Abstract
Development of the TEM-CCF2/4-AM FRET based system has enabled investigators to track translocation of effector proteins into mammalian cells during infection. This allows for separation of translocated and non-translocated cell populations for further study. Yersinia strains expressing translational Yop-TEM fusions, containing the secretion and translocation signals of a Yop with the TEM-1 portion of β-lactamase, are used to infect mice, tissues isolated from mice, or mammalian cells in culture. Infected and harvested mammalian cells are treated with either CCF2-AM or CCF4-AM, and cleavage of this fluorescent compound by TEM is detected by FACS analysis. A shift from green to blue emission spectra of individual cells is indicative of translocation of a given Yop-TEM fusion protein into the host cell during Yersinia infection due to a disruption in FRET between the two fluors of the compound. In Yersinia, this method has been used to understand Type III secretion dynamics and Yop functions in cells translocated by effectors during infection. Here, we describe how to generate Yop-TEM constructs, and how to detect, quantify, isolate, and study Yop-TEM containing cells in murine tissues during infection and in ex vivo tissues by cell sorting and flow cytometry analysis. In addition, we provide guidance for analyzing TEM-positive cells via a plate reader and fluorescent microscopy.
Keywords: β-lactamase, Bla, TEM, Yop translational fusions, Yersinia, Yops, translocation, neutrophils, CCF2, CCF4, FACS, T3SS, type III secretion, flow cytometry
1. Introduction
All three Yersinia species use a virulence plasmid-encoded Type III secretion system to translocate bacterial effector proteins (Yops) into eukaryotic cells (1–4). These Yops have wide-ranging effects on host cellular functions and allow bacteria to evade host immune responses to establish infection (4–6). For many years it had been challenging to identify and study the host cells injected with Yops during tissue infection, in part because Yersinia species are extracellular. Furthermore, it can be difficult to track Yop translocation into individual host cells by traditional fluorescence or epitope tagging methods without having to fix or lyse host cells. The development of the TEM-CCF2/4 system (7, 8) has allowed investigators to identify host cells targeted for injection by Yops by all three pathogenic Yersinia species and characterize both effector translocation and host responses to infection (9–11).
The Yop-TEM-CCF2/4 systems works by creating a translational fusion protein with the secretion and translocation signal sequences of Yops (contained within the first 100 amino acids of each Yop) (12) to the TEM-1 portion of β-lactamase. TEM-1 lacks the N-terminal signal sequence (23 amino acids) of β-lactamase (13) but retains its enzymatic ability to hydrolyze beta-lactam bonds in antibiotics and other molecules (13, 14). Upon translocation into mammalian cells, TEM cleaves the β-lactam ring of the cephalosporin core of the substrates CCF2 and CCF4. CCF2-AM and CCF4-AM are membrane-permeable substrates that contain two fluorophores linked by a cephalosporin ring. Once added to mammalian cells, CCF2-AM and CCF4-AM are cleaved by endogenous cytoplasmic esterases into their negatively charged forms and are retained in the cytosol. In the absence of TEM, excitation of the donor fluor (7-hydroxycoumarin) at 409 nm leads to the FRET excitation of the acceptor fluor (fluorescein) at 520 nm, and emission of green light. When TEM cleaves the cephalosporin β-lactam ring and separates the two fluors, excitation of 7-hydroxycoumarin at 409 nm leads to emission of blue light at 447 nm and the quenching of the acceptor fluor (8). Thus, after infection with a strain expressing a Yop-TEM fusion, shifts from green to blue emissions from mammalian cells containing CCF2/4-AM after infection with a strain expression a Yop-TEM fusion are a direct measurement of translocation of that effector protein into the host cells. Techniques used to detect shifts from blue to green fluorescence include Fluorescent Activated Cells Sorting (FACS), plate readers, and microscopy.
The Yop-TEM CCF2/4-AM FRET system is powerful because investigators can identify, separate, and study translocated cells in infected tissues from non-translocated cells in infected tissues. This method has been used in all three pathogenic Yersinia species to show the range of cells injected with Yops in different tissues during various types of infection including oral, intravenous, and intranasal infection (9–11, 15–17). Investigators have further exploited this method to study cells isolated from infected tissues and examine the consequences of Yop injection (18). Furthermore, bacterial and/or host mutants can be used to determine the contributions of specific Yop effectors, bacterial ligands and host factors in modulating Yersinia-host cell interactions in infected tissues (15, 17, 19–24). Finally, this method has been used in high-throughput screens to identify small molecule inhibitors of Yop translocation (25, 26).
In this chapter, we describe construction of Yop-TEM translational fusions which are expressed either on plasmids (9) or integrated into Yersinia genomic DNA (10). We also describe analytical FACS methods to identify and quantify cells translocated with Yops (9–11, 16, 17) and steps subsequent to FACS to analyze the effects of Yop translocation in mammalian cells from infected tissues (18). In addition, while not the primary focus of this chapter, we describe methods for plate reader assays for Yop-TEM translocation measurements (25, 26).
2. Materials:
2.1. Construction of Yop-TEM plasmids
Cloning plasmids: pBR322 (contains blaM gene), donor plasmid of interest (see Note 1)
Cloning reagents: restriction enzymes, CutSmart buffer
DNA oligonucleotides (see Note 2 for design)
PCR reagents: Phusion polymerase, 5x Phusion buffer, 50 mM MgCl2, 100 μM dNTPs
Ligation reagents: T4 DNA ligase, T4 DNA ligase buffer
SY327 or DH5α λpir competent cells
Luria Media Broth (LB) broth
LB agar plates
Kanamycin or antibiotic to select for plasmid
2.2. Bacterial conjugation
SY327 or DH5α λpir
E.coli strain with pRK600 donor plasmid
Yersinia recipient strain of interest
2xYT broth
LB agar plates
LB-irgasan/kanamycin plates
Toothpicks
Kanamycin
Chloramphenicol
Antibodies against β-lactamase
2.3. Verification of TEM constructs
Trichloroacetic acid (TCA)
Acetone
SDS sample buffer
SDS-PAGE gels and supplies for running western blots
Coomassie stain
Antibodies to beta lactamase
2.4. Preparation of Yersinia
Small culture tubes
2xYT media for overnight Yersinia cultures
Low Ca2+ 2xYT: 2xYT with 20 mM Na2C2O4, 20 mM MgCl2
Optical density spectrophotometer
Cuvettes
1X PBS
DMEM
RPMI 1640
Heat-inactivated Fetal calf serum (HI-FBS) (see Note 3)
LB-Irgasan plates
2.5. Preparation of 10X CCF2/4-AM
10X CCF2/4-AM for tissue culture experiments:1 μg/ml CCF2/4-AM, 1.5 mM probenecid, 100 μg/mL gentamicin (final concentrations). To make a 1 mM stock solution of CCF2/4-AM, add 185 μL of DMSO to 200 μg CCF2-AM or 182 μL of DMSO to 200 μg CCF4-AM. Store at −20°C in 6 μL aliquots. To prepare 600 μL of a 10X CCF2/4-AM stock mixture, add 36 μL solution B (provided with CCF2/4) to 6 μL 1 mM CCF2/4 in DMSO. Vortex, add 36 μl probenecid acid (250 mM stock); vortex. Add 462 μL PBS, vortex, add 60 μL gentamicin (10 mg/mL stock).
10X CCF2/4 for ex vivo and in vivo mouse infections: 1 μg/ml CCF2/4-AM, 1.5 mM probenecid (final concentrations). To make a 1 mM stock solution of CCF2/4-AM, add 185 μL of DMSO to 200 μg CCF2-AM or 182 μL of DMSO to 200 μg CCF4-AM. Store at −20°C in 6 μL aliquots. To prepare 600 μL of a 10X CCF2/4-AM stock mixture, add 36μl solution B (provided with CCF2/4) to 6 μL μl 1 mM CCF2/4 in DMSO. Vortex, add 36 μL probenecid acid (250 mM stock); vortex. Add 522 μL PBS, vortex.
Aluminum foil
2.6. In vitro infection of cultured cells
24-well plate
Confluent cultured cells
Tissue culture media (e.g. DMEM + 10% HI FBS)
0.25M Trypsin
1X PBS
5ml polystyrene for LSRII/FACS analysis tubes
LB-Irgasan plates
Optical density spectrophotometer
Cuvettes
70 μm filter
2.7. Ex vivo infection of splenocytes or lung cells:
Optical density spectrophotometer
Cuvettes
RPMI 1640
RPMI 1640 + 5% HI-FBS
2ml Eppendorf tubes
70-μm cell strainer
5ml syringes
6-well plate
1mg/ml of Collagenase D dissolved in PBS or water
Pharm Lyse 10X stock; dilute in sterile H2O for 1X
24-well plate
30-gauge needle and syringe
2.8. Mouse infection
Optical density spectrophotometer
Cuvettes
1x PBS
Anesthetics
Anesthesia chamber
50% glycerol
1X PBS, Ca2+- and Mg2+-free
5ml syringes
6-well plates
RPMI 1640
70-μm cell strainer
FACS buffer: 1% FBS in 1X PBS, Ca2+- and Mg2+-free
1x Fc blocking solution: 10μL Purified Rat α-mouse CD16/CD32 (0.5 mg/mL stock concentration) in 2ml of FACS buffer
Antibodies to surface markers
5ml polypropylene for MoFlo/FACS sorter tubes
5ml polystyrene for LSRII/FACS analysis tubes
LSRII (Becton Dickson) FACS machine
2.9. Preparation TEM+ and TEM− neutrophils (PMNs)
FACS buffer: 1% FBS in 1X PBS, Ca2+- and Mg2+-free
Sorting buffer: FACS buffer, 5mM EDTA, 50mM HEPES
1X Ca2+-, Mg2+-, phenol red-free Hank’s Balanced Salt Solution (HBSS)
1X PBS, Ca2+-, Mg2+-free
1X phenol red-free HBSS with Ca2+ and Mg2+
1M glucose
2.10. Analysis by microscopy:
Coumarin/Pacific Blue Longpass Filters (Chroma, Set Number: 19011)
10X or 40X Objective
Either epifluorescence or confocal microscope
2.11. RT-PCR
FACS buffer: 1% FBS in 1X PBS, Ca2+- and Mg2+-free
Sorting buffer: FACS buffer, 5mM EDTA, 50mM HEPES
TRIzol® Reagent
RNA extraction kits (e.g QIAGEN RNeasy Mini Kit)
2.12. Cytokine analysis:
RPMI 1640
Fetal bovine serum (FBS)
Brefeldin A (BFA) – Refer to manufacturer’s instructions, but stock concentration is typically at 10mg/ml. Store at −20°C.
BD Cytometic Bead Array or specific fluorescently labelled antibodies for cytokines of choice.
Buffers for intracellular staining – 4% Paraformaldehyde to fix cells; FACS buffer: 1% FBS in 1X PBS, Ca2+- and Mg2+-free and 0.1% saponin in FACS buffer for temporary permeabilization of cells
2.13. Analysis by plate reader:
Tissue culture media (ex. DMEM with 10% FBS or RPMI with 5% FBS
384-well plates
Gentamicin 10 mg/L stock in H2O
3. Methods:
3.1. Generation of Yop-TEM translational fusion construct plasmid
PCR amplify the blaM gene lacking the signal sequence (TEM) using primers containing restriction sites compatible with the donor plasmid (see Note 2).
Ligate TEM into donor plasmid using standard molecular biology techniques.
Transform into DH5α λpir and select on antibiotic agar plates.
PCR amplify the promoter and the desired portion of the Yop of interest containing the secretion and translocation signals (usually first 100 amino acids) from Yersinia DNA with primers containing restriction sites compatible with the MCS upstream from the TEM insertion into the donor plasmid (see Notes 4 and 5).
Ligate Yop promoter upstream of TEM in the donor plasmid standard molecular biology techniques.
Transform into DH5α λpir and select on antibiotic agar plates.
3.2. Generation Yersinia strains expressing Yop-TEM translational fusions
3.2.1. Conjugation of Yop-TEM translational fusion construct into Yersinia
Depending on the plasmid, the Yop-TEM translational fusion construct can be electroporated into Yersinia to be expressed in trans, or it can be integrated into the chromosome by allelic exchange as described here.
For allelic exchange, subclone the Yop-TEM translational fusion construct into a plasmid containing regions of homology to the desired Yersinia chromosomal insertion site to generate a merodiploid using standard molecular biology techniques (see Note 6).
Inoculate 2 mL cultures of three strains: recipient strain (Yersinia) in 2xYT, helper strain (DH5α with pRK600 with 40 μg/ml chloramphenicol) and donor strain (SY327 or DH5α λpir with plasmid of interest) in LB broth with appropriate antibiotics. Grow cultures overnight (see Note 7).
Transfer 500 μL of each overnight culture in new Eppendorf tubes, and spin at 17,000 x g for 2 minutes. Remove supernatant, add 500 μL antibiotic-free 2xYT and spin at 17,000 x g. Repeat twice.
Remove supernatant and resuspend the pellet in 500 μL 2xYT without antibiotics.
In 4 spots on an LB plate, place 5 μL of each strain on top of each other. Also spot each strain (5 μL) individually on the bottom part of the plate, which will serve as negative controls for selection on drug plates.
Invert once liquid is absorbed and leave at room temperature (RT) overnight.
In the morning use a toothpick, scoop up entire mating spot and smear on antibiotic selective plate (LB-Irgasan + antibiotic appropriate for plasmid selection: Kanamycin in case of pRS47), one spot per plate. Include individual strain spot controls.
In the afternoon, scoop up entire smeared field from each plate and streak for isolated colonies on new selective plates. Let Yersinia grow at RT for 2 days. Control colonies (individual strains) should not grow on selective plates.
Pick two isolated colonies from each mating and re-streak for isolated colonies on selective plates. Incubate at RT for 2 days.
Take an isolated colony from each streak and grow in 2xYT overnight at 26°C with antibiotics for Trichloroacetic acid (TCA) precipitation (see below).
3.2.2. Verification of secretion of Yop-TEM fusion protein
Test for Yop-TEM secretion by Trichloroacetic acid (TCA) precipitation assay of supernatants followed by Western Blot analysis with an antibody against Beta-lactamase.
Day 1 afternoon: Prepare overnight cultures of Yersinia for infection: Inoculate single colonies into individual sterile tubes containing 2 mL 2xYT plus antibiotics with Yersinia Yop-TEM translational fusion strain. Incubate overnight at 26°C with aeration. For screening colonies from mating, we recommend inoculating 8-10 single colonies.
Day 2: Dilute overnight Yersinia cultures 1:40 in 2 mL low Ca2+ 2xYT (see Note 8 and 9).
Grow Yersinia for 2 hours shaking at 26°C, and then shift to 2 hours shaking at 37°C
Transfer 900ul of bacterial culture to new Eppendorf tube. Add 100 μl TCA to each tube for a final concentration of 10%.
Incubate on ice for 15 minutes.
Spin down at max speed (17,000 x g) for 15 minutes.
Carefully decant supernatant; avoid aspirating pellet.
Wash pellet with ice cold acetone (500 μL), and spin down max speed for 5 minutes.
Carefully decant supernatant, and airdry pellet.
Resuspend pellet in 50 μl SDS sample buffer, boil samples at 95°C for 5 minutes, and run on 10 μl SDS-PAGE gel and stain with Coomassie to visualize secreted Yops.
Confirm the Yop-TEM fusion protein is secreted by Western blot using TEM specific antibodies.
Freeze down (−80°C) a single colony of the Yersinia Yop-TEM translational fusion strain that secretes Yops properly in 2xYT with 20% glycerol.
3.3. in vitro infection of cultured cells with Yersinia Yop-TEM translational fusion construct
3.3.1. Infection of cultured cells with Yersinia
Day 1: Plate cells in a 24 well plate (see Note 10).
Day 1: afternoon – Prepare overnight cultures of Yersinia for infection: Inoculate 2 mL 2xYT plus antibiotics with Yersinia Yop-TEM translational fusion strain. Incubate overnight at 26°C with aeration.
Day 2: Dilute overnight Yersinia cultures 1:40 in 2 mL low Ca2+ 2xYT (see Note 8 and 9).
Grow Yersinia for 2 hours shaking at 26°C, and then shift to 2 hours shaking at 37°C.
Take OD600 of cells to estimate culture concentration, and dilute in DMEM to desired concentration.
Plate serial dilutions of Yersinia used in infection on LB-Irgasan plates to determine MOI. Grow at 26°C for 2 days before counting CFUs.
Add 100 μL of Yersinia in DMEM at the desired MOI (based on starting cell concentration) to each well (see Note 11).
Spin 3 min at 20 x g at RT.
Incubate 1-2 hours in tissue culture incubator or until cells round up (see Note 12).
3.3.2. Preparation of samples for FACS analysis
Remove media from each well. Wash once with 1x PBS.
Add 100 μL 0.25M trypsin to each well, swirl, and incubate in tissue culture incubator for 3-4 minutes until cells lift off by gently tapping plate. Confirm by examination under a light microscope.
Add 400 μL DMEM + 10% HI-FBS to quench the trypsin. Pipette up and down vigorously to remove all cells from well and transfer all 500 μl to 5ml polystyrene for LSRII/FACS analysis tubes. If necessary, remove any clumps by filtering through a 70 μm filter to prevent clogging the machine.
Prepare 10X CCF2/4 master mix as written in Materials 2.4 and scale as necessary (see Notes 13 and 14).
Add 50 μL 10X CCF2/4 master mix to each FACS tube, incubate at RT in the dark for 20-40 minutes. Do not allow CCF2/4 incubation to continue for >1.5 hours (see Note 14). Store on ice until FACS analysis.
Run samples on FACS machine to determine populations of Yop-TEM+ and Yop-TEM− cells (see Method 3.6).
3.3.3. Preparation of samples for analysis by microscopy
Remove media from each well. Wash once with 1 mL 1x PBS.
Add 450 μL of PBS to each well and 50 μL CCF2/4 master mix.
Incubate at RT in the dark for 20-40 minutes.
Using Coumarin/Pacific Blue Longpass Filters (Chroma), view on microscope with 10X and 40X objectives (see Note 15).
3.4. Ex vivo infection with splenocytes or lung cells
3.4.1. Harvesting and infection splenocytes or lung cells with Yersinia
Day 1 afternoon: Prepare overnight cultures Yersinia for infection: Inoculate 2 mL 2xYT plus antibiotics with Yersinia Yop-TEM translational fusion strain. Incubate overnight at 26°C with aeration.
Day 2: Dilute overnight Yersinia cultures 1:40 in 2 mL low Ca2+ 2xYT (see Note 8 and 9).
Grow Yersinia for 2 hours shaking at 26°C, shift to 2 hours shaking at 37°C.
Meanwhile, aseptically harvest spleens or lungs into 1mL PBS in 2ml Eppendorf tubes. Keep on ice until all samples collected (see Note 16).
Prepare single cell suspension by passing organs through 70-μm cell strainer using a syringe plunger into well of 6 well plate. Add 5ml RPMI to wash cell strainer filter. Mix gently within each well. Transfer to 15 mL conical tube and incubate for 1 hour at 37°C (see Note 17).
Centrifuge single cell suspensions at 250 x g for 5 minutes at 4°C.
Treat with 5ml 1X Pharm Lyse for 5 minutes at 4°C.
Centrifuge 250 x g for 5 minutes at 4°C. Remove supernatant.
Resuspend in 10 ml RPMI supplemented with 5% HI-FBS. If there are cell clumps, passage a second time through 70-μm cell strainer into a 50ml conical tube, and centrifuge at 250 x g for 5 minutes at 4°C. Count cells.
Resuspend cells in RPMI + 5% HI-FBS at ~ 1-2×106 cells/mL.
Aliquot 200 μL of cell suspension in an appropriate number of wells in a 24-well plate depending on the number of experimental samples and controls needed. Typically, there are 2-3 replicates per experimental sample and controls for uninfected, CCF2/4-AM staining, and single antibody staining (see Table 1 for an example of plate set-up).
Take OD600 of diluted bacterial cultures to estimate culture (CFU) concentration, dilute in RPMI to desired concentration.
Plate serial dilutions of Yersinia used in infection on LB-Irgasan plates to determine MOI. Grow at 26°C for 2 days before counting CFUs.
Add Yersinia Yop-TEM translational fusion strain to wells at the desired MOI.
Spin 3 min at 20 x g at RT.
Incubate at 37°C for 1-2 hours.
Table 1:
An example of necessary controls and experimental samples for FACS
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| FACS gating Controls | Uninfected Cells Unstained | Uninfected Cells + Antibody 1 | Uninfected Cells + Antibody 2 | Uninfected Cells + Antibody 3 | Uninfected Cells + Antibody 4 | |
| FACS gating Controls | Uninfected cells + CCF2/4 | Infected cells WT ETEM + CCF2/4 | ||||
| Experimental samples | Uninfected cells + CCF2/4 + Antibody combination 1+2 | Uninfected cells + CCF2/4 + Antibody combination 3+4 | ||||
| Experimental samples | Infected Cells with WT ETEM + CCF2/4+ antibody combination 1+2 | Infected Cells with WT ETEM + CCF2/4 + antibody combination 3+4 | Infected cells Yersinia mutant 1 + CCF2/4 + Antibody combination 1+2 | Infected cells Yersinia mutant 1 + CCF2/4 + Antibody combination 3+4 | Infected cells Yersinia mutant 2 + CCF2/4 + Antibody combination 1+2 | Infected cells Yersinia mutant 2 + CCF2/4 + Antibody combination 3+4 |
Wells in bold should be infected and stained in either duplicate or triplicate.
3.4.2. Preparation of samples for FACS analysis
After infection, transfer cells by gently pipetting to an Eppendorf tube and centrifuge at 250 x g for 5 minutes at 4°C.
For FACS staining, depending on the number of antibody combinations to be used, resuspend in FACS buffer + 100 μg/ml gentamicin at a concentration of 0.5-2 × 106 cells/mL. Remove any clumps of cells by passing through a 70-μM cell strainer or physically with a pipet tip so as to not to clog the FACS machine.
Transfer 100 μL of each cell suspension to the appropriate number of 5 ml polypropylene (MoFlo/FACS sorter) or polystyrene (LSRII/FACS analysis) tubes depending on the number of staining combinations to be used. Typically, 50,000 cells are stained per antibody combination, but as few as 1 × 104 or as many as 2 × 105 may need to be stained (see Note 18). All of the following steps with CCF2/4-AM and antibody staining must be kept in the dark as much as possible. This includes keeping FACS samples in foil until it is time to FACS them.
Incubate cells with 10 μL 10X CCF2/4-AM mixture without gentamicin per 100μL of cells for 15 minutes at room temperature (22-24°C). Make sure to leave aside sufficient aliquots of cells that are not treated with CCF2/4-AM to serve as gating controls for antibodies (see Note 19 and Table 1).
Add 50 μL 1X Fc block per 100μL of cells. Incubate 10 minutes at 4°C.
Aliquot 50 μL of desired antibody combinations into each tube (see Notes 20 and 21).
Incubate for 15 minutes at 4°C. If in 96-well plate format, transfer to appropriate FACS tubes.
Add 200 μL of PBS into each tube and analyze immediately by FACS (Section 3.6).
3.5. Mouse infection
3.5.1. Intranasal infection of mice
Day 1 afternoon: From either a glycerol stock or a colony on a fresh plate, inoculate 2 mL of 2xYT with Yersinia. Grow overnight at 26°C with aeration.
Day 2 AM: Dilute overnight cultures 1:40 in 2 mL fresh 2xYT and allow cultures to grow for 8 h at 26°C with aeration.
Day 2 PM: Dilute 8 h cultures 1:100 into 2mL fresh 2xYT at 26°C and grow overnight with aeration (see Note 22).
Day 3: Take OD600 of overnight bacterial cultures to estimate culture concentration (CFU/ml), and dilute to appropriate CFU in 1x PBS. We normally infect mice with 500 CFU of wild-type Yersinia pseudotuberculosis IP2666 intranasally per mouse (see Note 23). Plate serial dilutions of Yersinia used in infection on LB-Irgasan plates to determine MOI. Grow at 26°C for 2 days before counting CFUs.
Administer anesthetics prior to intranasal infection (see Note 24). For isoflurane, set up anesthesia chamber with O2 at 1-2.5 psi and the isoflurane at 2.5-3 psi.
Place mice in chamber until their breathing slows and they remain completely limp upon stimulus such as rocking of the chamber.
Remove mouse from isoflurane chamber. Holding mouse at the scruff of the neck with nose pointed up, pipet 40-50 μL of bacteria in PBS slowly onto nose drop by drop (see Note 25). Make sure that the droplets are placed directly on the nostril openings. The mouse should inhale these droplets quickly. The speed of dropping the droplets should match that of the mouse’s inhalation and breathing.
Once the entire inoculum is inhaled, gently place the mouse in cage on area cleared of bedding. Monitor mouse until it regains consciousness and make sure it does not immediately cough up bacteria.
Monitor mice once or twice daily over the course of the infection.
3.5.2. Post-infection organ harvest
Collect organs in 1ml PBS in 2ml Eppendorf tubes. Keep on ice until single cell suspension is prepared (see Note 16).
Prepare single cell suspension by passing organs through 70-μm cell strainer using syringe stopper into well of 6 well plate. Add 5 ml RPMI to cell strainer and flush into well. Mix thoroughly.
Transfer 100 μL to Eppendorf tube for plating for CFU (see Note 26) and the rest to a 15ml conical tube containing a final concentration of 1mg/mL Collagenase D and 100 μg/mL gentamicin.
Incubate at 37°C for 30 minutes. Invert tubes 4-6 times.
Incubate at 37°C for another 30 minutes. At this point cells should appear dispersed within the suspension (see Note 27).
Centrifuge 250 x g at 4°C for 5 minutes. Remove supernatant. Resuspend the pellet in 5ml 1X Pharm Lyse. Incubate on ice for 5 minutes.
Centrifuge 250 x g at 4°C for 5 minutes. Remove supernatant.
Continue with step 11 of Section 3.4.2 for loading cells with CCF2/4-AM mixture and FACS staining with antibodies to surface markers.
3.6. Analytical FACS analysis
Keep all samples in the dark and on ice until they are ready to be analyzed by FACS.
Set up the FACS machine using the 409 nm laser and detection filters for 447 nm and 520 nm emission. Prepare to collect FSC vs SSC, 447nm vs 520nm, as well as all other fluor channels needed for antibody detection (see Note 28).
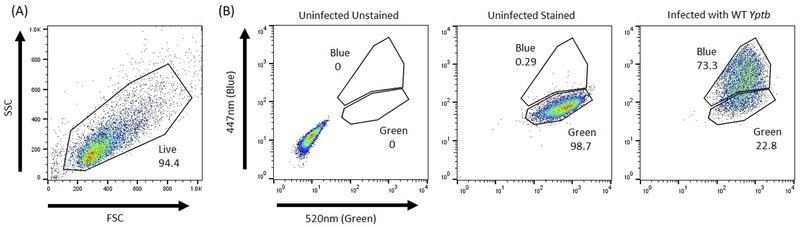
First run the unstained (non-CCF2/4-treated and no antibodies stained) control to set the voltages of the lasers (See Note 29–31, Table 1 and Figure 1a).
Run the CCF2/4-AM-treated uninfected control without antibody. This sample will define the “green-positive” or TEM− cells on the 520 nm axis as those cells that have shifted from the unstained sample (see Figure 1b).
Run the CCF2/4-AM-treated control cells infected with Yop-TEM expressing Yersinia but not treated with any antibodies (see Note 11). If Yop-TEM translocation occurred, there will be a shift of cells toward the 447nm axis, which are blue, or TEM+ cells. Cells that remain positive on only the 520nm axis are TEM−, which are green (See Figure 1b).
If staining cells for surface markers using antibodies, run single stained controls (See Note 20c for consideration of tandem-conjugated antibodies). Antibodies using fluorophores that are excited by the same laser may need compensation.
Run all additional samples using the voltages and channels set in steps 3-6.
After sample collection, export the data as FCS files.
Using a FACS analysis program of choice, analyze the data. See Figure 1 for an example of blue/green gating strategy.
Figure 1: Gating strategy for FACS.
(A) Representative image of using FSC vs SSC plot to gate on live cells. (B) HT29 cells were infected with WT IP2666 expressing YopE-TEM at an MOI of 10:1 for 1 hour or left uninfected. After infection, cells were harvested and stained with CCF4-AM for 20 minutes or left unstained. Samples were then analyzed by flow cytometry to determine green (520nm) and blue (447nm) cell populations, where green represents no translocation and blue represents translocation of the Yop-TEM. Left: Uninfected unstained samples are negative for both green and blue fluorescence. Middle: Uninfected stained samples are positive for green fluorescence, but negative for blue fluorescence. Right: Infected stained samples are positive for both green and blue fluorescence.
3.7. Using Yop-TEM+ and Yop-TEM− cells for functional and analytical assays
Using the same staining (see Section 3.4.2) and gating (see Section 3.6 and Figure 1) strategy, sort Yop-TEM+ and Yop-TEM− cells into sorting buffer in 15ml conical tubes (see Figure 1). Store on ice until all samples are sorted (see Note 32).
Sorted Yop-TEM+ and Yop-TEM− cells can be used in the following assays.
3.7.1. Preparation of PMNs for functional assays
For functional assays with PMNs, rest FACS-sorted cells at room temperature in sorting buffer for 1 hour.
Centrifuge at 250 x g, remove supernatant, and resuspend in 5 ml 1X Ca2+- and Mg2+-free PBS, or HBSS. Rest 30 minutes at room temperature.
Centrifuge at 250 x g, remove supernatant, and resuspend in 1X PBS or HBSS with Ca2+, Mg2+, and 5mM glucose at ~1×106 cells/ml. Rest 30 minutes at room temperature.
Shift cell suspension to 37°C for 15 minutes.
PMNs are now ready for functional assays such as production of reactive oxygen species.
3.7.2. Preparation of cells for western blotting
Spin down FACS sorted cells and lyse in SDS sample buffer for a final cell concentration of 0.25-1 × 108 cells/ml (see Note 33).
Boil samples at 95°C for 5 minutes. Run samples onto gel fresh or store in −20°C until analysis.
Load 2.5 × 105 - 1 × 106 cell equivalents on a SDS-PAGE gel - gel percentage is dependent on proteins of interest or use a 4-20% gradient gel.
Use transfer and western blotting protocols of your choice.
3.7.3. Preparation of cells for qRT-PCR
3.7.4. Preparation of cells for cytokine analysis
For in vivo mouse infections, harvest infected tissue in the presence of BD GolgiPlug according to manufacturer’s instructions, or 3 μg/mL Brefeldin A (BFA). All steps following tissue harvest must be in media containing either GolgiPlug or 3 μg/ml BFA (see Note 35).
Spin down FACS sorted cells and resuspend in RPMI + 10% FBS + 3 μg/mL BFA at a concentration of ~5 × 105 - 1 × 106 cells/ml (see Note 36).
Follow manufacturer’s instructions for BD Cytometic Bead Array (see Note 37).
Alternatively, follow intracellular staining protocol of choice. Most antibody manufacturers have an established protocol optimized for their reagents (see Note 38 for brief intracellular staining protocol, and Note 39 for consideration of cytokine antibodies).
Using gating strategy from Section 3.6, analyze by FACS.
3.8. High-Throughput Analysis by plate reader:
Day 1: Seed cultured cells in a 384-well plate: 1×104 cells/well in 25 μL RPMI with 5% HI-FBS or DMEM with 10% HI-FBS.
Day 1 afternoon: Prepare overnight cultures of Yersinia for infection: Inoculate 2 mL 2xYT plus antibiotics with Yersinia Yop-TEM translational fusion strain. Incubate overnight at 26°C with aeration.
Day 2: Dilute overnight Yersinia cultures 1:40 in 2 mL low Ca2+ 2xYT (see Note 8 and 9).
Grow Yersinia for 2 hours shaking at 26°C, and then shift to 2 hours shaking at 37°C.
Take OD600 of cells to estimate culture concentration, dilute in tissue culture media to desired concentration in 2 μL.
Plate serial dilutions of Yersinia used in infection on LB-Irgasan plates to determine MOI. Grow at 26°C for 2 days before counting CFUs.
Using robotic machine, add 3 μL of the 10X CCF2-AM mixture to each well of 384 well plates (see Note 40)
Incubate plate at 30°C for 30 minutes.
Add small molecule compound libraries with robotics, if desired.
Infect with Yersinia at the desired MOI. Include at least 8-12 wells/plate with each of the following controls: cells with CCF2/4-AM, cells alone, and CCF2/4-AM alone to control for background fluorescence signal. Use WT Yersinia expressing Yop-TEM as a positive control and a T3SS-defective Yersinia as a negative control on each plate.
Spin plate at 20 x g for 3 minutes.
Place plate in tissue culture incubator and allow infection to proceed for 1-2 hours.
To stop infection, add 100 μg/ml gentamicin to each well.
Use plate reader to measure blue fluorescence (447 nm) before green fluorescence (520nm) (see Note 41).
Acknowledgements
We thank all previous Mecsas laboratory members who used and optimized this technique in various procedures for their insights and notes. This work was supported by NIH AI R01 AI113166.
Notes:
Any compatible bacterial allelic exchange vector containing kanamycin resistance and a SacB gene can be used, for example pRS47 or derivatives of pCVD442 that have replaced the ampicillin resistance cassette with another drug resistance cassette (25). Alternatively, Yop-TEM fusion constructs can be generated on replication competent vectors, such as the pMMB series of vectors that are selectable with antibiotics other than ampicillin (9). If using replication competent plasmid, plasmid can be introduced by electroporation.
- TEM1F 5’ GAGAGAGCGGCCGCCACCCAGAAACGCTGGTG 3’
- TEM1R 5’ AGACAGAGCTCGCATGCTGAGTAAACTTGGTCTGACAGT 3’
To inactivate the complement system, and generate heat-inactivated (HI)-FBS, incubate thawed FBS at 55°C for 30 minutes. Complement factors can kill Yersinia, which can affect the multiplicity of infection (MOI) on the cells and skew CFU plating.
Instead of a native yop promoter, an inducible promoter such as IPTG, arabinose, or Tet promoter can be used if expression needs to be uncoupled from control of T3SS system
Ensure that Yop coding sequence is in frame with the TEM and that there are no stop codons between the two open reading frames.
The gene encoding the Yop-TEM construct can be inserted at a neutral site in the virulence plasmid or chromosome, if desired.
Yersinia should be grown at 26°C; E. coli can be grown at 37°C, but conjugation is performed at RT or 26°C overnight. When growing Yersinia in liquid culture, Irgasan should not be added to media. Irgasan is only added to agar plates.
Low Ca2+ 2xYT should always be made fresh from filter sterilized or autoclaved components.
The OD600 of overnight cultures should be measured to normalize the ODs before doing the 1:40 dilutions to best ensure comparable numbers of bacteria after 4 hours.
For adherent epithelial-like cell lines such as Hela, HEp-2, Caco-2 and HT-29 cells, 0.8-1.0 × 105 cells/well/24-well plate are added in 1 mL DMEM with 10% heat-inactivated (HI)-FBS. For adherent macrophage cell lines, such as RAW264.7 or J774, 2 × 105 cells/well/24-well plate can be seeded in 1 mL DMEM with 10% heat-inactivated (HI)-FBS. Depending on the growth rate of each cell line, adherent cells should be at ~80-90% confluency on the day of Yersinia infection. For adherent cells, adjust concentration based on surface area of 6, 12, 48, 96 or 384-well plates. For suspension cells, such as Jurkat cells and Raji B, cell concentrations can range from 2 × 105 – 1 × 106 cells/ml.
Leave at least two wells that are not infected with bacteria as negative controls. Add CCF2/4 to one of these wells. Also include WT Yersinia without Yop-TEM construct or a T3SS mutant expressing Yop-TEM as a negative control for translocation.
Epithelial cells, such as Hela and HEp-2 cells infected with T3SS-competent Yersinia expressing YopE should begin to round up after about 1 hour when infected at an MOI of 10:1 or greater, which will be visible under light microscopy. This is not apparent for most immune cells due to the fact that they are usually round.
All of the following CCF2/4-AM and antibody staining steps need to be kept in the dark as much as possible. It is also critical that CCF2/4 mixture be prepared fresh for each experiment and that it be kept in the dark. This includes keeping the samples that will be run on the FACS machine wrapped in foil until time to run the samples.
CCF2 vs. CCF4: CCF4 was engineered to have better solubility in aqueous solutions; however, our unpublished observations indicate that CCF4 is more rapidly turned over within cells compared to CCF2. Thus for applications where cells are loaded prior to infection (25), CCF2-AM is a better substrate because it lasts longer. In ex vivo and in vivo tissue infections, either CCF2-AM or CCF4-AM can be used as long as loading of cells with CCF2/4-AM occurs after infection and translocation of Yop-TEM.
This signal quenches within 30 seconds. Be quick!
To liberate all cells in spleens, including dendritic cells from spleen samples, perfuse tissues with 1 mg/mL Collagenase D for 30 minutes at 37°C prior to passing through filter. To perfuse spleens, inject several areas of the spleen with Collagenase D using a 30-gauge needle and syringe using about 50 μl of Collagenase D solution. Submerge the spleen in a small volume of the solution.
To help liberate cells from lungs, invert tubes 2-3 times every 15-20 minutes.
Alternatively, all the staining steps can be done in 96-well round, U, or V bottom plates. The resuspended cells in FACS buffer + 100 μg/mL gentamicin should be aliquoted to an appropriate number of wells in the plates. Typically, 50,000 cells should be stained per condition. Do not add more than 250 μl of the suspension to each well in the 96-well plate as larger volumes will contaminate adjacent samples (see Table 1).
At least one unstained (no antibodies/no CCF2/4-AM added) control and single antibody and CCF2/4-AM-only control samples are required. Also, set aside an aliquot from uninfected and infected CCF2/4-AM-treated cells to serve as gating controls (see Table 1).
- To titrate antibody, aseptically obtain cells (i.e. from the spleens or lungs) of a mouse. Prepare single cell suspension and lyse RBCs (see Method 3.5). Leave one unstained sample for gating control. Add a single antibody at 1:50, 1:100, 1:250, 1:500 dilution in FACS buffer to cell suspension.
- The emission spectra of fluorescent conjugated antibodies should not overlap with those of CCF2/4-AM, namely 447 and 520 nm. Good choices are PE, PE-Cy5, PE-Cy7, and APC, but not FITC and PacBlue.
- The tandem-conjugated antibodies such PE, PE-Cy5, and PE-Cy7 often require greater compensation than APC. Therefore, it is necessary to have single-stained controls to set laser intensity (voltages) and compensation.
- CD45: all WBCs
- CD11b: all phagocytes
- CD11b+ Ly6G+: PMNs only
- CD11bint CD11chi: alveolar macrophages
- CD11bint CD11chi: dendritic cells
- Gr1lo CD11b+: resident monocytes
- CD4+ TCRβ+: CD4 T cells
- CD8+ TCRβ+: CD8 T cells
- B220+ CD19+: B cells
Preparation of overnight cultures for infection of mice may differ between Yersinia strains and laboratories.
Required infectious doses will vary greatly depending on the Yersinia strain, mouse strain and route of infection. To detect TEM+ cells, overall translocation levels above 2-3% are required because the background CCF2/4+ staining in non-infected cells is around 0.5-1% of cells. Typically, translocation levels of 5-75% are optimal. To achieve these levels, the MOI and/or length of infection may need to be optimized.
General anesthetics can be administered to mice either as injectable (i.e. ketamine) or inhaled (i.e. isoflurane). Consideration for choice of anesthetics include strain, age, weight, disease model, and experimental protocol (27).
Intranasal delivery of bacteria requires 40-50μL of PBS to ensure that the inoculum will reach the lungs. A lower volume will more likely result in the infection of the nasopharynx.
A portion of the cell suspension can be diluted 1:1 in 50% glycerol and frozen for future repeated plating of CFU. Additionally, the cell suspension can be spun down at 17,000 x g, and the supernatant can be used for ELISAs assays or cells can be used for qRT-PCR.
If the cells are aggregating towards the bottom of the tube or appear to be clumping when tube is inverted, pellet the cell suspension at 250 x g for 5 minutes, and resuspend the pellet in fresh RPMI with 1mg/ml Collagenase D + 100 μg/ml gentamicin for an additional hour at 37C.
Both FACS and plate reader machines should be equipped with a 409nm laser for excitation, as well as 447nm and 520 nm filters for emission.
Make sure the unstained cells are visible on both the forward scatter control (FSC) and side scatter control (SSC). FSC measures the size of the cells, while SSC measures the granularity (internal complexity) of the cells.
The voltages of the lasers (i.e. 409 nm laser for CCF2/4) that will be used to analyze the FRET should be set to establish a negative population with the unstained control sample. Most cells emit a low level of fluorescence when excited (“autofluorescence”), and, therefore, will show up on the plot of 447 nm vs. 520 nm and relevant antibody fluors. This serves as the “negative” population for these channels. For all subsequent samples, cells that appears on the plot above this baseline “negative” population will be considered as positive cells (see Figure 1).
Each setup and gating strategy is subjective to the person running the samples. It is critical that these settings are not changed between samples within the same experiments. They may be different between experiments. The investigator must note all settings to compare data analysis appropriately across multiple experiments.
The rate of sorting will depend on the concentration of cells in each tube and FACS sorter. If a tube or series of tubes takes greater than 30 minutes, it is optimal to stagger your loading with CCF2/4 and staining of surface antibodies in order to reduce CCF2/4 turnover.
The number of Yop-TEM+ and Yop-TEM− cells can vary between mouse, organs, cell populations of interest, and strains used for infection. Thus, pooling cells from greater than one mouse is typically required so that at least 5 × 105 will be analyzed by western blot. A total of approximately 1-2 × 106 Yop-TEM+ and Yop-TEM− can be collected per wild-type Yersinia-infected mouse lung made up of 20-70% neutrophils, 1-4% alveolar macrophages, ~5% resident monocytes, ~5% dendritic cells, 2-10% CD8 T cells, 5-20% CD4 T cells, and 5-15% B cells.
Different cell types will have different RNA quantities and extraction efficiency; for example, efficiency of RNA extraction from neutrophils is significantly lower than other cells found in lungs or spleens. Ideally, 1 μg of RNA/condition is used as template for qRT-PCR, but smaller amounts can be used, if required. Frequently, we have found that we can use TEM+ and TEM−, PMNs and macrophages from one mouse lung or spleen for qRT-PCR analysis, in contrast to the number of mice required for Western Blot analysis and cytokine analysis.
Ex vivo addition of either GolgiPlug or 3 μg/ml BFA to cells blocks their intracellular protein transport processes preventing cytokines from being secreted into the supernatant during tissue processing. Optimal BFA concentration is dependent on cell types, and may need titration depending on manufacturer’s recommendation. Too low will not sufficiently block protein transport, and too high will cause cells to die during culture.
Pooling cells between multiple mice might be necessary. Generally, for intracellular staining, 50,000 cells are stained per antibody combination, but as few as 1 × 104 or as many as 2 × 105 may need to be stained. For intracellular staining, cells may need to be incubated and/or stimulated ex vivo for 1-6 hours (depending on cell types) for accumulation of the cytokines to reach a certain concentration (depending on the antibody and the cytokine) allowing for detection by flow cytometry
For BD Cytometic Bead Array, it might be necessary to dilute test samples of known concentration or assumed to contain high levels of a give cytokine by a desired dilution factor. For each test sample, we recommend making a couple of dilutions (i.e. 1:2, 1:10, or 1:50) to ensure that the median fluorescence values will fall within the generated standard curve.
Typical intracellular staining protocols for sorted cells may include 1) an initial wash with FACS buffer, 2) fixation with 1-4% paraformaldehyde (PFA) at room temperature for 10-20 minutes, 3) two washes with 0.1% saponin in FACS buffer for permeabilization, 4) intracellular stained with antibodies for cytokines in 0.1% saponin in FACS buffer, 6) a final wash with 1x PBS, and 7) suspension in 1x PBS for FACS analysis.
When choosing antibodies for cytokine analysis, avoid fluorophores that have excitations or emissions overlapping with CCF2 staining (excitation: 409nm/emission: 447nm and 520 nm), or any previously used surface antibodies. There will be residual CCF2 staining post-fixation, and antibodies to surface markers will still be detected.
CCF2-AM is preferred because it has a longer half-life and must be added before infection occurs. To ensure sterility of the robotics machine and prevent contamination with a BSL2 pathogen, all compounds are added before Yersinia.
To determine the values for green and blue fluorescence in each well, please see the Methods of: Harmon DE, Davis AJ, Castillo C, et al (2010) Identification and characterization of small-molecule inhibitors of Yop translocation in Yersinia pseudotuberculosis. Antimicrob Agents Chemother 54:3241–54 (25).
References:
- 1.Gemski P, Lazere JR, Casey T, et al. (1980) Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect Immun 28:1044–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Portnoy DA and Falkow S (1981) Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J Bacteriol 148:877–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornelis GR, Boland A, Boyd AP, et al. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev 62:1315–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewoody RS, Merritt PM, and Marketon MM (2013) Regulation of the Yersinia type III secretion system: traffic control. Front Cell Infect Microbiol 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung LK and Bliska JB (2016) Yersinia versus host immunity: how a pathogen evades or triggers a protective response. Curr Opin Microbiol 29:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinaud L, Sansonetti PJ, and Phalipon A (2018) Host Cell Targeting by Enteropathogenic Bacteria T3SS Effectors. Trends Microbiol [DOI] [PubMed] [Google Scholar]
- 7.Charpentier X and Oswald E (2004) Identification of the Secretion and Translocation Domain of the Enteropathogenic and Enterohemorrhagic Escherichia coli Effector Cif, Using TEM-1 nl-Lactamase as a New Fluorescence-Based Reporter. J Bacteriol 186:5486–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zlokarnik G, Negulescu PA, Knapp TE, et al. (1998) Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279:84–8 [DOI] [PubMed] [Google Scholar]
- 9.Marketon MM, DePaolo RW, DeBord KL, et al. (2005) Plague Bacteria Target Immune Cells During Infection. Science (80-) 309:1739–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand EA, Maldonado-Arocho FJ, Castillo C, et al. (2010) The presence of professional phagocytes dictates the number of host cells targeted for Yop translocation during infection. Cell Microbiol 12:1064–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köberle M, Klein-Günther A, Schütz M, et al. (2009) Yersinia enterocolitica targets cells of the innate and adaptive immune system by injection of Yops in a mouse infection model. PLoS Pathog 5:e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sory MP and Cornelis GR (1994) Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol 14:583–94 [DOI] [PubMed] [Google Scholar]
- 13.Sutcliffe JG (1978) Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A 75:3737–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Callaghan CH, Morris A, Kirby SM, et al. (1972) Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother 1:283–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maldonado-Arocho FJ, Green C, Fisher ML, et al. (2013) Adhesins and Host Serum Factors Drive Yop Translocation by Yersinia into Professional Phagocytes during Animal Infection. PLoS Pathog 9:e1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pechous RD, Sivaraman V, Price PA, et al. (2013) Early Host Cell Targets of Yersinia pestis during Primary Pneumonic Plague. PLoS Pathog 9:e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paczosa MK, Fisher ML, Maldonado-Arocho FJ, et al. (2014) Yersinia pseudotuberculosis uses Ail and YadA to circumvent neutrophils by directing Yop translocation during lung infection. Cell Microbiol 16:247–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolán HG, Durand EA, and Mecsas J (2013) Identifying Yersinia YopH-targeted signal transduction pathways that impair neutrophil responses during in vivo murine infection. Cell Host Microbe 14:306–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dewoody R, Merritt PM, and Marketon MM (2013) YopK controls both rate and fidelity of Yop translocation. Mol Microbiol 87:301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller B, Mühlenkamp M, Deuschle E, et al. (2015) Yersinia enterocolitica exploits different pathways to accomplish adhesion and toxin injection into host cells. Cell Microbiol 17:1179–1204 [DOI] [PubMed] [Google Scholar]
- 21.Autenrieth SE, Linzer T-R, Hiller C, et al. (2010) Immune evasion by Yersinia enterocolitica: differential targeting of dendritic cell subpopulations in vivo. PLoS Pathog 6:e1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Tam JW, Mena P, et al. (2015) CCR2+ Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8+ T Cell Response to the Protective YopE69-77 Epitope during Yersinia Infection. PLoS Pathog 11:e1005167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houppert AS, Kwiatkowski E, Glass EM, et al. (2012) Identification of chromosomal genes in Yersinia pestis that influence type III secretion and delivery of Yops into target cells. PLoS One 7:e34039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dewoody R, Merritt PM, Houppert AS, et al. (2011) YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol Microbiol 79:1445–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmon DE, Davis AJ, Castillo C, et al. (2010) Identification and characterization of small-molecule inhibitors of Yop translocation in Yersinia pseudotuberculosis. Antimicrob Agents Chemother 54:3241–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan MC, Wong WR, Dupzyk AJ, et al. (2014) An NF-κB-Based High-Throughput Screen Identifies Piericidins as Inhibitors of the Yersinia pseudotuberculosis Type III Secretion System. Antimicrob Agents Chemother 58:1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gargiulo S, Greco A, Gramanzini M, et al. (2012) Mice Anesthesia, Analgesia, and Care, Part I: Anesthetic Considerations in Preclinical Research. ILAR J 53:E55–E69 [DOI] [PubMed] [Google Scholar]