Abstract
Microbiomes play critical roles in ecosystems and human health, yet in most cases scientists lack standardized and reproducible model microbial communities. The development of fabricated microbial ecosystems, which we term EcoFABs, will provide such model systems for microbiome studies.
Microorganisms are ubiquitous on Earth, and their activities are critical to the health and survival of all life. Highly complex microbial communities, consisting of hundreds to hundreds of thousands of different taxa, inhabit essentially all natural ecosystems. Within each ecosystem, these dynamic communities exhibit tremendous compositional and functional heterogeneity over time and space[1, 2]. Although direct characterization of native microbiomes provides the most relevant data for understanding their biology and ecology, such studies are typically limited by their high cost, their limited replicability and the inability to use reductionistic technologies such as engineered microbes.
The identification, development and adoption of common model organisms, such as Escherichia coli, Saccharomyces cerevisiae, Arabidopsis thaliana, Drosophila melanogaster, Danio rerio and Mus musculus[3], has led to rapid advances in the understanding of molecular pathways and cellular life by enabling reproducible observations and targeted interventions. Laboratory experiments have also proven to be a powerful tool for microbiome science[4, 5], yielding many important insights into the ecology of microbes, including Dallinger’s studies of ecological specialization in the 1880s[6]; Gause’s examination of resource competition in the 1930s that led to the competitive exclusion principle[7]; important insights into the spatiotemporal assembly, architecture and communication in model flow-cell oral biofilms in the 1990s[8]; and many others[9, 10].
Critical insights into the importance of microbiomes for host health and disease have been obtained with defined microbiota and germ-free hosts, such as D. melanogaster, D. rerio and M. musculus[3]. Gnotobiotic mouse facilities[11] in particular have been used to show that gut microbes regulate host serotonin biosynthesis[12], that microbiota from obese mice have a greater capacity to harvest energy from diet compared with those from lean mice, and that gnotobiotic mice colonized by microbiota from obese mice have significantly more body fat than those colonized by microbes from lean mice[13]. However, gnotobiotic mouse facilities are expensive to operate and are not widely accessible. Further, standardized and reproducible systems are largely nonexistent in other areas of microbiome sciences[14].
We assert that there is an urgent need for the microbiome community to develop a few widely accepted standardized microbial ecosystems coupled with standardized workflows, computational tools, data standards and computational models (Fig. 1). Such reproducible fabricated ecosystems (EcoFABs; see http:\\eco-fab.org) will enable scientists to reproduce and build on each other’s research (Fig. 1). We propose that, following in the great tradition of model organisms[15], it will be possible to advance microbiome sciences through the reproducible interrogation of microbial community activities within relevant ecological frameworks. We envision that these microbial model systems will complement existing model organisms, clinical studies and field studies. Such model microbial communities should be developed to address the health or environmental research priorities outlined by the larger community, for example, studying plant–microbe interactions for agricultural research[16].
Fig. 1. Vision for fabricated model microbial ecosystems and their impact on microbiome science.
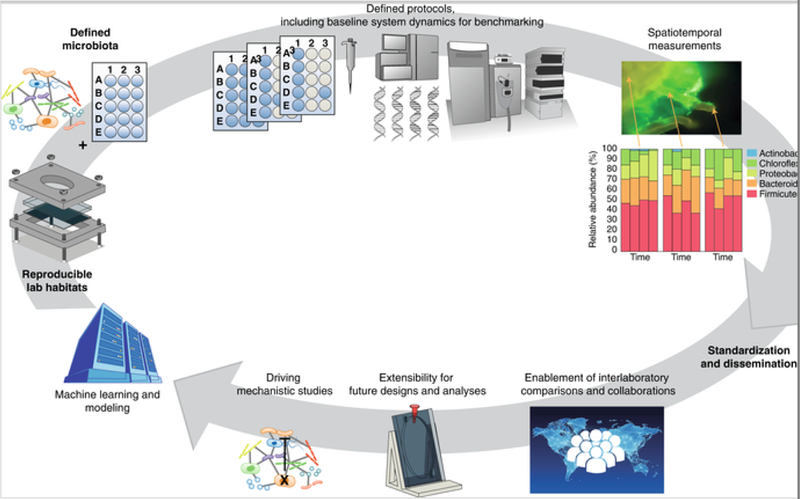
EcoFABs are composed of defined microbiota, laboratory habitats, and protocols for cultivation and spatiotemporal analysis. Once standardized and disseminated, these will enable multi-laboratory collaborations, adaptions, mechanistic studies to develop theory and predictive models.
Current experimental systems for host-microbe interactions
Important progress has been made both in individual laboratories and by collaborative groups to establish well-defined in vitro model systems for human-health-related research. Efforts include, for instance, microbial chemostat systems[17], the Lubbock chronic-wound biofilm model[18] and the development of organ-on-a-chip systems[19]. Such in vitro systems can be set up with or without the presence of immune cells and mechanical deformation, mimicking aspects of the intestinal environment (for example, refs. [17, 19]). Mechanical deformation that results in mixing, water-flow changes and exposure to oxygen through active contractions of the intestinal walls are critical in shaping the microbial community of the human gut. To investigate these processes in detail, a fluidic channel, termed the ‘minigut’, has been developed to determine the effect of flow rate and frequency of contractions on microbial community composition and cell density[20] (Fig. 2a–c). Notably, bacterial dynamics were captured by a simple reaction-diffusion model without adjustable parameters. Thus, in vitro model systems can provide mechanistic insights to guide the development of quantitative modeling frameworks for accurate predictions.
Fig. 2. Two example devices for analysis of microbial interactions.
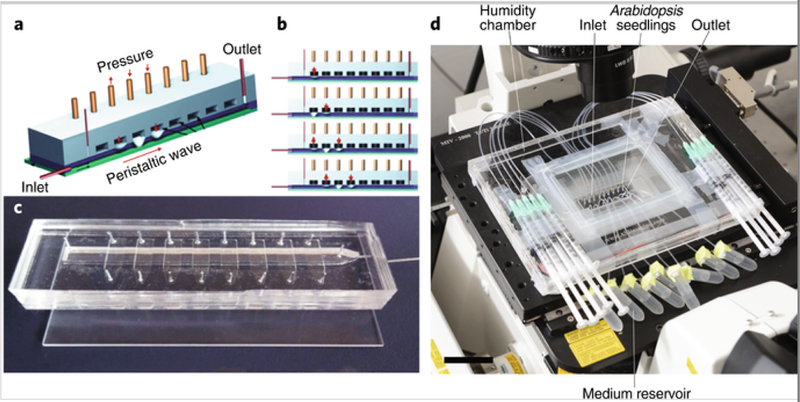
a,b, Minigut actuation design creates a peristaltic wave. c, Photograph of the minigut chip, which is approximately 8.4 cm × 2 cm. d, RootChip design for high-resolution imaging of plant–microbe interactions across controlled environmental conditions. Scale bar, 5 cm. Reproduced with permission from Cremer et al.[20] (a–c) and Massalha et al.[23] (d).
Progress has also been made in defining research priorities for plant-associated microbiota[16, 21] and developing experimental approaches for studying plant microbiomes[22, 23]. Although translation between the laboratory and the field is extremely challenging, recently an approach for developing core microbiomes has been proposed that provides a powerful framework for the selection of microbiota[24]. For example, an experimental approach based on laboratory screening of binary plant–bacteria association assays can inform the design of small synthetic communities with predictable host phenotypes[25]. Other experimental approaches, such as RootChip-type devices[26], allow control of plant–microbe cultivation for high-resolution imaging of the spatiotemporal dynamics of bacterial localization and competition on roots[23] (Fig. 2d). Similar devices have been developed for studying marine systems, for example, to investigate microbial interactions that support the base of the food web and drive biogeochemical cycles in the ocean[27].
However, there has been little standardization in cultivation systems to study complex microbial communities. Researchers typically use single strains or simple cocultures, which are generally poor models for the complex and metabolically diverse microbial communities found in nature[10]. Alternatively, researchers may use systems populated with complex mixtures of microbes obtained from the environment to capture the diversity of natural microbial communities. Although these approaches provide a better understanding of native microbiomes, they lack reproducibility (even within a single laboratory) in the absence of a microbial inoculum that is stable over time.
Standardized microbial ecosystems will advance microbiome science
Standardized EcoFABs should enable the addition or subtraction of system components (genes, pathways, microbes, environmental conditions and others) and analysis of microbial localization, activities and interactions across relevant temporal and spatial scales. EcoFABs will enable researchers to carry out large-scale analysis of gene function(s) in communities to dissect ecological questions with a high level of precision[28], and to define functional traits related to interspecies interactions[29, 30, 31]. Standardization of habitats, microbes and protocols (including workflows, computational tools and models, as well as data standards) will allow data from many laboratories to be compared and correlated for the testing and development of ecological theories[32] and generalizable principles[30]. Ultimately, concerted efforts would lead to a better understanding of community assembly, structure, interactions and emergent activities. Such findings will help guide the development of computational and theoretical models that accurately predict the trajectories of microbiomes to harness their activities.
The development of broadly applicable EcoFABs will require control over a diverse set of variables, such as microbial organisms, environmental conditions, and ecological forces that govern community assembly and succession. The function and composition of a microbiome are strongly influenced by its history, as are the activities of individual microbes. Hence, a standardized approach will be needed to ‘boot up’, monitor and maintain these systems. The necessity of robust, accessible and detailed protocols, and of tracking the providence of designs (for example, by using tools such as GitHub.com) and strains (possibly using genetic barcoding), will be critical to define valid comparisons across laboratories and over time. These protocols should define the inoculum (in terms of both organismal membership and absolute abundance), methods of viability assessment, environmental fabrication and setup, culture conditions, and acceptable dynamic ranges of experimental parameters (for example, temperature, pH, media composition, community stability and others).
The use of common experimental protocols and data reporting standards in EcoFABs will provide an opportunity to also standardize analytical approaches for microbiome research. This is important because different sample-processing protocols, instruments and bioinformatic analyses can produce divergent results. Although these challenges are widely acknowledged with respect to high-throughput sequencing, where small protocol changes can lead to different results, the problem is generalizable to other analysis modalities, such as phenotypic screens and omics analyses[33].
Balancing reproducibility with relevance
The development of EcoFABs must balance biological relevance with reproducibility, which has long been recognized as a considerable challenge[4]. The microbiota associated with EcoFAB systems must be experimentally tractable, broadly applicable to diverse biological questions and ideally composed of organisms known to coexist in natural settings. Given these design criteria, the development of even a handful of foundational EcoFABs will be a major effort for the scientific community. Most natural microbial communities will not constitute useful laboratory models, just as most individual species are not suitable as model organisms and not all questions can be addressed in a few model communities[31].
The goal of achieving reproducibility across laboratories should guide the construction of basic culture conditions, including the generation of a ‘minimal microbial community’ comprising the complete set of organisms that are sufficient and necessary to capture broadly applicable community function(s) or phenomena, in a defined spatiotemporal environmental context. At a minimum, these simplified systems must display reproducible dynamics and genotype–environment interactions. We propose defining simplified communities based on a ‘functional group paradigm’ (grouping of organisms based on similar characteristics within the community). In this paradigm, which is often used in ecology, particular ecosystem ecologies (habitats, tolerances and sensitivities) and metabolisms would be identified that possess similar habitats, biotic associations and functional dynamics (in terms of their tolerance and sensitivity to perturbations). Representative microbial communities from these functionally related systems could then be selected as the appropriate microbial study systems.
Establishing the field and clinical relevance of EcoFABs will require the demonstration that important native microbiome activities are reflected and that the results are reproducible both within and between laboratories[34]. We acknowledge that there are certainly many limitations to experiments using model systems versus natural ecosystems[9, 10, 31]. One particularly important focus for model ecosystems will be to support the development and refinement of generalizable principles and mechanisms that can then be tested in more relevant and complex systems.
It will take new communities of scientists to develop and disseminate EcoFABs
Historically, the development and establishment of model organisms have extended over several decades and often involved multiple generations of scientists[15]. Typically, these result from a few researchers generating extensive resources, attracting early adopters, which leads to additional resources and users, and eventually the formation of communities of scientists. To rapidly accelerate the development and acceptance of model microbiome ecosystems, it will be essential to learn from the failures and successes of previous model organism efforts. This requires the consideration of many factors, including the needs of large research communities, major societal challenges and scientific questions to be addressed, as well as the goals of funding agencies. We recommend that the scientific community develop the scientific basis and the necessary resources for a small number (three to four) of broadly applicable EcoFABs (animal, plant, soil and aquatic) in the next four years that can be further adapted and extended by individual groups of scientists (http://eco-fab.org/ecofab-summit-2017/) through the use of a set of common design principles (Box 1).
Box 1. EcoFAB design principles.
Keep it simple
Base EcoFABs and minimal analyses should be as simple and inexpensive as possible to enable widespread adoption, large numbers of replicates, and integrated studies across laboratories, with the recognition that some studies will require specialized EcoFABs.
Design for extensibility
Enabling extensions should address the needs of large groups of researchers from diverse fields.
Standardize system dynamics
EcoFABs should be standardized to ‘boot up’ and display reproducible dynamics.
Accurately recapitulate important aspects of natural systems
EcoFABs should capture key natural processes.
Enable mechanistic studies
Genetically tractable organisms should be used when possible to enable mechanistic insight at the molecular level.
Embrace complexity
EcoFABs should allow experimentation with minimal to complex microbiomes and habitats.
Build in simple analytics and sensors
Inclusion of inexpensive sensors and analytical measures should allow for debugging, comparison and standardization.
Space and time are important
Moving toward manipulation, imaging and analysis spanning relevant scales from individual microbes to whole communities over time are important.
Future-proof data
Data standards, analytical approaches, system tolerance and modeling approaches must be considered up front to enable future cross-EcoFAB meta-analyses. When possible, data should also be preserved in their original, raw form to enable future analyses.
Give it away
To maximize impact and usefulness, the EcoFAB resources should be freely accessible to the greater scientific community. This means that the base-level capabilities will need to be common to most research groups, and that we will embrace a culture of sharing and interlab cooperation. Open-access experimental design and standardized reporting methods will also aid the overall inclusiveness of EcoFAB efforts.
Because large networks of users and open access to resources and data will expedite scientific discovery, inclusiveness and collaboration will facilitate the development and acceptance of EcoFABs by a broad user community. To help organize these efforts, we have formed an EcoFAB Steering Committee (http://eco-fab.org/ecofab-steering-committee/) with four main responsibilities: (1) defining criteria for the design, dissemination and standards of data collection; (2) organizing multi-laboratory comparisons to evaluate reproducibility (see below); (3) broadly promoting the use of EcoFABs; and (4) preparing an annual white paper (or papers) identifying key challenges for the field that can be uniquely addressed with EcoFABs. Working groups focused on specific microbiomes will complement these efforts. For example, a working group has made major progress in developing a standardized plant rhizosphere EcoFAB following the design principles listed in Box 1. This EcoFAB will be complete with a defined microbiome (including mutant bacteria) and a standardized cultivation system[22, 34]. The working group plans to make these integrated capabilities available within the next year and to host a focused conference within the next two years. Our long-term vision is to create an EcoFAB community and a corresponding portal consistent with the highly successful approaches used by the Drosophila community (for example, the Drosophila Board and https://flybase.org) and Arabidopsis community (for example, Multinational Arabidopsis Steering Committee and https://araport.org).
Multi-laboratory comparisons will be essential for verifying that the designs, protocols and data types collected are sufficient for researchers in multiple laboratories to generate reproducible results[34]. Under the guidance of the steering committee, carefully designed studies that include computational analysis and quality metrics for models will be performed. We advocate the use of low-cost methods that are accessible to most scientists around the world. If successful, these experiments will define reference sets, thus enabling other researchers to benchmark their work against these results.
One of the most exciting opportunities for standardized microbial ecosystems is the ability to compare data across experiments and laboratories. This will require a community portal for open and uniform sharing of designs, protocols, models and data. It will also require that groups embrace an open-science culture: sharing data, following similar protocols and collecting a minimal set of standardized data. This will be critical to ensuring the utility of current results to future investigations and ultimately enabling cross-microbiome studies to elucidate general principles of microbiome structure and activities.
Conclusions
The success of standardized and community-accepted model systems warrants the development of analogous model microbiomes. By standardizing a few of these systems, along with common protocols and data standards, researchers will be able to build on each other’s work, test predictions, identify governing mechanisms and build predictive models. These systems will help microbiome science rapidly advance from often observational, correlative studies to reproducible mechanistic investigations. Comparison of findings and mechanisms across diverse model systems will facilitate the development of general principles that are applicable across microbiomes and ultimately can be validated in studies of natural systems. Advancing this vision will require effective community organization and participation to prioritize widely applicable EcoFABs, to set design and reporting standards, and to establish venues for exchanging findings and ideas that advance microbiome science.
Acknowledgements
Material is based on work supported by the National Science Foundation under grants 1332344 and 1804187 (to K.Z.), the Trial Ecosystem Advancement for Microbiome Science and the Microbial Community Analysis and Functional Evaluation in Soils Programs at Lawrence Berkeley National Laboratory by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research Awards DE-AC02-05CH11231 (to T.R.N.), DE-SC0018277 (to J.R.D.) and DE-SC0012658, DE-SC0012586, DE-SC00138344 (to K.Z.), DE-SC0013887 (to E.A.S.), the Center for Bioenergy Innovation (CBI), DE-AC05-000R22725 (to C.M.), by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research under contract DE-AC05-76RL01830 with Pacific Northwest National Laboratory through the iPASS and PREMIS Initiatives (to C.J.), and the Strategic Planning Support Activities Program of Lawrence Berkeley National Laboratory (to T.R.N.). Work at Lawrence Livermore National Laboratory (J.P.R.) was conducted under the auspices of Contract DE-AC52-07NA27344 and SCW1039. We thank P. Kim and Z. Rostomian for assistance in figure preparation, and D. Gilbert and J. Tanamachi for helpful comments.
Footnotes
Competing interests T.R.N. is an inventor on a related patent application. M.D.W. is a founder and shareholder of Growcentia, Inc.
References
- 1.Thompson LR et al. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zengler K & Zaramela LS Nat. Rev. Microbiol. 16, 383–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller B & Grossniklaus UJ Proteomics 73, 2054–2063 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Wimpenny JWT CRC Handbook of Laboratory Model Systems for Microbial Ecosystems (CRC Press, 1988). [Google Scholar]
- 5.Winogradsky S Arch. Sci. Biol. 3, 297–352 (1895). [Google Scholar]
- 6.Dallinger WD J. R. Microsc. Soc. 7, 184–199 (1887). [Google Scholar]
- 7.Gause GF The Struggle for Existence (The Williams & Wilkins Company, 1934). [Google Scholar]
- 8.Kolenbrander PE et al. Microbiol. Mol. Biol. R 66, 486–505 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jessup CM et al. Trends Ecol. Evol. 19, 189–197 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Wolfe BE Msystems 3, 00161–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welch JLM, Hasegawa Y, McNulty NP, Gordon JI & Borisy GG Proc. Natl. Acad. Sci. USA 114, E9105–E9114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yano JM et al. Cell 161, 264–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ et al. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Zhalnina K, Zengler K, Newman D & Northen TR MBio 9, e01175–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koornneef M & Meinke D Plant J. 61, 909–921 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Busby PE et al. PLoS Biol. 15, e2001793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald JA et al. J Microbiol. Methods 95, 167–174 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Agostinho AM et al. J. Appl. Microbiol. 111, 1275–1282 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Kim HJ, Li H, Collins JJ & Ingber DE Proc. Natl. Acad. Sci. USA 113, E7–E15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cremer J et al. Proc. Natl. Acad. Sci. USA 113, 11414–11419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blaser MJ et al. MBio 7, e00714–e00716 (2016). [Google Scholar]
- 22.Gao J et al. J. Vis. Exp. 134, e57170 (2018). [Google Scholar]
- 23.Massalha H, Korenblum E, Malitsky S, Shapiro OH & Aharoni A Proc. Natl. Acad. Sci. USA 114, 4549–4554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toju H et al. Nat. Plants 4, 247–257 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Herrera Paredes S et al. PLoS Biol. 16, e2003962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossmann G et al. Plant Cell 23, 4234–4240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert BS et al. Nat. Microbiol. 2, 1344–1349 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Handelsman J Microb. Biotechnol. 2, 138–139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green JL, Bohannan BJ & Whitaker RJ Science 320, 1039–1043 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Little AEF, Robinson CJ, Peterson SB, Raffa KE & Handelsman J Annu. Rev. Microbiol. 62, 375–401 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Ruby EG Nat. Rev. Microbiol. 6, 752–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prosser JI et al. Nat. Rev. Microbiol. 5, 384–392 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Sinha R, Abnet CC, White O, Knight R & Huttenhower C Genome Biol. 16, 276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasse J et al. New Phytol. (2018). [Google Scholar]


