Abstract
Long non-coding RNA (lncRNA) downregulated in hepatocellular carcinoma (DRHC) is a tumor suppressor in liver cancer. However, the role of this lncRNA in breast cancer has not been investigated. The present study revealed that lncRNA DRHC was downregulated and lncRNA Hox transcript antisense RNA (HOTAIR) was upregulated in tumor tissues compared with adjacent healthy tissues in patients with triple negative breast cancer (TNBC). Expression levels of lncRNA DRHC and lncRNA HOTAIR were negatively correlated in tumor tissues but not in adjacent healthy tissues. The lncRNA DRHC expression level was correlated with tumor size but not tumor metastasis. In vitro overexpression of lncRNA DRHC in TNBC cell lines resulted in decreased expression of lncRNA HOTAIR; however, lncRNA HOTAIR overexpression did not affect the expression level of lncRNA DRHC. Overexpression of lncRNA DRHC inhibited, while overexpression of lncRNA HOTAIR promoted the proliferation of the TNBC cell lines. In addition, lncRNA HOTAIR overexpression attenuated the inhibitory effects of lncRNA DRHC overexpression on cancer cell proliferation. The results obtained in the current study suggested that lncRNA DRHC may inhibit the proliferation of TNBC cells by downregulating the expression of lncRNA HOTAIR.
Keywords: triple negative breast cancer, long non-coding RNA, downregulated in hepatocellular carcinoma, lncRNA Hox transcript antisense RNA, proliferation
Introduction
As the most commonly diagnosed malignancy in females, breast cancer is the second leading cause of cancer-associated mortalities among female patients (1,2). Triple-negative breast cancer (TNBC) is characterized by the absence of the estrogen and progesterone receptors and erb-b2 receptor tyrosine kinase 2 (3). Chemotherapy remains the primary treatment option for patients with early and advanced TNBC (4). However, the lack of targeted therapies usually leads to poor prognosis (5). At present, the identification of a molecular target for the treatment of TNBC remains challenging.
Long non-coding RNAs (lncRNAs) are a group of non-protein coding RNA transcripts composed of >200 nucleotides (6). While messenger RNAs serve as mediators between DNA and protein, lncRNAs participate in cell growth and development directly in the form of RNA (7). lncRNAs encode no protein products but regulate gene expression at multiple levels (7). Therefore, regulation of lncRNA expression may indirectly regulate the expression of downstream genes that may serve roles in pathological or physiological processes (7–9). Previous studies demonstrated that lncRNAs are essential players in the pathogenesis of human diseases, particularly different types of human cancer (8,9). Therefore, lncRNAs may be promising therapeutic targets for the treatment of cancer (8,9). lncRNA Hox transcript antisense RNA (HOTAIR) is a well-characterized oncogenic lncRNA in several types of cancer (10). lncRNA DRHC has been recently reported to be a tumor suppressor in liver cancer (11); however, its role in other types of human cancer remains unknown. The present study revealed that lncRNA DRHC was downregulated in TNBC patients and inhibited the proliferation of TNBC cancer cells, possibly through downregulation of lncRNA HOTAIR.
Materials and methods
Patient samples
A total of 68 female patients with TNBC were enrolled in the Heilongjiang Farms and Land General Hospital (Haerbin, China) between April 2016 and July 2018. The inclusion criteria were as follows: i) Patients diagnosed by histopathological biopsies; ii) patients with complete medical records; and iii) patients who were willing to participate in the study. The exclusion criteria were as follows: i) Patients with other types of malignancies; ii) patients with chronic disease; and iii) patients who were treated within three months prior to admission. All patients received breast biopsies and tumor tissue as well as healthy adjacent tissue samples were obtained from each patient. The age of patients ranged between 32 and 66 years (median, 49 years; mean ± standard deviation, 48.6±5.7 years). The patients included 13 cases at stage I, 22 cases at stage II, 16 cases at stage III and 17 cases at stage IV. Tumor size was determined by performing magnetic resonance or X-ray imaging. Patients were divided into groups according to tumor size: i) 0–2 cm (n=19); ii) 2–5 cm (n=30); and iii) >5 cm (n=19). Patients were divided into groups according to the absence or presence of metastasis: i) Non-metastasis group (n=30); and ii) metastasis group (n=38). The Ethics Committee of Heilongjiang Farms & Land General Hospital approved the present study prior to enrolment of patients. All patients signed informed consent.
Cell lines
The present study included two TNBC cell lines, BT-549 and HCC70. The two cell lines were purchased from the American Type Culture Collection (ATCC). Cells were cultured using RPMI-1640 medium (ATCC) supplemented with 10% fetal bovine serum, 1% Penicillin and Streptomycin (all from Sangon Biotech Co., Ltd.) at 37°C with 5% CO2.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
In order to detect the expression of lncRNA DRHC and lncRNA HOTAIR in both in vitro cultivated TNBC cells and tissue samples from TNBC patients, the RNeasy Mini kit (Qiagen GmbH) was used to extract total RNA, according to the manufacturer's instructions. The Applied Biosystems™ High-Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to perform reverse transcription (25°C for 5 min, 55°C for 20 min and 75°C for 10 min). The qScript One-Step RT-qPCR kit (Quantabio) was used to perform qPCR, according to the manufacturer's protocol. All qPCR reactions were performed using an ABI 7500 system. lncRNA DRHC, lncRNA HOTAIR and GAPDH endogenous control primer pairs were designed and synthesized by Shanghai GenePharma Co., Ltd. The following primer sequences were used: lncRNA DRHC forward, 5′-GCTTAGAAATTTCTTCCACCTG-3′; lncRNA DRHC reverse, 5′-CTGCCCTAGCGATTTGTGAA-3′; lncRNA HOTAIR forward, 5′-CAGTGGGGAACTCTGACTCG-3′; lncRNA HOTAIR reverse, 5′-GTGCCTGGTGCTCTCTTACC-3′; GAPDH forward, 5′-GTCTCCTCTGACTTCAA-3′; and GAPDH reverse, 5′-ACCACCCTGTTGCTGTA-3′. The following thermocycling conditions were used: 95°C for 2 min, followed by 40 cycles of 95°C for 10 sec and 55°C for 30 sec. The expression of lncRNA DRHC and lncRNA HOTAIR was normalized to GAPDH endogenous control using the 2−ΔΔCq method (12). The sample with the lowest expression level was set to ‘1’ and all other samples were normalized to this sample.
Cell transfection
Vectors expressing lncRNA DRHC (11) or lncRNA HOTAIR (NR_047517.1) as well as the empty pcDNA3.1 vector were purchased from Sangon Biotech Co., Ltd. BT-549 and HCC70 cell lines were cultured overnight to reach 70–80% confluence. Vectors (10 nM) were transfected into cells using Lipofectamine® 2000 (cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific, Inc.). Cells treated with Lipofectamine 2000 but without vectors were used as control (C) cells. Cells transfected with empty vectors were used as the negative control (NC) cells. Cells were collected 24 h following transfection for subsequent experimentation. The transfection efficacy was determined using RT-qPCR.
Cell proliferation assay
Cell proliferation was assessed using transfected cells with lncRNA DRHC or lncRNA HOTAIR overexpression rates of 200%, as determined by RT-qPCR. Briefly, cells were harvested and 0.1 ml cell suspension (4×104 cells/ml) in RPMI-1640 medium was added into each well of a 96-well plate. A total of 10 µl Cell Counting Kit-8 (CCK-8, Sigma-Aldrich; Merck KGaA) solution was added to each well following 24, 48, 72 and 96 h of incubation (37°C, 5% CO2). The cells were cultured for an additional 4 h and the optical density values were measured at a wavelength of 450 nm.
Statistical analysis
All experiments in this study were repeated three times and the data are presented as the mean ± standard deviation (SD). All statistical analyses were performed using the GraphPad Prism software (version 6.0; GraphPad Software, Inc.). Comparison of the expression levels of lncRNA DRHC or lncRNA HOTAIR between tumor tissues and adjacent healthy tissues were performed using the paired t-test. Comparisons among three groups were performed by the one-way analysis of variance followed by the Tukey's test. Correlations between the expression levels of lncRNA DRHC and lncRNA HOTAIR in tumor tissues and adjacent healthy tissues were analyzed using Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA DRHC and lncRNA HOTAIR are dysregulated in TNBC tissues
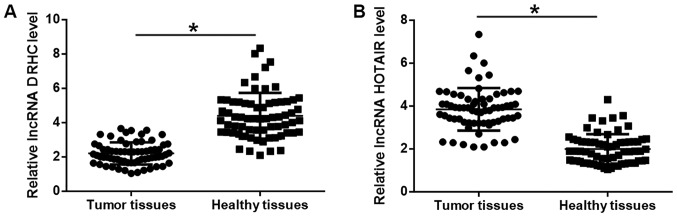
The expression levels of lncRNA DRHC and lncRNA HOTAIR in tumor and adjacent healthy tissues obtained from 68 patients with TNBC were measured using RT-qPCR. Results revealed that lncRNA DRHC was significantly downregulated (Fig. 1A), while lncRNA HOTAIR was significantly upregulated (Fig. 1B) in tumor tissues compared with adjacent healthy tissues (P<0.05).
Figure 1.
lncRNA DRHC and lncRNA HOTAIR expression levels were dysregulated in triple negative breast cancer tissues. Reverse transcription-quantitative polymerase chain reaction results revealed that (A) lncRNA DRHC was significantly downregulated while (B) lncRNA HOTAIR was significantly upregulated in tumor tissues compared with adjacent healthy tissues. The sample with the lowest expression level was set to ‘1’ and all other samples were normalized to this sample. *P<0.05. lncRNA, long non-coding RNA; DRHC, downregulated in hepatocellular carcinoma; HOTAIR, Hox transcript antisense RNA.
Expression levels of lncRNA DRHC and lncRNA HOTAIR are negatively correlated in TNBC tissues
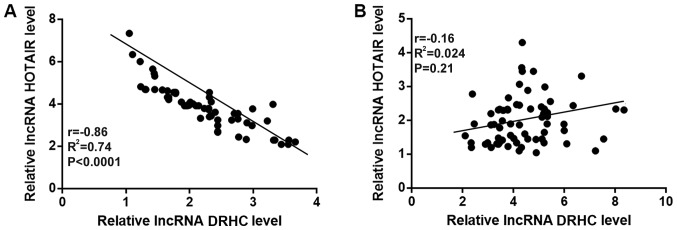
Correlations between the expression levels of lncRNA DRHC and lncRNA HOTAIR in tumor and adjacent healthy tissues were analyzed using Pearson's correlation coefficient. As shown in Fig. 2, the expression levels of lncRNA DRHC and lncRNA HOTAIR were negatively correlated in tumor tissues (P<0.0001; r, −0.86; Fig. 2A) but not in adjacent healthy tissues (P=0.21; r, −0.16; Fig. 2B).
Figure 2.
lncRNA DRHC and lncRNA HOTAIR were negatively correlated in TNBC tissues. Pearson's correlation coefficient revealed that the expression levels of lncRNA DRHC and lncRNA HOTAIR were negatively correlated in (A) tumor tissues but not in (B) adjacent healthy tissues. The sample with the lowest expression level was set to ‘1’ and all other samples were normalized to this sample. lncRNA, long non-coding RNA; DRHC, downregulated in HCC; HOTAIR, Hox transcript antisense RNA.
lncRNA DRHC is a potential upstream inhibitor of lncRNA HOTAIR expression in TNBC cells
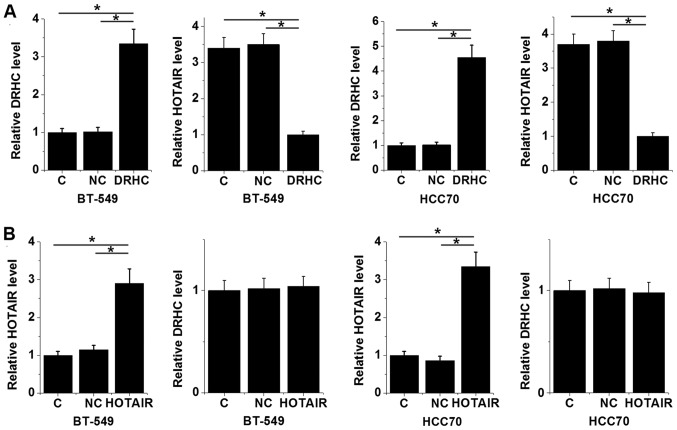
In vitro overexpression experiments were performed to further investigate the interactions between lncRNA DRHC and lncRNA HOTAIR. As shown in Fig. 3A, overexpression of lncRNA DRHC in the TNBC cell lines BT-549 and HCC70 decreased the expression level of lncRNA HOTAIR (P<0.05). However, lncRNA HOTAIR overexpression did not significantly affect the expression level of lncRNA DRHC (Fig. 3B).
Figure 3.
lncRNA DRHC is a potential upstream inhibitor of lncRNA HOTAIR expression in TNBC cells. Reverse transcription-quantitative polymerase chain reaction results revealed that (A) overexpression of lncRNA DRHC decreased the expression of lncRNA HOTAIR in the TNBC cell lines BT-549 and HCC70 while (B) lncRNA HOTAIR overexpression did not significantly affect the expression level of lncRNA DRH compared with C and NC groups. The sample with the lowest expression level was set to ‘1’ and all other samples were normalized to this sample. DRHC and HOTAIR on the x-axes represent transfection with vectors delivering DRHC and HOTAIR, respectively. *P<0.05. lncRNA, long non-coding RNA; DRHC, downregulated in HCC; HOTAIR, Hox transcript antisense RNA; TNBC, triple negative breast cancer; C, control; NC, negative control.
lncRNA DRHC expression level is correlated with tumor size but not tumor metastasis
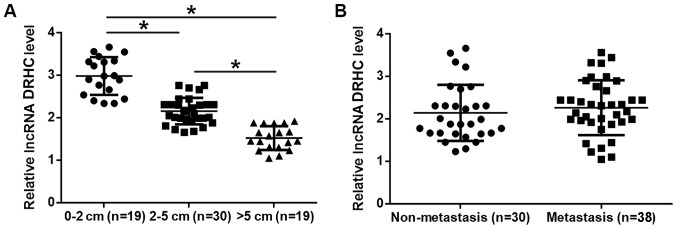
As shown in Fig. 4A, the expression levels of lncRNA DRHC in tumor tissues decreased significantly as the tumor diameter increased (P<0.05). However, no significant difference in the expression level of lncRNA DRHC was found between the metastasis and non-metastasis groups (Fig. 4B).
Figure 4.
lncRNA DRHC expression level is correlated with tumor size but not tumor metastasis. (A) Expression levels of lncRNA DRHC in tumor tissues decreased significantly as the tumor diameter increased. (B) A significant difference in the expression level of lncRNA DRHC was not observed between the metastasis and non-metastasis groups. The sample with the lowest expression level was set to ‘1’ and all other samples were normalized to this sample. *P<0.05. lncRNA, long non-coding RNA; DRHC, downregulated in HCC; HOTAIR, Hox transcript antisense RNA.
lncRNA DRHC may inhibit the proliferation of TNBC cell lines through lncRNA HOTAIR
The correlation between DRHC and tumor size indicated the possible involvement of DRHC in the proliferation of TNBC cells. Therefore, the CCK-8 assay was performed to investigate the role of DRHC in the proliferation of TNBC cell lines. Compared with the C and NC groups, the overexpression of lncRNA DRHC inhibited while the overexpression of lncRNA HOTAIR promoted the proliferation of TNBC cell lines (P<0.05; Fig. 5). In addition, lncRNA HOTAIR overexpression attenuated the inhibitory effects of lncRNA DRHC overexpression on cancer cell proliferation (P<0.05; Fig. 5).
Figure 5.
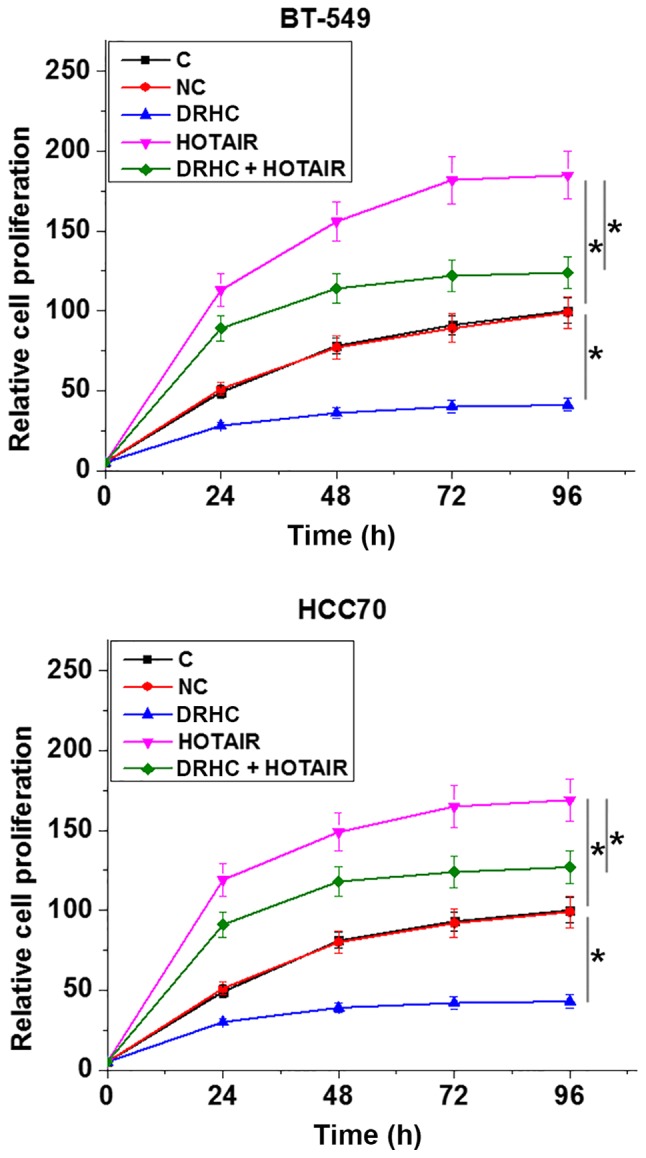
lncRNA DRHC inhibits the proliferation of TNBC cell lines potentially through lncRNA HOTAIR. The optical density value of the C group at 96 h was set to ‘100’ and all other groups were normalized to this group. Overexpression of lncRNA DRHC inhibited, while overexpression of lncRNA HOTAIR promoted the proliferation of TNBC cell lines. In addition, lncRNA HOTAIR overexpression attenuated the inhibitory effects of lncRNA DRHC overexpression on cancer cell proliferation. *P<0.05. lncRNA, long non-coding RNA; DRHC, downregulated in HCC; TNBC, triple negative breast cancer; HOTAIR, Hox transcript antisense RNA; C, control; NC, negative control.
Discussion
In a recent study, lncRNA DRHC was reported to be a tumor suppressor in liver cancer (11). However, the involvement of lncRNA DRHC in other human diseases remains unknown. The present study demonstrated that lncRNA DRHC was downregulated in TNBC tissues compared with healthy adjacent tissues. Furthermore, lncRNA DRHC may serve a role as tumor suppressor in TNBC by inhibiting tumor cell proliferation by decreasing the expression level of lncRNA HOTAIR.
lncRNA HOTAIR is a well-studied oncogenic lncRNA involved in different types of human cancer, such as liver cancer, breast cancer and lung cancer (10). lncRNA HOTAIR is upregulated during the development of TNBC (13). Consistent with the previous studies (10,13), the present study demonstrated significantly upregulated expression of lncRNA HOTAIR in TNBC tissues compared with adjacent healthy tissues. Additionally, the overexpression of lncRNA HOTAIR increased the proliferation of TNBC cell lines in vitro. In effect, decreasing the expression level of lncRNA HOTAIR may be a promising therapeutic target for TNBC.
As a tumor suppressor, lncRNA DRHC was downregulated in liver cancer (11). The present study was the first to report the downregulated expression pattern of lncRNA DRHC in TNBC. In addition, the present study revealed that lncRNA DRHC was correlated with tumor size but not tumor metastasis in patients with TNBC. Therefore, lncRNA DRHC may specifically participate in the proliferation of TNBC cells. Previous in vitro studies revealed that lncRNA DRHC inhibited the proliferation and invasion of liver cancer cells (13,14). The current study demonstrated that lncRNA DRHC overexpression inhibited the proliferation of TNBC cell lines.
lncRNAs achieve their biological functions through the interactions with multiple signaling molecules, including proteins (15) and microRNAs (16). However, no study on the investigation of the interactions between different lncRNAs in general was reported. The current study revealed that lncRNA DRHC is a potential upstream inhibitor of lncRNA HOTAIR expression, and that the inhibition of lncRNA HOTAIR expression by lncRNA DRHC may be involved in the regulation of TNBC cell proliferation. The results obtained in the current study provided new insights into the pathogenesis of TNBC.
The inhibition of lncRNA HOTAIR expression by lncRNA DRHC is likely indirect due to the lack of significant correlation between lncRNA HOTAIR and lncRNA DRHC expression levels in healthy tissues. In addition, lncRNA HOTAIR regulates cancer cell migration and invasion in different types of cancer, including breast cancer (17). Our future study will further investigate the roles of DRHC in regulating TNBC cell invasion and migration. Additionally, lncRNA DRHC may interact with multiple downstream effectors to achieve a fine regulation of cancer cell behaviors (18).
The present study investigated the interaction between lncRNA HOTAIR and lncRNA DRHC in TNBC, which to the best of the authors' knowledge had not been previously reported. Future experiments are required to investigate the interaction between DRHC and the classical cancer-related signaling pathways, such as PTEN and CDKs. In addition, preliminary data (not shown) suggests that DRHC may not be downregulated in other types of breast cancer, suggesting that DRHC may serve specific roles in TNBC. Future studies with a larger number of patients are required to further test this hypothesis.
In conclusion, the results obtained in the current study suggested that lncRNA DRHC may serve as a tumor suppressor in TNBC. lncRNA DRHC may inhibit the proliferation of TNBC cells by downregulating lncRNA HOTAIR.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FY designed the experiments. FY, LW and BZ conducted the experiments. FY analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committee of Heilongjiang Farms & Land General Hospital (Haerbin, China) approved the present study prior to the enrolment of patients. Written informed consent for the participation in this study was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ma J, Jemal A. Springer; New York, NY: 2013. Breast cancer metastasis and drug resistance; pp. 1–18. [Google Scholar]
- 2.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 3.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 4.Bartsch R, Ziebermayr R, Zielinski CC, Steger GG. Triple-negative breast cancer. Wien Med Wochenschr. 2010;160:174–181. doi: 10.1007/s10354-010-0773-6. [DOI] [PubMed] [Google Scholar]
- 5.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 8.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang R, Zhang X, Lu D, Wang J, Zhuo J, Wei X, Ling Q, Xie H, Zheng S, Xu X. lncRNA DRHC inhibits proliferation and invasion in hepatocellular carcinoma via c-Myb-regulated MEK/ERK signaling. Mol Carcinog. 2019;58:366–375. doi: 10.1002/mc.22934. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Collina F, Aquino G, Brogna M, Cipolletta S, Buonfanti G, De Laurentiis M, Di Bonito M, Cantile M, Botti G. LncRNA HOTAIR up-regulation is strongly related with lymph nodes metastasis and LAR subtype of Triple Negative Breast Cancer. J Cancer. 2019;10:2018–2024. doi: 10.7150/jca.29670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang YL, Overstreet AM, Chen MS, Wang J, Zhao HJ, Ho PC, Smith M, Wang SC. Combined inhibition of EGFR and c-ABL suppresses the growth of triple-negative breast cancer growth through inhibition of HOTAIR. Oncotarget. 2015;6:11150–11161. doi: 10.18632/oncotarget.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2015;17:106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W, Geng D, Li S, Chen Z, Sun M. Lnc RNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA 2 axis in breast cancer. Cancer Med. 2018;7:842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Hu W, Xu W, Shi Y, Dai W. lncRNA HOTAIR upregulates COX-2 expression to promote invasion and migration of nasopharyngeal carcinoma by interacting with miR-101. Biochem Biophys Res Commun. 2018;505:1090–1096. doi: 10.1016/j.bbrc.2018.09.190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.