Fetal hemoglobin (HbF) is the major modulator of the phenotype of sickle cell anemia and increased concentrations can reduce disease severity.(1, 2) HbF levels are controlled by genetic elements linked to the haplotype of the HBB gene cluster and by trans-acting quantitative trait loci (QTL). Variants in BCL11A and HBS1L-MYB were strongly associated with HbF levels in many populations of diverse race and ethnicity including cohorts of patients with sickle cell anemia, and the functional elements of these loci have been identified.(3) (4–6) ZBTB7A (LRF) suppresses γ-globin gene (HBG2, HBG1) expression.(7) HbF levels were 70% after ZBTB7A knockout in a human erythroid cell line (HUDEP-2) that normally expressed adult HbA on terminal differentiation.(7) Knocking out both ZBTB7A and BCL11A increased HbF to more than 90% of total hemoglobin. ZBTB7A and BCL11A acted independently as HBG silencers. In previous genome-wide association studies (GWAS) variants of ZBTB7A or in linkage disequilibrium (LD) with this gene were not associated with HbF.(8–12) Because of the profound effect of ZBTB7A on HbF expression we asked whether polymorphisms of this gene or its promoters and proximal enhancer elements, and in putative binding motifs for ZBTB7A in and adjacent to the HBB gene cluster are associated with HbF levels in sickle cell anemia.
Genetic data from GWAS and next generation sequencing from diverse patients who were homozygous for the sickle hemoglobin gene and had varying levels of HbF were available for analysis (Table 1). Included were 21 individuals studied by whole genome sequencing (WGS) and 15 Saudi Benin and 8 Saudi AI haplotype homozygotes studied using exome sequencing GWAS were available from 822 African American HbS homozygotes of diverse haplotypes (Cooperative Study of Sickle Cell Disease or CSSCD cohort), 104 Saudi AI haplotype homozygotes who originated from the Eastern Province and 71 Saudi Benin haplotype homozygotes who originated from the Southwestern Province. GWAS data were also imputed to the 1000 Genomes (Phase 3) reference. All studies were approved by the Institutional Review Boards of the participating institutions.
Table 1.
Cohorts analyzed. Fourteen Saudi and 3 Indian AI haplotype HbS homozygotes, 3 African Americans with the Benin haplotype (selected because of their unusually high HbF) and 1 African American with the Senegal haplotype had whole genome sequencing (WGS). Saudi AI patients who had WGS all had the same minor alleles for BCL11A and MYB. The other cohorts included African Americans of diverse HBB haplotypes who were participants in the Cooperative Study of Sickle Cell Disease (CSSCD) and Saudi patients with the AI and Benin haplotype. The approximate number of variants represented on the 2 Illumina arrays used for genome-wide association studies (GWAS) is shown in parentheses.
| Cohorts | n | Age (y) | HbF (%) |
|---|---|---|---|
| Saudi Benin Haplotype-Exome-Seq | 15 | 21.5±10.5 | 9.8±3.7 |
| Saudi Benin Haplotype-GWAS (700K) | 100 | 18.6±11.0 | 10.8±4.6 |
| African American Benin-WGS | 3 | 26.0±3.6 | 19.8±0.4 |
| African American Senegal- WGS | 1 | 5.9 | 16.0 |
| Saudi AI Haplotype-Exome-Seq | 8 | 32.3±11.1 | 12.7±5.5 |
| Saudi AI Haplotype cohort 1-GWAS(600K) | 42 | 26.4±11.1 | 17.6±5.018 |
| Saudi AI Haplotype cohort 2-GWAS(700K) | 62 | 23.9±9.6 | 18.8±7.5 |
| Saudi AI Haplotype-WGS | 7 | 25.9±6.8 | 23.5±2.6 |
| Saudi AI Haplotype-WGS | 7 | 34.1±10.3 | 8.2±1.3 |
| Indian AI haplotype-WGS | 3 | 22.7±5.5 | 26.0±4.5 |
| CSSCD-GWAS (600K) | 822 | 13.6±11.3 | 5.2±5.6 |
To detect variants in ZBTB7A and its promoters or putative proximal enhancers that might be associated with HbF we searched 100 kb upstream and 100 kb downstream of the ZBTB7A coding sequences using data from WGS. Using IMPUT2 the SNPs in this region were imputed to the 1000 Genomes (Phase 3) reference panel in the CSSCD and Saudi cohorts that were studied by GWAS in order to test associations of HbF with these variants using an additive genetic model. The most significant association with HbF in the CSSCD cohort did not pass the correction for multiple testing (rs114623325, p-value 0.002). Two of the 23 Saudi patients studied by exome sequencing with HbF levels of 6.9% and 14.3% were heterozygous for a CGC insertion polymorphism (transcript NM_015898, c.539_540insCGC (p.Ala181_Ser182insAla) that appeared to be a neutral variant. These data suggest that it is unlikely than common variants in ZBTB7A or its promoters and proximal enhancers accounted for HbF variation in sickle cell anemia.
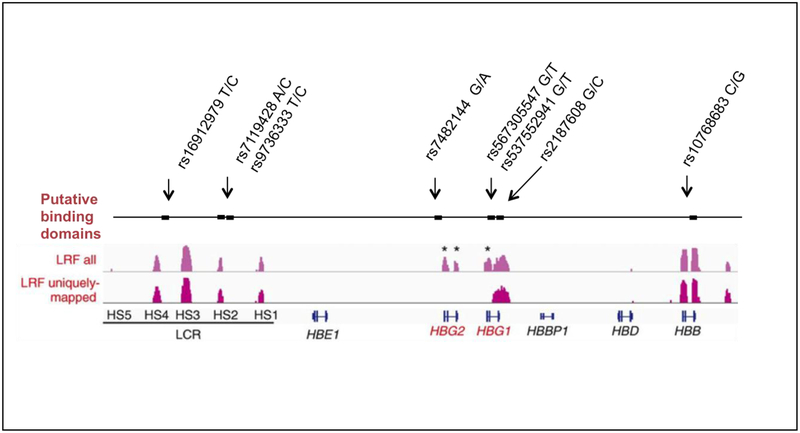
ZBTB7A affects HbF gene silencing through its binding in and about the HBB gene cluster. Accordingly, using the permissive motif ([GAC][ACG][GTAC][AC]CC[CAG][CTA]) as a target, we searched for variants in these putative binding motifs in the interval downstream of OR51V1 (5’ olfactory receptor gene cluster) and upstream of OR51B4 (3’ olfactory gene cluster) flanking the HBB-like genes and its locus control region (LCR) in chromosome 11p15.5. Within the putative binding motifs we found 8 motif-modifying SNPs. The binding domains identified by bioinformatic analysis overlapped some of the ZBTB7A binding occupancy data based on ChIPseq signals that were previously reported.(7) Figure 1 displays the position of these SNPs in putative binding motifs along with the experimentally defined ZBTB7A occupancy regions. Table 2 provides additional details of these SNPs. Rs16912979 is present in hypersensitive site (HS)-4 of the LCR; and rs7119428 and rs9736333, which are in perfect LD, are located in HS-2. The alternative allele (C) of both HS-2 SNPs, was present only in African American sickle cell anemia samples of diverse African-origin haplotypes and are in adjacent potential ZBTB7A binding sites at coordinates 11:5302080–5302087 and 11:5302057–11:5302064. HS-2 and HS-4 also contain GATA1 and TAL1 binding sites that were shown to be involved in hemoglobin switching. The alternative C allele of rs7119428 that was found in HS-2 had a minor allele frequency of about 0.22 in the HapMap Yoruban sample and 0.17 in another African population reported in dbSNP; the major A allele was monomorphic in Europeans, Asians and Indians as reported in dbSNP. Rs7119428, which was not represented on the Human610-Quad array (~600,000 SNPs), was imputed with very high confidence in cohorts studied with this chip (Table 1). The C allele had a frequency of 0.56 in 822 cases of the African American CSSCD cohort, in which all HBB haplotypes except for the AI haplotype were represented. This SNP was included on the Illumina Human Omni Express BeadChip (~700,000 SNPs). Its allele frequency was 0.88 in Saudis with the Benin haplotype who were studied using this chip. There was a significant association of this variant with HbF in the CSSCD cohort after adjusting for age and sex (beta coefficient = -0.05, p-value 0.02) but the effect was small. In 15 Saudi patients with the Benin haplotype who were heterozygous for the C allele, HbF was 13.7%; 85 C homozygotes had HbF of 10.2% however this difference was not significant (p-value 0.14). These preliminary results suggest that the C alleles of both rs7119428 and rs9736333 in HS-2, and also other SNPs in LD with these alleles might be play some role in the decreased HbF in sickle cell anemia of African descent compared with AI haplotype sickle cell anemia. Although rs7119428 was rare in Saudi AI sickle cell anemia we found 3 heterozygotes for the C allele and their HbF was about 10% compared with nearly 20% in homozygotes for the common A allele.
Figure 1.
Approximate locations of the putative binding motifs for ZBTB7A (first track) that contained polymorphisms (shown above track) and were located between OR51V1 in the 5’ olfactory receptor gene cluster and OR51B4 in the 3’ olfactory gene cluster flanking the HBBlike genes and its locus control region (LCR) on chromosome 11p15.5. SNPs in the binding motifs are shown (arrows) with their major/alternative alleles. ChIP-seq data (tracks 2 and 3) is taken from(7) along with the relative locations of globin genes and the LCR (track 4). Extensive homology between HBG2 and HBG1 makes mapping to these regions difficult and ChIP-seq data only showed uniquely-mapped binding in HBG1 (track 3)
Table 2.
SNPs in putative ZBTB7A binding motifs in the HBB gene cluster (RSID) along with their genomic locations (v37), reference sequence (ref-seq and variation according to haplotype (Benin, Senegal, AI). Underlined and bolded are the variant alleles. S denotes positive (+) or negative (-) strand location of the putative binding motif.
| site | location (v37) | S | Ref-seq | Benin | Senegal | AI | RSID | REF allele | ALT allele |
|---|---|---|---|---|---|---|---|---|---|
| HS-4 | 11:5309693–5309700 | + | GGTCCCCA | GGCCCCCA | GGCCCCCA | GGTCCCCA | rs16912979 | T | C |
| HS-2 | 11:5302080–5302087 | − | AGGGGCCT | CGGGGCCT | AGGGGCCT | AGGGGCCT | rs7119428 | A | C |
| HS-2 | 11:5302057–5302064 | − | GGGGGTGG | GGGGGCGG | GGGGGTGG | GGGGGTGG | rs9736333 | T | C |
| HBG2 5' | 11:5276167–5276174 | + | GGGACCGT | GGGACCGT | GGAACCGT | GGAACCGT | rs7482144 | G | A |
| HBG1 intron 1 | 11:5270900–5270907 | − | AGGGTCCT | AGTTTCCT | AGGGTCCT | AGGGTCCT | rs567305547 rs537552941 |
G | T |
| HBG1 intron 2 | 11:5269931–5269938 | + | GCCACCAT | GCCACCAT | CCCACCAT | CCCACCAT | rs2187608 | G | C |
| HBB intron 2 | 11:5247791–5247798 | + | CGTCCCAT | GGTCCCAT | GGTCCCAT | GGTCCCAT | rs10768683 | C | G |
A putative ZBTB7A binding motif was found 5’ to HBG2 although this region was devoid of uniquely mapped binding sites as reported in (7). This motif contained rs7482144 (G/A), the well-studied Xmn1 restriction site that is associated with high HbF in the African Senegal and AI haplotypes.(13) Variants were also present in HBG1 and HBB. The T allele of rs567305547 and rs537552941 in the small intron of HBG1, with an alternative allele frequency of about 0.40 in Africans, was found in 2 of 3 African Benin haplotype samples studied with WGS. All AI haplotype patients were monomorphic for the G allele, a result confirmed by Sanger sequencing in a subset of cases. These 2 intronic SNPs were not on the haplotype reference panel used for imputation and therefore they could not be imputed so any association with HbF could not be tested.
SNPs in putative ZBTB7A binding sites distinguish the high HbF AI haplotype from African origin haplotypes of sickle cell anemia where HbF is usually lower. They are present in sites with characteristics of active enhancers like transcription factor binding and epigenetic marks and are therefore candidates for the functional elements of this haplotype. Perhaps the variants of this haplotype alter looping of the LCR to globin gene promoters, however mechanistic studies are required to validate these genetic associations.(14, 15)
Acknowledgments:
Funded in part by the University of Dammam, SP 11/2011, Office of Collaboration and Knowledge Exchange, University of Dammam, and R01 HL 068970, RC2 HL 101212, R01 87681 (MHS), T32 HL007501 (EMS), T32 GM074905 (KB) from the NIH Bethesda, MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest disclosure:The authors declare no competing interests.
References
- 1.Alsultan A, Alabdulaali MK, Griffin PJ, Alsuliman AM, Ghabbour HA, Sebastiani P, et al. Sickle cell disease in Saudi Arabia: the phenotype in adults with the Arab-Indian haplotype is not benign. Br J Haematol. 2014; 164(4):597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330(23):1639–44. [DOI] [PubMed] [Google Scholar]
- 3.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013. 11;342(6155):253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae H, Baldwin CT, Sebastiani P, Telen MJ, Ashley-Koch A, Garrett ME, et al. Metaanalysis of 2040 sickle cell anemia patients: BCL11A and HMIP are the major genetic modifiers of HbF in African Americans. Blood. 2012;120:1961–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrell JJ, Sherva RM, Chen ZY, Luo HY, Chu BF, Ha SY, et al. A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood. 2011;117(18):4935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest. 2014. 1;124(4):1699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016; 351(6270):285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solovieff N, Milton JN, Hartley SW, Sherva R, Sebastiani P, Dworkis DA, et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5’ olfactory receptor gene cluster. Blood. 2010;115(9):1815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA. 2008;105(5):1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mtatiro SN, Singh T, Rooks H, Mgaya J, Mariki H, Soka D, et al. Genome wide association study of fetal hemoglobin in sickle cell anemia in Tanzania. PLoS One. 2014;9(11):e111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ngo D, Bae H, Steinberg MH, Sebastiani P, Solovieff N, Baldwin CT, et al. Fetal hemoglobin in sickle cell anemia: genetic studies of the Arab-Indian haplotype. Blood Cells Mol Dis. 2013;51(1):22–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA. 2008;105(33):11869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagel RL, Fabry ME, Pagnier J, Zohoun I, Wajcman H, Baudin V, et al. Hematologically and genetically distinct forms of sickle cell anemia in Africa. The Senegal type and the Benin type. N Engl J Med. 1984;312:880–4. [DOI] [PubMed] [Google Scholar]
- 14.Kim YW, Lee S, Yun J, Kim A. Chromatin looping and eRNA transcription precede the transcriptional activation of gene in the beta-globin locus. Biosci Rep. 2015;35(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun WJ, Kim YW, Kang Y, Lee J, Dean A, Kim A. The hematopoietic regulator TAL1 is required for chromatin looping between the beta-globin LCR and human gamma-globin genes to activate transcription. Nucl Acids Res. 2014;42(7):4283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]