Abstract
β-catenin regulates its target genes which are associated with proliferation, differentiation, migration and angiogenesis, and the dysregulation of Wnt/β-catenin signaling facilitates hallmarks of colorectal cancer (CRC). Identification of β-catenin targets and their potential roles in tumorigenesis has gained increased interest. However, the number of identified targets remains limited. The present study implemented a novel strategy, interrogating gene fitness profiles derived from large-scale RNA interference and CRISPR-CRISPR associated protein 9 screening data to identify β-catenin target genes in CRC cell lines. Using these data sets, pair wise gene fitness similarities were determined which highlighted a total of 13 genes whose functions were highly correlated with β-catenin. It was further demonstrated that the expression of these genes were altered in CRC, illustrating their potential roles in the progression of CRC. The present study further demonstrated that these targets could be used to predict disease-free survival in CRC. In conclusion, the findings provided novel approaches for the identification of β-catenin targets, which may become prognostic biomarkers or drug targets for the management of CRC.
Keywords: colon cancer, β-catenin target, gene fitness screening, prognostic biomarker
Introduction
The Wnt/β-catenin signaling pathway serves important roles in the tumorigenesis of colorectal cancer (CRC). In colon cancer cell lines, the disruption of the adenomatous polyposis coli protein, caused by loss of heterozygosity or mutation, leads to deregulation of the β-catenin protein (1). Subsequently, it is transported into the nucleus and activates its target genes by recruiting cofactors of the transcription factor/lymphoid enhancer binding factor family (2). It is crucial to identify β-catenin target genes, since they are involved in cellular processes that contribute to proliferation and migration in colorectal carcinoma (3–8). Based on a literature review, the Nusse group (9) revealed 28 β-catenin target genes in human colon cancer, which are presented on their website (https://web.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes). In an experimental approach, RNA interference (RNAi) of β-catenin was implemented by treating DLD1 and SW480 cells with small interfering RNA (siRNA) (10). By comparison with controls, in which samples were treated with mock siRNA, the identified differentially expressed genes are considered to be potential β-catenin target genes (10). The study then incorporated results from another study with a similar design, in which LS174T cells were treated with short hairpin RNA against β-catenin (11), and identified a total of 335 target genes. Recently, a multi-omics approach was used by Ewing et al (12) to decipher the oncogenic β-catenin network in HCT116 cells by comparing transcriptome, expression proteome and interactome data of wild type and β-catenin mutated samples. The results were subsequently integrated into a functional molecular network. However, target genes identified by these studies (10–12) lack functional information. Additionally, expression levels occasionally fail to reflect molecular dysfunction where post-translational modifications are involved (13).
Therefore, the present study aimed to identify β-catenin target genes by leveraging large-scale RNAi and CRISPR-CRISPR associated protein 9 (Cas9) genetic perturbation datasets, since they may provide an opportunity to derive gene-gene functional associations. Initially, in yeast, the gene-gene associations were revealed by epistatic analysis, where the phenotypic readout of a genetic perturbation depends on the status of a second gene (14). Based on this, genetic interaction networks could be constructed to reveal functionally associated genes (15). For human cell lines, the utility of this approach is limited due to the exponential increase in the combinatorial space (16,17). However, genome-wide single gene perturbation screens are more advanced and can be applied in a large collection of cell lines (18,19). For these screens, highly variable genetic dependencies for cellular fitness are observed in cancer cell lines, which may reflect diverse alterations during tumorigenesis (19). Upon perturbation of each gene, the genomic state of each cancer cell exhibits a unique overall fitness response. Notably, Project Acheilles achieved promising results by systematically elucidating genetic vulnerabilities across 501 cancer cell lines using RNAi (18) and in 342 cancer cell lines using CRISPR-Cas9 (19). The data could provide an opportunity to identify potential β-catenin target genes by deriving gene-gene functional associations.
The present study reanalyzed data generated from large-scale RNAi (18) and CRISPR-Cas9 (19) single gene perturbation screen studies and ranked gene candidates by their fitness correlation of β-catenin. The present study focused on genes that were highly correlated with β-catenin, and identified 13 β-catenin target genes. These genes were fitted into statistical models that could successfully predict CRC prognosis. Overall, the present study applied a novel strategy to identify β-catenin target genes, and the findings could establish the utility of this approach to reveal functional associations among Wnt/β-catenin signaling and deepen the understating of CRC pathogenesis.
Materials and methods
Filtering fitness data for analysis
Fitness screening data from RNAi (18) and CRISPR-Cas9 (19) experiments in CRC cell lines (Table SI) were downloaded from the DepMap website (depmap.org/portal/). Subsequently, genes were filtered based on the dependence scores upon genetic depletion across cell lines in both datasets. Therefore, genes that had a minimum dependency score >-0.3, for both the RNAi and CRISPR-Cas9 dataset, and were expressed in the cell lines with transcripts per kilobase million (TPM) >0, were selected for downstream analysis. Gene expression data (20) were retrieved from the Cancer Cell Line Encyclopedia (CCLE) project (portals.broadinstitute.org/ccle).
Computing the β-catenin fitness correlation landscape
Following filtering of the fitness data, the Pearson correlation coefficient (PCC) of fitness profiles of all other genes with β-catenin was computed to generate the fitness profile correlation landscape. The genes were ranked based on the PCC scores (between-1 and 1; Fig. 1A). It was hypothesized that potential targets could be among those genes exhibiting a positive or negative correlation with the fitness profile of β-catenin.
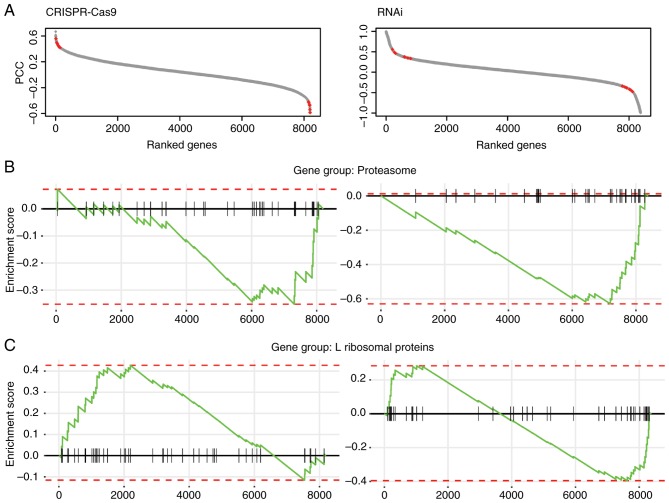
Figure 1.
Overview of fitness profile correlations between β-catenin and selected genes in the present study. (A) Scatter plot of ranked genes based on the PCC. Red dots represent β-catenin targets identified in the present study. The ranked genes were significantly enriched in (B) proteasome and (C) L ribosomal proteins. L ribosomal proteins, large ribosomal proteins; PCC, Pearson correlation coefficient; RNAi, RNA interference; Cas9, CRISPR associated protein 9.
Microarray data
All microarray data used in the present study were retrieved from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (ncbi.nlm.nih.gov/geo/) (21) using GEO accession numbers. The siRNA β-catenin and control treatment microarray dataset accession numbers were GSE44097 (10) for the DLD1 and SW480 cell lines, and GSE18560 (12) for Ls174T cells. The CRC dataset accession numbers were GSE68468 (22), GSE14333 (23), GSE17536 (24), GSE17537 (25), GSE24549 (26), GSE24550 (27), GSE31595 (28), GSE37892 (29) and GSE39582 (30). For the GSE24549 and GSE24550 datasets, gene expression profiles were generated using an Affymetrix Human Exon 1.0 ST array, and GSE68468 gene expression profiles were generated using an Affymetrix Human Genome U133A array. The remaining data were generated using an Affymetrix Human Genome U133 Plus 2.0 array. The disease-free survival outcome information was retrieved from the Prediction of Clinical Outcomes for Genomics database (https://precog.stanford.edu/index.php) (31) using the aforementioned NCBI GEO accession numbers.
Raw intensity files (*.CEL) were downloaded from the GEO database and then processed using an in-house bioinformatics pipeline. Briefly, the raw files were loaded into R version 3.5.2 (32) environment using the Bioconductor package oligo (version 1.46.0) (33). The rma algorithms from the oligo package were applied for background correction and normalization. Gene annotation was processed based on the custom chip definition files (version 22.0.0) (34) downloaded from the BrainArray website (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp).
RNA-sequencing (RNA-seq) data
RNA-seq data were retrieved from the NCBI Sequence Read Archive database using the following accession numbers: SRP029880 (35) for CRC samples and SRP101345 (36) for HCT116 cell lines with either mutant catenin b1 (CTNNB1)/β-catenin allele disrupted or wild-type CTNNB1/β-catenin allele disrupted. The raw reads files (*.fastq) were downloaded and processed using Salmon software (version 0.13.0) (37) to quantify the expression levels of transcripts. Reference transcriptome data (GRCh38 release 94) were downloaded from the Ensembl database (http://www.ensembl.org/)
The cancer genome Atlas (TCGA)-colon adenocarcinoma (COAD) data
RNA-seq expression profiles of colon cancer (n=521) were retrieved from the TCGA-COAD dataset deposited in the NCI Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov) using the Bioconductor package, TCGAbiolinks (version 2.10.5) (38). The raw fragments per kilobase million values were converted to TPM (transcripts per million) values for downstream analysis. The differences between the expression levels between primary solid tumors and normal solid tissues were tested using a Wilcoxon rank sum test.
Differential gene expression analysis
The limma (version 3.38.3) (39) package was implemented to conduct the differential expression analysis. The main strategy of this package is to use linear models to evaluate differential expression levels in the context of a multifactor designed experiment. The P-values were adjusted using Benjamini and Hochberg correction (40).
Cox proportional hazard modeling
The Cox proportional hazards modeling of CRC disease-free survival and log-rank tests was conducted using the R package, survival (version 2.44–1.1; CRAN.R-project.org/package=survival). Using this statistical model, gene expression values were summarized into the unified scores. The median value of these scores was chosen as a threshold, samples with scores above which were defined as high-risk groups and low-risk groups were determined. The Kaplan-Meier plot was generated using the R package, survminer (version 0.4.4; CRAN.R-project.org/package=survminer).
Gene set enrichment analysis (GSEA)
GSEA was conducted using a Bioconductor package, fgsea (version 1.8.0) (41), with gene family annotations from the HUGO database (https://www.genenames.org). In the present study, GSEA determined whether members of the gene families tended to be enriched at the top (positively correlated) or bottom (negatively correlated) of the ranked gene list and measured the probability of such distribution to be generated randomly. The NES (normalized enrichment score) was adjusted from the ES (enrichment score), which reflected to what extent the gene sets from the gene families were overrepresented in the ranked gene list (42).
Results
ß-catenin fitness correlated genes enriched in CRC associated signaling pathways
As demonstrated in a previous study (43), the correlated fitness effects across cancer cell lines can serve as a measure to reveal gene-gene functional associations. In other words, genes that share similar fitness profiles may be functionally associated, which makes them potential regulating targets. Therefore, the present study aimed to identify β-catenin target genes by computing its fitness correlation with other genes perturbed in CRC cell lines (CRISPR-Cas9, n=27; RNAi, n=45). The present study applied the stringent computational pipeline in parallel and retrieved a total of 8,200 genes from the CRISPR-Cas9 dataset and 8,381 genes from the RNAi dataset. From these genes, the present study computed the PCC of fitness profiles with β-catenin. As presented in Fig. 1A, the genes were ranked from the most positive to the most negative correlation with β-catenin and they were significantly enriched in the gene groups of proteasome (Fig. 1B; CRISPR-Cas9, NES=−1.45, P=0.05; RNAi, NES=−2.25, P=0.0024) and large ribosomal proteins (Fig. 1C; CRISPR-Cas9, NES=1.54, P=0.016; RNAi, NES=−1.47, P=0.04). The revealed associations between β-catenin and the proteasome and ribosomal proteins may highlight candidate β-catenin targets.
Identified β-catenin target genes are involved in CRC tumorigenesis
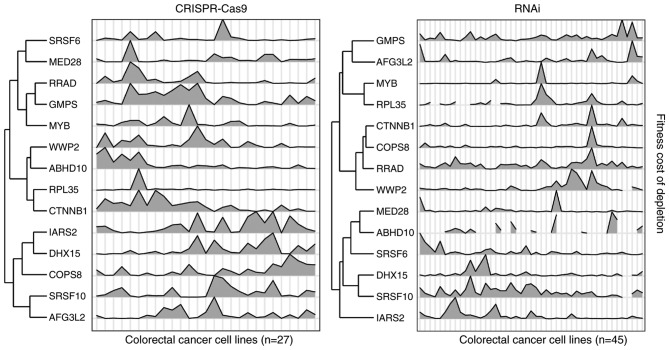
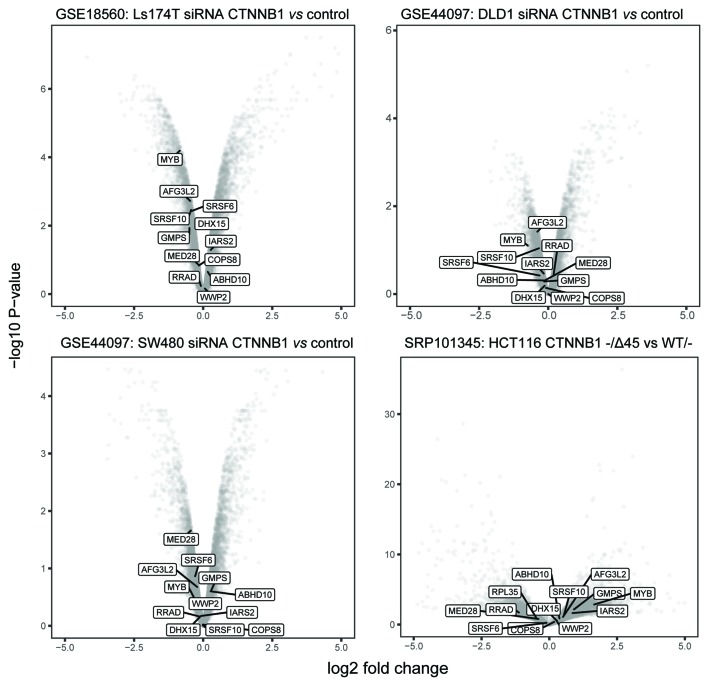
The correlated genes were filtered using stringent criteria (P≤0.03; Pearson correlation test), and 210 genes in the CRISPR-Cas9 dataset and 754 genes in RNAi dataset were identified as potential β-catenin targets, since their fitness profiles were significantly correlated with β-catenin in CRC cell lines. Since the resulting fitness profile correlations exhibited similar trends in terms of gene set enrichment analysis, the overlapping significantly correlated genes from both datasets were selected, and 13 genes were identified as high confidence β-catenin targets (Fig. 2; Table I). Notably, these genes seldom exhibited significant expression alterations in colon cancer cell lines that were treated with siRNA that inhibited β-catenin, or harbored β-catenin mutations (Fig. 3).
Figure 2.
Fitness profiles of β-catenin and 13 identified target genes in the CRISPR-Cas9 and RNAi datasets. Rows (genes) were hierarchically clustered. ABHD10, abhydrolase domain containing 10; AFG3L2, AFG3 like matrix AAA peptidase subunit 2; Cas9, CRISPR associated protein 9; COPS8, COP9 signalosome subunit 8; CTNNB1, catenin b1; DHX15, DEAH-box helicase 15; GMPS, guanine monophosphate synthase; IARS2, isoleucyl-tRNA synthetase 2; MED28, mediator complex subunit 28; MYB, MYB proto-oncogene, transcription factor; RNAi, RNA interference; RPL35, ribosomal protein L35; RRAD, RRAD, Ras related glycolysis inhibitor and calcium channel regulator; SRSF6, serine and arginine rich splicing factor 6; SRSF10, serine and arginine rich splicing factor 10; WWP2, WW domain containing E3 ubiquitin protein ligase 2.
Table I.
β-catenin target genes identified in the present study.
| CRISPR-Cas9 | RNAi | |||||
|---|---|---|---|---|---|---|
| Symbol | PCC | P-value | PCC | P-value | Chromosomal location | Description |
| ABHD10 | 0.430 | 0.025 | −0.477 | 0.016 | 3q13.2 | Abhydrolase domain containing 10 |
| AFG3L2 | −0.427 | 0.026 | 0.371 | 0.012 | 18p11.21 | AFG3 like matrix AAA peptidase subunit 2 |
| COPS8 | −0.465 | 0.014 | 0.336 | 0.024 | 2q37.3 | COP9 signalosome subunit 8 |
| DHX15 | −0.587 | 0.001 | −0.440 | 0.003 | 4p15.2 | DEAH-box helicase 15 |
| GMPS | 0.496 | 0.009 | 0.349 | 0.019 | 3q25.31 | Guanine monophosphate synthase |
| IARS2 | −0.421 | 0.029 | −0.341 | 0.022 | 1q41 | Isoleucyl-tRNA synthetase 2, mitochondrial |
| MED28 | −0.473 | 0.013 | −0.421 | 0.004 | 4p15.32 | Mediator complex subunit 28 |
| MYBa | 0.484 | 0.011 | 0.462 | 0.001 | 6q23.3 | MYB proto-oncogene, transcription factor |
| RPL35 | 0.455 | 0.017 | 0.559 | <0.001 | 9q33.3 | Ribosomal protein L35 |
| RRAD | 0.559 | 0.002 | 0.487 | 0.001 | 16q22.1 | RRAD, Ras related glycolysis inhibitor and calcium channel regulator |
| SRSF10 | −0.537 | 0.004 | −0.383 | 0.010 | 1p36.11 | Serine and arginine rich splicing factor 10 |
| SRSF6 | 0.420 | 0.029 | −0.366 | 0.013 | 20q13.11 | Serine and arginine rich splicing factor 6 |
| WWP2 | 0.423 | 0.028 | 0.373 | 0.013 | 16q22.1 | WW domain containing E3 ubiquitin protein ligase 2 |
P-values indicate the significance level of the PCC value.
Previously reported as known β-catenin target (51). Cas9, CRISPR associated protein 9; PCC, Pearson correlation coefficient; RNAi, RNA interference.
Figure 3.
Volcano plots revealing differentially expressed genes for siRNA-transfected vs. control samples and β-catenin mutant vs. wild type samples. β-catenin targets identified in the present study were highlighted. Data were obtained from the National Center for Biotechnology Information Gene Expression Omnibus database (accession numbers, GSE18560: top-left, GSE44097: top-right and bottom-left, and SRP101345: bottom-right). ABHD10, abhydrolase domain containing 10; AFG3L2, AFG3 like matrix AAA peptidase subunit 2; COPS8, COP9 signalosome subunit 8; CTNNB1, catenin b1; DHX15, DEAH-box helicase 15; GMPS, guanine monophosphate synthase; IARS2, isoleucyl-tRNA synthetase 2; MED28, mediator complex subunit 28; MYB, MYB proto-oncogene, transcription factor; RRAD, RRAD, Ras related glycolysis inhibitor and calcium channel regulator; siRNA, small interfering RNA; SRSF6, serine and arginine rich splicing factor 6; SRSF10, serine and arginine rich splicing factor 10; WWP2, WW domain containing E3 ubiquitin protein ligase 2.
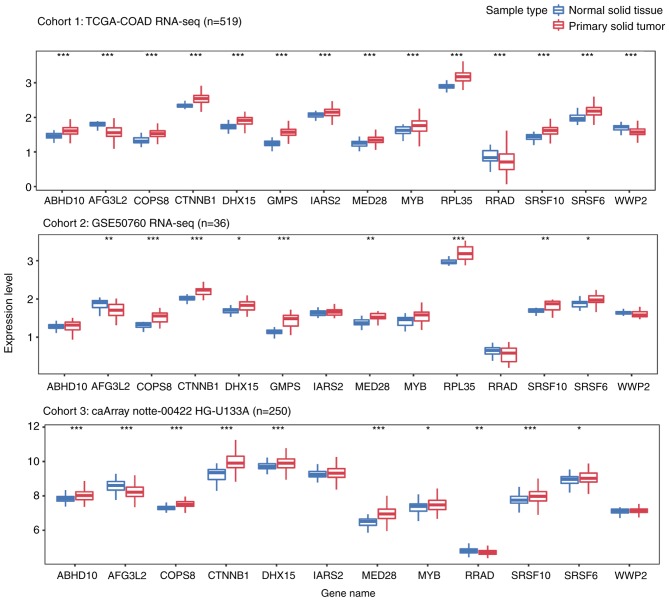
Additionally, the present study explored the expression levels of these genes in primary CRC and normal tissue samples from three independent cohorts, including a total of 805 samples. As presented in Fig. 4, all 13 genes were differentially expressed in solid tumor tissues from cohort 1. Among the genes, ten were upregulated and three were downregulated in CRC tumor samples. Although similar trends were observed in the other cohorts, certain genes (ABHD10, IARS2, MYB, RRAD, WWP2 in cohort2; IARS2 and WWP2 in cohor3), were not significantly differentially expressed between tumor and normal samples.
Figure 4.
Boxplots demonstrating gene expression levels of β-catenin targets in CRC primary solid tumors vs. normal solid tissues. Analysis was performed using the Wilcoxon rank sum test. *P<0.05, **P<0.01, ***P<0.001, Normal solid tissue compared with respective Primary solid tumor. ABHD10, abhydrolase domain containing 10; AFG3L2, AFG3 like matrix AAA peptidase subunit 2; COPS8, COP9 signalosome subunit 8; CTNNB1, catenin b1; DHX15, DEAH-box helicase 15; GMPS, guanine monophosphate synthase; IARS2, isoleucyl-tRNA synthetase 2; MED28, mediator complex subunit 28; MYB, MYB proto-oncogene, transcription factor; RNA-seq, RNA sequencing; RPL35, ribosomal protein L35; RRAD, RRAD, Ras related glycolysis inhibitor and calcium channel regulator; SRSF6, serine and arginine rich splicing factor 6; SRSF10, serine and arginine rich splicing factor 10; TCGA-COAD, The Cancer Genome Atlas-colon adenocarcinoma; WWP2, WW domain containing E3 ubiquitin protein ligase 2.
Modeling target genes as a CRC prognostic biomarker
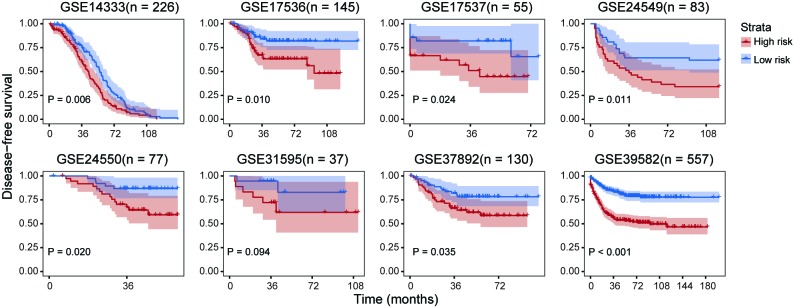
There is increasing interest to discover CRC prognostic biomarkers as patients could benefit from earlier diagnosis and a personalized treatment strategy. For this purpose, the present study aimed to establish gene signature prognostic models that could predict disease-free survival outcomes in CRC. The expression profiles of the 13 genes were fitted into a Cox proportional hazards model and a unified score was computed for each sample. For each cohort, the median value of the aforementioned unified score was selected to divide high- and low-risk groups. Log-rank tests indicated that the high- and low-risk groups exhibited significant differences in disease-free survival prognosis except for GSE31595 (Fig. 5).
Figure 5.
Kaplan-Meier plots demonstrating disease-free survival analysis of prognostic models based on expression profiles of β-catenin targets. P-values were calculated using the log-rank test.
Discussion
The present study explored large-scale RNAi and CRISPR-Cas9 screening data and identified β-catenin target genes based on fitness profile similarities generated from a large-scale study, CCLE (20) and the DepMap (18,19) project. As illustrated in a previous study, this fitness correlation strategy can recapitulate gene functional modularity by rewiring human protein complexes and establishing a human functional similarity network (43). A total of 79 CRC cell lines were considered in the present study. This set the CRC context, in which β-catenin regulates target genes. For both RNAi and CRISPR-cas9 datasets, the genes where the fitness data were highly correlated with β-catenin were significantly enriched in the proteasome and ribosomal families. Lack of proteasomal degradation of β-catenin facilitates its entry to the nucleus and targeting genes, including cyclin D1 (4) and AKT1 (44). This eventually leads to the proliferation and differentiation of cells. Furthermore, disturbances in ribosomal proteins have been observed in a variety of cancerous tissues, including glioblastoma (45), breast (46), esophagus (47), liver (48) and cervix tissues (49). For CRC, similar trends have been observed. For example, the expression levels of several ribosomal proteins are abnormally regulated in primary (50) and metastatic (51) CRC. It is worth noting that, in the previous strategy, the identified β-catenin targets from different cell lines merely overlapped (10). This limitation was overcome by computing gene fitness correlation and thus yielded reliable results.
Notably, among the 13 identified targets, the MYB proto-oncogene, transcription factor (MYB), has previously been reported as a known β-catenin target (52). Upregulated MYB levels and activated β-catenin may induce robust upregulation of MYC promoter activity in CRC (53). Previous studies have suggested that the dysregulation of MYB is associated with several rare types of cancer, including adenoid cystic cancer (54–60), spiradenocarcinoma (61) and cutaneous cylindroma (62). For the mediator complex subunit 28 gene, the same trend in expression alterations has been observed in epithelial-derived types of cancer (63). Although, to the best of our knowledge, there are no reports indicating the involvement of COP9 signalosome subunit 8 (COPS8) in CRC, a proteomic study revealed that COPS8 is upregulated in prostate cancer (64). The serine and arginine rich splicing factor 6 and serine and arginine rich splicing factor 10 genes are members of the serine-arginine family, which regulates RNA splicing. As a result, their roles in regulating alternative splicing may promote cancer pathogenesis (65–67). WW domain containing E3 ubiquitin protein ligase 2 modulates transforming growth factor b-dependent transcription and epithelial-mesenchymal transition (68,69). It should be noted that some of the identified targets seldom exhibited altered expressions in the siRNA β-catenin approaches. This suggests that the method used in the present study may serve as a complementary alternative for the identification of β-catenin targets, which may be potentially missed by other strategies.
The present study further revealed the associations of the 13 identified targets with CRC disease free survival outcomes in eight independent cohorts encompassing 1,310 individuals. Indeed, the prognostic potential of these genes has been revealed in previous studies (70,71). For instance, loss of the AFG3 like matrix AAA peptidase subunit 2 gene, located in the 18p11.32–21 region, have been associated with a significantly longer progression-free survival in patients with CRC (70). Additionally, MYB is associated with metastasis in pancreatic tumors (71).
However, it worth mentioning that the present study used a stringent criterion to filter the data and reported β-catenin targets with high confidence. While there are two datasets available, CRISPR-Cas9 and RNAi, the present study selected the overlap of both datasets for downstream analysis. This may lead to the misinterpretation of the remaining potential targets. For example, survivin, cyclin-D1 and axin-2 are well established β-catenin targets. The present study could not recapture them, since they exhibited less significant correlations with β-catenin comparing the identified 13 targets in this study. It was anticipated that a more robust strategy, such as machine learning, should be employed to explore the data. Additionally, the prognostic model requires improvements by statistical modeling, so that these findings can be applied to clinical practice using less complicated assays. Finally, experimental studies are required to verify the findings. The identified β-catenin target genes are of high confidence and the pathways are associated with CRC pathogenesis, which provides resources for the research community.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the present study are available in the figshare repository, doi.org/10.6084/m9.figshare.8872769.
Authors' contributions
HZ and BS conceived of and designed the study. HZ and LH performed the gene fitness correlation, gene set enrichment, differential gene experssion and prognostic modelling analysis. DY collected and re-formatted the data. HZ and BS wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Novellasdemunt L, Antas P, Li VS. Targeting Wnt signaling in colorectal cancer. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am J Physiol Cell Physiol. 2015;309:C511–C521. doi: 10.1152/ajpcell.00117.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 3.Tetsu O, McCormick F. Beta-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 4.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He TC, Chan TA, Vogelstein B, Kinzler KW. PPARδ is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–345. doi: 10.1016/S0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford HC, Fingleton BM, Rudolph-Owen LA, Goss KJ, Rubinfeld B, Polakis P, Matrisian LM. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 7.Brabletz T, Jung A, Dag S, Hlubek F, Kirchner T. Beta-Catenin regulates the expression of the Matrix Metalloproteinase-7 in human colorectal cancer. Am J Pathol. 1999;155:1033–1038. doi: 10.1016/S0002-9440(10)65204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hlubek F, Spaderna S, Jung A, Kirchner T, Brabletz T. Beta-Catenin activates a coordinated expression of the proinvasive factors laminin-5 gamma2 chain and MT1-MMP in colorectal carcinomas. Int J Cancer. 2004;108:321–326. doi: 10.1002/ijc.11522. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Göke B, Kolligs FT. Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genomics. 2014;15:74. doi: 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mokry M, Hatzis P, Schuijers J, Lansu N, Ruzius FP, Clevers H, Cuppen E. Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes. Nucleic Acids Res. 2012;40:148–158. doi: 10.1093/nar/gkr720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing RM, Song J, Gokulrangan G, Bai S, Bowler EH, Bolton R, Skipp P, Wang Y, Wang Z. Multiproteomic and transcriptomic analysis of oncogenic β-Catenin molecular networks. J Proteome Res. 2018;17:2216–2225. doi: 10.1021/acs.jproteome.8b00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Baryshnikova A, Costanzo M, Myers CL, Andrews B, Boone C. Genetic interaction networks: Toward an understanding of heritability. Annu Rev Genomics Hum Genet. 2013;14:111–133. doi: 10.1146/annurev-genom-082509-141730. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boettcher M, Tian R, Blau JA, Markegard E, Wagner RT, Wu D, Mo X, Biton A, Zaitlen N, Fu H, et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat Biotechnol. 2018;36:170–178. doi: 10.1038/nbt.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 2017;14:573–576. doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. Defining a cancer dependency map. Cell. 2017;170:564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. Computational correction of copy number effect improves specificity of CRISPRCas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2012;41(Database Issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, Liu H, Krier C, Stengel RF, Barany F, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 23.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhøffer M, et al. Metastasis-Associated gene expression changes predict poor outcomes in patients with dukes Stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman TJ, Smith JJ, Chen X, Washington MK, Roland JT, Means AL, Eschrich SA, Yeatman TJ, Deane NG, Beauchamp RD. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of β-catenin. Gastroenterology. 2012;142:562–571.e2. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sveen A, Agesen TH, Nesbakken A, Rognum TO, Lothe RA, Skotheim RI. Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Med. 2011;3:32. doi: 10.1186/gm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agesen TH, Sveen A, Merok MA, Lind GE, Nesbakken A, Skotheim RI, Lothe RA. ColoGuideEx: A robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
- 28.Thorsteinsson M, Kirkeby LT, Hansen R, Lund LR, Sørensen LT, Gerds TA, Jess P, Olsen J. Gene expression profiles in stages II and III colon cancers: Application of a 128-gene signature. Int J Colorectal Dis. 2012;27:1579–1586. doi: 10.1007/s00384-012-1517-4. [DOI] [PubMed] [Google Scholar]
- 29.Laibe S, Lagarde A, Ferrari A, Monges G, Birnbaum D, Olschwang S, COL2 Project A seven-gene signature aggregates a subgroup of stage II colon cancers with stage III. OMICS. 2012;16:560–565. doi: 10.1089/omi.2012.0039. [DOI] [PubMed] [Google Scholar]
- 30.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.RCoreTeam: R, corp-author. A language and environment for statistical computing. 2018 [Google Scholar]
- 33.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SK, Kim SY, Kim JH, Roh SA, Cho DH, Kim YS, Kim JC. A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol Oncol. 2014;8:1653–66. doi: 10.1016/j.molonc.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D, Skomorovska Y, Song J, Bowler E, Harris R, Ravasz M, Bai S, Ayati M, Tamai K, Koyuturk M, et al. ELF3 is an antagonist of oncogenic-signalling-induced expression of EMT-TF ZEB1. Cancer Biol Ther. 2019;20:90–100. doi: 10.1080/15384047.2018.1507256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 41.Sergushichev A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. 2016 Jun 20; doi: 10.1101/060012. doi: https://doi.org/10.1101/060012. [DOI] [Google Scholar]
- 42.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan J, Meyers RM, Michel BC, Mashtalir N, Sizemore AE, Wells JN, Cassel SH, Vazquez F, Weir BA, Hahn WC, et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst. 2018;6:555–568.e7. doi: 10.1016/j.cels.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dihlmann S, Kloor M, Fallsehr C, von Knebel Doeberitz M. Regulation of AKT1 expression by beta-catenin/Tcf/Lef signaling in colorectal cancer cells. Carcinogenesis. 2005;26:1503–1512. doi: 10.1093/carcin/bgi120. [DOI] [PubMed] [Google Scholar]
- 45.Lopez CD, Martinovsky G, Naumovski L. Inhibition of cell death by ribosomal protein L35a. Cancer Lett. 2002;180:195–202. doi: 10.1016/S0304-3835(02)00024-1. [DOI] [PubMed] [Google Scholar]
- 46.Henry JL, Coggin DL, King CR. High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Res. 1993;53:1403–1408. [PubMed] [Google Scholar]
- 47.Wang Q, Yang C, Zhou J, Wang X, Wu M, Liu Z. Cloning and characterization of full-length human ribosomal protein L15 cDNA which was overexpressed in esophageal cancer. Gene. 2001;263:205–209. doi: 10.1016/S0378-1119(00)00570-9. [DOI] [PubMed] [Google Scholar]
- 48.Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39:129–138. doi: 10.1002/hep.20017. [DOI] [PubMed] [Google Scholar]
- 49.Cheng Q, Lau WM, Chew SH, Ho TH, Tay SK, Hui KM. Identification of molecular markers for the early detection of human squamous cell carcinoma of the uterine cervix. Br J Cancer. 2002;86:274–281. doi: 10.1038/sj.bjc.6600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, Nita ME, Takagi T, Nakamura Y, Tsunoda T. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544–3549. [PubMed] [Google Scholar]
- 51.Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 52.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/S0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 53.Ciznadija D, Tothill R, Waterman ML, Zhao L, Huynh D, Yu RM, Ernst M, Ishii S, Mantamadiotis T, Gonda TJ, et al. Intestinal adenoma formation and MYC activation are regulated by cooperation between MYB and Wnt signaling. Cell Death Differ. 2009;16:1530–1538. doi: 10.1038/cdd.2009.94. [DOI] [PubMed] [Google Scholar]
- 54.Gao R, Cao C, Zhang M, Lopez MC, Yan Y, Chen Z, Mitani Y, Zhang L, Zajac-Kaye M, Liu B, et al. A unifying gene signature for adenoid cystic cancer identifies parallel MYB-dependent and MYB-independent therapeutic targets. Oncotarget. 2014;5:12528–12542. doi: 10.18632/oncotarget.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rettig EM, Tan M, Ling S, Yonescu R, Bishop JA, Fakhry C, Ha PK. MYB rearrangement and clinicopathologic characteristics in head and neck adenoid cystic carcinoma. Laryngoscope. 2015;125:E292–E299. doi: 10.1002/lary.25356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.North JP, McCalmont TH, Fehr A, van Zante A, Stenman G, LeBoit PE. Detection of MYB alterations and other immunohistochemical markers in primary cutaneous adenoid cystic carcinoma. Am J Surg Pathol. 2015;39:1347–1356. doi: 10.1097/PAS.0000000000000463. [DOI] [PubMed] [Google Scholar]
- 57.Bishop JA, Yonescu R, Epstein JI, Westra WH. A subset of prostatic basal cell carcinomas harbor the MYB rearrangement of adenoid cystic carcinoma. Hum Pathol. 2015;46:1204–1208. doi: 10.1016/j.humpath.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Argyris PP, Wetzel SL, Greipp P, Wehrs RN, Knutson DL, Kloft-Nelson SM, García JJ, Koutlas IG. Clinical utility of myb rearrangement detection and p63/p40 immunophenotyping in the diagnosis of adenoid cystic carcinoma of minor salivary glands: A pilot study. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;121:282–289. doi: 10.1016/j.oooo.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 59.Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJ, Kluk MJ, Carey CD, Rodig SJ, Sholl LM, Afrogheh AH, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet. 2016;48:265–272. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen TY, Keeney MG, Chintakuntlawar AV, Knutson DL, Kloft-Nelson S, Greipp PT, Garrity JA, Salomao DR, Garcia JJ. Adenoid cystic carcinoma of the lacrimal gland is frequently characterized by MYB rearrangement. Eye (Lond) 2017;31:720–725. doi: 10.1038/eye.2016.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Horst MP, Marusic Z, Hornick JL, Luzar B, Brenn T. Morphologically low-grade spiradenocarcinoma: A clinicopathologic study of 19 cases with emphasis on outcome and MYB expression. Mod Pathol. 2015;28:944–953. doi: 10.1038/modpathol.2015.48. [DOI] [PubMed] [Google Scholar]
- 62.Rajan N, Andersson MK, Sinclair N, Fehr A, Hodgson K, Lord CJ, Kazakov DV, Vanecek T, Ashworth A, Stenman G. Overexpression of MYB drives proliferation of CYLD-defective cylindroma cells. J Pathol. 2016;239:197–205. doi: 10.1002/path.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Maul RS, Rao J, Apple S, Seligson D, Sartippour M, Rubio R, Brooks MN. Expression pattern of the novel gene EG-1 in cancer. Clin Cancer Res. 2004;10:3504–3508. doi: 10.1158/1078-0432.CCR-03-0467. [DOI] [PubMed] [Google Scholar]
- 64.Lee EK, Cho H, Kim CW. Proteomic analysis of cancer stem cells in human prostate cancer cells. Biochem Biophys Res Commun. 2011;412:279–285. doi: 10.1016/j.bbrc.2011.07.083. [DOI] [PubMed] [Google Scholar]
- 65.Wei N, Cheng Y, Wang Z, Liu Y, Luo C, Liu L, Chen L, Xie Z, Lu Y, Feng Y. SRSF10 plays a role in myoblast differentiation and glucose production via regulation of alternative splicing. Cell Rep. 2015;13:1647–1657. doi: 10.1016/j.celrep.2015.10.038. [DOI] [PubMed] [Google Scholar]
- 66.Zhou X, Li X, Cheng Y, Wu W, Xie Z, Xi Q, Han J, Wu G, Fang J, Feng Y. BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat Commun. 2014;5:4581. doi: 10.1038/ncomms5581. [DOI] [PubMed] [Google Scholar]
- 67.Li H, Cheng Y, Wu W, Liu Y, Wei N, Feng X, Xie Z, Feng Y. SRSF10 regulates alternative splicing and is required for adipocyte differentiation. Mol Cell Biol. 2014;34:2198–2207. doi: 10.1128/MCB.01674-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soond SM, Smith PG, Wahl L, Swingler TE, Clark IM, Hemmings AM, Chantry A. Novel WWP2 ubiquitin ligase isoforms as potential prognostic markers and molecular targets in cancer. Biochim Biophys Acta 1832. 2013:2127–2135. doi: 10.1016/j.bbadis.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 69.Soond SM, Chantry A. Selective targeting of activating and inhibitory Smads by distinct WWP2 ubiquitin ligase isoforms differentially modulates TGFβ signalling and EMT. Oncogene. 2011;30:2451–2462. doi: 10.1038/onc.2010.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haan JC, Labots M, Rausch C, Koopman M, Tol J, Mekenkamp LJ, van de Wiel MA, Israeli D, van Essen HF, van Grieken NC, et al. Genomic landscape of metastatic colorectal cancer. Nat Commun. 2014;5:5457. doi: 10.1038/ncomms6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srivastava SK, Bhardwaj A, Arora S, Singh S, Azim S, Tyagi N, Carter JE, Wang B, Singh AP. MYB is a novel regulator of pancreatic tumour growth and metastasis. Br J Cancer. 2015;113:1694–1703. doi: 10.1038/bjc.2015.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available in the figshare repository, doi.org/10.6084/m9.figshare.8872769.