pearson's correlation coefficient analysis was performed in both TCGA KIRC cohort and the Korean ccRCC cohort. Prior to statistical analysis, significantly altered mRNA levels of miRNA biogenesis components were sorted from both cohorts (Table II). We found significant correlations between the mRNA levels of specific miRNA biogenesis components in both cohorts (Table II). Notably, significant correlations were observed not only between the mRNA levels of DICER1 and DROSHA, but also between those of DROSHA and DGCR8 in both cohorts (Fig. 4; Table II).
Table II.
Pearson's correlation analysis between individual components of microRNA biogenesis.
| A, TCGA KIRC cohort | ||
|---|---|---|
| Correlations between components | Pearson's correlation coefficient value | P-value |
| DICER1 and DROSHA | 0.311 | 0.008 |
| DICER1 and AGO1 | 0.265 | 0.024 |
| DICER1 and TNRC6B | 0.609 | <0.001 |
| DICER1 and TNRC6C | 0.452 | <0.001 |
| DICER1 and AGO3 | 0.543 | <0.001 |
| DROSHA and DGCR8 | 0.364 | 0.002 |
| DROSHA and AGO1 | 0.258 | 0.029 |
| DROSHA and TNRC6B | 0.599 | <0.001 |
| DROSHA and TNRC6C | 0.316 | 0.007 |
| DROSHA and AGO3 | 0.267 | 0.023 |
| DGCR8 and TNRC6B | 0.350 | 0.003 |
| DGCR8 and TNRC6C | 0.336 | 0.004 |
| AGO1 and TNRC6B | 0.270 | 0.022 |
| AGO1 and AGO3 | 0.529 | <0.001 |
| TNRC6B and TNRC6C | 0.558 | <0.001 |
| TNRC6B and AGO3 | 0.365 | 0.002 |
| DICER1 and DGCR8 | 0.168 | 0.157 |
| DGCR8 and AGO1 | 0.095 | 0.428 |
| DGCR8 and AGO3 | 0.065 | 0.587 |
| AGO1 and TNRC6C | 0.162 | 0.175 |
| TNRC6C and AGO3 | 0.025 | 0.836 |
| B, Korean ccRCC cohort | ||
| Correlations between components | Pearson's correlation coefficient value | P-value |
| DICER1 and DROSHA | 0.877 | <0.001 |
| DICER1 and DGCR8 | 0.954 | <0.001 |
| DICER1 and AGO2 | 0.774 | <0.001 |
| DROSHA and DGCR8 | 0.840 | <0.001 |
| DROSHA and AGO2 | 0.641 | <0.001 |
| DGCR8 and AGO2 | 0.837 | <0.001 |
TCGA KIRC, The Cancer Genome Atlas kidney clear cell carcinoma; ccRCC, clear cell renal cell carcinoma; DGCR8, DiGeroge syndrome critical region gene 8; TNR, trinucleotide repeat containing.
Figure 4.
Correlation between inter-individual mRNA levels of microRNA biogenesis components in TCGA KIRC cohort and the Korean ccRCC cohort. DICER1 and RNASEN (DROSHA) and DGCR8 and RNASEN (DROSHA) in (A) TCGA KIRC cohort and the (B) Korean ccRCC cohort. TCGA KIRC, The Cancer Genome Atlas kidney clear cell carcinoma; ccRCC, clear cell renal cell carcinoma; ST, adjacent non-neoplastic surround tissues; DGCR8, DiGeroge syndrome critical region gene 8; RNASEN (DROSHA), RNase III endonuclease DROSHA.
Relationship between mRNA levels of the four miRNA biogenesis components and their clinicopathological parameters in both cohorts
To investigate the clinicopathological effects of altered mRNA levels of the selected four miRNA biogenesis components in both cohorts, the correlations between mRNA levels of the miRNA biogenesis components and clinicopathological parameters were statistically evaluated. For TCGA KIRC cohort, downloaded data were processed as the ccRCC to ST ratio. Seventy-two patients with ccRCC, whose clinicopathological data were available, were classified according to each clinicopathological characteristic (Table III). Statistical analysis of the TCGA KIRC cohort data revealed that mRNA levels of DICER1 and DGCR8 were significantly associated with histologic grade, respectively (P=0.025, P=0.044; Table III). Moreover, the mRNA levels of DGCR8 were significantly associated with sex (P=0.024; Table III). Prior to statistical analysis of the Korean ccRCC cohort, raw qPCR data were processed using the 2−∆∆Cq method (17). Fifty-three patients with ccRCC were classified according to each clinicopathological parameter (Table IV). Altered mRNA levels of the analyzed components were not significantly associated with any clinical parameters, including sex, age, T stage, Fuhrman grade/The International Society of Urological Pathology (ISUP) grade, lymphovascular invasion, and peri-renal fat invasion (Table IV).
Table III.
DICER1, DROSHA and DGCR8 mRNA expression levels in relation to clinicopathological parameters of TCGA KIRC.
| DICER1 | DROSHA | DGCR8 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters | Low | High | P-value | Low | High | P-value | Low | High | P-value |
| Sex | 0.293a | 0.836a | 0.024a | ||||||
| Male | 28 | 24 | 30 | 22 | 31 | 21 | |||
| Female | 8 | 12 | 11 | 9 | 6 | 14 | |||
| Age, years | 0.477b | 0.640a | 0.057b | ||||||
| <60 | 22 | 18 | 21 | 18 | 25 | 15 | |||
| ≥60 | 14 | 18 | 19 | 13 | 12 | 20 | |||
| T stage | 0.803c | 0.916c | 0.983c | ||||||
| 1a,1b | 13 | 14 | 16 | 11 | 13 | 14 | |||
| 2 | 9 | 5 | 7 | 7 | 9 | 5 | |||
| 3a,3b | 13 | 16 | 17 | 12 | 14 | 15 | |||
| 4 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| M stage | 0.181a | 0.524a | 0.683a | ||||||
| No metastasis | 24 | 29 | 29 | 24 | 28 | 25 | |||
| Metastasis | 12 | 7 | 12 | 7 | 9 | 10 | |||
| Pathologic stage | 0.682a | 0.570a | 0.754a | ||||||
| Stage I | 11 | 14 | 12 | 13 | 11 | 14 | |||
| Stage II | 6 | 5 | 8 | 3 | 6 | 5 | |||
| Stage III | 7 | 9 | 9 | 7 | 8 | 8 | |||
| Stage IV | 12 | 8 | 12 | 8 | 12 | 8 | |||
| Histologic grade | 0.025c | 0.169c | 0.044c | ||||||
| G1 | 0 | 1 | 1 | 0 | 0 | 1 | |||
| G2 | 10 | 18 | 12 | 16 | 10 | 18 | |||
| G3 | 16 | 12 | 18 | 10 | 18 | 10 | |||
| G4 | 10 | 5 | 10 | 5 | 9 | 6 | |||
Pearson's chi-square test
Fisher's Exact test
Linear by linear association.
Table IV.
DICER1, DROSHA, DGCR8, and AGO2 mRNA expression levels in relation to clinicopathological parameters of Korean clear cell renal cell carcinoma.
| DICER1 | DROSHA | DGCR8 | AGO2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value |
| Sex | 0.560a | 0.077b | 0.810a | 0.728b | ||||||||
| Male | 20 | 7 | 15 | 12 | 20 | 7 | 21 | 6 | ||||
| Female | 21 | 5 | 21 | 5 | 20 | 6 | 22 | 4 | ||||
| Age, years | 0.941a | 0.168a | 0.379a | 0.293b | ||||||||
| <60 | 21 | 6 | 16 | 11 | 19 | 8 | 20 | 7 | ||||
| ≥60 | 20 | 6 | 20 | 6 | 21 | 5 | 23 | 3 | ||||
| T stage | 0.589c | 0.878c | 0.342c | 0.677c | ||||||||
| 1A,1B | 22 | 6 | 18 | 10 | 22 | 6 | 23 | 5 | ||||
| 2A,2B | 7 | 1 | 7 | 1 | 7 | 1 | 7 | 1 | ||||
| 3A,3B | 12 | 5 | 11 | 6 | 11 | 6 | 13 | 4 | ||||
| Fuhrman grade/ISUP grade | 0.250c | 0.280c | 0.105c | 0.305c | ||||||||
| 1 | 2 | 0 | 1 | 1 | 2 | 0 | 2 | 0 | ||||
| 2 | 16 | 1 | 15 | 12 | 16 | 1 | 16 | 1 | ||||
| 3 | 15 | 10 | 14 | 11 | 15 | 10 | 17 | 8 | ||||
| 4 | 8 | 1 | 6 | 3 | 7 | 2 | 8 | 1 | ||||
| Lymphovascular invasion | 0.183b | 0.667b | 0.053b | 0.604b | ||||||||
| (−) | 37 | 9 | 32 | 14 | 37 | 9 | 38 | 8 | ||||
| (+) | 4 | 3 | 4 | 3 | 3 | 4 | 5 | 2 | ||||
| Peri-renal fat invasion | 0.423b | 0.471b | 1.000b | 0.665b | ||||||||
| (−) | 32 | 11 | 28 | 15 | 32 | 11 | 34 | 9 | ||||
| (+) | 9 | 1 | 8 | 2 | 8 | 2 | 9 | 1 | ||||
Pearson's chi-square test
Fisher's Exact test
Linear by linear association. DGCR8, DiGeroge syndrome critical region gene 8; AGO2, Argonaute 2.
Discussion
Dysregulation of miRNA biogenesis components has been reported to be associated with carcinogenesis (18), cancer chemosensitivity (19), angiogenesis (13), metastasis (20), proliferation and migration (21), and the prognosis of patients with cancer (20,22) in various cancers, including ccRCC (12,13). Although the dysregulation and pathophysiological role of DICER1 in ccRCC has been investigated (12,13), studies of other miRNA biogenesis components, such as DROSHA, DGCR8, and AGO2 are needed. In the present study, we investigated the mRNA levels of miRNA biogenesis components and analyzed their clinicopathological relevance in the TCGA KIRC cohort and a Korean ccRCC cohort.
First, our hierarchical clustering results revealed considerable dysregulation of miRNA biogenesis-related genes in ccRCC tissues compared with ST from TCGA KIRC cohort. Among the dysregulated genes, DICER 1 was significantly downregulated in ccRCC tissues. Similarly, in our Korean ccRCC cohort, DICER 1 was significantly downregulated. These results are consistent with those of previous studies of ccRCC in which downregulated DICER1 mRNA levels were detected (12). Particularly, one study revealed that DICER1 was more downregulated in VHL-deficient (VHL mutation and methylation) ccRCCs compared with ST (13). Because nearly all samples (71 of 72 patients with ccRCC) were VHL-deficient (data not shown) in the dataset of 72 patients with ccRCC of TCGA KIRC cohort, we did not evaluate whether DICER1 mRNA levels were downregulated in VHL-deficient ccRCCs compared with wild-type VHL ccRCCs. Notably, we found for the first time that DROSHA and DGCR8 were significantly downregulated in the ccRCC tissues of both cohorts. On the other hand, AGO2 was only significantly downregulated in the Korean ccRCC cohort. Although we did not identify the exact cause underlying the downregulation of AGO2 in only the Korean ccRCC cohort, it is possible that racial differences between the two cohorts affected the AGO2 mRNA levels. These results demonstrate that the downregulated mRNA levels of DICER1, DROSHA, and DGCR8 are common in ccRCC samples and that downregulation of the three components is important in the pathophysiology of ccRCC.
Accumulating evidence indicates significant correlations between the inter-individual levels of miRNA biogenesis components in paired samples of cancer and ST: DGCR8 and AGO2 in colorectal cancer (23); DICER1 and DROSHA in triple negative breast cancer (24); and DICER1 and DROSHA, as well as DROSHA and DGCR8, in TCGA papillary thyroid carcinoma (25). Therefore, we investigated the correlation between the mRNA levels of the components in both cohorts. Our data revealed significant correlations between DICER1 and DROSHA as well as between DROSHA and DGCR8 in both cohorts.
Recent studies showed that aberrant regulation of miRNA biogenesis components is associated with specific clinicopathological parameters (10,26–29). For example, DICER1 mRNA levels were significantly correlated with overall TNM staging (12). Subsequent studies evaluated the clinicopathological implications of dysregulated miRNA biogenesis components in ccRCC. In contrast to a recent study (12), our results revealed that there was no significant correlation between the mRNA levels of any components and stages of T and M in TCGA KIRC cohort. Furthermore, DICER1 and DGCR8 mRNA levels were significantly correlated with histologic grade of ccRCC. However, we found no significant correlation between dysregulation of the miRNA biogenesis components and any clinicopathological parameters in the Korean ccRCC cohort, including sex, age, T stage, Fuhrman grade/ISUP grade (histologic grade), lymphovascular invasion, and peri-renal fat invasion. These differences between cohorts may be attributed to the small numbers of enrolled patients and racial differences. Because few studies have examined the clinicopathological implications of dysregulated the miRNA biogenesis components in ccRCC, further studies focusing on these issues in ccRCC are required.
Because RT-qPCR and RNA-Seq are different experimental assays utilizing different methodologies and show varying sensitivities, dynamic ranges of the results, and experimental principles, the results of the present study should be interpreted with caution. Nevertheless, we detected downregulation of DICER1, DROSHA, DGCR8, and AGO2 and significant correlations between DICER1 and DROSHA as well as between DROSHA and DGCR8 in ccRCC. Collectively, DICER1, DROSHA, DGCR8, and AGO2 were significantly downregulated and correlated with each other in ccRCC, suggesting that they serve important roles in the pathophysiology of ccRCC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ccRCC
clear cell renal cell carcinoma
- miRNA
microRNA
- TCGA KIRC
The Cancer Genome Atlas Kidney Clear Cell Carcinoma
- qPCR
quantitative PCR
- DGCR8
DiGeroge syndrome critical region gene 8
- AGO2
Argonaute 2
- ST
adjacent non-neoplastic surround tissues
Funding
The present study was supported by the National Research Foundation of Korea (grant funded by the Korea Government Ministry of Science, ICT and Future Planning; grant nos. 2017R1C1B5016670 and 2014R1A5A2010008).
Availability of data and materials
The datasets analyzed during the present study are available from The Cancer Genome Atlas (tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp) and the UCSC Xena (xena.ucsc.edu). The datasets generated in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
SSL and SK contributed to the conception and design of the study, analysis of the data, interpretation of the results, and the writing of the manuscript. HM and SK contributed to the acquisition of data. SSL, HM and SK performed the experiments. JYH, BHK and MSC contributed to the conception and design of the study, and assisted with the determination of the clinical implications of the study. SK reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy and integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The biospecimens for this retrospective study were provided by the Keimyung Human Bio-Resource Bank, a member of the National Biobank of Korea, which is supported by the Ministry of Health and Welfare. The experimental study was approved by the Institutional Review Board of Keimyung University and Dongsan Medical Center (approval no. 2015-10-021-004) and was performed in accordance with the principles of the Declaration of Helsinki. The requirement for written informed consent was waived due to the retrospective nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11:517–525. doi: 10.1038/nrurol.2014.194. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, Delahunt B, Eble JN, Fleming S, Ljungberg B, Medeiros LJ, et al. The Heidelberg classification of renal cell tumours. J Pathol. 1997;183:131–133. doi: 10.1002/(SICI)1096-9896(199710)183:2<131::AID-PATH931>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 4.Campbell D, Walker R, Mathew T, Craig J. Commentary on ‘Influence of industry on renal guideline development’. Clin J Am Soc Nephrol. 2007;2:211. doi: 10.2215/CJN.00300107. [DOI] [PubMed] [Google Scholar]
- 5.Papachristou DJ, Korpetinou A, Giannopoulou E, Antonacopoulou AG, Papadaki H, Grivas P, Scopa CD, Kalofonos HP. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011;459:431–440. doi: 10.1007/s00428-011-1119-5. [DOI] [PubMed] [Google Scholar]
- 6.Mockenhaupt S, Schurmann N, Grimm D. When cellular networks run out of control: Global dysregulation of the RNAi machinery in human pathology and therapy. Prog Mol Biol Transl Sci. 2011;102:165–242. doi: 10.1016/B978-0-12-415795-8.00006-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Song ML, Min H, Hwang I, Baek SK, Kwon TK, Park JW. miRNA biogenesis-associated RNase III nucleases Drosha and Dicer are upregulated in colorectal adenocarcinoma. Oncol Lett. 2017;14:4379–4383. doi: 10.3892/ol.2017.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hata A, Kashima R. Dysregulation of microRNA biogenesis machinery in cancer. Crit Rev Biochem Mol Biol. 2016;51:121–134. doi: 10.3109/10409238.2015.1117054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tchernitsa O, Kasajima A, Schäfer R, Kuban RJ, Ungethüm U, Györffy B, Neumann U, Simon E, Weichert W, Ebert MP, Röcken C. Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies gastric cancer progression. J Pathol. 2010;222:310–319. doi: 10.1002/path.2759. [DOI] [PubMed] [Google Scholar]
- 11.Shaikhibrahim Z, Lindstrot A, Ochsenfahrt J, Fuchs K, Wernert N. Epigenetics-related genes in prostate cancer: Expression profile in prostate cancer tissues, androgen-sensitive and -insensitive cell lines. Int J Mol Med. 2013;31:21–25. doi: 10.3892/ijmm.2012.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X, Fan Y, Gao Y, Zhang Y, Huang Q, Ai Q, Ni D, Chen W, Zhang P, Song E, et al. Dicer is down-regulated in clear cell renal cell carcinoma and in vitro Dicer knockdown enhances malignant phenotype transformation. Urol Oncol. 2014;32:46.e9–e17. doi: 10.1016/j.urolonc.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Fan Y, Li H, Ma X, Gao Y, Bao X, Du Q, Ma M, Liu K, Yao Y, Huang Q, et al. Dicer suppresses the malignant phenotype in VHL-deficient clear cell renal cell carcinoma by inhibiting HIF-2α. Oncotarget. 2016;7:18280–18294. doi: 10.18632/oncotarget.7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–1812. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edge S, Byrd DR, Compton CC, Fritz AG, Greene F, Trotti A, editors. Springer; New York: 2010. AJCC cancer staging handbook: From the AJCC cancer staging manual. [Google Scholar]
- 17.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Gurtner A, Falcone E, Garibaldi F, Piaggio G. Dysregulation of microRNA biogenesis in cancer: The impact of mutant p53 on Drosha complex activity. J Exp Clin Cancer Res. 2016;35:45. doi: 10.1186/s13046-016-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Li WF, Wu X, Zhang HC, Chen L, Zhang PY, Liu LY, Ma D, Chen T, Zhou L, et al. Dicer regulates non-homologous end joining and is associated with chemosensitivity in colon cancer patients. Carcinogenesis. 2017;38:873–882. doi: 10.1093/carcin/bgx059. [DOI] [PubMed] [Google Scholar]
- 20.To SKY, Mak ASC, Eva Fung YM, Che CM, Li SS, Deng W, Ru B, Zhang J, Wong AST. β-catenin downregulates Dicer to promote ovarian cancer metastasis. Oncogene. 2017;36:5927–5938. doi: 10.1038/onc.2017.185. [DOI] [PubMed] [Google Scholar]
- 21.Yu Z, Wang L, Wang C, Ju X, Wang M, Chen K, Loro E, Li Z, Zhang Y, Wu K, et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun. 2013;4:2812. doi: 10.1038/ncomms3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz-Garcia CV, Agudo-López A, Pérez C, López-Martín JA, Rodríguez-Peralto JL, de Castro J, Cortijo A, Martínez-Villanueva M, Iglesias L, García-Carbonero R, et al. DICER1, DROSHA and miRNAs in patients with non-small cell lung cancer: Implications for outcomes and histologic classification. Carcinogenesis. 2013;34:1031–1038. doi: 10.1093/carcin/bgt022. [DOI] [PubMed] [Google Scholar]
- 23.Kim B, Lee JH, Park JW, Kwon TK, Baek SK, Hwang I, Kim S. An essential microRNA maturing microprocessor complex component DGCR8 is up-regulated in colorectal carcinomas. Clin Exp Med. 2014;14:331–336. doi: 10.1007/s10238-013-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passon N, Gerometta A, Puppin C, Lavarone E, Puglisi F, Tell G, Di Loreto C, Damante G. Expression of Dicer and Drosha in triple-negative breast cancer. J Clin Pathol. 2012;65:320–326. doi: 10.1136/jclinpath-2011-200496. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Park WJ, Jeong KJ, Kang SH, Kwon SY, Kim S, Park JW. Racial differences in expression levels of miRNA machinery-related genes, Dicer, Drosha, DGCR8, and AGO2, in Asian Korean papillary thyroid carcinoma and comparative validation using the cancer genome atlas. Int J Genomics. 2017;2017:5789769. doi: 10.1155/2017/5789769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaksman O, Hetland TE, Trope' CG, Reich R, Davidson B. Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum Pathol. 2012;43:2062–2069. doi: 10.1016/j.humpath.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, London WB, Chang CH, Yu AL. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stratmann J, Wang CJ, Gnosa S, Wallin A, Hinselwood D, Sun XF, Zhang H. Dicer and miRNA in relation to clinicopathological variables in colorectal cancer patients. BMC Cancer. 2011;11:345. doi: 10.1186/1471-2407-11-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu GS, Yang ZL, Liu DC. Immunohistochemical study of Dicer and Drosha expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Pathol Res Pract. 2012;208:392–397. doi: 10.1016/j.prp.2012.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the present study are available from The Cancer Genome Atlas (tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp) and the UCSC Xena (xena.ucsc.edu). The datasets generated in the present study are available from the corresponding author upon reasonable request.