Abstract
Cervical cancer (CC) is one of the leading causes of cancer-associated mortalities in women from developing countries. Similar to other types of cancer, CC is considered to be a multifactorial disease, involving socioeconomic, cultural, immunological and epigenetic factors, as well as persistent human papilloma virus (HPV) infection. It has been well established that cancer stem cells (CSCs) play an important role in defining tumor size, the speed of development and the level of regression following treatment; therefore, CSCs are associated with a poor prognosis. CSCs have been detected in many types of cancer, including leukemia, pancreatic, colon, esophagus, liver, prostate, breast, gastric and lung cancer. In cervical cancer, CSCs have been associated with resistance to normally used drugs such as cisplatin. The present review summarizes the strategies that high-risk HPV viruses (HPV-16 and HPV-18) have developed to transform normal epithelial cells into cancer cells, as well as the cellular pathways and studies associated with the identification of cervical cancer stem cell biomarkers. In this sense, the present review provides state of the art information regarding CC development.
Keywords: cervical cancer stem cells, biomarkers of cancer stem cells, cervical cancer, human papillomavirus
1. Overview
Cancer is the leading cause of mortality from non-infectious diseases worldwide, while mortalities associated with infectious diseases have been declining (reported in 2014) (1). Cancer is defined as a multistep process involving the uncontrolled proliferation of genetically altered cells capable of invading adjacent and distant tissues, via utilization of the blood and lymphatic systems (2,3). Certain types of cancer are more prevalent in developing countries, mainly due to intrinsic factors such as poverty, late or incorrect diagnosis (1,4), infectious diseases, and other factors including obesity, physical inactivity, alcoholism and smoking (5).
In 2015, >8.8 million cancer-associated mortalities were reported worldwide; 70% of which occurred in low and middle-income countries and 25% of these were induced by viral infections, such as hepatitis virus or human papilloma virus (HPV) (6,7). HPV-16 and HPV-18 are the main biological factors associated with the development of cervical cancer (CC), and are the third most common cause of malignant tumors. According to the 2018 report of GLOBOCAN, CC remains the fourth most common cause of cancer-associated mortalities among women worldwide (6,7), and one of the most fatal types of cancer among the female population in developing countries (7–9). In Mexico, CC is the second most common cause of cancer-associated mortality in women (2018), primarily due to poor clinical diagnosis during the early stages of disease (7,10). Previous reports between 2015 and 2017 revealed that HPV-16 and HPV-18 are responsible for 65–75% of precancerous cervical lesions and are present in 99% of patients with CC worldwide (9,11–13). However, it has also been established that HPV infections do not necessarily trigger CC (9,12,13). In this sense, a significant number of factors are involved; contraceptives pills, multiple sexual partners, multiple births, obesity, smoking, alcoholism, poor diet, immunosuppressive cervical microenvironment, abnormal vaginal microbiota, co-infections with Chlamydia trachomatis or human immunodeficiency virus and the presence of cervical cancer stem cells (CCSCs) (9,12,14–19).
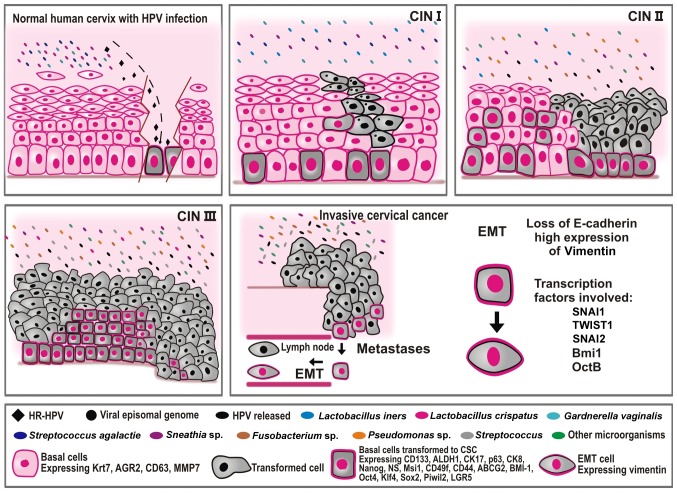
Regarding abnormal vaginal microbiota, certain published reports have indicated remarkable changes in the microenvironment of patients during the late stages of the disease. Therefore, patients positive for HPV exhibit more complex bacterial diversity than a healthy subject. Reports on patients with HPV frequently exhibit the presence of Lactobacillus iners; however, Sneathia spp., Megasphaera elsdenii and Shuttleworthia satelles were commonly found in cervical intraepithelial neoplasia (CIN) cases (9,19,20–22). Therefore, some of these microbes could be considered as a microbiological marker associated with the HPV-infection and CIN. Besides, Fusobacterium spp. is the most abundant microorganism found in CC cases (19). Figs. 1 and 2 illustrate the changes in the cervical microbiota through the progression of CIN I to CC.
Figure 1.
An overview of factors involved in the progression of CC. The figure illustrates cervical cancer progression as a consequence of the multiple factors and steps involved. CC is initiated by persistent infection with HR HPV (type 16 or type 18) that reaches the basal cells of the transformation zone, turning it into CCSCs. The microorganisms present during this stage of the recent infection in a healthy cervix are much less diverse and much less complex than those reported in more advanced stages of the disease, such as in CIN II, CIN III, and CC. Lactobacillus spp. are the principal microorganisms observed in a healthy cervix and in CIN stage I, and are the first line of defense against pathogenic microorganisms. As the CIN advances, more toxic microorganisms are observed in the cervix such as Fusobacterium, Sneathia and Streptococcus. HPV, human papilloma virus; CIN, cervical intraepithelial neoplasia; CC, cervical cancer; CCSCs, cervical cancer stem cells; CSCs, cancer stem cells; SNAI1, snail family transcriptional repressor 1; SNAI2, snail family transcriptional repressor 2; TWIST1, twist family bHLH transcription factor 1; BMI1, BMI1 polycomb ring finger proto-oncogene; Oct 4, octamer-binding transcription factor 4B; Krt7, keratin 7; AGR2, anterior gradient protein 2 homolog; Sox2, sex determining region Y-box 2; Klf4, Krüppel-like factor 4; ALDH1, aldehyde dehydrogenase 1; CD, cluster of differentiation; CK, cytokeratin; NS, Nucleostemin; MSI1, musashi RNA binding protein 1; ABCG2, adenosine triphosphate-binding cassette subfamily G member 2; PIWIL2, piwi like RNA-mediated gene silencing 2; LGR5, Leucine-rich repeat-containing G-protein-coupled receptor 5; HR, high-risk; EMT, epithelial-mesenchymal transition; MMP, matrix metalloproteinase; ABCG2, adenosine triphosphate-binding cassette subfamily G member 2.
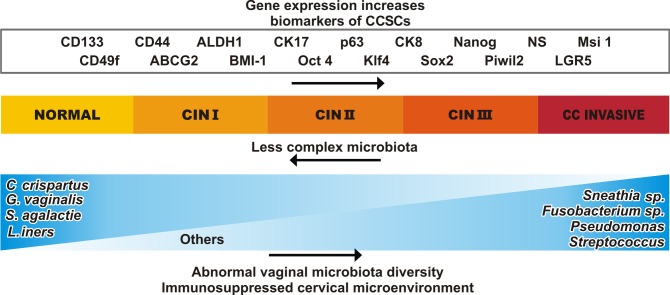
Figure 2.
Major changes occur in the expression of genes and the cervical microenvironment as CIN I progress to CC. It has been observed that certain biomarkers of CCSCs exhibit increased expression and concentration levels, such as NANOG, SOX2 and KLF4, and there is also an increase in markers that are associated with CCSCs such as, CD133, Cd44, ALDH1, CK17, p63, CK8, NS, MSI1, CD49f, ABCG2, BMI1, PIWIL2 and LGR5. Many of these markers have been reported in other types of CSCs. In the cervical microenvironment, the complexity and quantity of the microbiota are altered. As the disease progresses, the genus Lactobacillus tends to disappear and instead, microorganisms as Fusobacterium sp., Sneathia sp., Pseudomonas and Streptococcus appear. CIN, cervical intraepithelial neoplasia; CC, cervical cancer; CCSCs, cervical cancer stem cells; CD, cluster of differentiation; CK, cytokeratin; NS, Nucleostemin; MSI1, musashi RNA binding protein 1; PIWIL2, piwi like RNA-mediated gene silencing 2; LGR5, Leucine-rich repeat-containing G-protein-coupled receptor 5; BMI1, BMI1 polycomb ring finger proto-oncogene; ABCG2, adenosine triphosphate-binding cassette subfamily G member 2; KLF4, Krüppel-like factor 4.
Although a number of genetic and molecular events require further clarification in cervical carcinogenesis, it is currently accepted that HPV viruses depend on epithelial cell differentiation to establish their progeny and spread their viral genes (11–13). HPV has a more significant opportunity to infect basal cells with stem cell-like properties and integrate its viral DNA into the genome of these cells, driving the oncogenic transformation into CCSCs (13,23). The infection occurs where the ectocervix and endocervix join, known as the transformation zone (TZ) or the squamocolumnar junction (12,13,24). The TZ has a unique morphology and genetic expression profile, expressing keratin 7, anterior gradient 2, CD63 and matrix metalloproteinase 7, as well as other genes (24). Another essential factor in cervical carcinogenesis is the epithelial-mesenchymal transition (EMT). EMT is a crucial process for the generation of invasive cells and metastasis and it is regulated by snail family transcriptional repressors 1 (SNAI1) and 2 (SNAI2), and twist family bHLH transcription factor 1 (TWIST1) transcription factors. The EMT is a rich source of CSCs and its induction promotes metastasis, tumor cell invasion and drug resistance. EMT-specific changes include the loss of E-cadherin expression and upregulation of N-cadherin and Vimentin (25–28).
On the other hand, the presence of CCSCs is another factor that is equally important for triggering the development of CC, as they provide several malignant characteristics to the tumor, such as the proliferation and metastasis capacity, resistance to radiation and cisplatin-based chemotherapy. Currently there are no therapies capable of eliminating CCSCs, and their presence results in poor prognosis in CC. In this sense, development of dual therapies against CC could eliminate both primary cancer cells and CCSCs. Specific biomarkers from CSCs are being considered as promising targets for the development of new therapies in many types of cancer. In the present review, the leading factors involved in cervical cancer development were investigated, with a particular emphasis on specific biomarkers reported in CCSC.
2. HPV: A key factor in CC development
HPV consists of double-stranded and circularized DNA that contains non-coding, long control regions and eight open reading frames that encode viral proteins located in the ‘early’ and ‘late’ regions (8,29). The early stages of HPV DNA replication occur in proliferative basal epithelial cells, yielding low viral copy numbers (8). However, the progeny of basal cells replicates and move into the spinous cells layer where viruses mature; allowing the expression of late capsid proteins and the release of infectious virions within desquamated cells (8).
Role of viral proteins
Early HPV genes are involved in the synthesis of proteins associated with replication and maintenance of the viral genome. In addition, late genes encode proteins associated with the formation of the viral coat.
E1 is a DNA helicase required for viral replication in host cells. E1 establishes the viral genome as a multicopy episome in the nucleus of cells and can arrange protein-protein and protein-nucleic acid interactions. In this sense, p80, human SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1, histone H1, E1 binding protein and p56, are essential in order to link the DNA replication network of the host cell with the replication origin of the virus. Given the role that E1 serves in viral DNA replication and its immediate interaction with the host machinery, there has been speculation regarding its participation in the development of cervical cancer (30–33).
E2 is considered the central transcriptional regulator of the papilloma viruses as it interacts with E1 when DNA replication begins, and because this protein scatters the viral episomal genomes during the division of the infected cells. E2 mediates the interactions between the viral genome and chromatin adapter proteins at the point of mitotic division and indirectly regulates transcription of E6 and E7 producing several effects on the cell cycle that affect proliferation, differentiation, apoptosis and senescence (11,34–36).
E4 is associated with successful virus release and can also be used as a biomarker of active HPV infections. E4 is found between the early replication of HPV genes; however, this protein is also expressed during the late stages of the infection (37–40).
The E5 protein presents oncogenic activities in cultured cells and animals, and it is thought to play an important role in the productive virus cycle. The E5 protein has also been reported to modulate the activity of cellular proteins, interacting with targets such as Bax or NF-κB, which induce cell proliferation, apoptosis and senescence (41–43).
E6 is expressed when transformed cells migrate to the spinous cell layer. E6 stimulates protein synthesis by increasing translation through rapamycin (mTOR) complex 1, which enhances the 5′ mRNA cap translation initiation-complex. It was reported that loss of E6 resulted in poor maintenance of the HPV genome due to the failure in p53 degradation. E6 avoids apoptotic responses by binding to procaspase-8, and it is also able to inhibit the responses of interferon by downregulating multiple interferon response genes (44–47).
E7 induces HPV viral replication by reprograming the cellular environment; together with E6, it induces a potent transformation activity in the host cells. The influence of E7 is observed in many cellular processes including viral replication, transformation, cell cycle and cell death, through direct or indirect interactions with a large number of proteins (48–52). E6 and E7 induce the degradation of p53 tumor suppressor protein via the ubiquitin-proteasome pathway, triggering uncontrolled proliferation of the infected cell population (15,47,48).
L1 protein is comprised of the icosahedral surface of the HPV virions and it is an initial point of contact between host cells and the virus. L1 is flexible enough to release the viral genome into a new target cell; however, assembly of the new virions occurs only in fully differentiated keratinocytes that are ready to flake (53–56). Furthermore, L2 also participates in papillomavirus assembly, initiating the infection process. L2 helps HPV DNA encapsidation and it is a significant component of the late-stage proteins. Therefore, L1 and L2 proteins are involved in virion assembly and in early events of infection; for this reason, they could be used as potential vaccine antigens (29,57–59).
3. CSCs
The term ‘cell’, originally proposed in 1665 by the English scientist Robert Hooke, has been redefined (60). Currently, researchers recognize the existence of a group of cells with the capacity to renew with high plasticity. SCs differentiate into the most appropriate lineage, depending on the stimuli that they receive from the surrounding microenvironment. In addition to self-renewing, SCs can generate specialized cells with limited proliferative capacity (61).
The function of this subset of cells is to maintain tissue homeostasis during daily turnover and regenerate tissue injuries (62). With the discovery of humans SCs, even in adults, two new challenges have emerged: i) Developing laboratory protocols that allow the isolation of SCs in sufficient amounts; and ii) understanding the molecular mechanisms that define the fate of these cells (27,63,64). The identification and isolation of SCs have been performed taking advantage of molecular biomarkers consisting of differentially expressed proteins (13). However, the expression levels of these markers change significantly depending on the environmental conditions, particularly in in vitro cultures (64,65). Malignant SCs are called CSCs; these cells share some characteristics with normal SCs; such as self-renewal, differentiation, high expression of telomerase, apoptosis evasion and the ability to migrate (66). CSCs are also able to transport substances like drugs throughout the membrane, via the membrane transport protein adenosine triphosphate-binding cassette subfamily G member 2 (ABCG2), which leads to the recurrence of disease in patients following surgery and chemotherapy (13,23,24,27,52).
Theories regarding the origin of CSCs
Given the similarities between SCs and CSCs, several theories exist regarding the origin of CSCs. DNA mutations can affect somatic cells and SCs; therefore, essential alterations can target SCs and produce CSCs with high tumorigenic capacity. Another possibility is regarding the DNA mutations that target stemness genes in malignant progenitor cells, transforming progenitor cells into CSC (13,24). However, the possibility exists that CSCs could be present in a dormant state until the initiation of carcinogenesis (23,24,67).
CSC and metastases
As aforementioned, CSCs are involved in promoting cancer metastasis due to their migratory and invasion capacity to distant organs. Previous studies have demonstrated that tumor-associated CSCs are associated with adverse outcomes and high rates of metastasis (23,25,26,68,69). EMT is considered to be the primary source of metastatic cells. The transformation of this cell is induced by transcription factors, such as SNAI1, SNAI2, TWIST1 and BMI1 polycomb ring finger proto-oncogene (BMI1), which are highly expressed in CSCs. The EMT produces a loss of epithelial adhesion and apical-basal polarity, allowing the release of transformed cells with CSC characteristics into the circulatory system (25–27). Fig. 1 illustrates the cells undergoing EMT, resulting in metastasis.
4. Biomarkers in CCSCs
Certain specific markers have been reported for CSCs, including CD44, CD90, CD133, CD271, epithelial cell adhesion molecule and aldehyde dehydrogenase 1 (ALDH1). However, there is no set of universal biomarkers to identify and isolate CSCs (23,27,65–74). Therefore, the main method to study CSCs is through the ‘side population’ (SP), which is a small subpopulation within the tumor mass present in some cases (up to 20%) (75). The SP exhibits CSC-like characteristics, such as the ability to initiate tumors, development of resistance to chemotherapeutic drugs, and its potential as a predictor of patient outcome. Therefore the SP could be an alternative source for studying CSCs with unknown biomarkers (75,76). Analyses on the basis of the ability of the cells to efflux the fluorescent dye Hoechst and provide a system to identify multipotent SCs (76). The spheroid cell formation assay can also be used as it is based on the capacity of CSCs to grow and form spheres in non-adherent conditions. The principal focus is to analyze and compare the expression of surface markers via fluorescence-activated cell sorting (FACS), confocal microscopy, immunohistochemistry (IHC), reverse transcription (RT)-quantitative PCR and the isolation of CSCs for tumorigenic efficiency tests and tumor subpopulations analysis in animal models (25,27,65,67).
Therefore, CCSC populations exhibit characteristic expression profiles and cell surface markers that (on tumor tissues of patients) make their isolation possible in vitro and in vivo, as well as their evaluation, and research into the progression of cancer will make creating dual therapies directed towards specific targets of the CCSC possible in the near future. Among the main proteins expressed in CCSC are cytokeratin (CK) and CDs.
CKs −5, −8, −13, −17, −18 and −19 are proteins expressed in reserve cells and the immature squamous metaplastic cells of the cervix. CK19 was described by Wang et al (77), where it was revealed that the expression of CK19 in CC was significantly higher than that observed in patients with benign lesions. In addition, high levels of CK19 expression were identified by RT-PCR in the CCSCs of sentinel lymph nodes from patients with CC (77). In a study by Ikeda et al (78), CK8 and CK17 were investigated by performing IHC on the tissues of patients with different grades of CIN and CC. Therefore, the authors concluded that CK8 and CK17 were expressed in CIN and CC tissues, and that CK17 was associated with metastatic processes and the development of highly malignant diseases (78). Thus, CK17 and CK19 could be considered as biomarkers of CCSCs via the primary cultivation of CC clinical samples (79). In addition, analysis of main and side population using Hoechst 33342 dye, flow cytometry sorting method, as well as tumor formation in nude mice revealed that the side population presented higher tumorigenicity and CSC characteristics (79).
CD44 and CD133 proteins have been broadly accepted as general CSC markers in many types of tumor. CD44 and CD133 transmembrane glycoproteins are involved in normal cellular processes (13) and also in cancer cell migration, aggregation and tumor development (13,24,72). Therefore, these glycoproteins can be used as surface markers to isolate several types of CSCs including breast, prostate, pancreas, colorectal, gastric and cervical cancer (13,72,80–82). On the other hand, CD49f is a highly expressed protein in CCSC and it is advantageous in its identification and isolation.
López et al (74) analyzed the presence of certain surface markers in sphere cells derived from HeLa, SiHa, CaSki and C-41 cell lines grown at low density (1,000 cells/ml) in serum-free medium and discovered an increase in CD49f and CD133 positive cells when compared with the monolayer cells.
Ortiz-Sánchez et al (69) reported characteristic phenotypes of putative CCSC including, CD49f+, CK17, p63+, AII+ and ALDH; the sphere culture exhibited a stemness state characterized by the presence of OCT4, NANOG and β-catenin. In addition, it was observed that the presence of CD49f and AII was associated with the possibility that HR-HPV can infect normal cervical cells (SCs). Ortiz-Sánchez et al (69) also demonstrated the high tumorigenic capacity of ALDHbright cells when compared with ALDHlow cells.
Tyagi et al (83) demonstrated the role of viral oncoprotein E6 in SC signaling and the maintenance of stemness in CC. CSCs that expressed a set of phenotypic markers including CD49f, ABCG2, CD71 and CD133 were isolated from primary cervical tumors and cancer-derived cell lines grown as spheres. In addition, transcripts of self-renewal and stemness markers including OCT4, SOX2, NANOG, leucine-rich repeat-containing G protein-coupled receptor 1 (LGR1) and CD133 were identified, along with the overexpression of E6, Hes family bHLH transcription factor 1, a protein involved in enhancing self-renewal properties and the ability of tumorsphere formation (83).
Hou et al (84) reported that there was a correlation between CCSC markers in patient tissue samples and the prediction of CIN prognosis. The authors analyzed paraffin-embedded surgical samples through IHC, snap-frozen CC samples and normal cervical samples using RT-PCR for CD49f, SOX2, ALDH1 and musashi RNA binding protein 1 (MSI1). In this sense, patients with tumors classified as high MSI1 and low CD49f expression had the poorest prognosis, whereas tumors without MSI1 and CD49f upregulation had the best prognosis in CC. Hou et al (84) reported, for the first time, clinical evidence regarding CCSC markers associated with the prognosis of patients with CC.
Other proteins differentially expressed in CCSC are NANOG, Nucleostemin (NS), MSI1, TWIST, nestin, ALDH1, BMI1, piwi-like RNA-mediated gene silencing 2 (PIWIL2), TIMP metallopeptidase inhibitor 4 (TIMP4), LGR5, OCT and SOX.
Ye et al (85) reported the role of NANOG, NS and MSI1 in cervix carcinogenesis and progression of cervical carcinoma by performing IHC analysis on 235 paraffin-embedded samples with normal cervical epithelia, CIN-I, -II and -III; and CC. The CINs were staged as follows (86): I, low-grade lesion with mildly atypical cellular changes in the lower third of the epithelium (mild dysplasia); CIN II, high-grade lesion with moderately atypical cellular changes confined to the basal two-thirds of the epithelium (moderate dysplasia); and CIN III, high-grade lesion with severely atypical cellular changes encompassing the full thickness of the epithelium (severe dysplasia). Ye et al (85) observed high levels of expression of the three proteins in CC, CIN -II and -III, and low expression levels in CIN I and normal cervical epithelia. However, there was no correlation among NANOG, NS and MSI1 expression levels and the prognosis of CC. In addition, NANOG, NS and MSI1 also have a critical role in the carcinogenesis of glioma, liver, gastric and other types of cancer (85).
In 2011, Li and Zhou (87) revealed the activation of Wnt/β-catenin and Akt signaling pathways in TWIST-overexpressing cells that had CSC characteristics, such as tumorsphere formation and ALDH1 and CD44 expression. The study reported that TWIST is an inducer of morphological changes associated with EMT. In addition, the spheroid cells gained expression of human actin α-cardiac muscle 1 (also known as α-smooth muscle actin) and Vimentin mesenchymal markers. Functional analysis demonstrated that spheroid cells are more resistant than monolayer cells to paclitaxel (87). On the other hand, knockdown of β-catenin expression by small interfering RNA transfection and Akt signaling pathway inhibition by the PI3K/Akt inhibitor wortmannin, suppressed the expression of CD44 (87).
Sato et al (88), determined the role of Nestin in CIN and CC via IHC and in situ hybridization analysis. Nestin is usually expressed in the brain; however, it was also identified in CSCs, several metastasized carcinomas and in types of cancer with poor prognosis. In this study, Sato et al (88) analyzed tissue samples from 26 cases of each stage of CIN and 55 of CC. The results revealed low Nestin expression levels in the basal squamous epithelium of CIN I; however, in CIN II Nestin was present in 65% of the cases. Nestin was found in most of the cases of CIN III localized in the squamous epithelium.
Furthermore, Nestin was detected in all of the invasive CC samples. In addition, Sato et al (88) analyzed the effects of Nestin overexpression via transfection of ME-180 cells; a metastatic cancer cell line derived from the cervix. Overexpression of Nestin induced a higher capacity to form spheres, produce a CD44high/CD24low pattern associated with the biomarkers found in breast CSCs, increased expression of ALDH, NANOG and OCT4. According to these results, the authors concluded that Nestin may be involved in the progression of CIN to CC and could be associated with the regulation of CSCs due to its ability to stimulate sphere formation in vitro (88).
ALDH1 is a cytosolic isoenzyme normally involved in retinol oxidation to produce retinoic acid. Liu and Zheng (89) reported high levels of ALDH1 in CCSCs. The authors observed that patients exhibited high ALDH1 activity level in a subpopulation of CC cases with a great capacity for self-renewal, high differentiation potential, and high tumorigenicity, just like CSCs. The authors used tools such as FACS and functional assays in xenografted NOD/SCID mice, as well as cultures of CC cells expressing high and low levels of ALDH1 in serum-free media to promote tumor sphere cell formation. The authors concluded that ALDH1 may function as a CSCC marker. In the study by Liu and Zheng (89), cells with high ALDH1 activity were resistant to cisplatin and produced a high expression of OCT4, NANOG, Krüppel-like factor 4 (KLF4) and BMI1.
Liu et al (90) demonstrated that SOX2-positive CC cells share all the characteristics with CSCs including self-renewal, differentiation and tumor-initiating properties. Additionally, SOX2-positive cervical cells increased the levels of OCT4, BMI1 and ALDH1 stemness markers, as well as Vimentin, SNAI1 and β-catenin (mesenchymal SCs markers) (91). For this reason, the authors concluded that SOX2 could be a critical factor in self-renewal, pluripotency and also a stemness factor needed for SCs and CCSCs differentiation (90).
Lizarraga et al (92) described the role of TIMP4 in the stemness of CC cells. TIMP4 is a tissue inhibitor that has been overexpressed in a number of cancer cell lines and in nude mice, to evaluate its function in carcinogenesis. The results by Lizarraga et al (92) revealed a faster tumor formation in nude mice that overexpressed TIMP4 in CC cells. Notably, activation of NF-κB signaling pathway and the increasing CSC population led to a high expression of pluripotency markers as OCT3/4 and SOX2, the EMT markers SNAI1 and Vimentin and the drug efflux transporters markers ABCG1 and ABCG2 (92).
Feng et al (93) demonstrated the role of PIWIL2 in CC tumorigenesis. They observed that PIWIL2 expression was present in the HPV+ CC cell lines HeLa, SiHa and CaSki, and was undetectable in HPV− cancer cell line C33A. Knockdown of PIWIL2 by short hairpin RNA in HeLa and SiHa cells decreased the tumorigenic, proliferation and chemoresistant capacity of these cells. On the other hand, overexpression of PIWIL2 in HaCat cells activated tumor-initiating capabilities and cMyc, KLF4, NANOG, OCT4 and SOX2 cell reprogramming factors were upregulated (93). Feng et al (93) also demonstrated that PIWIL2 reactivation by E6 and E7 oncoproteins is essential in the transformation of cervical epithelial cells into CSCs. PIWIL2 was highly expressed in CIN II, CIN III and CC, but its expression in healthy tissue and CIN I was low. PIWIL2 also suppressed the expression of P53 and P21 in CC cell lines, inducing cervical carcinogenesis (93).
Cao et al (94) previously reported the principal role of LGR5 in CC for the activation of Wnt/β-catenin signaling pathway. In addition, this study reported the role of LGR5 in CCSCs via overexpressing and silencing its effect in CC cell lines. Thus, they showed that overexpression of LGR5 induces CSC characteristics including tumorsphere formation, increased tumorigenic capacities in vivo, chemoresistance to cisplatin, augmented cell migration and invasion, and upregulate the expression of stem cell-associated transcription factors in vitro. LGR5-overexpression in HeLa and SiHa cells was revealed as being correlated with higher expression levels of BMI1, NANOG, OCT4 and KLF4, when compared with control or silenced cells (94). Figs. 1 and 2 summarize leading factors involved in the progression of CC, the significant changes in genetic expression from CCSC as well as the changes in microbiota population, throughout CC development and progression.
5. CSC signaling pathway
SCs and CSCs share Hedgehog (Hh), Notch, Wnt, NF-κB and PI3K/Akt/mTOR signaling pathways. Hh is an essential pathway for self-renewal and cell fate; it is associated with tumorigenesis, development and the progression of certain types of cancer including the maintenance of CSCs (95). The Hh signaling pathway drives stemness in CSCs via the genetic regulation of OCT4, SOX2 and BMI1 (96,97). Regarding CC, Hh has been associated with poor outcomes in irradiated patients and evidence has suggested that Hh is involved in the repopulation of cervical cells following chemoradiation (98).
The Notch signaling pathway regulates proliferation, stem cell maintenance, cell fate specification, differentiation and angiogenesis (99). In addition, the Notch signaling pathway is involved in cell-cell communication through transmembrane ligands and receptors (100). The canonical pathway involves five canonical Notch ligands: ∆-like canonical Notch ligand (DLL) 1, DLL3, DLL4, Jagged1 and Jagged2, as well as four receptor paralogs. Cancer cells express different Notch receptors and ligands (100) and the noncanonical pathway may also have some relevance in cancer. Thus, both pathways can control several types of tumor-associated cells, including CSCs or immune cells. Furthermore, expression of different Notch paralogs varies between different types of tumor (101). In CC, Notch can exhibit different roles depending on the disease progression: i) Maintaining immature epithelium by preventing terminal differentiation; ii) increasing NOTCH expression during the progress of CIN into CC; iii) regulation of Notch signaling pathway by E6 in CC cell lines; and iv) silencing Jagged1 in CaSki cells inhibits its tumorigenic capacity (101,102).
The Wnt signaling pathway is involved in cell proliferation and differentiation during embryogenesis. From Wnt signaling cascades, the canonical path is the best studied for its involvement in cancer development. There are many reports regarding CSC contribution to the maintenance of these cells through the Wnt-β-catenin signaling pathway (103–106). In CC, apoptosis is induced and tumor growth is inhibited if the Wnt signaling pathway is repressed. On the other hand, overactivation of Wnt/β-catenin signaling pathway is associated with cervical tumorigenesis with HPV infection (105,107). In the basal layer, Wnt ligands are required for sustaining the undifferentiated state of SCs. The majority of genetic mutations in colorectal cancer activate the Wnt signaling pathway, and CSCs are the most susceptible to transformation by these mutations (28,108).
NF-κB serves a pivotal role in HPV infected cells. This signaling pathway is involved in cancer development by regulating several oncogenic genes (109). The NF-κB signaling pathway has two routes: The canonical pathway that depends on the inhibitor of NF-κB kinase complex (IκB), and the non-canonical pathway, which is activated when the homodimer of IkB, inhibitor of nuclear factor kappa-B kinase subunit α (IKKα) is phosphorylated. IKKα is part of the IκB complex, and is associated with the growth, metastases and stemness of several types of cancer (110,111).
In the PI3K/Akt/mTOR signaling pathway, PI3K and mTOR serve a critical role in cell proliferation, angiogenesis, metabolism, differentiation and survival (112). This signaling pathway is usually activated when mTOR is not correctly regulated in cancer conditions. PI3K overexpression has been reported in ovarian cancer and CC (113,114). PI3K/Akt/mTOR is an essential signaling pathway to regulate self-renewal and the maintenance of stemness in SCs and CSCs. The role of CSCs is well recognized in prostate cancer (66,115); however, the mechanism through PI3K/Akt/mTOR signaling regulates CSC is unknown.
6. Conclusions
A large number of studies have sought to expand the current knowledge of the molecular pathogenesis of CC and the progression of viral infections leading to this invasive cancer. The majority of these studies have attempted to verify that the cause of CC is the HPV infection; however, recent studies have aimed to determine the factors and changes at the molecular level that are involved with the stemness and development of CC. In this sense, a growing amount of evidence has identified the CCSCs as a novel, fundamental and strategic key factor to be considered in cancer development, chemotherapy resistance and cancer regression. Regarding the origin of CCSCs, the best-accepted hypothesis involves the transformation of SCs, via HPV infection by using their E6 and E7 oncoproteins. However, there are a large number of proteins whose natural activity is the maintenance of stemness in healthy cells. Nevertheless, SOX2, NANOG, OCT4, Klf4 and Nestin proteins are also involved in maintaining the stemness of CSCs.
ABCG2, SNAI1, Vimentin and LGR5 are other proteins that are also involved in maintenance, although it is not their primary role. This evidence confirms the existence of a complex network of regulation in CC development and progression. Thus, searching biomarkers at each disease stage is essential to improve our current knowledge for the development and application of novel therapies and to allow for an accurate CC diagnoses.
Currently, several research groups are making a great effort to identify new targets involved in the stemness of cervical cancer cells. Therefore, genes, proteins and signaling pathways are under consideration. In this sense, the information described in the present review collates some potential candidates to accomplish the difficult task of controlling CCSC development. Thus, CD44, CD133 and CD49f could be an excellent target to direct the therapeutic efforts and block Hedgehog, PI3K/Akt/mTOR, Wnt or Notch signaling pathways in CCSC.
Acknowledgements
The authors would like to thank Dr Michelle Quezada from The University of Newcastle (Newcastle, UK) and Mr Yael Vargas for their assistance in editing the manuscript, and also Mr Humberto Vallejo for his support with the figure design.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
GMA, EOS, LRZ, CRS, EEI and JO were all responsible for reviewing the cited literature and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Barondess JA. Scanning the chronic disease terrain: Prospects and opportunities. Trans Am Clin Climatol Assoc. 2014;125:45–56. [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahasrabuddhe V, Luhn P, Wentzensen N. Human papillomavirus and cervical cancer: Biomarkers for improved prevention efforts. Future Microbiol. 2011;6:1083–1098. doi: 10.2217/fmb.11.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latino-Martel P, Cottet V, Druesne-Pecollo N, Pierre FH, Touillaud M, Touvier M, Vasson MP, Deschasaux M, Le Merdy J, Barrandon E, Ancellin R. Alcoholic beverages, obesity, physical activity and other nutritional factors, and cancer risk: A review of the evidence. Crit Rev Oncol Hematol. 2016;99:308–323. doi: 10.1016/j.critrevonc.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 7.International agency for Research on Cancer. http://gco.iarc.fr Global Cancer Observatory.
- 8.Scheurer ME, Tortolero-Luna G, Adler-Storthz K. Human papillomavirus infection: Biology, epidemiology, and prevention. Int J Gynecol Cancer. 2015;15:727–746. doi: 10.1111/j.1525-1438.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 9.Kyrgiou M, Mitra A, Moscicki AB. Does the vaginal microbiota play a role in the development of cervical cancer? Transl Res. 2017;179:168–182. doi: 10.1016/j.trsl.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Secretariat of Health, Mexico, corp-author. Statistics of breast cancer and uterine cervical cancer. https://www.gob.mx/salud/acciones-y-programas/informacion-estadistica. 2015
- 11.Domínguez-Catzín V, Reveles-Espinoza AM, Sánchez-Ramos J, Cruz-Cadena R, Lemus-Hernández D, Garrido E. HPV16-E2 protein modifies self-renewal and differentiation rate in progenitor cells of human immortalized keratinocytes. Virol J. 2017;14:65. doi: 10.1186/s12985-017-0736-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chhabra R. Cervical cancer stem cells: Opportunities and challenges. J Cancer Res Clin Oncol. 2015;141:1889–1897. doi: 10.1007/s00432-014-1905-y. [DOI] [PubMed] [Google Scholar]
- 13.Yao T, Lu R, Zhang Y, Zhang Y, Zhao C, Lin R, Lin Z. Cervical cancer stem cells. Cell Prol. 2015;48:611–625. doi: 10.1111/cpr.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ncube B, Bey A, Knight J, Bessler P, Jolly PE. Factors associated with the uptake of cervical cancer screening among women in portland, Jamaica. N Am J Med Sci. 2015;7:104–113. doi: 10.4103/1947-2714.153922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jungbauer F, Aderhold C, Birk R, Hoermann K, Kramer B, Kuhlin B, Thorn C, Umbreit C, Lammert A. Communicate or Die-A Model for HPV+ and HPV- CSCs and their interactions with SDF-1α. Anticancer Res. 2017;37:4827–4836. doi: 10.21873/anticanres.11890. [DOI] [PubMed] [Google Scholar]
- 16.American Cancer Society. What are the risk factors for cervical cancer? http://www.cancer.org/cancer/cervicalcancer/moreinformation/cervicalcancerpreventionandearlydetection. 2014
- 17.Alfaro KM, Gage JC, Rosenbaum AJ, Ditzian LR, Maza M, Scarinci IC, Miranda E, Villalta S, Felix JC, Castle PE, Cremer ML. Factors affecting attendance to cervical cancer screening among women in the Paracentral Region of El Salvador: A nested study within the CAPE HPV screening program. BMC Public Health. 2015;15:1058. doi: 10.1186/s12889-015-2360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngugi CW, Boga H, Muigai AW, Wanzala P, Mbithi JN. Factors affecting uptake of cervical cancer early detection measures among women in Thika, Kenya. Health Care Women Int. 2012;33:595–613. doi: 10.1080/07399332.2011.646367. [DOI] [PubMed] [Google Scholar]
- 19.Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, López-Estrada G, Delgado-Romero K, Burguete-García AI, Cantú D, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PLoS One. 2016;11:e0153274. doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra A, Maclntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, Bhatia R, Lyons D, Paraskevaidis E, Li JV, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep. 2015;5:16865. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra A, Maclntyre DA, Marchesi JR, Lee YS, Benett PR, Kyrgiou M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: What do we know and where are we going next? Microbiome. 2016;4:58. doi: 10.1186/s40168-016-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Da M, Zhang W, Qi Q, Zhang C, Han S. Role of Lactobacillus in cervical cancer. Cancer Manag Res. 2018;10:1219–1229. doi: 10.2147/CMAR.S165228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang R, Rofstad E. Cancer stem cells (CSCs), cervical CSCs and targeted therapies. Oncotarget. 2017;8:35351–35367. doi: 10.18632/oncotarget.10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao QX, Yao TT, Zhang BZ, Lin RC, Chen ZL, Zhou H, Wang LJ, Lu HW, Chen Q, Di N, Lin Z. Expression and functional role of ALDH1 in cervical carcinoma cells. Asian Pac J Cancer Prev. 2012;13:1325–1331. doi: 10.7314/APJCP.2012.13.4.1325. [DOI] [PubMed] [Google Scholar]
- 25.López J, Ruíz G, Organista-Nava J, Gariglio P, García-Carrancá A. Human papillomavirus infections and cancer stem cells of tumors from the uterine cervix. Open Virol J. 2012;6:232–240. doi: 10.2174/1874357901206010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Liu X, Ding D. Evidence for epithelial-mesenchymal transition in cancer stem-like cells derived from carcinoma cell lines of the cervix uteri. Int J Clin Exp Pathol. 2015;8:847–855. [PMC free article] [PubMed] [Google Scholar]
- 27.Yang MH, Imrali A, Heeschen C. Circulating cancer stem cells: The importance to select. Chin J Cancer Res. 2015;27:437–449. doi: 10.3978/j.issn.1000-9604.2015.04.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 29.Wang JW, Roden RB. L2, the minor capsid protein of papillomavirus. Virology. 2013;445:175–186. doi: 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo SR, Liu JS, Broker TR, Chow LT. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J Biol Chem. 1994;269:24058–24065. [PubMed] [Google Scholar]
- 31.Sanders CM, Kovalevskiy OV, Sizov D, Lebedev AA, Isupov MN, Anston AA. Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res. 2007;35:6451–6457. doi: 10.1093/nar/gkm705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egawa N, Nakahara T, Ohno S, Narisawa-Saito M, Yugawa T, Fujita M, Yamato K, Natori Y, Kiyono T. The E1 protein of human papillomavirus type 16 is dispensable for maintenance replication of the viral genome. J Virol. 2012;86:3276–3283. doi: 10.1128/JVI.06450-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergvall M, Melendy T, Archambault J. The E1 proteins. Virology. 2013;445:35–56. doi: 10.1016/j.virol.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin MT, Hirochika R, Hirochika H, Broker TR, Chow LT. Regulation of human papillomavirus type 11 enhancer and E6 promoter by activating and repressing proteins from the E2 open reading frame: Functional and biochemical studies. J Virol. 1988;62:2994–3002. doi: 10.1128/jvi.62.8.2994-3002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou SY, Wu SY, Zhou T, Thomas MC, Chiang CM. Alleviation of human papillomavirus E2-mediated transcriptional repression via formation of a TATA binding protein (or TFIID)-TFIIB-RNA polymerase II-TFIIF preinitiation complex. Mol Cell Biol. 2000;20:113–125. doi: 10.1128/MCB.20.1.113-125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride AA. The Papillomavirus E2 proteins. Virology. 2013;445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology. 2007;368:219–226. doi: 10.1016/j.virol.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borgogna C, Zavattaro E, de Andrea M, Griffin HM, Dell'Oste V, Azzimonti B, Landini MM, Peh WL, Pfister H, Doorbar J, et al. Characterization of beta papillomavirus E4 expression in tumours from Epidermodysplasia Verruciformis patients and in experimental models. Virology. 2012;423:195–204. doi: 10.1016/j.virol.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 39.Griffin H, Wu Z, Marnane R, Dewar V, Molijin A, Quint W, Van Hoof C, Struyf F, Colau B, Jenkins D, Doorbar J. E4 antibodies facilitate detection and type-assignment of active HPV infection in cervical disease. PLoS One. 2012;7:e49974. doi: 10.1371/journal.pone.0049974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80–98. doi: 10.1016/j.virol.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Lehman JM, Petti LM. Apoptosis of mortal human fibroblasts transformed by the bovine papillomavirus E5 oncoprotein. Mol Cancer Res. 2002;1:122–136. [PubMed] [Google Scholar]
- 42.Venuti A, Paolini F, Nasir L, Corteggio A, Roperto S, Campo MS, Borzacchiello G. Papillomavirus E5: The smallest oncoprotein with many functions. Mol Cancer. 2011;10:140. doi: 10.1186/1476-4598-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Maio D, Petti LM. The E5 proteins. Virology. 2013;445:99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. 2003;22:5938–5945. doi: 10.1038/sj.onc.1206894. [DOI] [PubMed] [Google Scholar]
- 45.Ansari T, Brimer N, Vande Pol SB. Peptide interactions stabilize and restructure human papillomavirus type 16 E6 to interact with p53. J Virol. 2012;86:11386–11391. doi: 10.1128/JVI.01236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanier K, ould M'hamed ould Sidi A, Boulade-Ladame C, Rybin V, Chapelle A, Atkinson A, Kieffer B, Travé G. Solution structure analysis of the HPV16 E6 oncoprotein reveals a self-association mechanism required for E6-mediated degradation of p53. Structure. 2012;20:604–617. doi: 10.1016/j.str.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaughlin-Drubin ME, Bromberg-White JL, Meyers C. The role of the human papillomavirus type 18 E7 oncoprotein during the complete viral life cycle. Virology. 2005;338:61–68. doi: 10.1016/j.virol.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 49.McLaughlin-Drubin ME, Huh KW, Münger K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J Virol. 2008;82:8695–8705. doi: 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLaughlin-Drubin ME, Münger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384:335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLaughlin-Drubin ME, Crum CP, Münger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLaughlin-Drubin ME, Meyers J, Munger K. Cancer associated human papillomaviruses. Curr Opin Virol. 2012;2:459–466. doi: 10.1016/j.coviro.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schäfer F, Florin L, Sapp M. DNA binding of L1 is required for human papillomavirus morphogenesis in vivo. Virology. 2002;295:172–181. doi: 10.1006/viro.2002.1361. [DOI] [PubMed] [Google Scholar]
- 54.Doorbar J. The papillomavirus life cycle. J Clin Virol. 2005;32(Suppl 1):S7–S15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Day PM, Lowy DR, Schiller JT. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J Virol. 2008;82:12565–12568. doi: 10.1128/JVI.01631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buck CB, Day PM, Trus BL. The papillomavirus major capsid protein L1. Virology. 2013;445:169–174. doi: 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirnbauer R, Chandrachud LM, O'Neil BW, Wagner ER, Grindlay GJ, Armstrong A, McGarvie GM, Schiller JT, Lowy DR, Campo MS. Virus-like particles of bovine papillomavirus type 4 in prophylactic and therapeutic immunization. Virology. 1996;219:37–44. doi: 10.1006/viro.1996.0220. [DOI] [PubMed] [Google Scholar]
- 58.Rubio I, Seitz H, Canali E, Sehr P, Bolchi A, Tommasino M, Ottonello S, Müller M. The N-terminal region of the human papillomavirus L2 protein contains overlapping binding sites for neutralizing, cross-neutralizing and non-neutralizing antibodies. Virology. 2001;409:348–359. doi: 10.1016/j.virol.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 59.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 60.Kutschera U. Founding fathers: The cell was defined 150 years ago. Nature. 2011;480:457. doi: 10.1038/480457c. [DOI] [PubMed] [Google Scholar]
- 61.Nava MM, Raimondi MT, Pietrabissa R. Controlling self-renewal and differentiation of stem cells via mechanical cues. J Biomed Biotechnol 2012. 2012:797410. doi: 10.1155/2012/797410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ge Y, Fuchs E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet. 2018;19:311–325. doi: 10.1038/nrg.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maruyama T. Stem/progenitor cells and the regeneration potentials in the human uterus. Reprod Med Biol. 2009;9:9–16. doi: 10.1007/s12522-009-0032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zapata AG, Alfaro D, García-Ceca J. Biology of stem cells: The role of microenvironments. Adv Exp Med Biol. 2012;741:135–151. doi: 10.1007/978-1-4614-2098-9_10. [DOI] [PubMed] [Google Scholar]
- 65.Yang B, Lu Y, Zhang A, Zhou A, Zhang L, Zhang L, Gao L, Zang Y, Tang X, Sun L. Doxycycline induces apoptosis and inhibits proliferation and invasion of human cervical carcinoma stem cells. PLoS One. 2015;10:e0129138. doi: 10.1371/journal.pone.0129138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aponte PM, Caicedo A. Stemness in cancer: Stem cells, cancer stem cells, and their microenvironment. Stem Cells Int 2017. 2017:5619472. doi: 10.1155/2017/5619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clevers H. The cancer stem cell: Premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 68.Kobayashi NC, Noronha SM. Cancer stem cells: A new approach to tumor development. Rev Assoc Med Bras (1992) 2015;61:86–93. doi: 10.1590/1806-9282.61.01.086. [DOI] [PubMed] [Google Scholar]
- 69.Ortiz-Sánchez E, Santiago-López L, Cruz-Domínguez VB, Toledo-Guzmán ME, Hernández-Cueto D, Muñiz-Hernández S, Garrido E, Cantú De León D, García-Carra A. Characterization of cervical cancer stem celllike cells: Phenotyping, stemness, and human papillomavirus co-receptor expression. Oncotarget. 2016;7:31943–31954. doi: 10.18632/oncotarget.8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, Chou SH, Chien CS, Ku HH, Lo JF. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14:4085–4095. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 71.Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Q, Shi S, Yen Y, Brown J, Ta JQ, Le AD. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett. 2010;289:151–160. doi: 10.1016/j.canlet.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 73.Murillo-Sauca O, Chung MK, Shin JH, Karamboulas C, Kwok S, Jung Y, Oakley R, Tysome JR, Farnebo LO, Kaplan MJ, et al. CD271 is a functional and targetable marker of tumor-initiating cells in head and neck squamous cell carcinoma. Oncotarget. 2014;5:6854–6866. doi: 10.18632/oncotarget.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.López J, Poitevin A, Mendoza-Martínez V, Pérez-Plasencia C, García-Carrancá A. Cancer-initiating cells derived from established cervical cell lines exhibit stem-cell markers and increased radioresistance. BMC Cancer. 2012;12:48. doi: 10.1186/1471-2407-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu C, Alman BA. Side population cells in human cancers. Cancer Lett. 2008;268:1–9. doi: 10.1016/j.canlet.2008.03.048. [DOI] [PubMed] [Google Scholar]
- 76.Richard V, Nair MG, Santhosh Kumar TR, Pillai MR. Side population cells as prototype of chemoresistant, tumor-initiating cells. Biomed Res Int. 2013 doi: 10.1155/2013/517237. 2013:517237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang HY, Sun JM, Lu HF, Shi DR, Ou ZL, Ren YL, Fu SQ. Micrometastases detected by cytokeratin 19 expression in sentinel lymph nodes of patients with early-stage cervical cancer. Int J Gynecol Cancer. 2006;16:643–648. doi: 10.1111/j.1525-1438.2006.00381.x. [DOI] [PubMed] [Google Scholar]
- 78.Ikeda K, Tate G, Suzuki T, Mitsuya T. Coordinate expression of cytokeratin 8 and cytokeratin 17 immunohistochemical staining in cervical intraepithelial neoplasia and cervical squamous cell carcinoma: An immunohistochemical analysis and review of the literature. Gynecol Oncol. 2008;108:598–602. doi: 10.1016/j.ygyno.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y, Wang M, Zeng Q, Lv Y, Bao B. Isolation and biological characteristics of human cervical cancer side population cells. Int J Clin Exp Pathol. 2017;10:869–876. [Google Scholar]
- 80.Takaishi S, Okumura T, Tu S, Wang S, Shibata W, Vingneshwaran R, Gordon SA, Shimada Y, Wang TC. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Su YJ, Lai HM, Chang YW, Chen GY, Lee JL. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 2011;30:3186–3199. doi: 10.1038/emboj.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hiraga T, Ito S, Nakamura H. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res. 2013;73:4112–4122. doi: 10.1158/0008-5472.CAN-12-3801. [DOI] [PubMed] [Google Scholar]
- 83.Tyagi A, Vishnoi K, Mahata S, Verma G, Srivastava Y, Masaldan S, Roy BG, Bharti AC, Das BC. Cervical cancer stem cells selectively overexpress HPV oncoprotein E6 that controls stemness and self-renewal through upregulation of HES1. Clin Cancer Res. 2016;22:4170–4184. doi: 10.1158/1078-0432.CCR-15-2574. [DOI] [PubMed] [Google Scholar]
- 84.Hou T, Zhang W, Tong C, Kazobinka G, Huang X, Huang Y, Zhang Y. Putative stem cell markers in cervical squamous cell carcinoma are correlated with poor clinical outcome. BMC Cancer. 2015;15:785. doi: 10.1186/s12885-015-1826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye F, Zhou C, Cheng Q, Shen J, Chen H. Stem-cell-abundant proteins nanog, nucleostemin and musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer. 2008;8:108. doi: 10.1186/1471-2407-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.The 1988 Bethesda System for reporting cervical/vaginal cytological diagnoses. National Cancer Institute Workshop. JAMA. 1989;262:931–934. doi: 10.1001/jama.262.7.931. [DOI] [PubMed] [Google Scholar]
- 87.Li J, Zhou BP. Activation of β-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer. 2011;11:49. doi: 10.1186/1471-2407-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sato A, Ishiwata T, Matsuda Y, Yamammoto T, Asakura H, Takeshita T, Naito Z. Expression and role of nestin in human cervical intraepithelial neoplasia and cervical cancer. Int J Oncol. 2012;41:441–448. doi: 10.3892/ijo.2012.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu SY, Zheng PS. High aldehyde dehydrogenase activity identifies cancer stem cells in human cervical cancer. Oncotarget. 2013;4:2462–2475. doi: 10.18632/oncotarget.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu XF, Yang WT, Xu R, Liu JT, Zheng PS. Cervical cancer cells with positive Sox2 expression exhibit the properties of cancer stem cells. PLoS One. 2014;9:e87092. doi: 10.1371/journal.pone.0087092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mei W, Lin X, Kapoor A, GU Y, Zhao K, Tang D. the contributions of prostate cancer stem cells in prostate cancer initiation and metastasis. Cancers (Basel) 2019;11(pii):E434. doi: 10.3390/cancers11040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lizarraga F, Espinosa M, Ceballos-Cancino G, Vazquez-Santillan K, Bahena-Ocampo I, Schwarz-Cruz Y Celis A, Vega-Gordillo M, Garcia Lopez P, Maldonado V, Melendez-Zajgla J. Tissue inhibitor of metalloproteinases-4 (TIMP-4) regulates stemness in cervical cancer cells. Mol Carcinog. 2016;55:1952–1961. doi: 10.1002/mc.22442. [DOI] [PubMed] [Google Scholar]
- 93.Feng D, Yan K, Zhou Y, Liang H, Liang J, Zhao W, Dong Z, Ling B. Piwil2 is reactivated by HPV oncoproteins and initiates cell reprogramming via epigenetic regulation during cervical cancer tumorigenesis. Oncotarget. 2016;7:64575–64588. doi: 10.18632/oncotarget.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cao HZ, Liu XF, Yang WT, Chen Q, Zheng PS. LGR5 promotes cancer stem cell traits and chemoresistance in cervical cancer. Cell Death Dis. 2017;8:e3039. doi: 10.1038/cddis.2017.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Batsaikhan BE, Yoshikawa K, Kurita N, Iwata T, Takasu C, Kashihara H, Shimada M. Cyclopamine decreased the expression of Sonic Hedgehog and its downstream genes in colon cancer stem cells. Anticancer Res. 2014;34:6339–6344. [PubMed] [Google Scholar]
- 97.Cochrane CR, Szczepny A, Watkins DN, Cain JE. Hedgehog signaling in the maintenance of cancer stem cells. Cancers (Basel) 2015;7:1554–1585. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rofstad EK, Sundfør K, Lyng H, Tropé CG. Hypoxia-induced treatment failure in advanced squamous cell carcinoma of the uterine cervix is primarily due to hypoxia-induced radiation resistance rather than hypoxia-induced metastasis. Br J Cancer. 2000;83:354–359. doi: 10.1054/bjoc.2000.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 101.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Venkatesh V, Nataraj R, Thangaraj G, Karthikeyan M, Gnanasekaran A, Kaginelli SB, Kuppanna G, Kapalla CG, Basalingappa KS. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Invest. 2018;5:5. doi: 10.21037/sci.2018.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu AJ, Watt FM. Beta-catenin signalling modulates proliferative potential of human epidermal keratinocytes independently of intercellular adhesion. Development. 1999;126:2285–2298. doi: 10.1242/dev.126.10.2285. [DOI] [PubMed] [Google Scholar]
- 104.Andrade AC, Nilsson O, Barnes KM, Baron J. Wnt gene expression in the post-natal growth plate: Regulation with chondrocyte differentiation. Bone. 2007;40:1361–1369. doi: 10.1016/j.bone.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: Turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 107.Ji J, Wei X, Wang Y. Embryonic stem cell markers Sox-2 and OCT4 expression and their correlation with WNT signal pathway in cervical squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:2470–2476. [PMC free article] [PubMed] [Google Scholar]
- 108.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van de Born M, Danenberg E, Clarke AR, Sanson OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 109.Gonzalez-Torres C, Gaytan-Cervantes J, Vazquez-Santillan K, Mandujano-Tinoco EA, Ceballos-Cancino G, Garcia-Venzor A, Zampedri C, Sanchez-Maldonado P, Mojica-Espinosa R, Jimenez-Hernandez LE, Maldonado V. NF-κB participates in the stem cell phenotype of ovarian cancer cells. Arch Med Res. 2017;48:343–351. doi: 10.1016/j.arcmed.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 110.Affara NI, Coussens LM. IKKalpha at the crossroads of inflammation and metastasis. Cell. 2007;129:25–26. doi: 10.1016/j.cell.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 111.Lüningschrör P, Kaltschmidt B, Kaltschmidt C. Knockdown of IKK1/2 promotes differentiation of mouse embryonic stem cells into neuroectoderm at the expense of mesoderm. Stem Cell Rev Rep. 2012;8:1098–1108. doi: 10.1007/s12015-012-9402-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 114.Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, Liu JM, Yang DM, Yang WK, Shen CY. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–2744. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 115.Xia P, Xu X. PI3K/Akt/mTOR signaling pathway in cancer stem cells: From basic research to clinical application. Am J Cancer Res. 2015;5:1602–1609. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.