Abstract
Abnormal methylation of secreted frizzled-related proteins (SFRPs) has been observed in various human cancer types. The loss of SFRP gene expression induces the activation of the Wnt pathway and is a vital mechanism for tumorigenesis and development. The aim of the present systematic review was to assess the association between SFRP methylation and cancer risk. A meta-analysis was systematically conducted to assess the clinicopathological significance of SFRP methylation in cancer risk. The Cochrane Library, PubMed and Web of Science databases were comprehensively searched, and 83 publications with a total of 21,612 samples were selected for the meta-analysis. The pooled odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were calculated to evaluate the degree of associations between SFRP promoter methylation and cancer risk. Subgroup analysis, meta regression and sensitivity analysis were used to identify the potential sources of heterogeneity. SFRP1, SFRP2, SFRP4 and SFRP5 hypermethylation was significantly associated with cancer risk, with ORs of 8.48 (95% CI, 6.26–11.49), 8.21 (95% CI, 6.20–10.88), 11.41 (95% CI, 6.42–20.30) and 6.34 (95% CI, 3.86–10.42), respectively. SFRP2 methylation was significantly associated with differentiation in colorectal cancer (OR, 2.16; 95% CI, 1.02–4.56). The results of the present study demonstrated that SFRP methylation may contribute to carcinogenesis, especially in certain cancer types, including hepatocellular carcinoma and colorectal cancer.
Keywords: secreted frizzled-related proteins, methylation, meta-analysis, systematic review, cancer
Introduction
Epigenetic regulation is described as a heritable DNA modification without changes in the DNA sequence (1). In cancer, epigenetic modifications involve DNA methylation of tumor suppressor gene (TSG) promoters, which inhibits gene transcription (2). Aberrant DNA methylation often occurs early in carcinogenesis in a number of cancer types, including breast cancer (3), colorectal cancer (4) and gastric cancer (5). Currently, the main techniques used in methylation research include third generation high-throughput sequencing, second generation high-throughput sequencing, whole genome bisulfite sequencing based on second generation sequencing, methylated DNA immunization co-precipitation sequencing, reduced representation bisulfite sequencing, gene chip detection technology and mass spectrometry detection. In addition, other techniques include methylation-specific PCR (MSP), bisulfite-treated sequencing and methylation-sensitive high-resolution melting curve analysis. These techniques offer the potential to screen highly methylated promoter genes to identify important biomarkers for tumors.
The Wnt signaling pathway is involved in cell proliferation, differentiation and fate determination (6). However, the abnormal activity of the Wnt pathway can lead to tumorigenesis (7). Previous studies have discovered that hepatocellular carcinoma (HCC) and >90% of colorectal cancer (CRC) tumors exhibit abnormal activation of the Wnt signal pathway and changes in the downstream components of the pathway (8,9).
Secreted frizzled-related proteins (SFRPs) are tumor suppressor genes involved in the Wnt signaling pathway by binding to Wnt ligands, forming a non-functional complex, and subsequently preventing the initiation of the signaling cascade (Fig. 1). To date, four mammalian SFRPs (SFRP1, 2, 4 and 5) have been identified that exhibit CpG promoter hypermethylation (10,11). The downregulation of SFRP genes by promoter methylation has been demonstrated in various cancer types, including cervical cancer (12), leukemia (13) and lung cancer (14). In the study by Sui et al (15), a meta-analysis was performed that identified an association between SFRP2 in tissue and feces and the risk of CRC. In another meta-analysis, SFRP2 methylation was identified as a new biomarker for screening early CRC by detecting stool-based DNA methylation (16). In addition, aberrant methylation of the SFRP1 promoter has been demonstrated to contribute to colorectal carcinogenesis (17). Despite numerous investigations, the association between SFRP methylation in CRC and clinicopathological significance still needs to be clarified, and the association between SFRP promoter methylation and multiple tumors remains controversial. For example, Kloten et al (18) reported that SFRP1 and SFRP2 methylation had no association with breast cancer, whereas Suzuki et al (19) reported that SFRP1 and SFRP2 methylation was associated with breast cancer. However, the association of methylated SFRP4 and SFRP5 in tumors has not been assessed in a meta-analysis. Therefore, the aim of the present study was to conduct a meta-analysis to further analyze the association between different types of cancer and SFRP methylation.
Figure 1.
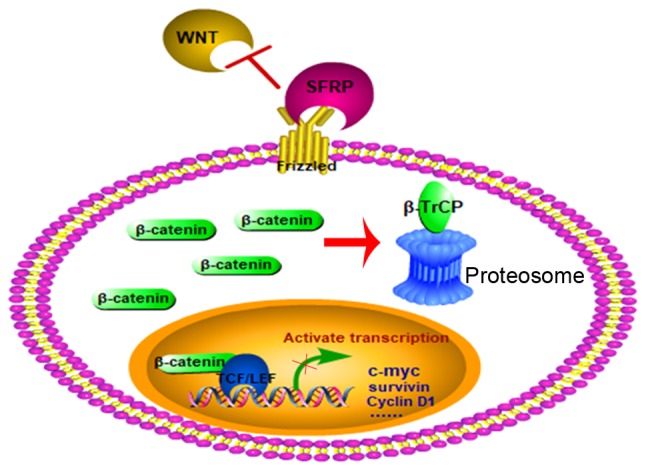
Pattern Diagram of SFRP blocking the WNT pathway. SFRP, secreted frizzled-related protein. β-TrCP, β-transducin repeats-containing proteins; TCF/LEF, T-cell factor/lymphocyte enhancer factor.
Materials and methods
Search strategy
A literature search was performed independently by two investigators using data recorded in the PubMed, Web of Science and Cochrane Library databases prior to December 2017, and was restricted to English language publications. Combinations of the following terms were used in the search strategy: ‘Frizzled-related protein’ OR ‘frizzled-related proteins’ OR ‘FRZB proteins’ OR ‘SFRPs’ OR ‘SFRP’ OR ‘SFRP1’ OR ‘SFRP2’ OR ‘SFRP4’ OR ‘SFRP5’ and ‘methy*’ OR ‘methylation’ OR ‘methylated’ and ‘neoplasm’ OR ‘tumor’ OR ‘cancer’ OR ‘neoplasia’ OR ‘carcinomas’. All eligible articles were retrieved, and their reference lists were further checked for potential additional articles.
Inclusion and exclusion criteria
Data extraction was performed independently by two reviewers. The inclusion criteria were as follows: i) The data were independent; ii) the case-control or cohort studies assessed the associations between SFRP methylation status and any type of human cancer or their clinicopathological features; iii) studies had sufficient data to calculate an odds ratio (OR) and 95% confidence interval (CI); iv) an explicit method of methylation detection was reported; v) studies were written in English; vi) only the most recent or detailed publication with a large sample size was selected to avoid duplicated publications; and vii) the number of subjects in the control groups was >5. The exclusion criteria for the meta-analysis were as follows: i) Reviews, letters, abstracts, case reports or expert opinions; ii) reports with insufficient data for calculation of OR; iii) studies regarding in vitro experiments or animal experiments; or iv) duplications of previous publications or replicated samples.
Data extraction and quality assessment
Two investigators extracted the following information independently from eligible studies according to the aforementioned inclusion and exclusion criteria: The first author, year of publication, region, names of genes, source of controls, detection method of methylation, clinicopathological characteristics, sample material and number of methylated (M) and unmethylated (U) samples in cases and controls (Table SI). The meta-analysis was performed based on the Preferred Reporting Items for Systematic Reviews and Meta-Analysis statement (20) The quality of studies was assessed according to the Newcastle-Ottawa Scale assessment for case-control or cohort studies (21). The scores ranged from 0 to 9 points; a score >6 points was considered to indicate a high-quality study.
Statistical analysis
The ORs and 95% CIs were calculated to examine the associations between SFRP methylation status and different types of human cancer. In all statistical tests, P<0.05 was considered to indicate a statistically significant difference.
The heterogeneity across studies was assessed by the Cochran's Q test and I2 (22). P≤0.1 or I2>50% indicated significant heterogeneity across studies, and the pooled OR was calculated using the random-effects model. Otherwise, the fixed-effects model was performed. A subgroup analysis was performed to evaluate the source of the heterogeneity. Meta-regression was performed based on regions, method, cancer types, publication year (year <2010 or ≥2010), case sample size (n<50 or n≥50) and sample materials to further explore the potential sources of heterogeneity.
Sensitivity analyses were conducted to assess the stability of the pooled OR and heterogeneity by sequentially omitting each study. Publication bias was assessed quantitatively by Egger's test and qualitatively by Begg's funnel plots (23). A combined Egger's of P-value<0.05 with an asymmetric funnel plot suggested the presence of publication bias. Meta-analysis was conducted using Review Manager 5.2 (Cochrane Collaboration) and STATA 12.0 (StataCorp LP).
Results
Identification of relevant studies
The article selection process is presented as a flow chart in Fig. 2. Based on the search strategies, a total of 824 potentially relevant articles were identified, and 83 articles were included in the final analysis following screening. The majority of the excluded abstracts and titles were reviews or studies with insufficient data.
Figure 2.
Flow chart of the literature search strategy. SFRP, secreted frizzled-related protein.
Study characteristics
In the present analysis, 21,612 samples from 83 articles were used to study SFRP promoter hypermethylation in 22 types of human tumors. Among the 83 articles, there were 79 case-control articles and 4 cohort articles. A total of 14 articles among the 79 case-control articles also contained cohort analyses. In addition, 46 articles focused on the methylation of a single gene, and the remaining 37 articles involved the methylation of multiple genes. SFRP1 was the focus of 61 studies (3 studies from cohort articles) (3,4,8,10–13,18,19,24–71), 56 studies focused on SFRP2 (2 studies from cohort articles) (4,5,8–10,12,13,18,19, 28,29,31–33,35,37,38,42–45,53, 57–59,62,65–67,71–90), 19 studies on SFRP4 (4,10,12,31,36,42–44,57,62,63,65–67,71,85,86,91) and 29 studies on SFRP5 (1 study from cohort articles) (4,8,10,12,19,24,29,31,42–45,49,53,57–59,62,63,65–67,71,78,85,86,92). In addition, of these 83 articles, 24 articles focused on CRC (4,5,9,30–35,72–85,93), 4 on leukemia (13,55–57), 4 on lung cancer (58,59,61,68), 8 on HCC (8,49–54,88), 6 on esophageal cancer (EC) (37–42,87), 9 on gastric cancer (GC) (5,39,43–47,84,92), 5 on BC (3,18,19,28), 4 on renal cell carcinoma (RCC) (68–71) and the remaining 21 on other types of cancer. A single article evaluated SFRP methylation levels in esophageal cancer and GC (39), and 1 article evaluated SFRP methylation levels in GC and CRC (5). The information on SFRP1, SFRP2, SFRP4 and SFRP5 methylation was collected from eligible studies and presented in Table SI.
Association between SFRP1 promoter methylation and cancer risk
The results of the meta-analysis demonstrated that the frequency of SFRP1 methylation was significantly higher in patients with cancer compared with that in control samples. The pooled OR from 58 studies on SFRP1, which included 6,358 samples with various cancer types, was 8.48 (95% CI, 6.26–11.49; Table I; Fig. 3). In the analysis by cancer type, SFRP1 methylation was associated with HCC (OR, 5.00; 95% CI, 2.74–9.11; P<0.001), GC (OR, 10.27; 95% CI, 5.14–20.50; P<0.001), CRC (OR, 7.86; 95% CI, 4.87–12.68; P<0.001), EC (OR, 16.18; 95% CI, 3.77–69.47; P<0.001), RCC (OR, 12.18; 95% CI, 5.66–6.21; P<0.001), CC (OR, 60.61; 95% CI, 7.10–517.42; P<0.001), leukemia (OR, 12.85; 95% CI, 3.64–45.31; P<0.001), lung cancer (OR, 10.68; 95% CI, 5.94–19.20; P<0.001), bladder cancer (OR, 8.20; 95% CI; 3.23–20.76; P<0.001), ovarian cancer (OR, 22.19; 95% CI, 10.54–46.72; P<0.001) and endometrial carcinoma (OR, 3.07; 95% CI, 1.03–9.12; P=0.04), but not BC (OR, 10.70; 95% CI, 0.82–140.26; P=0.07). In the analysis based on method, significantly increased cancer risk was associated with SFRP1 methylation according to the MSP method (OR, 9.95; 95% CI, 7.26–13.64; P<0.001), COBRA method (OR, 5.48; 95% CI, 1.89–15.85; P=0.002), MethyLight (OR, 8.92; 95% CI, 1.10–72.12; P=0.04) and methylation-sensitive restriction endonuclease digestion and quantitative PCR (OR, 3.26; 95% CI, 1.33–7.95; P=0.01), but not by QMSP (OR, 5.01; 95% CI, 0.62–40.67; P=0.13). Stratified analysis by sample material revealed that significantly increased cancer risk was associated with SFRP1 methylation in tissue (OR, 9.46; 95% CI, 6.93–12.92; P<0.001), stool (OR, 9.33; 95% CI, 3.09–28.15; P<0.001) and blood (OR, 9.30; 95% CI, 3.60–24.03; P<0.001) samples, but not in bone marrow samples (OR, 2.01; 95% CI, 0.35–11.69; P=0.44). Moreover, subgroup analysis based on region demonstrated that SFRP1 methylation was associated with cancer in patients from Asia (OR, 7.83; 95% CI, 5.70–10.76; P<0.001), Europe (OR, 7.58; 95% CI, 2.74–20.98; P<0.001) and North America (OR, 18.21; 95% CI, 9.28–35.71; P<0.001; Table I).
Table I.
Pooled OR and 95% CI for the association between SFRP1, SFRP2, SFRP 4 and SFRP5 promoter hypermethylation and cancer risk.
| SFRP1 | SFRP2 | SFRP4 | SFRP5 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | n | OR (95% CI) | P-value | I2, % | n | OR (95% CI) | P-value | I2, % | n | OR (95% CI) | P-value | I2, % | n | OR (95% CI) | P-value | I2, % |
| Total | 58 | 8.48 (6.26–11.49) | <0.001 | 62 | 54 | 8.21 (6.20–10.88) | <0.001 | 77 | 19 | 11.41 (6.42–20.30) | <0.001 | 63 | 28 | 6.34 (3.86–10.42) | <0.001 | 79 |
| Cancer type | ||||||||||||||||
| Hepatocellular carcinoma | 8 | 5.00 (2.74–9.11) | <0.001 | 50 | 3 | 1.91 (1.20–3.03) | 0.006 | 0 | 3 | 4.11 (1.95–8.67) | <0.001 | 0 | ||||
| Gastric cancer | 7 | 10.27 (5.14–20.50) | <0.001 | 51 | 5 | 6.97 (2.66–18.25) | <0.001 | 71 | 2 | 7.66 (0.87–67.08) | 0.070 | 89 | 4 | 3.36 (0.47–23.83) | 0.220 | 94 |
| Colorectal cancer | 6 | 7.86 (4.87–12.68) | <0.001 | 39 | 23 | 8.41 (5.91–11.97) | <0.001 | 73 | 3 | 11.04 (2.50–48.76) | 0.002 | 67 | 4 | 6.37 (2.18–18.62) | <0.001 | 66 |
| Esophageal cancer | 5 | 16.18 (3.77–69.47) | <0.001 | 80 | 4 | 7.28 (1.72–30.79) | 0.007 | 76 | ||||||||
| Breast cancer | 4 | 10.70 (0.82–140.26) | 0.070 | 88 | 3 | 30.81 (0.52–1837.06) | 0.100 | 91 | ||||||||
| Renal cell carcinoma | 5 | 12.18 (5.66–26.21) | <0.001 | 0 | 2 | 13.48 (5.37–33.79) | <0.001 | 0 | 2 | 7.39 (3.22–16.98) | <0.001 | 0 | 2 | 9.75 (4.29–22.12) | <0.001 | 4 |
| Cervical cancer | 2 | 60.61 (7.10–517.42) | <0.001 | 0 | 2 | 93.72 (29.05–302.32) | <0.001 | 0 | 3 | 215.02 (40.42–1143.79) | <0.001 | 0 | 2 | 11.89 (0.50–282.90) | 0.130 | 87 |
| Ovarian cancer | 2 | 22.19 (10.54–46.72) | <0.001 | 0 | 2 | 19.00 (6.54–55.16) | <0.001 | 77 | 2 | 22.01 (10.49–46.19) | <0.001 | 42 | 2 | 59.70 (23.59–151.07) | <0.001 | 0 |
| Endometrial carcinoma | 2 | 3.07 (1.03–9.12) | 0.040 | 0 | 2 | 5.97 (2.06–17.33) | 0.001 | 0 | 2 | 0.71 (0.16–3.19) | 0.660 | 47 | 2 | 2.09 (0.44–10.04) | 0.360 | 61 |
| Bladder cancer | 3 | 8.20 (3.23–20.76) | <0.001 | 0 | ||||||||||||
| Lung cancer | 3 | 10.68 (5.94–19.20) | <0.001 | 0 | ||||||||||||
| Leukemia | 4 | 12.85 (3.64–45.31) | <0.001 | 27 | 2 | 14.07 (1.84–107.61) | 0.010 | 0 | ||||||||
| Sample material | ||||||||||||||||
| Tissue | 42 | 9.46 (6.93 −12.92) | <0.001 | 58 | 36 | 8.29 (5.79–11.87) | <0.001 | 79 | 16 | 12.12 (6.50–22.59) | <0.001 | 69 | 23 | 6.63 (3.85–11.42) | <0.001 | 83 |
| Blood | 11 | 9.30 (3.60–24.03) | <0.001 | 62 | 7 | 7.50 (2.30–24.42) | <0.001 | 86 | 2 | 9.15 (1.18–70.81) | 0.030 | 0 | 2 | 17.36 (2.29–131.85) | 0.006 | 0 |
| Feces | 2 | 9.33 (3.09–28.15) | <0.001 | 0 | 10 | 9.20 (7.06–11.99) | <0.001 | 42 | ||||||||
| Bone marrow | 2 | 2.01 (0.35–11.69) | 0.440 | 0 | ||||||||||||
| Region | ||||||||||||||||
| Asia | 41 | 7.83 (5.70–10.76) | <0.001 | 56 | 42 | 8.69 (6.40–11.81) | <0.001 | 77 | 13 | 11.69 (5.86–23.32) | <0.001 | 61 | 21 | 6.54 (3.51–12.21) | <0.001 | 81 |
| Europe | 10 | 7.58 (2.74–20.98) | <0.001 | 77 | 6 | 5.73 (1.76–18.72) | 0.004 | 81 | 2 | 2.08 (1.22–3.55) | 0.007 | 28 | ||||
| North America | 6 | 18.21 (9.28–35.71) | <0.001 | 0 | 5 | 6.99 (2.53–19.33) | <0.001 | 72 | 4 | 9.39 (5.25–16.79) | <0.001 | 0 | 4 | 11.70 (6.60–20.74) | <0.001 | 37 |
| Method | ||||||||||||||||
| MSP | 48 | 9.95 (7.26–13.64) | <0.001 | 55 | 41 | 8.82 (6.21–12.54) | <0.001 | 77 | 17 | 11.25 (6.63–19.11) | <0.001 | 52 | 25 | 7.21 (4.32–12.03) | <0.001 | 79 |
| COBRA | 2 | 5.48 (1.89–15.85) | 0.002 | 67 | 5 | 9.74 (5.59–16.98) | <0.001 | 74 | ||||||||
| MethyLight | 2 | 8.92 (1.10–72.12) | 0.04 | 0 | 3 | 12.19 (6.92–21.48) | <0.001 | 0 | ||||||||
| MSRE-qPCR | 2 | 3.26 (1.33–7.95) | 0.01 | 0 | ||||||||||||
| QMSP | 3 | 5.01 (0.62–40.67) | 0.13 | 83 | 2 | 5.00 (0.60–41.83) | 0.140 | 89 | 2 | 25.86 (0.18–3638.77) | 0.20 | 93 | ||||
| Reverse hybridization method | 2 | 2.46 (0.45–13.58) | 0.300 | 68 | ||||||||||||
COBRA, combined bisulfite restriction analysis; MSP, MSP, methylation-specific PCR; MSRE-qPCR, methylation-sensitive restriction endonucleases; OR, odds ratio; QMSP, real-time quantitative methylation-specific polymerase chain reaction; SFRP, secreted frizzled-related protein.
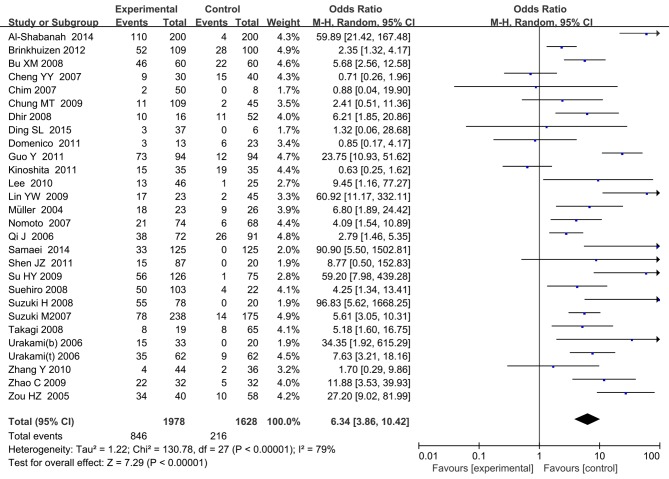
Figure 3.
Forest plots of secreted frizzled-related protein 1 promoter methylation between cancer and control groups. b, blood; CI, confidence interval; M-H, Mantel-Haenszel; t, tissue.
However, heterogeneity was identified across the included studies (I2=62%; P<0.001). Subgroup analysis was used based on sample materials, regions, cancer types and assay method to explain the sources of heterogeneity. Moderate or extensive heterogeneity still remained in the majority of the subgroups (Table I).
Association between SFRP2 promoter methylation and cancer risk
In total, 54 studies with 8,577 samples were included in the meta-analysis to assess the association between SFRP2 methylation status and cancer risk. A significant association was identified between SFRP2 promoter hypermethylation and increased cancer risk, with an OR of 8.21 (95% CI, 6.20–10.88; P<0.001) (Table I; Fig. 4). Analysis by cancer type revealed that significantly increased cancer risk was associated with SFRP2 methylation in HCC (OR, 1.91; 95% CI, 1.20–3.03; P=0.006), CRC (OR, 8.32; 95% CI, 5.88–11.78; P<0.001), GC (OR, 6.97; 95% CI, 2.66–18.25; P<0.001), EC (OR, 7.28; 95% CI, 1.72–30.79; P=0.007), leukemia (OR, 14.07; 95% CI, 1.84–107.61; P=0.01), CC (OR, 93.72; 95% CI, 29.05–302.32; P<0.001), ovarian cancer (OR, 19.00; 95% CI, 6.54–55.16; P<0.001), endometrial carcinoma (OR, 5.97; 95% CI, 2.06–17.33; P=0.001) and RCC (OR, 13.48; 95% CI, 5.37–33.79; P=0.001), but not BC (OR, 30.81; 95% CI, 0.52–1,837.06; P=0.10). In the subgroup analysis based on method, significantly increased cancer risk was associated with SFRP2 methylation as determined by the MSP method (OR, 8.82; 95% CI, 6.21–12.54; P<0.001), COBRA method (OR, 9.74; 95% CI, 5.59–16.98, P<0.001) and MethyLight (OR, 12.19; 95% CI, 6.92–21.48; P<0.001), but not according to the QMSP (OR, 5.00; 95% CI, 0.60–41.83; P=0.14) or reverse hybridization (OR, 2.46; 95% CI, 0.45–13.58; P=0.30) methods. In addition, subgroup analysis based on region reported that SFRP2 methylation was associated with cancer in patients from Asia (OR 8.69; 95% CI, 6.40–11.81; P<0.001), Europe (OR, 5.73; 95% CI, 1.76–18.72; P=0.004) and North America (OR, 6.99; 95% CI, 2.53–19.33; P<0.001). Stratified analysis by sample material revealed that significantly increased cancer risk was associated with SFRP2 methylation in tissue (OR, 8.29; 95% CI, 5.79–11.87; P<0.001), stool (OR, 9.20; 95% CI, 7.06–11.99; P<0.001) and blood sample (OR, 7.50; 95% CI, 2.30–24.42; P<0.001; Table I).
Figure 4.
Forest plots of secreted frizzled-related protein 2 promoter methylation between cancer and control groups. b, blood; CI, confidence interval; CRC, colorectal cancer; GC, gastric cancer; M-H, Mantel-Haenszel; s, stool; t, tissue.
Due to significant heterogeneity among the included studies (I2=77%; P<0.001), subgroup analysis was performed. However, extensive heterogeneity remained in the majority of the subgroups (Table I).
Association between SFRP4 promoter methylation and cancer risks
In total, 19 studies with 2,440 samples were included in the meta-analysis to determine the effects of SFRP4 promoter hypermethylation on cancer risk. A significant association was identified between SFRP4 promoter hypermethylation and increased cancer risk with an OR of 11.41 (95% CI, 6.42–20.30; P<0.001) (Table I; Fig. 5). Analysis by cancer type revealed that significantly increased cancer risk was associated with SFRP4 methylation in CRC (OR, 11.04; 95% CI, 2.50–48.76; P=0.002), ovarian cancer (OR, 22.01, 95% CI, 10.49–46.19; P<0.001), CC (OR, 215.02; 95% CI, 40.42–1143.79; P<0.001 and RCC (OR, 7.39; 95% CI, 3.22–16.98; P<0.001), but not endometrial carcinoma (OR, 0.71; 95% CI, 0.16–3.19; P=0.66) or GC (OR, 7.66; 95% CI, 0.87–67.08; P=0.07). In the subgroup analysis based on method, significantly increased cancer risk was associated with SFRP1 methylation as assessed by the MSP method (OR, 11.25; 95% CI, 6.63–19.11; P<0.001), but not by QMSP (OR, 25.86; 95% CI, 0.18–3638.77; P=0.20). In addition, subgroup analysis based on region exhibited that SFRP4 methylation was associated with cancer in patients from Asia (OR, 11.69; 95% CI, 5.86–23.32; P<0.001), and North America (OR, 9.39; 95% CI, 5.25–16.79; P<0.001). Stratified analysis by sample material revealed that significantly increased cancer risk was associated with SFRP4 methylation in tissue (OR, 12.12; 95% CI, 6.50–22.59; P<0.001) and blood sample (OR, 9.15; 95% CI, 1.18–70.81; P<0.001; Table I).
Figure 5.
Forest plots of secreted frizzled-related protein 4 promoter methylation between cancer and control groups. b, blood; CI, confidence interval; M-H, Mantel-Haenszel; t, tissue.
However, evidence of heterogeneity was identified across the studies (I2=63%; P<0.001). When subgroup analysis was performed, heterogeneity was reduced in several subgroups but remained high (Table I).
Association between SFRP5 promoter methylation and cancer risk
In total, 28 studies including 3,606 samples were analyzed to evaluate the association of SFRP5 methylation status with various cancer types. The pooled OR of SFRP5 hypermethylation was 6.34 (95% CI, 3.86–10.42; P<0.001) (Table I; Fig. 6). Analysis by cancer type revealed that significantly increased cancer risk was associated with SFRP5 methylation in HCC (OR, 4.11; 95% CI, 1.95–8.67; P<0.001), CRC (OR, 6.37; 95% CI, 2.18–18.62; P<0.001), ovarian cancer (OR, 59.70; 95% CI, 23.59–151.07; P<0.001) and RCC (OR, 9.75; 95% CI, 4.29–22.12; P<0.001), but not endometrial carcinoma (OR, 2.09; 95% CI, 0.44–10.04; P=0.36), GC (OR, 3.36; 95% CI, 0.47–23.83; P=0.22) and CC (OR, 11.89; 95% CI, 0.50–282.90; P=0.13). In the stratified analysis based on method, significantly increased cancer risk was associated with SFRP5 methylation as determined by the MSP method (OR, 7.21; 95% CI, 4.32–12.03; P<0.001). Additionally, subgroup analysis based on region demonstrated that SFRP5 methylation was associated with cancer in patients from Asia (OR, 6.54; 95% CI, 3.51–12.21; P<0.001), Europe (OR, 8.69; 95% CI, 6.40–11.81; P<0.001) and North America (OR, 2.08; 95% CI, 1.22–3.55; P<0.007). Stratified analysis by sample material revealed that significantly increased cancer risk was associated with SFRP5 methylation in tissue (OR, 6.63; 95% CI, 3.85–11.42; P<0.001) and blood sample (OR, 17.36; 95% CI, 2.29–131.85; P<0.006; Table I).
Figure 6.
Forest plots of secreted frizzled-related protein 5 promoter methylation between cancer and control groups. b, blood; CI, confidence interval; M-H, Mantel-Haenszel; t, tissue.
High levels of heterogeneity were identified among the included studies (I2=79%; P<0.001). When subgroup analysis was performed, moderate or extensive heterogeneity remained in most of the subgroups (Table I).
Association between SFRP1 and SFRP2 promoter methylation and clinicopathological features of multiple cancer types
Overall, 2 studies of SFRP2 in BC, 3 studies of SFRP1 in lung cancer, 5 studies of SFRP2 in GC and 11 studies of SFRP2 in CRC provided sufficient data to assess the association between the methylation status and clinicopathological features. No associations were observed between SFRP1 methylation and certain factors, including sex, smoking habit or pathological type in lung cancer. SFRP2 methylation was not associated with any reported clinicopathological features in BC and GC. However, SFRP2 gene promoter hypermethylation was associated with CRC tumor differentiation (‘poor or other’ vs. ‘well or moderate’: OR, 2.16; 95% CI, 1.02–4.56; P=0.04; Table SII).
Sensitivity and meta-regression analysis
Sensitivity analysis was performed by omitting each study in turn to evaluate its effect on the pooled OR and to identify heterogeneous studies. To analyze the association between SFRP2 methylation and tumor differentiation in CRC, deletion of the study by Takeda et al (93) increased the pooled OR from 2.16 (95% CI, 1.02–4.56) to 2.70 (95% CI, 1.39–5.25), whereas the heterogeneity was reduced from 59% (P=0.02) to 39% (P=0.30).
Meta-regression analysis was conducted to explore the potential sources of heterogeneity for SFRP1, SFRP2, SFRP4 and SFRP5. The results demonstrated that sample materials may be a contributor to heterogeneity (P=0.099) for SFRP1, whereas publication year, case sample size, cancer type, region and method were not (P=0.103–0.903). In addition, publication year, sample size, region, method, cancer type and sample materials did not contribute to the heterogeneity for SFRP2 (P=0.488–0.993), SFRP4 (P=0.262–0.760) and SFRP5 (P=0.102–0.923; Table SIII).
Publication bias
Publication bias was evaluated using Begg's funnel plots and Egger's test. Begg's funnel plots did not reveal asymmetry (Fig. S1). However, publication bias was detected in the analysis of SFRP1 in various cancer types (Egger's test, P=0.044; Table SIII). Therefore, a trim and fill analysis was performed to identify and amend the bias. A total of 12 adjusted studies were added to the original meta-analysis of SFRP1 with various cancer types (Fig. S1). The OR remained significant for the association between SFRP1 methylation and risk of various cancer types (OR, 1.872; 95% CI, 1.568–2.176). Egger's tests indicated that no significant publication bias was identified for SFRP2 (P=0.386), SFRP4 (P=0.992) or SFRP5 (P=0.254; Table SIV).
Discussion
Hypermethylation of SFRP gene promoter is an important mechanism of Wnt signaling pathway activation, and it is also key to tumor formation and development (7). In the present study, the association between SFRP gene promoter methylation and tumor risk was analyzed. The results demonstrated that the frequency of SFRP1, SFRP2, SFRP4 and SFRP5 promoter methylation was 8.48-, 8.21-, 11.41- and 6.34-fold higher, respectively, in patients with cancer compared with that in healthy controls. Additionally, SFRP2 promoter methylation was significantly associated with poor differentiation in CRC.
A subgroup analysis based on cancer type was conducted to further investigate SFRP promoter methylation in specific tumor types. The results demonstrated that SFRP1 promoter methylation was significantly associated with HCC, GC, CRC, EC, RCC, CC, ovarian cancer, endometrial carcinoma, bladder cancer, lung cancer and leukemia. SFRP2 promoter methylation was associated with HCC, GC, CRC, EC, RCC, CC, ovarian cancer and endometrial carcinoma. Additionally, SFRP4 promoter methylation was associated with CRC, ovarian cancer, CC and RCC, and SFRP5 promoter methylation was associated with HCC, CRC, ovarian cancer and RCC. SFRP1 and SFRP2 methylation were not associated with BC. SFRP4 and SFRP5 promoter methylation was not associated with endometrial carcinoma and GC, and SFRP5 promoter methylation was not associated with CC. Therefore, despite the limited number of studies that described other specific types of cancer, SFRPs gene hypermethylation was associated with the majority of cancer types.
In the present study, SFRP methylation was significantly associated with cancer risk in tissue, blood and stool samples. The level of SFRP2 promoter methylation detected in stool samples was higher compared with that in tissue samples. A similar frequency of SFRP1 promoter methylation was detected in stool and tissue samples. The results suggested that the detection of fecal methylation biomarkers may represent a new non-invasive method to screen for malignancies. To date, studies of fecal methylation detection have mainly focused on the early diagnosis and screening of colorectal cancer (7). Stool samples contain a large number of colorectal cancer cells that escape from tumors, as well as normal colorectal epithelial cells and cell-free DNA that originates from cell degradation. The alkaline environment in the intestinal tract is conducive to DNA preservation (94). Thus, fecal DNA may be a good specimen for the molecular detection of cancer.
Subgroup analysis based on methylation detection method revealed significant associations between SFRP methylation and cancer using MSP, COBRA and MethyLight, but not QMSP. However, heterogeneity between studies remained moderately high in the MSP and COBRA subgroups. For MSP, primers based on different loci and PCR conditions used in different studies contributed to heterogeneity. COBRA can only obtain the methylation status at specific restriction sites, and primer issues similar to those for MSP contributed to heterogeneity. Subgroup analysis based on different regions revealed that SFRP methylation was associated with cancer in patients from all covered regions. This finding indicated that although the lifestyles, environments and genetic factors were different, the relevance of SFRP methylation remained stable. However, whether geographical differences exist between specific cancer types and SFRP methylation requires further clarification.
In the subgroup analyses, the I2 value was reduced in some stratified analyses to an extent, but it was insufficient to conclude that test method, sample material, cancer type and region may cause heterogeneity. Meta-regression was performed to explain the sources of heterogeneity; the results demonstrated that the sample material contributed to the bias among studies that investigated the association between SFRP1 methylation and multiple cancer types. For other studies, region, publication year, sample material, case sample size, cancer type and assay method did not significantly contribute to the heterogeneity.
Heterogeneity was observed in the majority of the analyses in the present study, even when the results were pooled and assessed using the random-effects model and stratification analyses based on sample material, methylation test method, cancer type and region. However, none of the previously mentioned factors contributed to the heterogeneity among the studies on SFRP2, SFRP4 and SFRP5. Publication bias was observed among the studies concerning SFRP1, but not SFRP2, SFRP4 or SFRP5 methylation. A trim and fill analysis was performed to amend the bias, and SFRP1 methylation still exhibited a significant association with cancer risk. The sensitivity analysis did not alter the significance of the association, which further supported the stability of the results.
The present study has several potential limitations. First, although the analysis was conducted with precise data extraction and strict criteria for study inclusion, heterogeneity in the subgroup analyses remained high. The possibility of unidentified confounders and selection biases could not be completely avoided. Second, the inclusion of articles only in English may have led to selection bias, as studies with potentially high-quality data published in other languages may pose difficulties in obtaining an accurate medical translation. Third, the primers based on specific locations of CpG islands and PCR conditions used to detect the status of SFRP methylation were not uniform. Fourth, some analyses were based on a limited number of studies, which led to inevitable bias. Publication bias was not identified for these studies, which subsequently influenced the gene-based analysis. However, the complete literature search identified a certain number of negative results to minimize the publication bias.
In conclusion, the results of the present meta-analysis suggested that SFRP promoter methylation may be associated with cancer risk. In addition, SFRP2 methylation was associated with CRC differentiation. Well-designed studies with larger sample sizes are needed in the future to strengthen these observations and confirm the association between SFRP promoter methylation and other cancer types.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- SFRP
secreted frizzled-related protein
- CRC
colorectal cancer
- GC
gastric cancer
- HCC
hepatocellular carcinoma
- BC
breast cancer
- CC
cervical cancer
- EC
esophageal cancer
- RCC
renal cell carcinoma
- MSP
methylation-specific PCR
- QMSP
quantitative real-time MSP
- COBRA
combined bisulfite restriction analysis
- MethyLight
methylation-specific multiplex ligation-dependent probe amplification
Funding
The present study was supported by the Chinese National Science Foundation Projects (grant no. 81101868) and the Applied Basic Research Programs of Wuhan Science and Technology Department (grant no. 2015061701011642).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JY and JQ conceived and designed the research. YX, ML and FW performed data acquisition, data analysis and manuscript preparation. FZ, ZZ and YL assisted with data acquisition, data analysis and statistical analysis. JY, YX and JQ contributed in writing the manuscript. JY, YX and JQ read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Davies PF, Manduchi E, Jimenez JM, Jiang YZ. Biofluids, cell mechanics and epigenetics: Flow-induced epigenetic mechanisms of endothelial gene expression. J Biomech. 2017;50:3–10. doi: 10.1016/j.jbiomech.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeshima H, Wakabayashi M, Hattori N, Yamashita S, Ushijima T. Identification of coexistence of DNA methylation and H3K27me3 specifically in cancer cells as a promising target for epigenetic therapy. Carcinogenesis. 2015;36:192–201. doi: 10.1093/carcin/bgu238. [DOI] [PubMed] [Google Scholar]
- 3.Agostini M, Enzo MV, Bedin C, Belardinelli V, Goldin E, Del Bianco P, Maschietto E, D'Angelo E, Izzi L, Saccani A, et al. Circulating cell-free DNA: A promising marker of regional lymphonode metastasis in breast cancer patients. Cancer Biomark. 2012;11:89–98. doi: 10.3233/CBM-2012-0263. [DOI] [PubMed] [Google Scholar]
- 4.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7117. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagasaka T, Tanaka N, Cullings HM, Sun DS, Sasamoto H, Uchida T, Koi M, Nishida N, Naomoto Y, Boland CR, et al. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009;101:1244–1258. doi: 10.1093/jnci/djp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T, Clevers H. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Surana R, Sikka S, Cai W, Shin EM, Warrier SR, Tan HJ, Arfuso F, Fox SA. Secreted frizzled related proteins: Implications in cancers. Biochim Biophys Acta 1845. 2014:53–65. doi: 10.1016/j.bbcan.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, Shinomura Y. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–389. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Zhu YQ, Wu YQ, Zhang P, Qi J. Detection of promoter hypermethylation of Wnt antagonist genes in fecal samples for diagnosis of early colorectal cancer. World J Gastroenterol. 2014;20:6329–6335. doi: 10.3748/wjg.v20.i20.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung MT, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Chu TY, Lai HC, Lin YW. Promoter methylation of SFRPs gene family in cervical cancer. Gynecol Oncol. 2009;112:301–306. doi: 10.1016/j.ygyno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Uhm KO, Lee ES, Lee YM, Kim HS, Park YN, Park SH. Aberrant promoter CpG islands methylation of tumor suppressor genes in cholangiocarcinoma. Oncol Res. 2008;17:151–157. doi: 10.3727/096504008785114110. [DOI] [PubMed] [Google Scholar]
- 12.Lin YW, Chung MT, Lai HC, De Yan M, Shih YL, Chang CC, Yu MH. Methylation analysis of SFRP genes family in cervical adenocarcinoma. J Cancer Res Clin Oncol. 2009;135:1665–1674. doi: 10.1007/s00432-009-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghasemi A, Rostami S, Chahardouli B, Alizad Ghandforosh N, Ghotaslou A, Nadali F. Study of SFRP1 and SFRP2 methylation status in patients with de novo acute myeloblastic leukemia. Int J Hematol Oncol Stem Cell Res. 2015;9:15–21. [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi YH, Iwadate M, Umeyama H. SFRP1 is a possible candidate for epigenetic therapy in non-small cell lung cancer. BMC Med Genomics. 2016;9(Suppl 1):S28. doi: 10.1186/s12920-016-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui C, Ma J, Chen Q, Yang Y. The variation trends of SFRP2 methylation of tissue, feces, and blood detection in colorectal cancer development. Eur J Cancer Prev. 2016;25:288–298. doi: 10.1097/CEJ.0000000000000185. [DOI] [PubMed] [Google Scholar]
- 16.Yang Q, Huang T, Ye G, Wang B, Zhang X. Methylation of SFRP2 gene as a promising noninvasive biomarker using feces in colorectal cancer diagnosis: A systematic meta-analysis. Sci Rep. 2016;6:33339. doi: 10.1038/srep33339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YZ, Liu D, Zhao YX, Wang HT, Gao Y, Chen Y. Aberrant promoter methylation of the SFRP1 gene may contribute to colorectal carcinogenesis: A meta-analysis. Tumour Biol. 2014;35:9201–9210. doi: 10.1007/s13277-014-2180-x. [DOI] [PubMed] [Google Scholar]
- 18.Kloten V, Becker B, Winner K, Schrauder MG, Fasching PA, Anzeneder T, Veeck J, Hartmann A, Knüchel R, Dahl E. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15:R4. doi: 10.1186/bcr3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. J Clin Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 22.Zintzaras E, Ioannidis JP. HEGESMA: Genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinkhuizen T, van den Hurk K, Winnepenninckx VJ, de Hoon JP, van Marion AM, Veeck J, van Engeland M, van Steensel MA. Epigenetic changes in basal cell carcinoma affect SHH and WNT signaling components. PLoS One. 2012;7:e51710. doi: 10.1371/journal.pone.0051710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoehr R, Wissmann C, Suzuki H, Knuechel R, Krieg RC, Klopocki E, Dahl E, Wild P, Blaszyk H, Sauter G, et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84:465–478. doi: 10.1038/labinvest.3700068. [DOI] [PubMed] [Google Scholar]
- 26.Sun J, Chen Z, Zhu T, Yu J, Ma K, Zhang H, He Y, Luo X, Zhu J. Hypermethylated SFRP1, but none of other nine genes ‘informative’ for western countries, is valuable for bladder cancer detection in Mainland China. J Cancer Res Clin Oncol. 2009;135:1717–1727. doi: 10.1007/s00432-009-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Wang H, Bu R, Fei X, Zhao C, Song Y. Methylation and aberrant expression of the Wnt antagonist secreted Frizzled-related protein 1 in bladder cancer. Oncol Lett. 2012;4:334–338. doi: 10.3892/ol.2012.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, Galm O, Camara O, Dürst M, Kristiansen G, et al. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25:3479–3488. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Yang B, Du Z, Gao YT, Wang YJ, Jing X, Bai T. Identification and validation of specific methylation profile in bile for differential diagnosis of malignant biliary stricture. Clin Biochem. 2010;43:1340–1344. doi: 10.1016/j.clinbiochem.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Caldwell GM, Jones C, Gensberg K, Jan S, Hardy RG, Byrd P, Chughtai S, Wallis Y, Matthews GM, Morton DG. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–888. doi: 10.1158/0008-5472.CAN-03-1346. [DOI] [PubMed] [Google Scholar]
- 31.Dhir M, Montgomery EA, Glöckner SC, Schuebel KE, Hooker CM, Herman JG, Baylin SB, Gearhart SL, Ahuja N. Epigenetic regulation of WNT signaling pathway genes in inflammatory bowel disease (IBD) associated neoplasia. J Gastrointest Surg. 2008;12:1745–1753. doi: 10.1007/s11605-008-0633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patai ÁV, Valcz G, Hollósi P, Kalmár A, Péterfia B, Patai Á, Wichmann B, Spisák S, Barták BK, Leiszter K, et al. Comprehensive DNA methylation analysis reveals a common ten-gene methylation signature in colorectal adenomas and carcinomas. PLoS One. 2015;10:e0133836. doi: 10.1371/journal.pone.0133836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salehi R, Mohammadi M, Emami MH, Salehi AR. Methylation pattern of SFRP1 promoter in stool sample is a potential marker for early detection of colorectal cancer. Adv Biomed Res. 2012;1:87. doi: 10.4103/2277-9175.105169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Bauer M, Croner RS, Pelz JO, Lodygin D, Hermeking H, Stürzl M, Hohenberger W, Matzel KE. DNA stool test for colorectal cancer: Hypermethylation of the secreted frizzled- related protein-1 gene. Dis Colon Rectum. 2007;50:1618–1627. doi: 10.1007/s10350-007-0286-6. [DOI] [PubMed] [Google Scholar]
- 35.Rawson JB, Manno M, Mrkonjic M, Daftary D, Dicks E, Buchanan DD, Younghusband HB, Parfrey PS, Young JP, Pollett A, et al. Promoter methylation of Wnt antagonists DKK1 and SFRP1 is associated with opposing tumor subtypes in two large populations of colorectal cancer patients. Carcinogenesis. 2011;32:741–747. doi: 10.1093/carcin/bgr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Domenico M, Santoro A, Ricciardi C, Iaccarino M, Iaccarino S, Freda M, Feola A, Sanguedolce F, Losito S, Pasquali D, et al. Epigenetic fingerprint in endometrial carcinogenesis: The hypothesis of a uterine field cancerization. Cancer Biol Ther. 2011;12:447–457. doi: 10.4161/cbt.12.5.15963. [DOI] [PubMed] [Google Scholar]
- 37.Clement G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett's esophagus. Oncogene. 2006;25:3084–3092. doi: 10.1038/sj.onc.1209338. [DOI] [PubMed] [Google Scholar]
- 38.Ishii T, Murakami J, Notohara K, Cullings HM, Sasamoto H, Kambara T, Shirakawa Y, Naomoto Y, Ouchida M, Shimizu K, et al. Oesophageal squamous cell carcinoma may develop within a background of accumulating DNA methylation in normal and dysplastic mucosa. Gut. 2007;56:13–19. doi: 10.1136/gut.2005.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C, Li N, Lu H, Wang Z, Chen C, Wu L, Liu J, Lu Y, Wang F. Circulating SFRP1 promoter methylation status in gastric adenocarcinoma and esophageal square cell carcinoma. Biomed Rep. 2015;3:123–127. doi: 10.3892/br.2014.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu JB, Qiang FL, Dong J, Cai J, Zhou SH, Shi MX, Chen KP, Hu ZB. Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World J Gastroenterol. 2011;17:4917–4921. doi: 10.3748/wjg.v17.i44.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng Y, Wang QG, Wang JX, Zhu ST, Jiao Y, Li P, Zhang ST. Epigenetic inactivation of the SFRP1 gene in esophageal squamous cell carcinoma. Dig Dis Sci. 2011;56:3195–3203. doi: 10.1007/s10620-011-1734-7. [DOI] [PubMed] [Google Scholar]
- 42.Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, Burgart LJ, Ahlquist DA. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett's esophagus. Int J Cancer. 2005;116:584–591. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- 43.Cheng YY, Yu J, Wong YP, Man EP, To KF, Jin VX, Li J, Tao Q, Sung JJ, Chan FK, Leung WK. Frequent epigenetic inactivation of secreted frizzled-related protein 2 (SFRP2) by promoter methylation in human gastric cancer. Br J Cancer. 2007;97:895–901. doi: 10.1038/sj.bjc.6603968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y, Guo W, Chen Z, Kuang G, Yang Z, Dong Z. Hypermethylation and aberrant expression of Wnt-antagonist family genes in gastric cardia adenocarcinoma. Neoplasma. 2011;58:110–117. doi: 10.4149/neo_2011_02_110. [DOI] [PubMed] [Google Scholar]
- 45.Kinoshita T, Nomoto S, Kodera Y, Koike M, Fujiwara M, Nakao A. Decreased expression and aberrant hypermethylation of the SFRP genes in human gastric cancer. Hepatogastroenterology. 2011;58:1051–1056. [PubMed] [Google Scholar]
- 46.Liu JB, Wu XM, Cai J, Zhang JY, Zhang JL, Zhou SH, Shi MX, Qiang FL. CpG island methylator phenotype and Helicobacter pylori infection associated with gastric cancer. World J Gastroenterol. 2012;18:5129–5134. doi: 10.3748/wjg.v18.i36.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao CH, Bu XM, Zhang N. Hypermethylation and aberrant expression of Wnt antagonist secreted frizzled-related protein 1 in gastric cancer. World J Gastroenterol. 2007;13:2214–2217. doi: 10.3748/wjg.v13.i15.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rettori MM, de Carvalho AC, Longo AL, de Oliveira CZ, Kowalski LP, Carvalho AL, Vettore AL. TIMP3 and CCNA1 hypermethylation in HNSCC is associated with an increased incidence of second primary tumors. J Transl Med. 2013;11:316. doi: 10.1186/1479-5876-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2015;21:6317–6328. doi: 10.3748/wjg.v21.i20.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Q, Stern JE, Hawes SE, Lu H, Jiang M, Kiviat NB. DNA methylation changes in normal liver tissues and hepatocellular carcinoma with different viral infection. Exp Mol Pathol. 2010;88:287–292. doi: 10.1016/j.yexmp.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hua D, Hu Y, Wu YY, Cheng ZH, Yu J, Du X, Huang ZH. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol. 2011;91:455–460. doi: 10.1016/j.yexmp.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Liu JB, Zhang YX, Zhou SH, Shi MX, Cai J, Liu Y, Chen KP, Qiang FL. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J Gastroenterol. 2011;17:4718–4724. doi: 10.3748/wjg.v17.i42.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomoto S, Kinoshita T, Kato K, Otani S, Kasuya H, Takeda S, Kanazumi N, Sugimoto H, Nakao A. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer. 2007;97:1260–1265. doi: 10.1038/sj.bjc.6604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih YL, Shyu RY, Hsieh CB, Lai HC, Liu KY, Chu TY, Lin YW. Promoter methylation of the secreted frizzled-related protein 1 gene SFRP1 is frequent in hepatocellular carcinoma. Cancer. 2006;107:579–590. doi: 10.1002/cncr.22023. [DOI] [PubMed] [Google Scholar]
- 55.Uhm KO, Lee ES, Lee YM, Park JS, Kim SJ, Kim BS, Kim HS, Park SH. Differential methylation pattern of ID4, SFRP1, and SHP1 between acute myeloid leukemia and chronic myeloid leukemia. J Korean Med Sci. 2009;24:493–497. doi: 10.3346/jkms.2009.24.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pehlivan M, Sercan Z, Sercan HO. sFRP1 promoter methylation is associated with persistent Philadelphia chromosome in chronic myeloid leukemia. Leuk Res. 2009;33:1062–1067. doi: 10.1016/j.leukres.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 57.Shen JZ, Xu CB, Fu HY, Wu DS, Zhou HR, Fan LP. Methylation of secreted frizzled related protein gene in acute leukemia patients in China. Asian Pac J Cancer Prev. 2011;12:2617–2621. [PubMed] [Google Scholar]
- 58.Suzuki M, Shigematsu H, Nakajima T, Kubo R, Motohashi S, Sekine Y, Shibuya K, Iizasa T, Hiroshima K, Nakatani Y, et al. Synchronous alterations of Wnt and epidermal growth factor receptor signaling pathways through aberrant methylation and mutation in non small cell lung cancer. Clin Cancer Res. 2007;13:6087–6092. doi: 10.1158/1078-0432.CCR-07-0591. [DOI] [PubMed] [Google Scholar]
- 59.Yoshino M, Suzuki M, Tian L, Moriya Y, Hoshino H, Okamoto T, Yoshida S, Shibuya K, Yoshino I. Promoter hypermethylation of the p16 and Wif-1 genes as an independent prognostic marker in stage IA non-small cell lung cancers. Int J Oncol. 2009;35:1201–1209. doi: 10.3892/ijo_00000437. [DOI] [PubMed] [Google Scholar]
- 60.Zhang YW, Miao YF, Yi J, Geng J, Wang R, Chen LB. Transcriptional inactivation of secreted frizzled-related protein 1 by promoter hypermethylation as a potential biomarker for non-small cell lung cancer. Neoplasma. 2010;57:228–233. doi: 10.4149/neo_2010_03_228. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Wang R, Song H, Huang G, Yi J, Zheng Y, Wan J, Chen L. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303:21–28. doi: 10.1016/j.canlet.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 62.Lee CH, Hung YJ, Lin CY, Hung PH, Hung HW, Shieh YS. Loss of SFRP1 expression is associated with aberrant beta-catenin distribution and tumor progression in mucoepidermoid carcinoma of salivary glands. Ann Surg Oncol. 2010;17:2237–2246. doi: 10.1245/s10434-010-0961-z. [DOI] [PubMed] [Google Scholar]
- 63.Chim CS, Pang R, Fung TK, Choi CL, Liang R. Epigenetic dysregulation of Wnt signaling pathway in multiple myeloma. Leukemia. 2007;21:2527–2536. doi: 10.1038/sj.leu.2404939. [DOI] [PubMed] [Google Scholar]
- 64.Chang Q, Pang JC, Li KK, Poon WS, Zhou L, Ng HK. Promoter hypermethylation profile of RASSF1A, FHIT, and sFRP1 in intracranial primitive neuroectodermal tumors. Hum Pathol. 2005;36:1265–1272. doi: 10.1016/j.humpath.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 65.Al-Shabanah OA, Hafez MM, Hassan ZK, Sayed-Ahmed MM, Abozeed WN, Alsheikh A, Al-Rejaie SS. Methylation of SFRPs and APC genes in ovarian cancer infected with high risk human papillomavirus. Asian Pac J Cancer Prev. 2014;15:2719–2725. doi: 10.7314/APJCP.2014.15.6.2719. [DOI] [PubMed] [Google Scholar]
- 66.Su HY, Lai HC, Lin YW, Chou YC, Liu CY, Yu MH. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int J Cancer. 2009;124:387–393. doi: 10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- 67.Bu XM, Zhao CH, Zhang N, Gao F, Lin S, Dai XW. Hypermethylation and aberrant expression of secreted frizzled-related protein genes in pancreatic cancer. World J Gastroenterol. 2008;14:3421–3424. doi: 10.3748/wjg.14.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Awakura Y, Nakamura E, Ito N, Kamoto T, Ogawa O. Methylation-associated silencing of SFRP1 in renal cell carcinoma. Oncol Rep. 2008;20:1257–1263. [PubMed] [Google Scholar]
- 69.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 70.Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F, Maher ER. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29:2104–2117. doi: 10.1038/onc.2009.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, et al. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–6997. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- 72.Babaei H, Mohammadi M, Salehi R. DNA methylation analysis of secreted frizzled-related protein 2 gene for the early detection of colorectal cancer in fecal DNA. Niger Med J. 2016;57:242–245. doi: 10.4103/0300-1652.188357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bagci B, Sari M, Karadayi K, Turan M, Ozdemir O, Bagci G. KRAS, BRAF oncogene mutations and tissue specific promoter hypermethylation of tumor suppressor SFRP2, DAPK1, MGMT, HIC1 and p16 genes in colorectal cancer patients. Cancer Biomark. 2016;17:133–143. doi: 10.3233/CBM-160624. [DOI] [PubMed] [Google Scholar]
- 74.Chang E, Park DI, Kim YJ, Kim BK, Park JH, Kim HJ, Cho YK, Sohn CI, Jeon WK, Kim BI, et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: A preliminary report in Korean patients. Hepatogastroenterology. 2010;57:720–727. [PubMed] [Google Scholar]
- 75.Huang ZH, Li LH, Yang F, Wang JF. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol. 2007;13:950–954. doi: 10.3748/wjg.v13.i6.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leung WK, To KF, Man EP, Chan MW, Hui AJ, Ng SS, Lau JY, Sung JJ. Detection of hypermethylated DNA or cyclooxygenase-2 messenger RNA in fecal samples of patients with colorectal cancer or polyps. Am J Gastroenterol. 2007;102:1070–1076. doi: 10.1111/j.1572-0241.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 77.Lu H, Huang S, Zhang X, Wang D, Zhang X, Yuan X, Zhang Q, Huang Z. DNA methylation analysis of SFRP2, GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in fecal DNA. Oncol Lett. 2014;8:1751–1756. doi: 10.3892/ol.2014.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Müller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Mühlthaler M, Ofner D, Margreiter R, Widschwendter M. Methylation changes in faecal DNA: A marker for colorectal cancer screening? Lancet. 2004;363:1283–1285. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 79.Park SK, Song CS, Yang HJ, Jung YS, Choi KY, Koo DH, Kim KE, Jeong KU, Kim HO, Kim H, et al. Erratum: Field cancerization in sporadic colon cancer. Gut Liver. 2016;10:981. doi: 10.5009/gnl10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pehlivan S, Artac M, Sever T, Bozcuk H, Kilincarslan C, Pehlivan M. Gene methylation of SFRP2, P16, DAPK1, HIC1, and MGMT and KRAS mutations in sporadic colorectal cancer. Cancer Genet Cytogenet. 2010;201:128–132. doi: 10.1016/j.cancergencyto.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 81.Tang D, Liu J, Wang DR, Yu HF, Li YK, Zhang JQ. Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin Invest Med. 2011;34:E88–E95. doi: 10.25011/cim.v34i1.15105. [DOI] [PubMed] [Google Scholar]
- 82.Wang DR, Tang D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol. 2008;14:524–531. doi: 10.3748/wjg.14.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang W, Wang X, Li X, Wang M, Chen X, Wu X, Wang Y, Fan Y, Jin H. The specific methylation characteristics of cancer related genes in Chinese colorectal cancer patients. Tumour Biol. 2014;35:8267–8279. doi: 10.1007/s13277-014-2100-0. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, Song YF, Lu HN, Wang DP, Zhang XS, Huang SL, Sun BL, Huang ZG. Combined detection of plasma GATA5 and SFRP2 methylation is a valid noninvasive biomarker for colorectal cancer and adenomas. World J Gastroenterol. 2015;21:2629–2637. doi: 10.3748/wjg.v21.i9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samaei NM, Yazdani Y, Alizadeh-Navaei R, Azadeh H, Farazmandfar T. Promoter methylation analysis of WNT/β-catenin pathway regulators and its association with expression of DNMT1 enzyme in colorectal cancer. J Biomed Sci. 2014;21:73. doi: 10.1186/s12929-014-0073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suehiro Y, Okada T, Okada T, Anno K, Okayama N, Ueno K, Hiura M, Nakamura M, Kondo T, Oga A, et al. Aneuploidy predicts outcome in patients with endometrial carcinoma and is related to lack of CDH13 hypermethylation. Clin Cancer Res. 2008;14:3354–3361. doi: 10.1158/1078-0432.CCR-07-4609. [DOI] [PubMed] [Google Scholar]
- 87.Hao XW, Zhu ST, He YL, Li P, Wang YJ, Zhang ST. Epigenetic inactivation of secreted frizzled-related protein 2 in esophageal squamous cell carcinoma. World J Gastroenterol. 2012;18:532–540. doi: 10.3748/wjg.v18.i6.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao C, Wang L, Zhu L, Zhang C, Zhou J. Secreted frizzled- related protein 2 is epigenetically silenced and functions as a tumor suppressor in oral squamous cell carcinoma. Mol Med Rep. 2014;10:2293–2298. doi: 10.3892/mmr.2014.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perry AS, O'Hurley G, Raheem OA, Brennan K, Wong S, O'Grady A, Kennedy AM, Marignol L, Murphy TM, Sullivan L, et al. Gene expression and epigenetic discovery screen reveal methylation of SFRP2 in prostate cancer. Int J Cancer. 2013;132:1771–1780. doi: 10.1002/ijc.27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brebi P, Hoffstetter R, Andana A, Ili CG, Saavedra K, Viscarra T, Retamal J, Sanchez R, Roa JC. Evaluation of ZAR1 and SFRP4 methylation status as potentials biomarkers for diagnosis in cervical cancer: Exploratory study phase I. Biomarkers. 2014;19:181–188. doi: 10.3109/1354750X.2013.867535. [DOI] [PubMed] [Google Scholar]
- 92.Zhao C, Bu X, Zhang N, Wang W. Downregulation of SFRP5 expression and its inverse correlation with those of MMP-7 and MT1-MMP in gastric cancer. BMC Cancer. 2009;9:224. doi: 10.1186/1471-2407-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takeda M, Nagasaka T, Dong-Sheng S, Nishie H, Oka T, Yamada E, Mori Y, Shigeyasu K, Morikawa T, Mizobuchi S, Fujiwara T. Expansion of CpG methylation in the SFRP2 promoter region during colorectal tumorigenesis. Acta Med Okayama. 2011;65:169–177. doi: 10.18926/AMO/46628. [DOI] [PubMed] [Google Scholar]
- 94.Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.