Abstract
The aim of the present study was to identify genes associated with and the underlying mechanisms in nasopharyngeal carcinoma (NPC) using microarray data. GSE12452 and GSE34573 gene expression profiles were obtained from the Gene Expression Omnibus (GEO) database. GEO2R was utilized to obtain differentially expressed genes (DEGs). In addition, the Database for Annotation, Visualization and Integrated Discovery was used to perform pathway enrichment analyses for DEGs using the Gene Ontology (GO) annotation along with the Kyoto Encyclopedia of Genes and Genomes (KEGG). Furthermore, Cytoscape was used to perform module analysis of the protein-protein interaction (PPI) network and pathways of the hub genes were studied. A total of 298 genes were ascertained as DEGs in the two datasets. To functionally categorize these DEGs, we obtained 82 supplemented GO terms along with 7 KEGG pathways. Subsequently, a PPI network consisting of 10 hub genes with high degrees of interaction was constructed. These hub genes included cyclin-dependent kinase (CDK) 1, structural maintenance of chromosomes (SMC) 4, kinetochore-associated (KNTC) 1, kinesin family member (KIF) 23, aurora kinase A (AURKA), ATAD (ATPase family AAA domain containing) 2, NDC80 kinetochore complex component, enhancer of zeste 2 polycomb repressive complex 2 subunit, BUB1 mitotic checkpoint serine/threonine kinase and protein regulator of cytokinesis 1. CDK1, SMC4, KNTC1, KIF23, AURKA and ATAD2 presented with high areas under the curve in receiver operator curves, suggesting that these genes may be diagnostic markers for nasopharyngeal carcinoma. In conclusion, it was proposed that CDK1, SMC4, KNTC1, KIF23, AURKA and ATAD2 may be involved in the tumorigenesis of NPC. Furthermore, they may be utilized as molecular biomarkers in early diagnosis of NPC.
Keywords: nasopharyngeal carcinoma, Gene Expression Omnibus, bioinformatics analysis, diagnosis, microarray
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor of the nasopharyngeal epithelium with an uneven geographical distribution, including 81% of novel cases occurring in Asia; however, only 9% in Africa (1). Regarding the number of newly diagnosed patients, the top 5 countries include China, Indonesia, Vietnam, India and Malaysia, accounting for 67% of patients with NPC worldwide, in 2012 (1). Despite the therapeutic enhancements in the treatment of NPC, late diagnosis, poor prognosis and metastasis influence survival (2). Therefore, it is crucial to investigate novel diagnostics and therapeutics intended for patients with NPC. Advances in radiotherapy and comprehensive chemotherapy strategies have enhanced outcomes in patients with primary NPC (3,4). Furthermore, NPC presents with highly malignant recurrence with local tissue invasion and distant metastasis, the main reason for radiotherapy failure (5).
In the course of tumorigenesis and development, various types of genetic and epigenetic abnormalities, such as methylation and allele imbalance contribute to the pathogenesis of NPC (5,6). Previous studies have reported that the molecular mechanism of NPC has provided an important understanding and revealed the etiology of NPC (7,8). Nasopharyngeal carcinoma is caused by multiple genes and different environmental factors, such as mutations in cyclin D1 and X-ray repair cross complementing 1, and an excess intake of salted fish and alcohol (8–10).
Microarray technology has been widely applied in the examination of general genetic abnormalities (11–13). Furthermore, the Gene Expression Omnibus (GEO) database is increasingly recognized by researchers with an increasing interest in data mining and bioinformatics and a number of tumor-associated genes, including for NPC, being discovered (14,15). However, only a limited number of studies integrate these microarray datasets in identifying essential genes and pathway regulatory networks in NPC (6,14).
In the present study, differentially expressed genes (DEGs) were screened based on gene expression profiling data obtained from GEO. Consequently, two datasets comparing gene expression in tumor and non-tumor tissues were used and DEGs considered to be involved in NPC development were identified. Gene Ontology (GO) term enrichment, Kyoto Encyclopedia of Genes and Genomes (KEGG), protein-protein interaction (PPI) network and gene co-expression network analyses were performed to uncover the underlying mechanisms of NPC occurrence and development.
Materials and methods
Microarray data
Two gene expression profiles, GSE12452 (16) and GSE34573 (17), were downloaded from the GEO database using the following criteria: i) Containing NPC and non-tumor samples; ii) based on the Illumina HiSeq 2000 (Homo sapiens; GPL11154) chip platform; and iii) sample size >30.
The array data for GSE12452 contains 41 samples, including 31 NPC and 10 normal nasopharyngeal tissues (accessed on December 23, 2017) (16). The array data for GSE34573 includes 64 samples, of which, 18 samples included expression profiles from chip scan (platform: GPL570) and used for further analysis in the present study, which included 15 NPC tumor and 3 normal control (NC) tissues (accessed on December 23, 2017) (17).
Data processing
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), an interactive web tool, is used to compare two sets of data and is capable of analyzing GEO array data (18). GEO2R was applied in screening differentially expressed mRNAs in NPC and normal tissues. P<0.05 and |log2fold-change|≥1 were set as cut-off standards. A DEG heat map was subsequently generated using the online tool Morpheus (https://software.broadinstitute.org/morpheus/).
Analysis of functional and pathway enrichment
The GO database denotes a large number of gene annotation terms (19). KEGG is used to identify functional and metabolic pathways (20). Analyses were accessed from Database for Annotation, Visualization and Integrated Discovery (DAVID; v6.8; http://david.ncifcrf.gov/). The cut-off criterion was P<0.05.
Analysis of PPI network and hub proteins
Another online software, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; v11.0; http://string-db.org/), provides an important integration and assessment of the PPI (21). Cytoscape (v3.70) is a common open-source software tool that is useful in the visual evaluation of biomolecule interaction networks comprising of protein, gene and other forms of interactions (22). DEGs were mapped using STRING to assess the PPI information prior to visualization with Cytoscape. Nodes that are usually linked to the proteins are described as hub proteins and are crucial in the network (23).
Statistical analysis
The receiver operating characteristic (ROC) curve was constructed using SPSS 19.0 (IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs
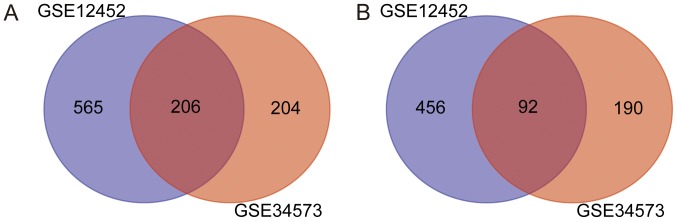
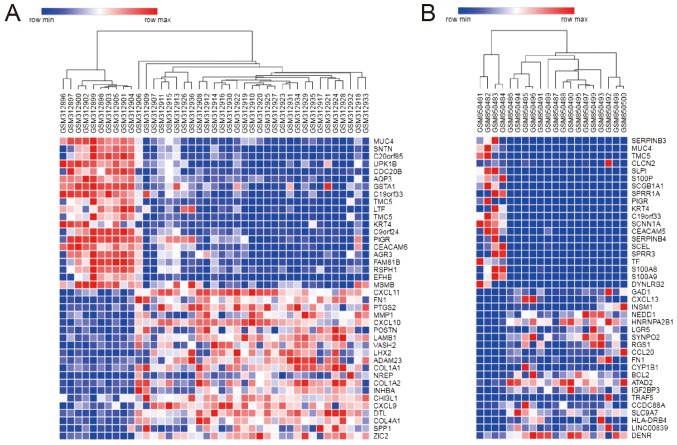
In the present study, two gene expression profiles were evaluated, including 46 NPC and 13 NC samples. On the basis of the GEO2R analysis, a total of 1,319 and 692 genes were identified for GSE12452 and GSE34573, respectively. Venn Diagram online software (http://bioinformatics. psb.ugent.be/webtools/Venn/) was used to determine the intersection of the two datasets as presented in Fig. 1. A total of 298 genes were identified as DEGs in the two sets, with 206 up- and 92 downregulated genes. Fig. 2 illustrates the DEGs expression heat map, including the top 40 genes.
Figure 1.
Overlapping DEGs from the GSE12452 and GSE34573 microarray datasets. Venn diagram of (A) up- and (B) downregulated genes from the GSE12452 and GSE34573 datasets. DEGs, differentially expressed genes.
Figure 2.
Heat map of the top 40 differentially expressed genes. Heat map for the (A) GSE12452 and (B) GSE34573 datasets. Each column represents a sample and each row a gene. Red indicates upregulated and blue downregulated genes.
Analysis of GO term enrichment
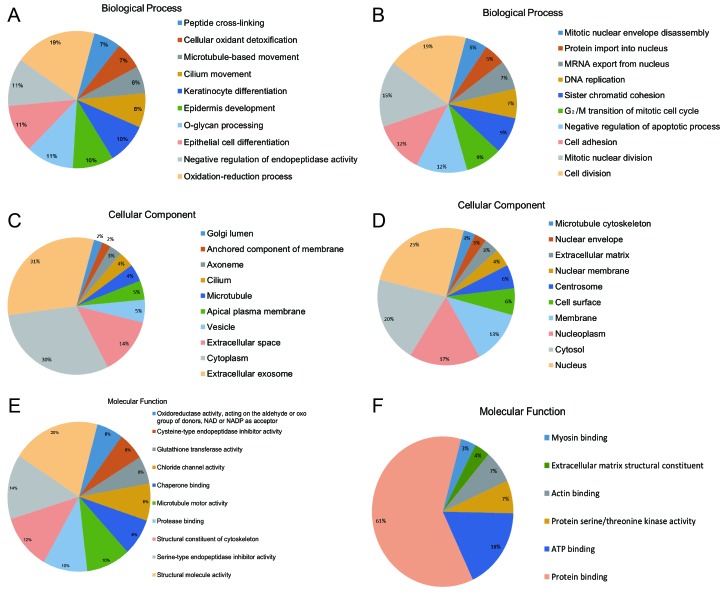
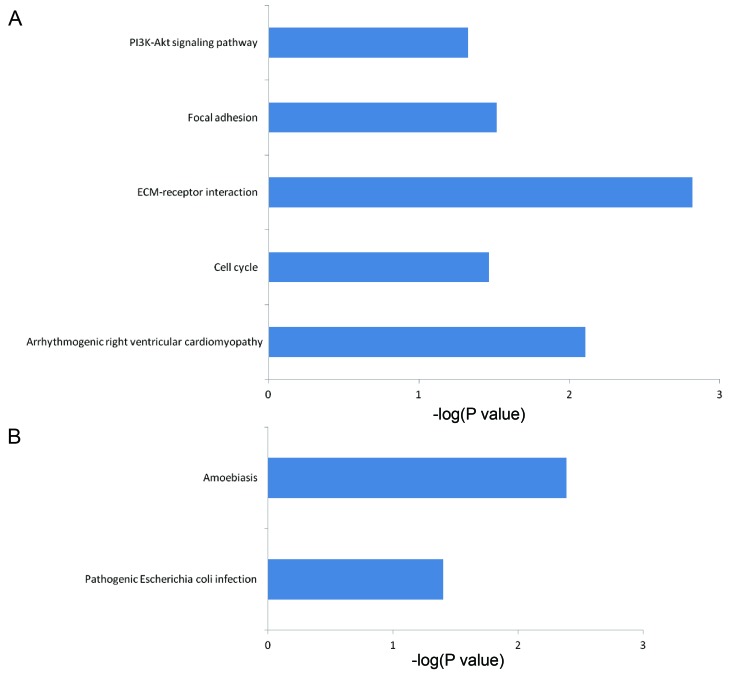
To functionally categorize the 298 DEGs, 82 enhanced GO terms and 7 KEGG pathways were obtained. Tables I and II summarize the enhanced top 10 GO terms by P-value. These genes may play a crucial role in the development of nasopharyngeal carcinoma. These GO terms were classified based on their association with biological processes, including response to epithelial cell differentiation, positive regulation of the intrinsic apoptotic signaling pathway, keratinocyte differentiation, cell division, mitotic nuclear division, G2/M transition of the mitotic cell cycle, mRNA export from the nucleus and DNA replication. In addition, there were numerous enriched GO terms associated with protein binding, activities of protein serine or threonine kinases, extracellular matrix (ECM) structural constituent and cell surface as presented in Fig. 3, divided by biological process, cellular component and molecular function. Fig. 4 illustrates the enriched KEGG pathways, including the phosphoinositide 3-kinase (PI3K)-protein kinase B (Akt) signaling pathway, ECM-receptor interactions, the cell cycle, arrhythmogenic right ventricular cardiomyopathy and focal adhesion pathways.
Table I.
Kyoto Encyclopedia of Genes and Genomes and GO functional annotation for upregulated genes (top 10).
| Category | Term | Count | P-value |
|---|---|---|---|
| GOTERM_CC_DIRECT | Extracellular exosome | 76 | <0.001 |
| GOTERM_CC_DIRECT | Cytoplasm | 74 | <0.001 |
| GOTERM_CC_DIRECT | Extracellular space | 33 | <0.001 |
| GOTERM_CC_DIRECT | Vesicle | 13 | <0.001 |
| GOTERM_BP_DIRECT | Oxidation-reduction process | 12 | 0.0206 |
| GOTERM_CC_DIRECT | Apical plasma membrane | 11 | <0.001 |
| GOTERM_CC_DIRECT | Microtubule | 10 | 0.0036 |
| GOTERM_MF_DIRECT | Structural molecule activity | 10 | <0.001 |
| GOTERM_CC_DIRECT | Cilium | 9 | <0.001 |
| GOTERM_BP_DIRECT | O-glycan processing | 7 | <0.001 |
GO, Gene Ontology.
Table II.
Kyoto Encyclopedia of Genes and Genomes and GO functional annotation for downregulated genes (top 10).
| Category | Term | Count | P-value |
|---|---|---|---|
| GOTERM_MF_DIRECT | Protein binding | 52 | <0.001 |
| GOTERM_CC_DIRECT | Nucleus | 36 | <0.001 |
| GOTERM_CC_DIRECT | Cytosol | 29 | <0.001 |
| GOTERM_CC_DIRECT | Nucleoplasm | 24 | 0.0018 |
| GOTERM_CC_DIRECT | Membrane | 18 | 0.0154 |
| GOTERM_MF_DIRECT | ATP binding | 15 | 0.0056 |
| GOTERM_BP_DIRECT | Cell division | 11 | <0.001 |
| GOTERM_BP_DIRECT | Mitotic nuclear division | 9 | <0.001 |
| GOTERM_CC_DIRECT | Cell surface | 9 | 0.0028 |
| GOTERM_CC_DIRECT | Centrosome | 8 | 0.0029 |
GO, Gene Ontology.
Figure 3.
GO terms associated with the identified target genes. GO analysis of (A) up- and (B) downregulated target genes of the intersection of GSE12452 and GSE34573 according to biological process. GO analysis of (C) up- and (D) downregulated target genes of the intersection of GSE12452 and GSE34573 according to cellular component. GO analysis of (E) up- and (F) downregulated target genes of the intersection of GSE12452 and GSE34573 according to molecular function. GO, Gene Ontology; NADP, nicotinamide adenine dinucleotide phosphate.
Figure 4.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of the identified target genes. Pathways corresponding to the (A) up- and (B) downregulated genes. ECM, extracellular matrix; PI3K, phosphoinositide 3-kinase; Akt, protein kinase B.
Module screening in the PPI network
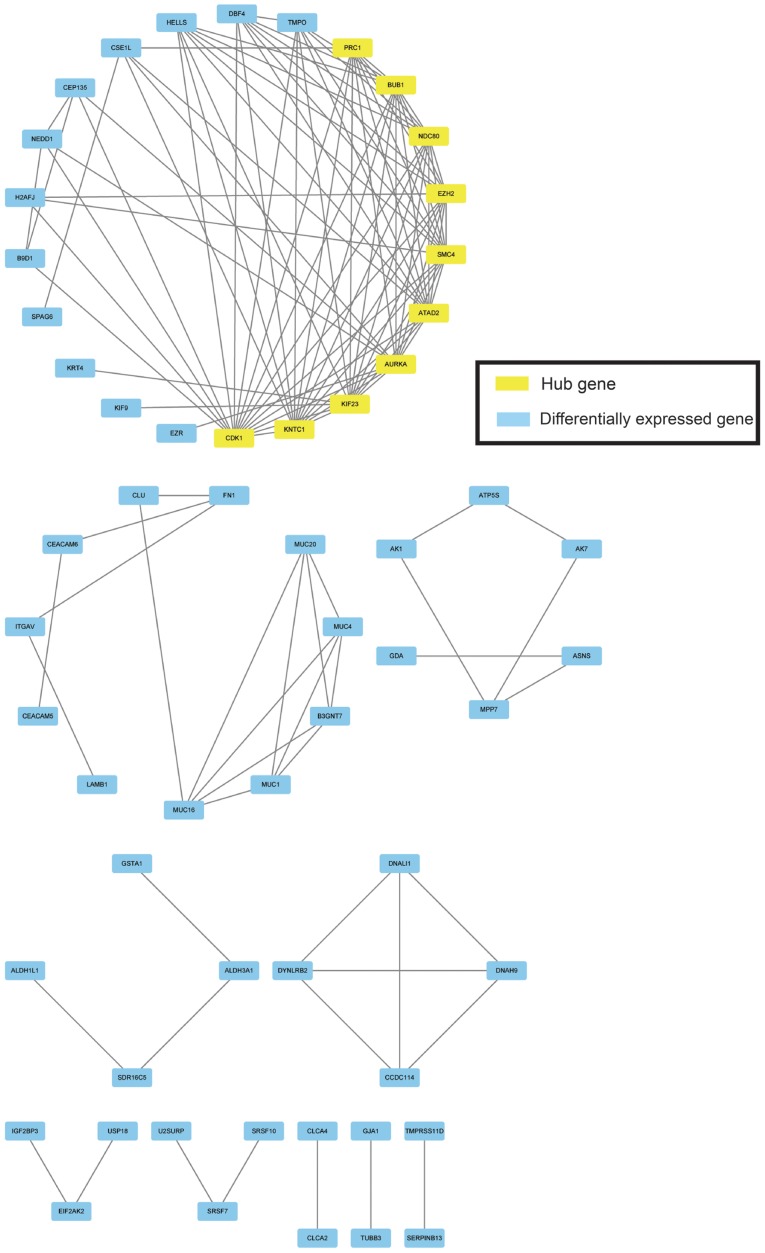
Based on the STRING output, the top 10 hub nodes with higher degrees of interaction were screened, including cyclin-dependent kinase (CDK) 1, structural maintenance of chromosomes (SMC) 4, kinetochore-associated (KNTC) 1, kinesin family member (KIF) 23, aurora kinase A (AURKA), ATPase Family AAA domain containing 2 (ATAD2), NDC80 kinetochore complex component (NDC80), enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), BUB1 mitotic checkpoint serine/threonine kinase (BUB1) and protein regulator of cytokinesis 1 (PRC1) (Fig. 5). The hub genes that were discovered in the PPI network analysis could play important roles in the aberrant signaling pathways and are potential targets for future research.
Figure 5.
Protein-protein interaction network of the 298 differentially expressed genes. Yellow colored nodes represent hub genes.
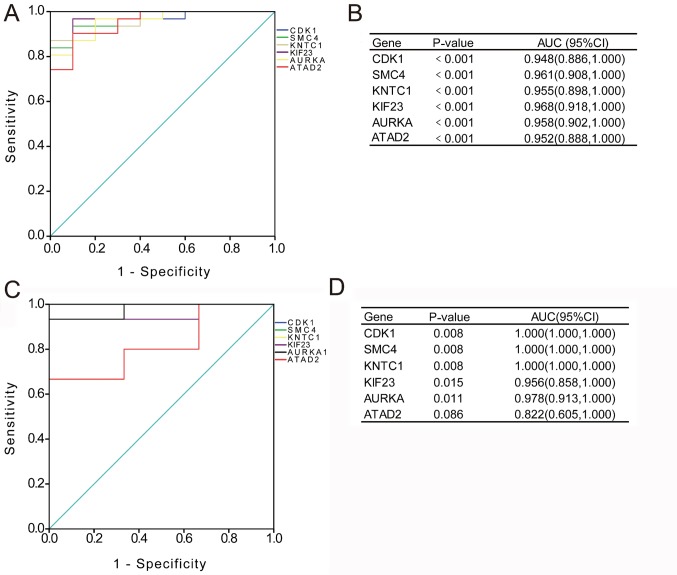
Diagnostic values of CDK1, SMC4, KNTC1, KIF23, AURK and ATAD2 expression in NPC tissues
To verify the diagnostic value of the hub genes, expression levels in NPC tissues were evaluated using ROC curves. As presented in Fig. 6A and B, the area under the curve (AUC) for CDK1, SMC4, KNTC1, KIF23, AURKA and ATAD2 in NPC and non-cancerous nasopharyngeal tissue determined for the GSE12452 dataset were 0.948 [95% confidence interval (CI), 0.866–1.000; P<0.001], 0.961 (95% CI, 0.908–1.000; P<0.001), 0.955 (95% CI, 0.898–1.000; P<0.001), 0.968 (95% CI, 0.918–1.000; P<0.001), 0.958 (95% CI, 0.902–1.000; P<0.001) and 0.952 (95% CI, 0.888–1.000; P<0.001), respectively. In addition, as indicated in Fig. 6C and D, the AUC for CDK1, SMC4, KNTC1, KIF23, AURKA and ATAD2 in NPC and non-cancerous nasopharyngeal tissue for the GSE34573 dataset were 1.000 (95% CI, 1.000–1.000; P=0.008), 1.000 (95% CI, 1.000–1.000; P=0.008), 1.000 (95% CI, 1.000–1.000; P=0.008), 0.956 (95% CI, 0.858–1.000; P=0.015), 0.978 (95% CI, 0.913–1.000; P=0.011) and 0.822 (95% CI, 0.605–1.000; P=0.086), respectively. These results suggest that these genes may be diagnostic markers for nasopharyngeal carcinoma.
Figure 6.
Diagnostic value of HUB genes. (A) ROC curves and (B) AUC values for the selected genes from the GSE12452 dataset. (C) ROC curves and (D) AUC values for the selected genes from the GSE34573 dataset. ROC, receiver operating characteristic; AUC, area under ROC curve; CI, confidence interval; DEG, differentially expressed gene; CDK1, cyclin-dependent kinase 1; SMC4, structural maintenance of chromosomes 4; KNTC1, kinetochore-associated 1; KIF23, kinesin family member 23; AURKA, aurora kinase A; ATAD2, ATPase Family AAA domain containing 2.
Discussion
In Southern China, NPC is a common malignant tumor (24,25). Sihui City in Southern China has the highest overall incidence rate, with 27.2 per 100,000 men and 11.3 per 100,000 women in 2003, while the average incidence rates in other parts of China are 4.31 per 100,000 men and 1.81 per 100,000 women, respectively (24,25). Advances in treatment, including radio- and chemotherapy, had an impact on clinical results, improving the long-term survival of patients with NPC (26). Despite the aforementioned, 70% of patients with NPC are diagnosed at mid to late stages and the 5-year survival rates for these patients are 40–70% (27). To gain insight into the pathogenesis of NPC, this study focused on DEGs in NPC compared with normal tissues based on two gene expression datasets obtained from the GEO database. A total of 206 up- and 92 downregulated DEGs were identified in the two datasets. It was indicated that DEGs, including CDK1, SMC4, KNTC1, KIF23, AURKA, ATAD2, NDC80, EZH2, BUB1 and PRC1, were hub genes in the PPI network analysis.
CDK1 was indicated as one of the most important genes in NPC compared with normal tissues. CDK1 is a well-preserved serine/threonine kinase, which forms multiplexes with its cyclin partners, contributing in the progression and regulation of the cell cycle (26). One previous study has reported that the CDK1-cyclin B1 signaling pathway is a target in NPC therapy (23). In previous research, overactivity of the CDKs were reported to be due to different genetic and epigenetic events in human types of cancer and inhibition causes cell cycle arrest and apoptosis (28). In a published study, CDK1 selectively targets human breast cancer cells, suggesting that further research into CDK1 suppression is required for a potential application in breast cancer treatment (29). In addition, CDK1 indicated to have a high AUC in the present study. Therefore, it was concluded that CDK1 may promote growth of NPC cells by regulating the cell cycle and it may serve as a biomarker.
SMC4 is a member of the SMC gene family (30). A study reported that SMC4 expression is substantially associated with tumor size, degree of tumor differentiation, progressive stages and vascular invasion of hepatocellular carcinoma (31). Clinical and experimental data propose that SMC4 stimulates the formation of colorectal cancer (32). In addition, SMC4 had a high AUC in the present study. Therefore, it is suggested that SMC4 may promote the development of tumors and may serve as a biomarker.
The ATAD2 gene is located on human chromosome 8q24.13 (33). ATAD2 is a member of the ATPase family. Caron et al (34) reported that ATAD2 is highly expressed in mouse testis and in many tumor cell lines, including leukemia, lymphoma, osteosarcoma, breast cancer, lung cancer and cervical cancer. Other studies have verified that high expression of ATAD2 is significantly associated with poor prognosis in patients with hepatocellular carcinoma, as well as ovarian cancer (35,36).
Furthermore, KNTC1, KIF23 and AURKA were reported to be the important hub proteins in the PPI network analysis, whereas KIF23 and AURKA had been implicated in tumor progression in the present study. The study by Takahashi et al (37) indicated that KIF23 may be a novel therapeutic target in malignant glioma. A previous study has examined and affirmed that KIF23 is a novel transcriptional target gene of the tumor suppressor protein p53 (38). It was reported that p53 decreases KIF23 mRNA and protein expression levels in various cell types. Previous studies have further reported that AURKA is a central kinase-encoding gene and overexpression causes amplification of centrosome, chromosomal instability, as well as transformation in mammalian cells (39,40). AURKA is associated with different types of cancer, including NPC (41), colorectal cancer (42), gastric cancer (43), esophageal cancer (44), multiple myeloma (45), breast cancer (46,47) and bladder cancer (48). These studies suggest that AURKA may be considered as a potential therapeutic target for NPC treatment. KNTC1 serves an important role in the mitotic checkpoint (49). It regulates the main mechanism that prevents chromosome instability usually observed in human tumors (50).
The present study indicated that a number of processes identified in the GO enrichment analysis were associated with NPC, including the physiological processes of cell growth and proliferation. These processes were associated with numerous cancers including lung cancer (51), breast cancer (52) and skin cancer (53), and any abnormality in these processes may cause cancer development. A total of 7 KEGG pathways were enriched and with P<0.05 in the current study. The ECM-receptor interaction pathway has been demonstrated to be involved in the tumorigenesis of various cancers (54–56) and in the current study, the KEGG pathway analysis indicated that these DEGs were also enriched in the ECM-receptor interaction pathway. The PI3K-Akt signaling pathway contributes to the regulatory function in cell survival, proliferation, metastasis, metabolism, angiogenesis and apoptosis (57–60). It is aberrantly expressed in many human tumors, including NPC, colon, breast and ovarian cancer (57–60). In the current study, it was established that the focal adhesion and the cell cycle were associated with the occurrence and development of tumors.
The development of bioinformatics tools has led to the development of various models and the establishment of databases (61–65). A number of them effectively predict potential miRNA-disease associations with high precision (61–65). These will provide novel directions for the diagnosis and treatment of patients with cancers, including NPC.
However, there are some limitations in the present study. First, different datasets in the GEO database microarray and platform were not unified. Second, it should be emphasized that the regulatory network and the potential mechanisms examined in the current study were solely based on bioinformatics analyses and were not verified by in vitro or in vivo experiments. Lastly, the datasets used in this study come from a single database and require further validation.
In summary, this study provided a bioinformatics analysis of DEGs, which may be involved in the development of NPC. Critical node proteins were identified, indicating an association with other proteins in the network, including CDK1, SMC4, KNTC1, KIF23, AURKA and ATAD2. These proteins may be used in NPC tumorigenesis through modulation of the cell cycle and nucleic acid metabolic processes, and may also be utilized as molecular biomarkers in the early diagnosis of NPC. There is a requirement for further experimental studies to validate the results of the present study.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81460460, 81360405 and 81760542), the China Postdoctoral Science Foundation (grant no. 2016M602918XB) and the Research Foundation of the Science and Technology Department of Guangxi Province, China (grant nos. 2016GXNSFAA380252 and 2014GXNSFBA118114).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available in the GEO repository, (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
RW designed the study. RW, KL and MK analyzed data and wrote the manuscript. ZZ and WQ analyzed data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lam KO, Lee AW, Choi CW, Sze HC, Zietman AL, Hopkins KI, Rosenblatt E. Global pattern of nasopharyngeal cancer: Correlation of outcome with access to radiation therapy. Int J Radiat Oncol Biol Phys. 2016;94:1106–1112. doi: 10.1016/j.ijrobp.2015.11.047. [DOI] [PubMed] [Google Scholar]
- 2.Peng H, Chen L, Zhang J, Li WF, Mao YP, Zhang Y, Liu LZ, Tian L, Lin AH, Sun Y, Ma J. Induction chemotherapy improved Long-term outcomes of patients with locoregionally advanced nasopharyngeal carcinoma: A propensity matched analysis of 5-year survival outcomes in the Era of intensity-modulated radiotherapy. J Cancer. 2017;8:371–377. doi: 10.7150/jca.16732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan WL, Tan EH, Lim DW, Ng QS, Tan DS, Jain A, Ang MK. Advances in systemic treatment for nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5:21. doi: 10.21037/cco.2016.03.03. [DOI] [PubMed] [Google Scholar]
- 4.Kang M, Zhou P, Liao X, Xu M, Wang R. Prognostic value of masticatory muscle involvement in nasopharyngeal carcinoma patients treated with intensity-modulated radiation therapy. Oral Oncol. 2017;75:100–105. doi: 10.1016/j.oraloncology.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Ren XY, Zhou GQ, Jiang W, Sun Y, Xu YF, Li YQ, Tang XR, Wen X, He QM, Yang XJ, et al. Low SFRP1 expression correlates with poor prognosis and promotes cell invasion by activating the Wnt/β-catenin signaling pathway in NPC. Cancer Prev Res (Phila) 2015;8:968–977. doi: 10.1158/1940-6207.CAPR-14-0369. [DOI] [PubMed] [Google Scholar]
- 6.Tao Q, Chan AT. Nasopharyngeal carcinoma: Molecular pathogenesis and therapeutic developments. Expert Rev Mol Med. 2007;9:1–24. doi: 10.1017/S1462399407000312. [DOI] [PubMed] [Google Scholar]
- 7.Sun R, Zhang Q, Guo L, Chen MY, Sun Y, Cao B, Sun J. HGF stimulates proliferation through the HGF/c-Met pathway in nasopharyngeal carcinoma cells. Oncol Lett. 2012;3:1124–1128. doi: 10.3892/ol.2012.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low SY, Tan BS, Choo HL, Tiong KH, Khoo AS, Leong CO. Suppression of BCL-2 synergizes cisplatin sensitivity in nasopharyngeal carcinoma cells. Cancer Lett. 2012;314:166–175. doi: 10.1016/j.canlet.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 9.Lakhanpal M, Singh LC, Rahman T, Sharma J, Singh MM, Kataki AC, Verma S, Chauhan PS, Singh YM, Wajid S, et al. Contribution of susceptibility locus at HLA class I region and environmental factors to occurrence of nasopharyngeal cancer in Northeast India. Tumour Biol. 2015;36:3061–3073. doi: 10.1007/s13277-014-2942-5. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Miao XP, Huang MY, Deng L, Hu LF, Ernberg I, Zeng YX, Lin DX, Shao JY. Polymorphisms of XRCC1 genes and risk of nasopharyngeal carcinoma in the Cantonese population. BMC Cancer. 2006;6:167. doi: 10.1186/1471-2407-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vercauteren SM, Sung S, Starczynowski DT, Lam WL, Bruyere H, Horsman DE, Tsang P, Leitch H, Karsan A. Array comparative genomic hybridization of peripheral blood granulocytes of patients with myelodysplastic syndrome detects karyotypic abnormalities. Am J Clin Pathol. 2010;134:119–126. doi: 10.1309/AJCPH27ZIZEJLORF. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Guo Z, Wang W, Sun Y, Zhang C, Wang X, Zhang L, Wang C. Application of single nucleotide polymorphism microarray and fluorescence in situ hybridization analysis for the prenatal diagnosis of a case with Pallister-Killian syndrome. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2018;35:232–235. doi: 10.3760/cma.j.issn.1003-9406.2018.02.019. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 13.Liu K, Kang M, Li J, Qin W, Wang R. Prognostic value of the mRNA expression of members of the HSP90 family in non-small cell lung cancer. Exp Ther Med. 2019;17:2657–2665. doi: 10.3892/etm.2019.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F, Shen C, Wang X, Wang H, Liu Y, Yu C, Lv J, He J, Wen Z. Identification of genes and pathways in nasopharyngeal carcinoma by bioinformatics analysis. Oncotarget. 2017;8:63738–63749. doi: 10.18632/oncotarget.19478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng C, Shen X, Jiang W. Potential biomarkers of HCC based on gene expression and DNA methylation profiles. Oncol Lett. 2018;16:3183–3192. doi: 10.3892/ol.2018.9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodd LE, Sengupta S, Chen IH, den Boon JA, Cheng YJ, Westra W, Newton MA, Mittl BF, McShane L, Chen CJ, et al. Genes involved in DNA repair and nitrosamine metabolism and those located on chromosome 14q32 are dysregulated in nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:2216–2225. doi: 10.1158/1055-9965.EPI-06-0455. [DOI] [PubMed] [Google Scholar]
- 17.Hu C, Wei W, Chen X, Woodman CB, Yao Y, Nicholls JM, Joab I, Sihota SK, Shao JY, Derkaoui KD, et al. A global view of the oncogenic landscape in nasopharyngeal carcinoma: An integrated analysis at the genetic and expression levels. PLoS One. 2012;7:e41055. doi: 10.1371/journal.pone.0041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41(Database Issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34(Database Issue):D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic acids Res. 2015;43(Database Issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47:8.13.11–24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai X, Yang Y, Wan J, Zhu R, Wu Y. Inhibition of LDH-A by oxamate induces G2/M arrest, apoptosis and increases radiosensitivity in nasopharyngeal carcinoma cells. Oncol Rep. 2013;30:2983–2991. doi: 10.3892/or.2013.2735. [DOI] [PubMed] [Google Scholar]
- 24.Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer. 2011;30:114–119. doi: 10.5732/cjc.010.10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei KR, Zheng RS, Zhang SW, Liang ZH, Li ZM, Chen WQ. Nasopharyngeal carcinoma incidence and mortality in China, 2013. Chin J Cancer. 2017;36:90. doi: 10.1186/s40880-017-0257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown NR, Korolchuk S, Martin MP, Stanley WA, Moukhametzianov R, Noble MEM, Endicott JA. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat Commun. 2015;6:6769. doi: 10.1038/ncomms7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 28.Johnson N, Li YC, Walton ZE, Cheng KA, Li D, Rodig SJ, Moreau LA, Unitt C, Bronson RT, Thomas HD, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat Med. 2011;17:875–882. doi: 10.1038/nm.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang J, Sergio CM, Sutherland RL, Musgrove EA. Targeting cyclin-dependent kinase 1 (CDK1) but not CDK4/6 or CDK2 is selectively lethal to MYC-dependent human breast cancer cells. BMC Cancer. 2014;14:32. doi: 10.1186/1471-2407-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei-Shan H, Amit VC, Clarke DJ. Cell cycle regulation of condensin Smc4. Oncotarget. 2019;10:263–276. doi: 10.18632/oncotarget.26467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, Yuan T, Liu M, Liu H, Xie J, Shen Y, Chen P. Overexpression of the structural maintenance of chromosome 4 protein is associated with tumor de-differentiation, advanced stage and vascular invasion of primary liver cancer. Oncol Rep. 2012;28:1263–1268. doi: 10.3892/or.2012.1929. [DOI] [PubMed] [Google Scholar]
- 32.Feng XD, Song Q, Li CW, Chen J, Tang HM, Peng ZH, Wang XC. Structural maintenance of chromosomes 4 is a predictor of survival and a novel therapeutic target in colorectal cancer. Asian Pac J Cancer Prev. 2014;15:9459–9465. doi: 10.7314/APJCP.2014.15.21.9459. [DOI] [PubMed] [Google Scholar]
- 33.Morozumi Y, Boussouar F, Tan M, Chaikuad A, Jamshidikia M, Colak G, He H, Nie L, Petosa C, de Dieuleveult M, et al. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J Mol Cell Biol. 2016;8:349–362. doi: 10.1093/jmcb/mjv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E, et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010;29:5171–5181. doi: 10.1038/onc.2010.259. [DOI] [PubMed] [Google Scholar]
- 35.Wu G, Liu H, He H, Wang Y, Lu X, Yu Y, Xia S, Meng X, Liu Y. miR-372 down-regulates the oncogene ATAD2 to influence hepatocellular carcinoma proliferation and metastasis. BMC Cancer. 2014;14:107. doi: 10.1186/1471-2407-14-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan WN, Zhang YX, Wang XM, Liu YJ, Zhang YQ, Que YH, Zhao WJ. ATAD2 is highly expressed in ovarian carcinomas and indicates poor prognosis. Asian Pac J Cancer Prev. 2014;15:2777–2783. doi: 10.7314/APJCP.2014.15.6.2777. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi S, Fusaki N, Ohta S, Iwahori Y, Iizuka Y, Inagawa K, Kawakami Y, Yoshida K, Toda M. Downregulation of KIF23 suppresses glioma proliferation. J Neurooncol. 2012;106:519–529. doi: 10.1007/s11060-011-0706-2. [DOI] [PubMed] [Google Scholar]
- 38.Fischer M, Grundke I, Sohr S, Quaas M, Hoffmann S, Knörck A, Gumhold C, Rother K. p53 and cell cycle dependent transcription of kinesin family member 23 (KIF23) is controlled via a CHR promoter element bound by DREAM and MMB complexes. PLoS One. 2013;8:e63187. doi: 10.1371/journal.pone.0063187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 40.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Hong MJ, Chow JP, Man WY, Mak JP, Ma HT, Poon RY. Co-inhibition of polo-like kinase 1 and Aurora kinases promotes mitotic catastrophe. Oncotarget. 2015;6:9327–9340. doi: 10.18632/oncotarget.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuß S, Burfeind P, Aigner A, Wordeman L, Bastians H. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol. 2014;16:779–791. doi: 10.1038/ncb2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsha A, Soutto M, Sehdev V, Peng D, Washington MK, Piazuelo MB, Tantawy MN, Manning HC, Lu P, Shyr Y, et al. Aurora kinase A promotes inflammation and tumorigenesis in mice and human gastric neoplasia. Gastroenterology. 2013;145:1312–1322.e1-e8. doi: 10.1053/j.gastro.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang XX, Liu R, Jin SQ, Fan FY, Zhan QM. Overexpression of Aurora-A kinase promotes tumor cell proliferation and inhibits apoptosis in esophageal squamous cell carcinoma cell line. Cell Res. 2006;16:356–366. doi: 10.1038/sj.cr.7310046. [DOI] [PubMed] [Google Scholar]
- 45.Dutta-Simmons J, Zhang Y, Gorgun G, Gatt M, Mani M, Hideshima T, Takada K, Carlson NE, Carrasco DE, Tai YT, et al. Aurora kinase A is a target of Wnt/beta-catenin involved in multiple myeloma disease progression. Blood. 2009;114:2699–2708. doi: 10.1182/blood-2008-12-194290. [DOI] [PubMed] [Google Scholar]
- 46.Li JJ, Weroha SJ, Lingle WL, Papa D, Salisbury JL, Li SA. Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc Natl Acad Sci USA. 2004;101:18123–18128. doi: 10.1073/pnas.0408273101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou Z, Yuan Z, Zhang Q, Long Z, Chen J, Tang Z, Zhu Y, Chen S, Xu J, Yan M, et al. Aurora kinase A inhibition-induced autophagy triggers drug resistance in breast cancer cells. Autophagy. 2012;8:1798–1810. doi: 10.4161/auto.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park HS, Park WS, Bondaruk J, Tanaka N, Katayama H, Lee S, Spiess PE, Steinberg JR, Wang Z, Katz RL, et al. Quantitation of Aurora kinase A gene copy number in urine sediments and bladder cancer detection. J Natl Cancer Inst. 2008;100:1401–1411. doi: 10.1093/jnci/djn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarfraz S, Hamid S, Ali S, Jafri W, Siddiqui AA. Modulations of cell cycle checkpoints during HCV associated disease. BMC Infect Dis. 2009;9:125. doi: 10.1186/1471-2334-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR, III, Tagaya M, Cleveland DW. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Xiao D, Wang Z, Zou Y, Huang L, Lin W, Deng Q, Pan H, Zhou J, Liang C, He J. MicroRNA-26a/b regulate DNA replication licensing, tumorigenesis, and prognosis by targeting CDC6 in lung cancer. Mol Cancer Res. 2014;12:1535–1546. doi: 10.1158/1541-7786.MCR-13-0641. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Bierie B, Li AG, Pathania S, Toomire K, Dimitrov SD, Liu B, Gelman R, Giobbie-Hurder A, Feunteun J, et al. BRCA1/FANCD2/BRG1-Driven DNA repair stabilizes the differentiation state of human mammary epithelial cells. Mol Cell. 2016;63:277–292. doi: 10.1016/j.molcel.2016.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu JH, Liao WT, Lee CH, Chang KL, Ke HL, Yu HS. ΔNp63 promotes abnormal epidermal proliferation in arsenical skin cancers. Toxicol In Vitro. 2018;53:57–66. doi: 10.1016/j.tiv.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Wu Z, Wang S, Jiang F, Li Q, Wang C, Wang H, Zhang W, Xue P, Wang SL. Mass spectrometric detection combined with bioinformatic analysis identified possible protein markers and key pathways associated with bladder cancer. Gene. 2017;626:407–413. doi: 10.1016/j.gene.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 55.Zhang HJ, Tao J, Sheng L, Hu X, Rong RM, Xu M, Zhu TY. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther. 2016;9:1801–1812. doi: 10.2147/OTT.S96535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu M, Bower KA, Chen G, Shi X, Dong Z, Ke Z, Luo J. Ethanol enhances the interaction of breast cancer cells over-expressing ErbB2 with fibronectin. Alcohol Clin Exp Res. 2010;34:751–760. doi: 10.1111/j.1530-0277.2010.01147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ni Z, Yi J. Oxymatrine induces nasopharyngeal cancer cell death through inhibition of PI3K/AKT and NF-κB pathways. Mol Med Rep. 2017;16:9701–9706. doi: 10.3892/mmr.2017.7822. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Takano C, Shimizu T, Yokobe S, Abe-Kanoh N, Zhu B, Nakamura T, Munemasa S, Murata Y, Nakamura Y. Inhibition of phosphatidylinositide 3-kinase ameliorates antiproliferation by benzyl isothiocyanate in human colon cancer cells. Biochem Biophys Res Commun. 2017;491:209–216. doi: 10.1016/j.bbrc.2017.07.078. [DOI] [PubMed] [Google Scholar]
- 59.Kaushik S, Shyam H, Sharma R, Balapure AK. Dietary isoflavone daidzein synergizes centchroman action via induction of apoptosis and inhibition of PI3K/Akt pathway in MCF-7/MDA MB-231 human breast cancer cells. Phytomedicine. 2018;40:116–124. doi: 10.1016/j.phymed.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Huang Y, Hua K, Zhou X, Jin H, Chen X, Lu X, Yu Y, Zha X, Feng Y. Activation of the PI3K/AKT pathway mediates FSH-stimulated VEGF expression in ovarian serous cystadenocarcinoma. Cell Res. 2008;18:780–791. doi: 10.1038/cr.2008.70. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief Bioinform. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Yan GY. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Huang L. LRSSLMDA: Laplacian regularized sparse subspace learning for MiRNA-disease association prediction. PLoS Comput Biol. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X, Xie D, Wang L, Zhao Q, You ZH, Liu H. BNPMDA: Bipartite Network projection for MiRNA-disease association prediction. Bioinformatics. 2018;34:3178–3186. doi: 10.1093/bioinformatics/bty333. [DOI] [PubMed] [Google Scholar]
- 65.Chen X, Wang L, Qu J, Guan NN, Li JQ. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the present study are available in the GEO repository, (https://www.ncbi.nlm.nih.gov/geo/).