Abstract
The first steps in phage lysis involve a temporally controlled permeabilization of the cytoplasmic membrane followed by enzymatic degradation of the peptidoglycan. For Caudovirales of Gram-negative hosts, there are two different systems: the holin-endolysin and pinholin-SAR endolysin pathways. In the former, lysis is initiated when the holin forms micron-scale holes in the inner membrane, releasing active endolysin into the periplasm to degrade the peptidoglycan. In the latter, lysis begins when the pinholin causes depolarization of the membrane, which activates the secreted SAR endolysin. Historically, the disruption of the first two barriers of the cell envelope was thought to be necessary and sufficient for lysis of Gram-negative hosts. However, recently a third functional class of lysis proteins, the spanins, has been shown to be required for outer membrane disruption. Spanins are so named because they form a protein bridge that connects both membranes. Most phages produce a two-component spanin complex, composed of an outer membrane lipoprotein (o-spanin) and an inner membrane protein (i-spanin) with a predominantly coiled-coil periplasmic domain. Some phages have a different type of spanin which spans the periplasm as a single molecule, by virtue of an N-terminal lipoprotein signal and a C-terminal transmembrane domain. Evidence is reviewed supporting a model in which the spanins function by fusing the inner membrane and outer membrane. Moreover, it is proposed that spanin function is inhibited by the meshwork of the peptidoglycan, thus coupling the spanin step to the first two steps mediated by the holin and endolysin.
1. INTRODUCTION
1.1. Scope
Except for cell division, phage lysis is the most common cell fate on the planet. Lysis of ocean bacteria by marine phages accounts for the largest carbon flux within the oceans, estimated to be 150 gigatons annually (Suttle, 2005). Nevertheless, systematic study of the lysis process came late to phage biology. Textbooks in the late 1980s expressed the simplest possible scenario for lysis as being initiated when phage maturation was completed and effected in a single step by the production of a phage “lysozyme” (Watson et al., 1987). In fact, timing of lysis is a tightly regulated and temporally programmed event that occurs independently of phage morphogenesis, and the lysozyme is but one of multiple phage-encoded lysis proteins (Fig. 1). This review will summarize the current understanding of phage lysis systems, focusing exclusively on the large dsDNA phages of Gram-negative bacteria, where the most progress has been made. In particular, we will focus on the newest findings of the lysis pathway, the requirement for outer membrane disruption after the degradation of the cell wall, and the interesting proteins that phages use for this purpose.
Fig. 1.
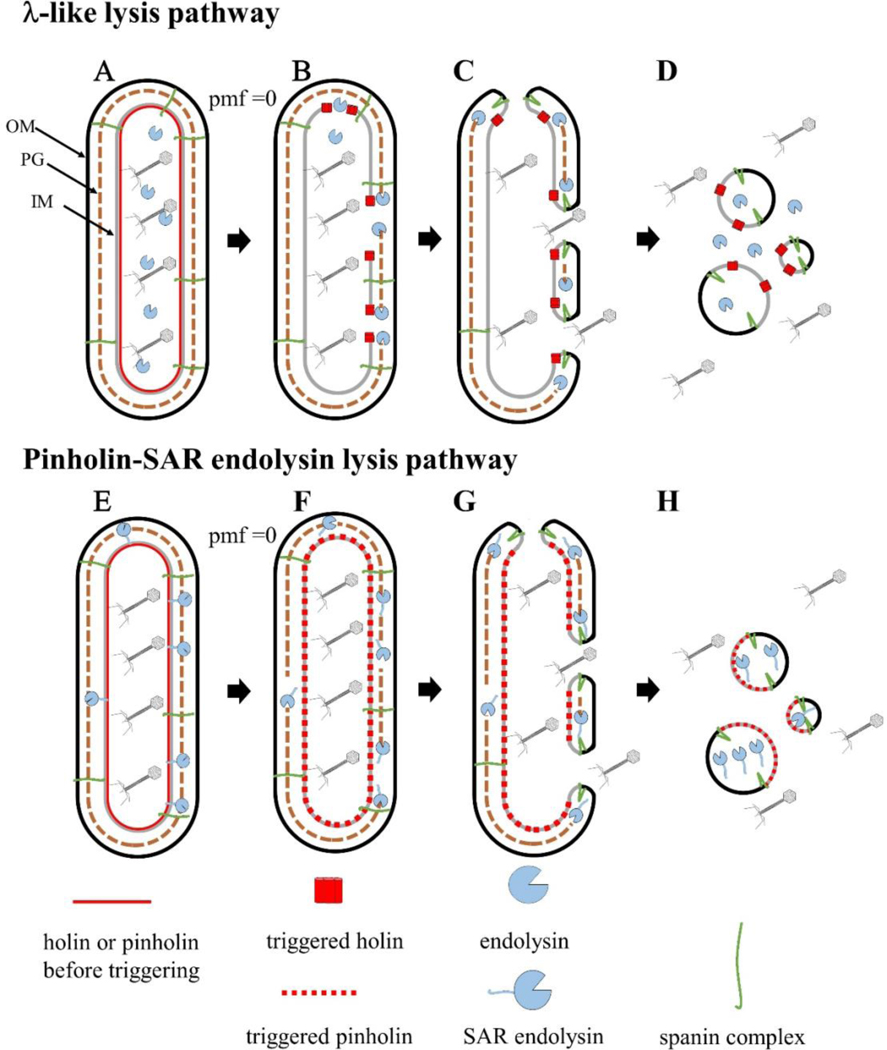
Cartoon model of the two pathways of phage lysis of Gram-negative hosts. During the late morphogenesis period, the endolysin or SAR endolysin accumulates in the cytoplasm (A) or inner membrane (E), respectively. Upon reaching a critical concentration, holin triggering results in micron-scale holes (B) or small heptameric pinholes (F), which release the endolysin into the periplasm (B), or release the SAR end- olysin from the inner membrane into the periplasm;peptidoglycan degradation occurs at this step for both pathways. PG degradation results in spanin activation, which removes the topological barrier of the outer membrane by fusing both membranes (C and G). This results in the release of phage progeny and cytoplasmic content (D and H).
1.2. Context for Lysis
Phages must overcome the barrier of the cell envelope at two points in the viral lifecycle. During infection, phages transfer nucleic acids across the cell envelope. By definition, the infection process must minimize damage to the envelope, whereas lysis is always a destructive process. Remarkably, time lapse microscopy has shown that lysis occurs as sudden explosive burst with little or no time during which the host morphology is changed (Movie 1) (Berry et al., 2012). This contrasts sharply with the gradual deformation that leads to lysis of cells exposed to antibiotics (Yao et al., 2012). Moreover, normal host physiology is maintained before this bursting event, as shown by Grundling et al. (2001) for the well-studied lysis pathway of phage lambda. Using a non-invasive assay based on flagella rotation, these authors showed that the proton motive force (PMF) is unaffected until a few seconds before lysis. The rate at which cells transition from the state of normal physiology to lysis is significant, considering the physical strength of the cell envelope, which can be inferred from studies demonstrating proliferation of E. coli under centrifugal forces >400,000 ×g (Deguchi et al., 2011).
1.3. Overview: The Establishment of the Holin-Endolysin-Spanin Lysis Model
Phage biology was long dominated by the study of a few paradigm coliphages, including the famous “Seven Dwarves,” T1 through T7, and phage lambda. Typically, it was found that lysates of these phages contained an activity that mimicked the ability of egg white lysozyme to cause lysis of E. coli cells treated to disrupt the outer membrane (OM). Jacob and Fuerst first designated this activity as “endolysin,” to indicate that it arises from within infected cells (Jacob et al., 1957; Jacob and Fuerst, 1958). It is now understood that endolysins are muralytic enzymes; i.e., enzymes produced by phages to degrade the murein or peptidoglycan (PG) layer. PG is comprised of disaccharidepeptide repeat units linked by glycosidic bonds that form glycan strands. These strands are cross-linked through pentapeptides by 4–3 and 3–3 linkages (Glauner et al., 1988). The PG confers a majority of structural protection to internal osmotic pressure of the cell (Holtje, 1998). Endolysins destroy cells by targeting linkages essential to the integrity of the PG (discussed below). In each system, the gene for the endolysin activity was usually among the first essential genes to be discovered in nonsense mutant screens and also to be associated with an absolute lysis-defective phenotype. It turned out that this uniform muralytic capacity did not imply a common biochemical mechanism. For example, in phages T4, T5, T7, and lambda, the endolysins comprised four different enzymatic activities: glycosylase (true lysozyme), endopeptidase, amidase, and transglycosylase, respectively (Oliveira et al., 2013). Holins were first discovered in T4 and lambda as genetic loci for nonsense mutations conferring not only an absolute lysis defect in liquid infections (like the endolysin genes) but also, uniquely, an intracellular hyper-accumulation of both progeny virions and endolysin activity (Josslin, 1970, 1971; Reader and Siminovitch, 1971). This led to the simple notion that the holin functioned to allow the endolysin to cross the cytoplasmic (inner) membrane (IM) and attack the peptidoglycan (PG) (Reader and Siminovitch, 1971; Wilson, 1982). Early on, it was shown that holins and endolysins from heterologous phages were able to cross-complement indicating that the membrane pores would have to be non-specific (Rennell and Poteete, 1985; Young, 1992). That, and the fact that the term porin was already in use, ultimately led to the definition of “holin” as a phage lysis protein that made non-specific “holes” in the membrane (Young, 1992). For many years, it was thought that the holin-endolysin system was necessary and sufficient for host lysis, with the additional detail that in some phages, a third protein, the antiholin, was elaborated, expressly for the purpose of regulating holin function (Bläsi et al., 1989, 1990). There were annoying problems with this broad scheme for some of the paradigm phage systems, including T1, P1, and Mu, where the endolysin was easily determined but no essential holin gene had been identified in thorough conditional mutant hunts. The sufficiency of the “two-step” model, in which the holin forms a hole in the IM, allowing the endolysin to escape and destroy the PG, resulting directly in lysis, was finally disproven in experiments involving phase contrast video microscopy of induced lambda lysogens (Berry et al., 2012). Mutations in genes Rz and/or Rz1, bizarre nested cistrons located adjacent to the S holin and R endolysin genes (Fig. 2), were shown to completely block lysis of the induced cells. Instead, the rod-shaped cells were rapidly converted to spherical forms suggesting that the PG had been destroyed (Berry et al., 2012; Cahill et al., 2017b). This implied the somewhat heretical notion that the OM was capable of withstanding the internal osmotic pressure in the absence of the PG. Later other investigators were able to conclude the same thing, based on studies of single cells undergoing lysis as a result of β-lactam antibiotic activity (Yao et al., 2012). The term “spanin” was invented for the Rz and Rz1 proteins because they were IM and OM proteins, respectively, thought to form a complex that spanned the entire periplasm (Berry et al., 2008; Summer et al., 2007). Thus was born the “three-step” model for lysis, in which three functional types of proteins...holins, endolysins, and spanins...act sequentially on the IM, PG, and OM components of the cell envelope to effect lysis.
Fig. 2.
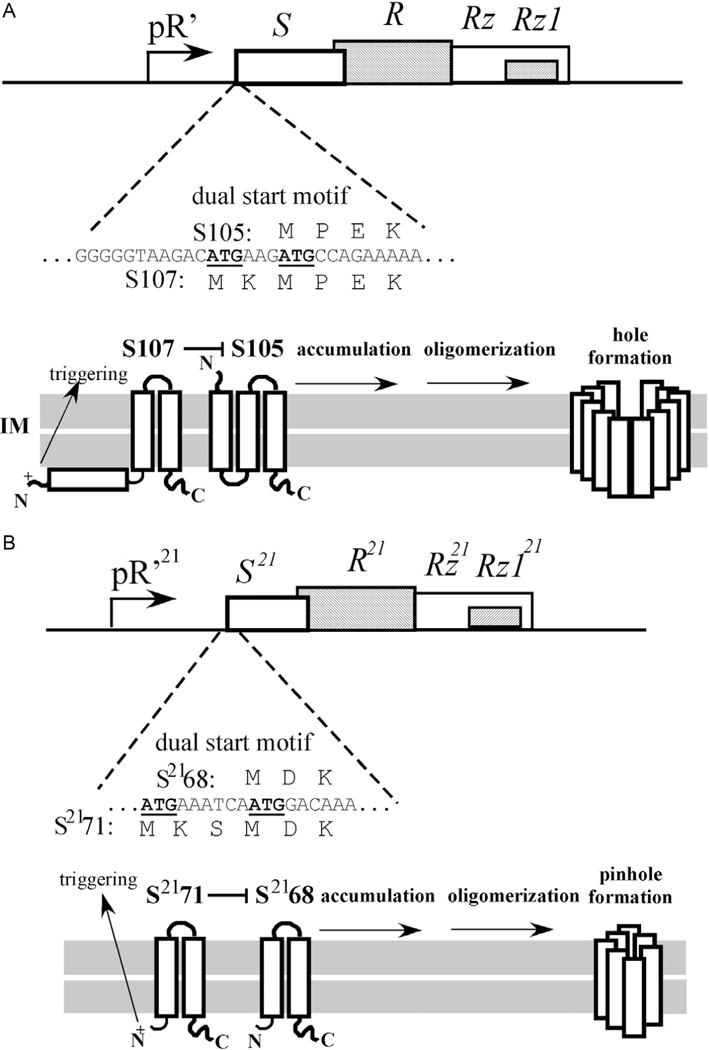
Comparison of two different holms with dual start motifs. (A) The lysis cassette of λ and the dual start motif of S (above). Below: The topology of S107 and S105. In this configuration, S107 negatively regulates S105. At the time of triggering the S107 TMD1 flips its N-terminus into the periplasm (see arrow). (B) The lysis cassette of phage 21 and the dual start motif of S21’ (above). Below: The topology of S2171 and S2168. In this configuration, S2171 negatively regulates S2168. At the time of triggering the S2171 TMD1 exits the bilayer and enters the periplasm (arrow).
1.4. Overview of Kinetic Regulation in Three-Step Lysis
The main players for lysis in most phages of Gram-negative hosts have been determined to be holins, endolysins, and spanins. The next question is how these proteins work, how their function is made sequential, and how the precise timing of lysis is determined. A general model addressing these issues has emerged (Young, 2013, 2014). First, during the entire segment of the infection cycle devoted to virion morphogenesis (usually called “late gene expression”), the lysis proteins accumulate harmlessly in their respective compartments (Fig. 1A). This continues until, at a genetically programmed time, the holin forms its lethal IM lesions, or holes, an event called “triggering.” Triggering is thus obligately associated with collapse of the PMF and cessation of macromolecular synthesis. In turn, hole formation activates the endolysin, which uses its muralytic activity for destruction of the PG (Fig. 1B). Next, the spanins are activated by degradation of the PG and cause disruption ofthe OM (Fig. 1C). The entire process occurs on a seconds timescale after triggering and is seen as an explosive event by video microscopy. Holin function thus initiates the lysis pathway and defines the length and thus the fecundity, in terms of released viable phage particles, of the infection cycle.
1.5. Three Steps But Two Choices (at Least) at Each Step
For each functional class of lysis proteins, two distinct sub-classes have been identified that have sharply different features in terms of membrane topology. Holins are either canonical holins or pinholins. As the names imply, they differ in the size and number of the membrane lesions formed upon triggering. The canonical holins form only a few micron-scale holes (Savva et al., 2014), through which the endolysins, which accumulate in the cytoplasm, escape to the periplasm and degrade the PG. In contrast, the pinholins form ~103nm scale holes (Pang et al., 2009), resulting in the sudden collapse of the PMF (compare Fig. 1B–F). Pinholins are always associated with what are termed SAR endolysins. These enzymes are secreted by the host translocon and accumulate in an inactive, membrane-tethered form in the periplasm. The membrane tether exits the membrane when the PMF is collapsed, earning the N-terminal domain the “SAR” acronym, for “signal anchor release” (Xu et al., 2004, 2005). Release from the membrane causes refolding of the SAR endolysin to an enzymatically active conformation (Fig. 1F). Thus both types of holins control endolysin-mediated degradation of the PG but do so by entirely different mechanisms.
The last step of the lysis process, disruption of the OM, also offers two completely different molecular solutions: two-component spanins, which are complexes formed from an OM lipoprotein (o-spanin) and a type II integral IM protein (i-spanin) (Fig. 3); and the unimolecular spanins (u-spanins) (Summer et al., 2007). The latter protein is unique in having both an OM lipoprotein determinant and a C-terminal transmembrane domain (TMD) that is embedded in the IM. As noted above, the functions of both types of holins and endolysins are fairly well established at the operational level, and our current understanding is described below. It is still unclear how the spanins of either class effect outer membrane disruption. Therefore, the function ofthese unusual proteins will be covered in more detail than for holins and endolysins. In particular, a model is advanced that the spanins disrupt the OM by molecular mechanisms that mimic the membrane-entry fusion proteins of mammalian viruses; that is, spanins cause IM-OM fusion as the last step in phage lysis (Rajaure et al., 2015).
Fig. 3.
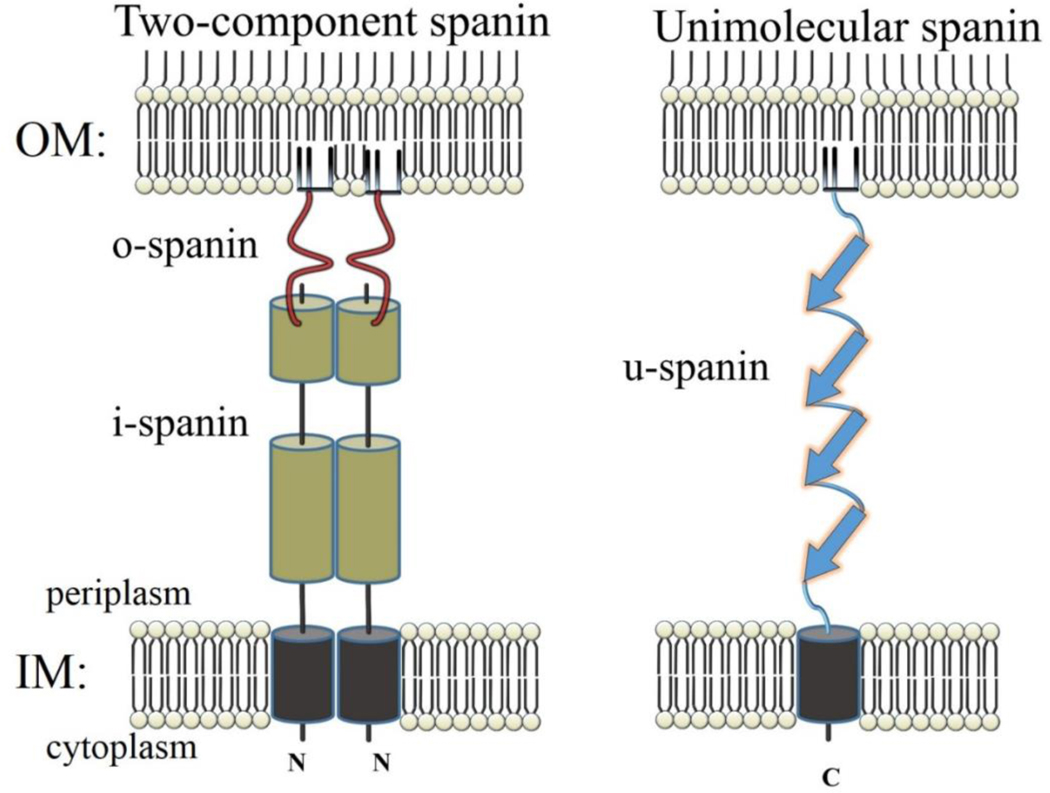
Cartoon comparing topology of two-component spanins to u-spanins, the inner membrane (IM) and outer membrane (OM). The N-terminus of the i-spanin and the C-terminus of the o-spanin are labeled. The peptidoglycan is not depicted.
In the following sections, six fundamentally different types of lysis proteins are discussed, two for each component of the envelope:
IM: (a) canonical holins and (b) pinholins.
PG: (a) canonical endolysins and (b) SAR endolysins.
OM: (a) two-component (2C) spanins and (b) unimolecular spanins.
Given the mosaic architecture and high recombination characteristics of the Caudovirales, there could be 2 × 2 × 2 = 8 possible combinations of these functional types, and in fact the combinations a-a-a, a-a-b, b-b-a, b-b-b, and a-b-a have been found in nature or constructed in the laboratory (Young, 2014). As will be developed below, the two combinations “b-a-a” and “b-a-b” turn out to be incompatible due to mechanistic reasons. We have already documented that endolysins come in at least four enzymatic flavors, but they turn out to be the least diverse of the lysis proteins. Holins, in particular, are almost unimaginably diverse and appear to have evolved many times. We will return to the diversity issue at the end and consider what it means to phage evolution and the fitness value of the lysis pathway.
2. HOLINS OF GRAM-NEGATIVE HOSTS
The best-studied holin is the S105 protein encoded by the lambda S gene, so named because a second product, S107, is also produced, from translational starts two codons upstream (Fig. 2A). S105 has three TMDs, with N-out topology (Fig. 2A) and a short, highly basic cytoplasmic domain. Two of the TMDs, 1 and 3, have moderately hydrophilic faces (Fig. 4). A detailed model for S105-mediated timing of lysis is described below. As is the case for many complex processes in membranes of living cells, the evidence for various aspects of the model is often circumstantial, indirect, and amalgamated from the complete range of biological study, from physiology to in vitro biochemistry with liposomes to structural biology (Deaton et al., 2004; Garrett and Young, 1982; Gründling et al., 2001; Savva et al., 2008; Smith et al., 1998). This mass of evidence will not be considered in detail in this work, which is more focused on the final step of the lysis process, OM disruption. Reviews on the role and mechanisms of holins and endolysins (Loessner, 2005; Oliveira et al., 2013; Wang et al., 2000) as well as more holin-centric broad overviews (Young, 2013, 2014) are available.
Fig. 4.
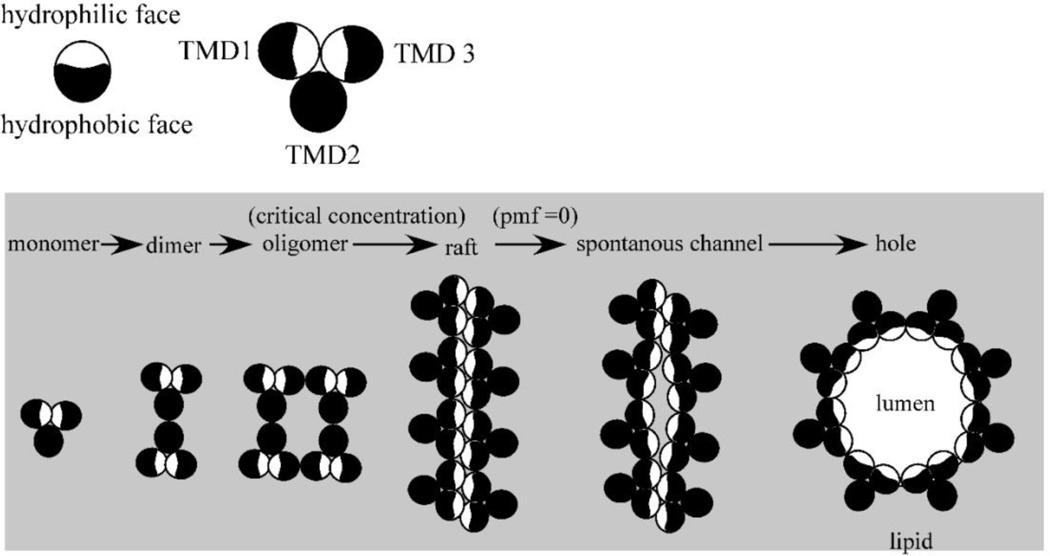
Model for the lambda holin function. A top-down view, each circle representing a TMD of S105. In the “death raft” model, the holin accumulates as dimers in which the more hydrophilic faces of TMD1 and TMD are sequestered intramolecularly. At a critical concentration, holin rafts form; here for simplicity, they are depicted as linear arrays, but the key feature is that in the rafts, many or all of the hydrophilic faces are now sequestered intermolecularly. Rafts cause local depolarization of the membrane, which in turn causes conformational changes in the holins leading to hole formation. The hydrophilic faces of TMD1 and TMD3 face the aqueous lumen of the hole (not drawn to scale).
2.1. The Lysis Pathway of the Lambda S105 Holin: How Is Lysis Timing Determined?
From the onset of late gene expression at about 8min after the beginning of the infection cycle, S105 accumulates as a homodimer in the membrane, with the hydrophilic faces sequestered against each other (Fig. 4). At some point, the homodimers reach a critical concentration, after which two-dimensional aggregates form, as was originally invoked by Krebs and Isenbarger (2000) for the assembly of bacteriorhodopsin into the “purple membrane.” It is thought that these S105 aggregates, or “death rafts,” are lipid-depleted, due to the intimate helical packing of the holins, and are thus poor insulators, leading to local reduction or collapse of the PMF. This in turn leads to alterations in the orientation of one or more of the TMDs, presumably leading to more leakage of protons, and so on. A massive and concerted reorganization of the raft into the final hole follows, resulting in a micron-scale lesion lined by a single layer of holin molecules, now with the hydrophilic faces of TMDs 1 and 3 bordering the lumen of the hole (Fig. 4). The saltatory expansion of a raft into a hole is a key feature of the model, as it solves the vexing problem for all pore-forming systems of how to displace the lipids from the pore. In this case, it is raft formation itself that does the job. Also, in this scheme, the triggering time is directly linked to a particular two-dimensional concentration of holins and thus a particular time after onset of late protein synthesis. Moreover, the triggering time/critical concentration is organically linked to the primary structure ofS105, in that even conservative changes in residues in any of the three TMDs result in dramatic changes in the timing of lysis, presumably because helix-helix packing is especially sensitive to steric clashes in the side chains of the TMDs. As discussed below, this allows lambda to explore “lysis time space” rapidly, since nearly every change in S either advances or retards triggering time and thus the length and fecundity of the infection cycle (Gründling et al., 2000; Raab et al., 1986; Zheng et al., 2008).
A key element of the S105 model is the sensitivity to the PMF. Experiments have shown that the PMF is maintained at its normal level until triggering occurs, which is why the model features the feed-forward element in that a raft of a certain size will start to short-circuit the local PMF, leading to “micro-holes,” further accelerating the loss of the PMF and rapidly resulting in the final micron-scale hole. Ryan and Rutenberg (2007) proposed a mathematical model for S105-mediated lysis timing that featured two nucleation events: first, the nucleation of a supersaturated pool of mobile holin monomers into rafts, and then an intra-raft nucleation of holes (Ryan and Rutenberg, 2007). Although crosslinking experiments suggest that the mobile holin is a homodimer, the behavior of the Ryan-Rutenberg model matches physiological observables remarkably well, especially in relating the effects of modest changes in holin-holin interaction affinities to the predicted lysis times. This model could be developed further if kinetic parameters describing the actual hole-forming process could be obtained, possibly by super-resolution fluorescence microscopy.
This model also explains an often-repeated observation dating from experiments of the Delbrück phage group in the 1940s, originally called “premature lysis” (Doermann, 1948). It was found that addition of energy poisons (e.g., cyanide or the protonophore dinitrophenol (DNP)) to an infected culture results in immediate lysis. Indeed, as phage biologists have often discovered, infected cultures begin to lyse as the result of any physical manipulation that would temporarily depress the PMF by sudden imposition of anerobiosis, like stopping the shaker and trying to collect the infected cells by filtration or centrifugation. The linkage to the current model for timed triggering is obvious; any exogenous event that causes a depression of the PMF during the holin pathway will artifactually cause triggering. There may be real biological consequence in that the holin pathway serves as the “canary in the cage” for the primary infecting phage. Superinfection by most other phages would usually cause a short-term collapse of the PMF concomitant with ejection of the gDNA from the new phage into the cytoplasm (Boulanger and Letellier, 1992; Cumby et al., 2015; Labedan and Letellier, 1981; Letellier and Labedan, 1985). The result would be instant premature triggering, which effectively “kills” the superinfecting phage and causes immediate liberation of the particles of the primary phage that have already been assembled, if any. This phage-vs-phage aspect of the holin-endolysin-spanin mode of lysis may have stimulated the evolution of the T7-like superfamily of Caudovirales, in that these very successful phages transport the gDNA into the host slowly and without collapsing the PMF (Kemp et al., 2004). This means that lambda would not be able to detect a superinfection by T7, which, considering its violent (i.e., immediate solubilization of the cellular DNA) and very short life cycle, would likely overwhelm the original infection.
2.2. Something Completely Different: The Pinholins
After its formal presentation in 1992 (Young, 1992), the holin-endolysin model did not last long as a universally applicable scheme. The first indication that another pathway existed was the fact that, despite syntenic arrangement of lysis genes, the holin gene S21 of the well-studied lambdoid phage ϕ21 did not complement the lysis defect of a null S allele in phage lambda lysis (Park et al., 2007). This led to the detailed genetic and biochemical characterization of S21, the first “pinholin” gene to be investigated. The holin, designated S2168 because it is 68 aa in length, has two TMDs, with N-in, C-in topology, and, like lambda S105, accumulates as homodimers in the bilayer (Pang et al., 2009). Unlike lambda, however, the native topology, with two TMDs in the bilayer, is not capable of triggering. Instead, TMD1 is not only non-essential for hole formation but also acts as a negative regulator of TMD2, which is the real hole-forming domain. Indeed, the TMD1 domains of both molecules of S2168 must exit the bilayer to license the resulting homodimer, stabilized by both intramembrane TMD2-TMD2 interactions and periplasmic TMD1-TMD1 interactions, to proceed down a pathway to triggering (Fig. 2B, 5A and B). Further mutational analyses, studies with S2168-GFP fusions and cysteine-accessibility experiments using single-Cys mutants revealed a markedly different triggering pathway (Pang et al., 2009,2013). Although the timing is still based on a critical concentration for triggering, the rafts that are formed are much smaller and more numerous, and ~103 “pinholes” are formed, instead of a few micron-scale holes. Cysteine-accessibility studies and cryo-electron microscopy of purified pinholes indicate that the pinhole is a homoheptamer, with the hydrophilic face ofTMD2 facing the lumen of the ~2nm hole (Panget al., 2009). Pinhole formation clearly would not permit the release of a pre-folded endolysin, so pinholins invariably require secreted endolysins, the SAR endolysins, described in the next section. Thus, the S21 and lambda pathways are operationally parallel, with the pinholin and canonical holins controlling the timing of lysis; it is just the molecular mechanisms are completely different.
Fig. 5.
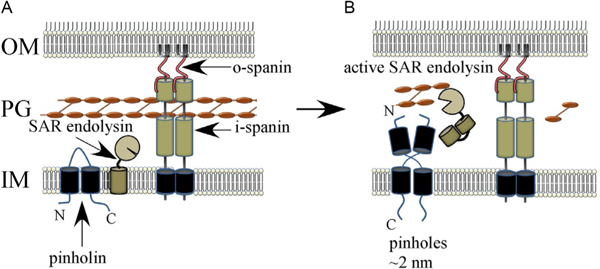
Model for pinholin and SAR endolysin function. (A) The SAR endolysin accumulates in an inactive form, tethered to the IM. The TMD1 of the pinholin is in the IM. The N-terminal domain of the pinholin exits the bilayer gradually, which leads to the formation of dimers (B) that oligomerize (not shown) and trigger to form a functional pinholin. This causes the loss of the PMF, which results in the release of the SAR endolysin from the IM and its refolding to an enzymatically active form.
2.3. Dual Starts and Intragenic Antiholins
The genes for both the canonical holin and pinholin prototypes, lambda S and ϕ21 S21, have dual translational starts and, in both cases, the upstream start serves for the translation of a second gene product, S107 and S2171, respectively, that acts as a specific antiholin (Bläsi et al., 1989; Bonovich and Young, 1991). In both cases, the ratio of holin to antiholin form is about 2:1, with a stem-loop structure in the mRNA determining the relative initiation frequency (Barenboim et al., 1999). In both cases, an extra positively charged residue in the N-terminal extension of the antiholin product blocks TMD1 from a topological change, but in opposite ways. TMD1 in S107 is unable to enter the bilayer whereas TMD1 in S2171 is unable to exit the bilayer (compare Fig. 2A and B) (Graschopf and Blasi, 1999; Park et al., 2006). In both cases, the heterodimer between the holin and antiholin is incompetent for participation in the triggering pathway, and in both cases, triggering depolarizes the membrane and thus removes the topological blockage, allowing the antiholin to assume the topology and lytic function of the holin product. Consequently, since only ~1/3 of the total holin gene products accumulate as holin-form homodimers but ~2/3 accumulate as holin-antiholin heterodimers, at the instant of triggering the amount of hole-forming molecules triples. This is a beautiful example of all-or-nothing regulation; that is, the lysis “clock” runs on a reduced amount of holin product but hole formation is amplified by the recruitment of the antiholin form.
2.4. Annotation, Diversity, and Evolution of Holins and Antiholins
Generally, in the annotation of a new phage genome, an endolysin gene can be detected by homology with one of the four classes of endolysin activities. Commonly, a nearby gene encoding a small membrane protein with two or more TMDs is designated as the holin. Lysis gene clusters are most easily recognized in phages of Gram-negative hosts since in most such “lysis cassettes,” the spanin genes with their distinctive embedded or overlapping architecture are adjacent or nearby. This is the main basis for the fact that, at the time of this review, the NIH Identical Protein database has >44 × 103 unique entries annotated as holins. In 2013, Reddy and Saier identified 52 families of holins (now 65, according to their Transporter Classification Database (TCD)) (Reddy and SaierJr, 2013; SaierJr et al., 2006), made up of small integral membrane proteins with 1–4 TMDs. Using HMM and statistical methods, these authors were able to cluster 21 of these families into 7 superfamilies. However, only a few of the genes encoding these holins have been subjected to experimental confirmation of holin function. Among the well-characterized pha ges, where both physiological and genetic methods can be brought to bear, holin function, including allele-specific triggering time, premature triggering by energy poisons, and the formation of either pinholes or micron-scale holes, has been thoroughly demonstrated only for a few phages. This includes the coliphages lambda and ϕ21, as discussed above, and also P2 (protein Y) and T4 (protein T) (Pang et al., 2010; Raab et al., 1986; Ramanculov and Young, 2001; Savva et al., 2014). P2 Y has three TMDs with the same topology as S105 and is grouped with S105 in the TCD as part of holin superfamily II. Interestingly, gene Y does not have a dual start; instead, there is suggestive physiological evidence the nearby gene lysA, which has four TMDs, may be an antiholin. This illustrates the annotation problem neatly. Imagine the difficulty in properly annotating the holin and antiholin if the P2 genome was novel. The gene cluster Y-K-lysA would show two membrane protein genes, corresponding to Y and lysA, flanking the gene for the endolysin K, which is easily identifiable based on homology to lambda R. Assignment of holin or antiholin function to either of the integral membrane proteins Y or LysA would be arbitrary, in the absence of genetic or physiological information.
2.5. T4: Signal Transduction in Lysis Control
After the lambda and ϕ21 systems, the best-studied lysis pathway is that of phage T4, where both the holin, gpi, and the endolysin, gpe, were discovered as essential lysis genes at different times during the long history of T4 genetics (Josslin, 1970; Mukai et al., 1967). Gpt is unusual among holins because of its single TMD and also because it has substantial N- and C-terminal soluble domains in the cytoplasm and the periplasm, respectively. Gpt-like holins are found only in T4-like and T5-like phages of enteric hosts. However, the timing and pathway of T4 lysis have all the same characteristics as that of lambda, including the signature critical concentration triggering, the sensitivity to energy poisons, and the formation of micron-scale holes (Ramanculov and Young, 2001; Savva et al., 2014). The major difference is that T4 lysis can be inhibited in real time. If a T4-infected cell is superinfected >5min after the initial infection, the gpt-mediated lysis clock is temporarily suspended. This is the LIN (lysis-inhibited) state, which can last indefinitely as long as superinfections continue to occur, ultimately resulting in a cell that has accumulated > 10-fold more virions than the normal ~200 (Hershey, 1946). It is not clear how the information of superinfection is conducted to the holin, but it is known that the contents of the phage head, including the 170kb gDNA and ~103 protein molecules, are ejected ectopically into the periplasm. Although the details are not known, the LIN signal flows through the RI protein of T4. RI has a SAR domain (see SAR endolysins below) that mediates its insertion into and then release from the cytoplasmic membrane and also confers extreme proteolytic instability via the periplasmic chaperone/protease DegP (Tran et al., 2007). RI blocks lysis by forming a complex with the periplasmic domain of gpt, so the LIN state reflects a competition between proteolytic degradation of RI or formation of RI-T complexes. Presumably the LIN signal affects the balance between these fates. Recently, a second T4 protein, RIII, has been shown to stabilize RI-mediated inhibition of the gpt holin (Chen and Young, 2016). RIII is a small, positively charged cytoplasmic polypeptide that binds the N-terminal cytoplasmic domain of gpt. Thus RI and RIII act together as periplasmic and cytoplasmic antiholins to impose LIN when the superinfection signal is detected. The logic of LIN is clear: superinfection is a signal reflecting the paucity of other host cells in the environment. In light of this environmental information, continuing the lysis pathway to liberate more progeny phage to face the same-host poor environment would obviously be suboptimal. Instead, involving LIN allows the phage makes optimal use of the prey at hand (to make more virions), extending the infection cycle until such time that superinfection signals cease.
2.6. Are Holins the Same in the Gram-Positive and Mycolata Domains?
The TCD classification scheme for the most part separates holins from ph ages that infect cells in the Gram-negative, Gram-positive, and Mycolata (Reddy and Saier Jr, 2013). In some cases, holins from phages of Gram-positive and Mycolata hosts have been shown to complement an S null in lambda lysis, indicating that these holins do at least form large holes (Catalao et al., 2011; Tedin et al., 1995). Although careful physiological studies have been done in some systems, detailed genetics and biochemistry has not been applied to holins outside of the classical coliphage systems. Indeed, it appears that some phages of Gram-positive and Mycolata hosts have two proteins that are both required for holin function, or even unprecedented chaperone-mediated lysis protein secretion (Catalão et al., 2010; Fernandes and São-José, 2017; Guo et al., 2018; Krogh et al., 1998). The reader is directed to several excellent and comprehensive reviews that describe these systems (Catalao et al., 2013), and, although it seems likely that holins still control the timing of lysis for all Caudovirales, interesting operational and mechanistic differences are bound to emerge if these holins receive the attention they deserve.
3. ENDOLYSINS OF GRAM-NEGATIVE HOSTS
3.1. Canonical Endolysins
For canonical holin-endolysin lysis, a key feature is that the nature of the micron-scale hole makes the molecular details of the endolysin irrelevant. This notion is reinforced by comparing the lysis cassettes of lambda and P22, two of the best-studied paradigm phages, in which the holin genes are ~95% identical but the endolysins are completely different enzymes: lambda R is a transglycosylase whereas P22 gp 14 is a true lysozyme (Bienkowska-Szewczyk and Taylor, 1980; Weaver et al., 1985). However, the ability of the diverse phage holins and endolysins to cross-complement has only been studied in bulk liquid culture. Significant differences might be revealed in single cell experiments with high-resolution phase contrast and fluorescence microscopy. For example, holins always have a small cytoplasmic domain, usually at the C-terminus and usually highly hydrophilic with basic character. It is possible that the endolysin is not distributed uniformly within the cytoplasm; weak interactions with the holin may bias its localization to the vicinity of the rafts and thus potentiate more rapid attack on the PG after hole formation. It should be noted that canonical endolysins are usually produced in great excess, as is evident from the fact that double nonsense mutants are needed to obtain a clean lysis-defective phenotype for the lambda R endolysin gene (Ry Young, Unpublished results). Phage endolysins in general have been recently been the subject of heightened interest and investigation as novel bacteriocides, or “enzybiotics” (Fischetti, 2010). This exciting development is based on the fact that most ph ages of Gram-positive hosts have endolysins that have separate cell wall binding (CWB) and enzymatic domains, usually N- and C-terminal, respectively (Fischetti, 2005). The CWB domain confers genus-specificity on the endolysin, an attractive feature for development as a useful antibiotic. Endolysins of nearly all phages with Gram-negative hosts have a single domain combining PG-binding and enzymatic activity (Briers et al., 2007). This difference probably originates from the fact that in Gram-positive cells the PG is a thick, multilayer barrier PG exposed to the medium. Among other considerations, unrestrained release of the endolysin from a cell undergoing phage lysis might cause unacceptable collateral damage to nearby cells that are potential prey. There are interesting exceptions in the case of two giant ph ages of Pseudomonas, phiKZ and EL (Briers et al., 2007), which have endolysins with an N-terminal CWB and C-terminal catalytic domain. Both enzymes were shown to have extraordinarily high enzymatic activity, ~ 100-fold more than egg white lysozyme, in cell wall degradation assays. However, nothing is known about the lysis pathway in these enormous ph ages, and the genome complexity has so far precluded identification of other lysis genes, so the significance of this unusual modularity is unknown.
3.2. SAR Endolysins
As described above, pinholins do not cause formation of macroscopic holes but, at least in the case of S2168, but instead form pinholes with nm diameters, serving only to depolarize the IM (Pang et al., 2009). Lytic function thus depends on the function of a SAR endolysin, which has a special N-terminal signal sequence that engages the host sec translocon but is not removed by leader peptidase. Accordingly, the SAR endolysin accumulates in the periplasm in a membrane-tethered form throughout the late gene expression period. This form is enzymatically inactive and its integration in the membrane is both metastable and PMF-sensitive. Depolarization of the membrane, usually mediated by pinholin triggering, results in release of the SAR endolysin from the bilayer, hence the acronym SAR corresponding to the three key topological features (signal anchor release) (Park et al., 2006). Upon release, the endolysin refolds to an enzymatically active form and attacks the PG (Fig. 5B). Several SAR endolysins have been subjected to detailed investigation, including crystallographic studies in the cases of Lyz and R21, the endolysins of the classic coliphages P1 and ϕ21, both of which are of the T4 gpe “true lysozyme” type of muramidase. Highresolution structures and in vitro biochemistry revealed that the activation pathways in these two SAR endolysins are completely different. The activation of Lyz requires dramatic refolding and disulfide bond isomerization involving a Cys residue in the newly-released SAR domain and a Cys residue in the catalytic triad (Xu et al., 2005). In R21, the active site is distorted by proximity to the membrane in the tethered form, so that the refolding to the active form is less dramatic than in Lyz (Sun et al., 2009). Genetic and biochemical analysis of a third SAR endolysin, from Erwinia phage ERA103, indicates that it also requires disulfide bond isomerization but in this case, the covalent switching relieves caging of the active site (Kuty et al., 2010).
Clearly, the key element common to the three well-characterized SAR endolysins is the SAR domain itself. Little is known about what distinguishes SAR domains from other TMDs. Sequence analysis of R21, Lyz, and other SAR endolysins revealed that the SAR domains have an enriched content of uncharged but rather hydrophilic residues Ala, Gly, Ser, and Thr residues at the expense of the hydrophobic residues like Leu and Ile that typically dominate TMDs (Oliveira et al., 2013; Xu et al., 2004). How this difference in residue content is involved in the ability of the SAR domain to exit the membrane quantitatively upon de-energization is unknown. The SAR endolysins that have been studied to date share the capacity for holin-independent lysis; that is, when expressed alone, there is a protein-specific basal level of release from the bilayer (Krupovič and Bamford, 2008; Kuty et al., 2010; Xu et al., 2004). This spontaneous release and activation are sufficiently high in phage P1 infections that in the absence of the holin, lysis still occurs, although it is significantly delayed, which explains why the holin gene in P1, lydA, is not essential for either lysis or plaque formation (Schmidt et al., 1996; Xu et al., 2004).
4. SPANINS
4.1. Studies on the Paradigm Lambda Spanin Complex
The lambda spanins Rz and Rz1 are unique in that both genes share the same DNA sequence (Fig. 6A) and are required for the same biological function (Berry et al., 2008). The Rz gene was discovered in 1979 by a transposon insertion into the lambda chromosome (Young et al., 1979). The transposon mutant was defective for lysis only in conditions where magnesium or other divalent cations were added to the medium. Although Rz was predicted to have an N-terminal TMD and lacked any recognizable enzymatic motifs, it was suggested that it might be an endopeptidase activity that had been detected in lambda lysates (Taylor, 1971) and incorrectly assigned to the R gene product (Kedzierska et al., 1996). Later, an unexpected 6.5kDa protein product was detected from expression of a DNA fragment containing a portion of the Rz gene, leading to the definition of the Rz1 cistron. Rz1 is entirely embedded in the +1 reading frame within Rz, with its own translational initiation signals (Taylor et al., 1996). The product was predicted to be a lipoprotein targeted to the OM; this was borne out by subcellular fractionation experiments (Berry et al., 2008; Kedzierska et al., 1996).
Fig. 6.
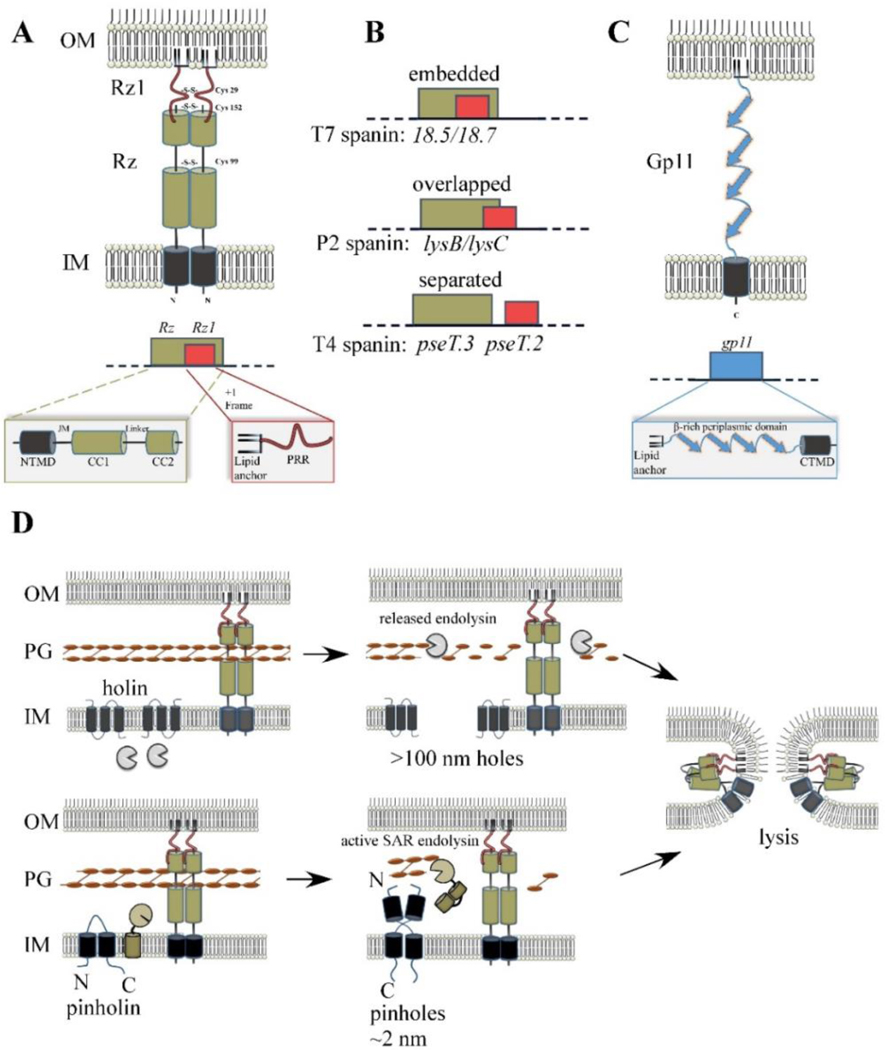
Spanin products, genes, and model for function. (A) Cartoon of λ spanin complex, comprised of Rz and Rz1, is shown within the cell envelope. The homotypic intermolecular disulfide linkages are denoted. Below: The Rz1gene is embedded within the +1 reading frame of Rz. Functional domains of the spanin complex that has been identified by genetic analysis are denoted in at relative positions respective to the predicted structural features. NTMD = N-terminal transmembrane domain. JM = juxtamembrane region, CC1 = alpha helix with predicted coiled-coil domains, CC2 = distal coiled-coil domain, PRR = proline-rich region. (B) Examples of different spanin genetic architectures, classified on the degree of overlap of the o-spanin gene (red) within the i-spanin (green). (C) The u-spanin gene product, exemplified by gp11 of T1. Below: Predicted domains of the gp11 product. (D) Model for spanin function following the canonical holin-endolysin (top) or pinholin-SAR endolysin pathway (bottom). For both pathways, spanins activate after PG degradation and undergo a conformational change that causes fusion of the IM and OM. This removes the topological barrier of the OM at the last step of lysis, which results in the release of phage progeny.
Although bioinformatic analysis showed that Rz and Rz1 equivalents are found in most phages of Gram-negative hosts (Summer et al., 2007), the two proteins remained mysterious for years and were generally referred to as auxiliary lysis functions. Finally, in a landmark 2012 study, video microscopy was used to show that, in the absence of Rz or Rz1 function, host cells convert from rod to sphere shapes instead of undergoing explosive lysis (Movie 2) (Berry et al., 2012). CryoEM imaging revealed that these spherical cells lack the PG layer and are bounded by the intact OM (Dewey et al., 2010). These data showed that indeed spanins played an essential role in lysis downstream of the holin-endolysin steps, presumably at the level of disruption of the OM. This essential role had been missed because the shearing forces attendant to the vigorous aeration used for lysis physiology experiments destroy the fragile spherical forms, thus in effect mechanically complementing the absolute lysis-defective phenotype. The term spanin was coined because in almost any perspective, the function of Rz-Rz1 would feature a complex between the two proteins linking the IM and OM; any such complex would necessarily span the entire periplasm.
4.1.1. Rz
Rz is a 153 aa type II integral membrane protein that extends ~130 residues into the periplasm. The periplasmic domain is comprised of two predicted alpha helices with a propensity for coiled-coil structure, denoted CC1 and CC2. Rz accumulates as a dimer that is linked by two homotypic intermolecular disulfide bonds (Fig. 6A) (Berry et al., 2013), although the disulfide bonds are not required for Rz dimerization in vitro (Berry et al., 2010). This was a surprising feature, in that intermolecular disulfide bonds are very unusual for bacteria; moreover, lytic function has an unusual dependency on these disulfide linkages, as will be discussed below.
The TMD of Rz was shown to be essential for lytic function but only as a membrane anchor, in that function was unaffected by replacement of the native TMD with the TMD of FtsI, a heterologous type II IM protein, or an artificial TMD comprised of random hydrophobic residues (Berry et al., 2008; Cahill et al., 2017b). Genetic analysis (discussed below) has highlighted the importance of the CC1 and CC2 domains for Rz function (Cahill et al., 2017b).
4.1.2. Rz1
Rz1 encodes a 60 aa prolipoprotein; mature Rz1 is anchored in the inner leaflet of the OM (Berry et al., 2008), as predicted from the 39 residues immediately distal to the lipoylated Cys (Kedzierska et al., 1996). Like Rz, Rz1 accumulates as a dimer, linked by homotypic intermolecular disulfide bonds at position Cys29 (Berry et al., 2013); again, see below for the role of this linkage in lytic function. In contrast to Rz, the periplasmic domain of Rz1 does not dimerize in vitro without this covalent linkage. Notably, Rz1 has high proline content: 10 of the 40 aa in the mature Rz1 sequence are proline. Genetic analysis showed that the proline-rich region (denoted PRR in Fig. 6A), including five consecutive Pro residues, plays an essential role in Rz1 function (Cahill et al., 2017b). Substitution of any of the five Pro residues except the middle one eliminates lytic function without affecting accumulation or spanin complex formation, suggesting that pentaproline sequence was directly involved in OM disruption. The essentiality of the pentaproline sequence is strikingly suggestive of a functional requirement for a PPAPP pentapeptide sequence in the reovirus p15 “fusion-associated small transmembrane” (FAST) membrane fusion protein (Top et al., 2012). The PPAPP motif in p15 forms a polyproline helix and, interestingly, the middle position in this stretch was not sensitive to missense substitutions. In the FAST fusion system, it was suggested that polyproline helix forces the exposure of neighboring hydrophobic side chains, which may play a role in the membrane fusion reaction.
4.2. The Spanin Complex
The topology and dimensions of Rz and Rz1 support the notion of a periplasm-spanning complex: 130 aa from the periplasmic domain of Rz, largely alpha-helical, could span ~20nm and the 40 aa from Rz1, iflargely in extended conformation, could span ~10nm. This is a good match to the 25nm width of the periplasm measured by cryoEM (Berry et al., 2010). Formation of a complex between Rz and Rz1 was demonstrated by co-immunoprecipitation using a functional oligohistidine-tagged Rz1 (denoted Rz1-His) (Berry et al., 2008). In a later study, similar experiments were done with mutant spanins lacking lytic function (Cahill et al., 2017b). Mutations were introduced into either Rz or Rz1-his and expressed in separate cultures. To test whether complexes were formed in vitro, lysates were solubilized in detergent, mixed, and bound to cobalt beads. In almost every case, there was co-elution of Rz and Rz1, indicating that the defect imposed by the mutations blocked spanin function at a later step, i.e., after complex formation.
Further evidence for Rz-Rz1 interaction in vivo comes from three sources. The first is the observation that expression of Rz without Rz1 results the generation of a ~12kDa proteolysis product in Rz immunoblots (Berry et al., 2013). This product was attributed to proteolysis of Rz in the region predicted to be a linker between the two coiled-coil domains (Fig. 6A); this cleavage is not detected in the presence of Rz1. Fluorescence microscopy using a functional GFP-Rz allele provided additional evidence for Rz-Rz1 interaction (Berry et al., 2012). GFP signal appears to uniformly label the IM when GFP-Rz is expressed alone but becomes punctate when co-expressed with Rz1. This provides evidence that the spanin complexes oligomerize with both Rz and Rz1 subunits are present. Recently, the E150G mutation near the C-terminus of Rz was shown to block complex formation with Rz1 in vitro (Cahill et al., 2017b). Moreover, Rz E150 and Rz1 R59 exhibited a covariant relationship in homologous protein pairs from other phages. Accordingly, expression of Rz1R59E restored function in the lysis-defective background of RzE150R, strongly suggesting that there is required salt bridge formed between these residues. However, a truncation of the Rz1 product to position 55 is functional, suggesting that the C-terminus-C-terminus interaction is complex and perhaps over-determined.
Additional evidence for complex formation comes from CD and TEM single-particle analysis of the purified periplasmic domains of Rz and Rz1 (Berry et al., 2010). The truncated proteins designated as sRz (residues 25–153) and sRzl (residues 21–60), lacking the TMD and signal sequence, respectively, were purified. CD analysis of sRz indicated less helical structure than predicted (27% vs 73% predicted), whereas sRz1 was only 6% helical, which was not surprising given its high proline content. Importantly, helicity increased significantly when the two truncated proteins were mixed in vitro, indicating that complex formation imparts structure to the Rz-Rz1, probably to the Rz segments predicted to be coiled-coil domains. When sRz and sRz1 were mixed and negatively stained, TEM imaging indicated the formation of rod-shaped structures, ~24nm in length and ~6nm in diameter, which is in agreement with measurements of the periplasm (Berry et al., 2010) and the width of the lacunae in the PG (Demchick and Koch, 1996). The width of these rods is consistent with 8–10 sRz molecules, if they are bundled like α-keratin protofilaments (Watts et al., 2002).
A potential caveat to the significance of the rod-shaped bundles has recently arisen. To avoid complications with oxidative disulfide bond formation during purification, both the sRz and sRz1 proteins were created with Ser substitutions at all the periplasmic Cys positions: Cys99 and Cys152 in Rz and Cys29 in Rz1. However, as noted above, in vivo, both Rz and Rz1 accumulate as covalent homodimers linked by disulfides at each Cys position, making the spanin complex a heterotetramer (Fig. 6A) (Berry et al., 2013). Moreover, serine substitutions are tolerated at each of the Cys positions, as long as either RzC152 or Rz1C29 is left unchanged, suggesting that a covalent intermolecular linkage is required near the C-terminal interface. This ambiguous requirement is odd, in the sense that the requirement for a covalent linkage would make more sense if it were between the Rz and Rz1 subunits, implicating the complex in a process that exerted stress on the intermembrane bridge. Instead, it appears that covalent stabilization of either the Rz or Rz1 homodimer is important and required at the heterotypic interface. In any case, the significance of the rod-shaped bundles formed from purified but Cys-less spanin subunits is uncertain.
4.3. Membrane Fusion Model for Spanin Function
The genetic, biochemical, and EM experiments described above established that the heterotetrameric spanin complex accumulates in the envelope, spanning the entire periplasm, and threaded through the PG. Inevitably, this picture evoked comparison to class I viral fusion systems and SNARE proteins and led to the proposition the spanins disrupt the OM by fusion to the IM (Berry et al., 2008). In the fusion model, spanin complexes accumulate in a metastable membrane-spanning conformation (Fig. 6A). A conserved mechanism of all membrane fusion systems is that a hairpin-like conformational change is required to drive opposing membranes close together (Martens and McMahon, 2008). In this perspective, the spanin complexes, once formed, are trapped within a single lacuna of the PG, preventing unrestrained lateral diffusion, oligomerization, and conversion to the putative hairpin conformation (Rajaure et al., 2015). This provides the necessary sequential character to the lysis pathway: PG degradation by the endolysin occurs only after holin function and in turn licenses the spanins for oligomerization and conformational changes that cause IM-OM fusion (Fig. 6D).
The membrane fusion model is supported by recent experiments with spheroplasts (Rajaure et al., 2015), which are spherical living cells created by enzymatic removal of the OM and PG from bacterial cells, with sucrose present for osmotic stabilization (Osborn and Munson, 1974). A mutant Rz1 allele, imRz1 was created in which positions 21 and 22 were converted to Asp residues; these changes block lipoproteins from Lol-dependent sorting to the OM and accordingly, redirect Rz1 to the IM. This allowed spheroplasts to be prepared from cells expressing wt Rz or the mutant imRz1, and labeled by different fluorescence proteins (Fig. 7). These spheroplasts, which have the periplasmic domains of Rz or Rz1 projecting from the surface, were shown to undergo efficient fusion requiring both subunits (Rajaure et al., 2015). The demonstration of fusogenic behavior for Rz/Rz1 is unprecedented for phage proteins. However, caution is warranted in interpreting these results, especially noting that the OM is an asymmetric bilayer that may behave very differently from the IM.
Fig. 7.
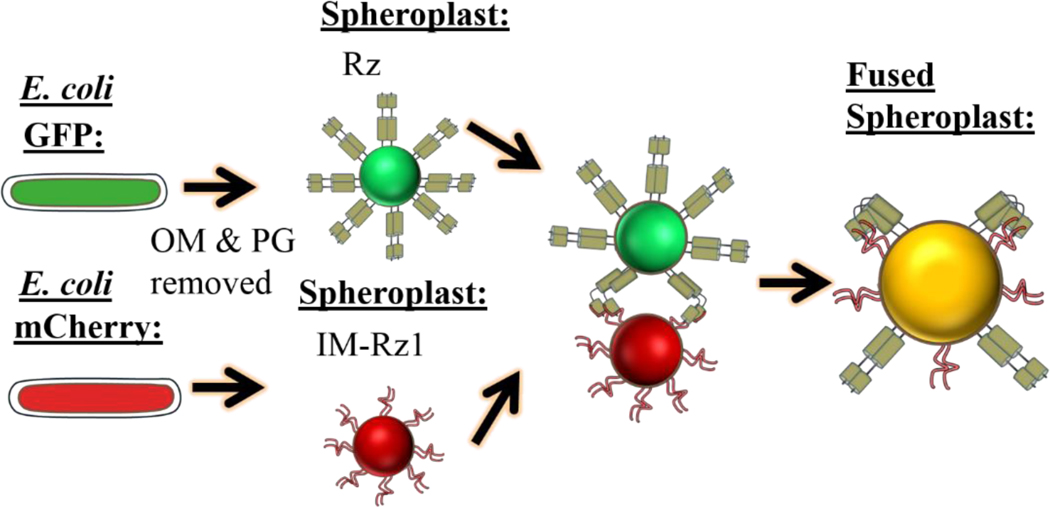
Spheroplast fusion assay. E. coli cells are induced for coexpression of GFP and Rz (green cell) or mCherry and Rz1 (red cell). Spheroplasts are formed by treating the cells with EDTA and lysozyme. IM-Rz1 is a mutant with an IM retention signal.
Further support for the membrane fusion model was derived from experiments with strains carrying mutations that confer D-Ala auxotrophy (Rajaure et al., 2015). Removal of D-Ala from the medium results in the cessation of net PG biosynthesis, so that the PG is gradually depleted. In these PG-depleted cells, induction of lambda carrying a null mutation in the holin gene resulted in lysis in a spanin-dependent manner, indicating that the spanins are causing membrane disruption directly, without participation of the holin or endolysin. The simplest notion is that upon extended limitation of PG precursors, the lacunae in the PG become sufficiently large to allow the oligomerization and concerted action of the spanin complex.
4.4. Genetic Analyses of the Spanins
Recently, two genetic analyses were conducted in order to address the mechanism of spanin function. In the first study, Rz and Rz1 were mutagenized by PCR and then cloned into separate plasmids (Cahill et al., 2017b). The plasmids were then transformed into a lysogen in which the prophage carried a null mutation in either Rz or Rz1. Induction of either library results in the intracellular retention of plasmids that fail to complement the lysis defect, generating a nearly saturated collection of mutant alleles with absolute lysis-defective phenotypes. Overall, mutations clustered within identifiable domains, including a proline-rich region in Rz1 (PRR) and coiled-coil domains CC1 and CC2 in Rz (Fig. 6A). These are domains that have been shown to be important in well-characterized membrane fusion systems, further supporting the membrane fusion model. In a follow-up study, suppression analysis was performed to look for evidence of intramolecular functional contacts that would be involved in the conformational changes predicted by the fusion model (Cahill et al., 2017a). This approach had been successful for the Kielian group in studies on the alphavirus fusogen (Liu et al., 2010). Starting with a collection of alleles with tight defects in plaque formation, a number of second-site suppressor mutations were identified, nearly all clustering within a membrane-proximal segment of Rz, denoted the juxtamembrane region (JM) (Fig. 6A) (Cahill et al., 2017a). However, these were not allele-specific, as would be expected for mutations affecting critical intramolecular contacts. Instead, the rescuing changes were polar substitutions in the hydrophobic interface of the coiled-coil in the JM region and thus were highly likely to be destabilizing to the coil. Western blotting indicated a laddering of the revertant Rz product, which is consistent with proteolytic degradation of Rz. How this rescues spanin function is not completely clear. The simplest notion is that the original missense changes block a step in the ordered conversion to the hairpin conformation of the complex to the hairpin configuration. In this view, destabilizing the structure of the JM helical region resulted in increased degrees of freedom for the complex, which could allow the hairpin form to be accessed by a bypass pathway.
4.5. Studies on the T1 gp11 Unimolecular Spanin
Rz and Rz1 constitute the prototype for the two-component spanin system, but the original bioinformatic analysis that identified analogous gene pairs in many phages of Gram-negative hosts also found that some phages have a single component system, or “u-spanin,” predicted to be single peptide connected to both membranes (Summer et al., 2007). The prototype u-spanin, gp11 of phage T1, complemented a spanin null for lambda lysis (Fig. 6C). Recently, the first genetic and biochemical characterization of gp11 was conducted (Kongari et al., 2018b). Site-directed mutagenesis studies showed that both the N-terminal OM lipoprotein signal and the C-terminal TMD of gp 11 were required. Like Rz, the TMD could be replaced with an artificial TMD sequence, indicating that the role of this domain is an IM anchor. Furthermore, a gp11-GFP chimera with the GFP moiety attached to the short cytoplasmic tail of the u-spanin was shown to be functional. Surprisingly, fluorescence microscopy of cells in which the chimera was expressed in the absence of other lysis proteins showed that the GFP signal was punctate in the membrane, indicating some oligomerization of gp11 even with the PG intact. The punctate signal distribution was dependent on the OM attachment, because mutant alleles lacking the N-terminal lipoylation signal exhibited a uniform GFP signal across the cell. The simplest interpretation of these data is that oligomers of the extended complex are threaded through the lacuna of the PG. Like the two-component spanins, premature u-spanin function is thought to be restrained by an intact PG meshwork. Supporting this idea, a strong mid-cell localization signal was observed when gp 11-GFP was co-expressed with lambda kil, which inhibits FtsZ ring formation. The authors suggested that the arrested septation might lead to regions at the mid-cell with less PG crosslinking. If so, the mid-cell signal could be evidence of the pathway of gp11 function. That is, once PG restraint is removed, there would be further oligomerization of gp 11 before gp 11-mediated fusion of the IM and OM. This idea was further supported by inductions of gene 11 in the same D-Ala auxotroph strains described above; as in the case of Rz-Rz1, the u-spanin was found to cause lysis in the PG-depleted conditions, in the absence of holin function and endolysin (Kongari et al., 2018b). The most straightforward interpretation of these data is that the PG regulates u-spanin function, and either depletion or degradation of the PG results in activation of the spanins and subsequent fusion of the IM and OM.
Despite these similarities, u-spanins have significant differences from two-component spanins. The secondary structure of the gp11 ectodomain is predicted to be dominated by beta-sheet structure and devoid of cysteine, which contrasts with the coiled-coil structure of Rz and requirement of disulfide bonds for Rz/Rz1 function (Berry et al., 2013; Kongari et al., 2018a). Regardless of the profound structural differences, there is mounting evidence for a similar endpoint for function in both spanin systems, i.e., IM/OM fusion during lysis. Therefore, u-spanin systems have been compared to class II viral fusion systems, based on the predicted beta-sheet structure, apparent oligomerization, and evidence of membrane fusion by gp 11.
Finally, an important distinction of gp 11 from Rz/Rz1 is that expression of gp11 at physiological levels is toxic, resulting in a decrease in OD ~50min after induction (Kongari et al., 2018b). Like the PG-depletion experiments described above, gp11-killed cells were rod-shaped ghosts, which indicated that gp 11 caused leakage of cytoplasmic contents. The simplest explanation for the toxicity of gp 11 is that the long interval of induction results in enough gp11 oligomerization to overwhelm the PG damping mechanism, causing gp 11-mediated IM/OM fusion. It is important to note that although gp11 is capable of causing release of cytoplasmic content independent of the holin and endolysin, employing gene 11 as a sole lysis gene would not be a viable strategy for phage release. The sacculi of these cells remain intact, which would prevent the release of larger cytoplasmic particles, such as phage. Another important point is that there may be a fitness cost for phages that use u-spanins. The eventual toxicity ofgp11 would limit the lysis clock function of the holin. In other words, the data above suggest that the holins would be unable to evolve programming for a longer latent period because early lysis would be triggered by gp 11 toxicity. This could explain why the u-spanins are less abundant than two-component systems (see below).
4.6. Diversity and Evolution of Spanins
A recent bioinformatic analysis investigated the diversity and conservation of spanin genes within 677 genomes of phages with Gram-negative hosts (Kongari et al., 2018a). Most (528 genomes) had two-component spanins that could be identified by a variety of approaches but mostly by a positional strategy requiring a type II integral IM protein to be located adjacent or very close to a predicted OM lipoprotein. Within the two-component class, three types of architectures are observed, categorized by whether the o-spanin gene was embedded within the i-spanin gene like lambda and T7 are overlapped like the P2 spanins, or completely separated like T4 spanins pseT.2 and pseT.3 (Fig. 6B). Within these, the most common genetic architecture was overlapped (Fig. 6B), with 228 genomes, whereas 182 and 118 genomes had embedded or separated architectures, respectively. The sequence diversity of the spanins was assessed by grouping spanins into families based on the amino acid identity. Families were defined so that every member shared ≥40% identity over ≥40% of the length of their sequence; these parameters would be expected to identify homologous relationships between proteins (Pearson, 2013a, b; Rost, 1999). The TMDs and OM lipoprotein signals were excluded from this analysis as non-specific localization signals. On this basis, there were 157 i-spanin families and 136 o-spanin families. U-spanins were found in 58 genomes and could be similarly grouped into 13 families. The number of singletons (one gene being the sole representative for its family) underscores the staggering diversity of each family: The i-spanin, o-spanin, and u-spanin families had 99 (out of 157 total), 65 (out of 136), and 6 (out of 13) singletons, respectively. Rz was the prototype of the largest i-spanin family with 83 members. The largest o-spanin and u-spanin families were represented by T4 pseT.3 and gp11 ofT1, respectively. Thus, as daunting as this diversity is, it would appear that the best-studied spanin systems are good exemplars for spanins in general.
The strange architecture of the two-component spanin genes is especially provocative in terms of evolution. The most parsimonious idea is that i-spanin and o-spanin genes arose as distinct genes, as in T4, but gradually contracted into the more compact overlapping and embedded architectures. Two evolutionary pressures can be invoked: first, phage genomes are always under pressure for coding space, given the packaging limitations of the phage capsid; second, the overlapped and embedded arrangements eliminate or minimize the potential for segregation of the interaction domains by recombination events. This idea for spanin evolution was supported by the finding that several families had members that cross the overlapped and separated architectural classes. In other words, T5 has an overlapped gene arrangement, whereas the spanin genes of phage Shivani are separated; however, both T5 and Shivani have spanin genes in the same family (Kongari et al., 2018a). It could be inferred that these genes have been “caught in the act” of merging. The most plausible model for the merger mechanism would be for the entire spanin locus to be duplicated and then a series of deletions and mutations on one locus would lead to the merger of one overlapped spanin pair. Then, if the nascent overlapped pair was functional, the pressure to maximize coding density would lead to deletion of the original locus.
4.7. Other Ways to Disrupt the OM?
A striking finding from the spanin bioinformatic analysis is that in 13% of the genomes, no-spanin genes could be identified. For most of these, the confidence for the absence of spanin genes is high, since both types of spanin systems have a protein with a signal peptidase II signal, or lipobox, and thus the absence of predicted lipoproteins rules out the presence of a spanin (Kongari et al., 2018a, b). It was suggested that there may be conditions where phages do not need to disrupt the OM for lysis. This may explain why a number of cyanobacterial phages are among the no-spanin group. However, some phages in this group have hosts for which many spanin-encoding phages are known. For example, Petty is a phage of Acinetobacter, which has many phages with annotated spanins. Moreover, Petty is a member of the ϕKMV-like podophage family with a small (40 kb) genome with all genes arranged on one strand. No reading frame annotated or otherwise has a potential lipobox. All but six of the late genes have unambiguous annotations, including a standard class I canonical holin and a soluble endolysin. Only two late genes have signals that would locate the protein somewhere in the envelope, one being the holin and the other a hypothetical protein with two predicted TMDs. Nevertheless, Petty infections terminate in explosive lysis events (Hernandez-Morales et al., 2018), suggesting that it uses a novel OM disruption mechanism. Experiments are underway to test the hypothetical genes for complementation of the lysis defect of λRzam Rz1am, based on the assumption that the OM disruption protein is not specific to the OM of Acinetobacter. However, the lack of genetic tools available for Acinetobacter complicates the search for the OM disruption function. Recently we identified phage PhiKT, which infects the strain E. coli 4 s, isolated from horse feces (Knirel et al., 2015). PhiKT is also a simple PhiKT-like podophage. Preliminary results indicate that like Petty, PhiKT causes explosive lysis and studies are underway to identify the mechanism responsible for OM disruption (Jesse Cahill et al., Unpublished results 2018).
5. EVOLUTION OF MULTIGENE LYSIS SYSTEMS
5.1. Why So Many Genes When One Is Enough for Lysis?
Gram-negative hosts also are prey to small single-strand nucleic acid phages, both ssDNA and ssRNA, including phages with grand roles in the history of molecular biology like ssDNA phage phiX174 and the ssRNA phages Qβ and MS2. All of these phages accomplish lysis by a single gene, each producing a single gene lysis protein (Sgl): E for phiX174 (Hutchison III and Sinsheimer, 1966), A2 for Qβ (Karnik and Billeter, 1983; Winter and Gold, 1983), and L for MS2 (Beremand and Blumenthal, 1979). E and L are small membrane proteins (91 and 75 aa, respectively) whereas A2 is the 45kDa virion maturation protein that is present in one copy per virion but in Qβ, doubles as the Sgl. E and A2 effect lysis by inhibiting essential enzymes steps in the PG biosynthesis pathway: MurA and MraY, respectively, and have thus been called “protein antibiotics” (Bernhardt et al., 2001). The mechanism of L lysis is still unknown but it causes dyscrasia of cell wall growth without affecting net PG synthesis. More recently, the lysis protein of ssRNA phage M has been shown to inhibit MurJ, the flippase that exports Lipid II to the periplasm (Chamakura et al., 2017). In each case, the Sgl is encoded by a reading frame embedded in one of the essential genes of the phage, or in the case of Qβ, is an added function of a structural protein. In contrast, the Caudovirales use multiple genes to accomplish lysis, including the multiple functional classes and sub-classes described above and also regulatory proteins including antiholins. Superficially, one would expect a fitness advantage for phages that could reduce coding space. If it was this simple, single gene lysis systems would be the dominant lysis strategy for both large and small phages. Since no known Caudovirales have single gene lysis systems, it raises the question of what advantages are provided by a more complex lysis system?
5.2. Temporal Regulation Provides a Fitness Advantage to the Holin-Based Lysis Systems
Regulation of the timing of lysis is arguably the most important feature of the holin-dependent lysis systems. The longer the morphogenesis period, the more progeny virions phage accumulate per cell (compare Fig. 8A and B). Therefore, a holin allele with an early lysis time will have a reduced burst size (average number of plaque-forming units released per cell) (Fig. 8B). Shortening the morphogenesis period would be advantageous in a host-dense environment, where the progeny can rapidly establish new infection cycles, thereby approaching exponential increase in the population (Fig. 8C) (Abedon, 1989; Abedon et al., 2001). Conversely, delaying the onset oflysis results in a larger burst size, which would be preferable in a host-depleted environment (Fig. 8A) (Hadas et al., 1997; Wang et al., 1996, 2000). From the perspective of evolution, the extraordinary mutational sensitivity of the holin clearly provides an advantage in terms of rapid adaptation. Any particular holin allele is a single base change away from a dramatically different temporal program. In contrast, the Sgl systems are much more difficult to change in terms of timing. To date, the only mutations that affect the timing of lysis in Sgl systems are those that change the synthesis rate of the Sgl protein; most mutations have no effect or inactivate the lysis protein (Bernhardt et al., 2002; Reed et al., 2013; Zheng et al., 2008).
Fig. 8.
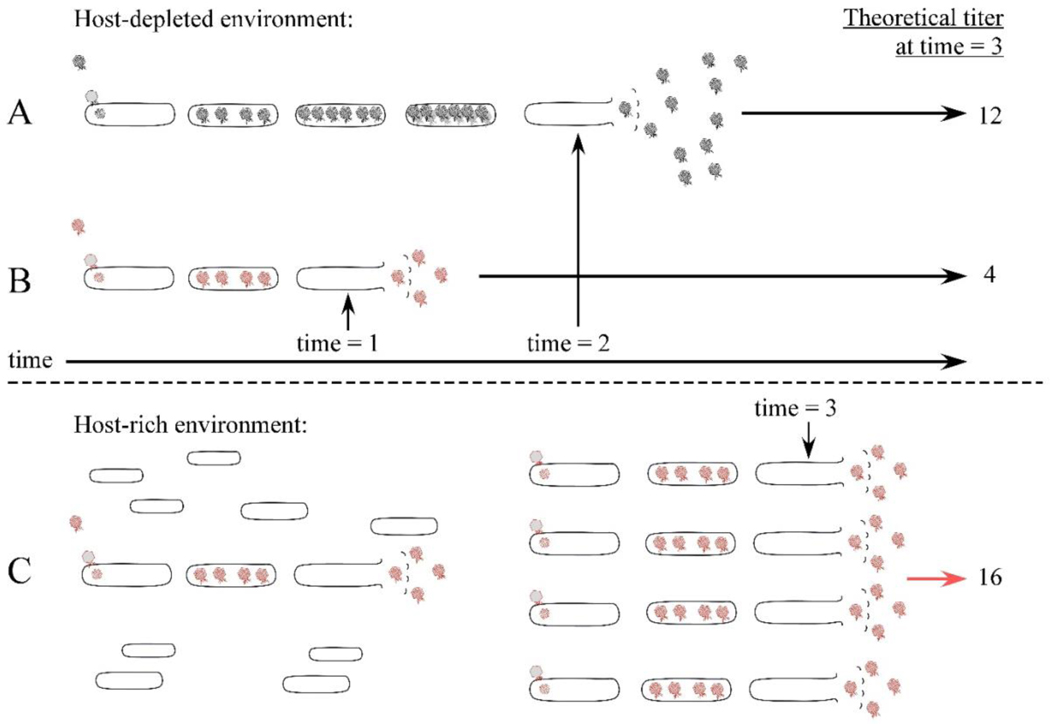
Model showing that phages with a shorter morphogenesis period have an advantage in a host-rich environment. Phages with late or early programmed lysis times are shown in black or red, respectively. The programmed lysis time for the red phage is time = 1 and time = 3 for the first and second generation, and the black phage lyses at time = 2. The late temporal program (A) has the advantage of releasing more phages in a single burst in the host-poor environment, compared to the shorter program (B). Below the dashed line: A host-rich environment will confer advantage to the early temporal program (C), since the cycles of exponential growth are shortened. Advantage is judged as the total free phage at time = 3, indicated as theoretical titer. It should be noted that for simplicity, variables such as the rate of adsorption or injection are not considered in this model.
The physiological state of the host prior to lysis might also be important in comparing the fitness value of holin-based and Sgl lysis. As described above, the holin-based programs preserve the physiological integrity of the entire cell envelope until just seconds before fatal hole formation, thus maximizing the fecundity of the infective cycle. In contrast, the Sgl systems work by inhibition or misdirection of cell wall synthesis. Therefore, there is an interval of increasing dyscrasia of the cell envelope prior to bursting and the release of the progeny. Although this has not been addressed by experiments, it seems likely that the increasing insult to the cell wall results in cellular stress responses that detract from the fecundity. In addition, at least for the three known cases where the Sgl protein inhibits an enzyme in the PG biosynthesis pathway, lysis does not occur until the infected cell attempts cell division. Thus even if mutant alleles of the Sgl protein do arise, the effect on lysis timing is masked by the progression of the cell cycle.
5.3. Evolution of Phage Lysis Systems
Some aspects of the TCD holin superfamily classification scheme cited above (Saier Jr. and Reddy, 2015) are intriguing in an evolutionary perspective. For example, the ϕ21 pinholin, which is the prototype of a very large family of similar proteins, is grouped in the same superfamily with canonical, large-hole forming holins. This suggests that pinholins and canonical holins are evolutionarily related. The most parsimonious perspective would likely be that pinholins evolved first, establishing the critical concentration- triggering time-PMF sensitivity features that underpin the ability of the holin to serve as the clock of the infection cycle. Interestingly, the membrane topology, heptameric structure, and 2.5-nm pore size of the well- studied MscS mechanosensitive channel of E. coli are all strikingly reminiscent features of the current model for the S2168 pinhole (Bass et al., 2002). The similarities extend beyond the structural level, in that the pinhole and the MscS channel both are regulated by physical parameters, the PMF and the tensile stress on the membrane, respectively, with the important difference that the mechanosensitive channel has a reversible pore opening cycle, whereas the pinholin hole formation is irreversible and lethal. The notion that large-hole holins evolved from pinholins also suggests that the first endolysins were SAR endolysins.
From this perspective, an attractive evolutionary scenario for lysis system evolution arises. Presumably, the primordial Caudovirales lacked lysis genes entirely and relied on eventual “rotting” of the dead host, consequent to the deleterious effects of intracellular phage development pathways. Acquisition of secretory muralytic enzymes from the host would give a phage a strong fitness advantage; i.e., active catalysis of host murein degradation, accelerating the release of progeny virions. However, late gene turn-on is the last transcriptional switch that occurs in phage infections, so the secreted endolysins would be accumulating in active form in the periplasm throughout the morphogenesis period. Mutations converting the signal sequence to an uncleaved SAR domain would provide a time delay between secretion and activation and presumably provide another boost in fitness. Further adaptation to an environment where accelerating or retarding the time of lysis was advantageous would be problematic. Likely the only way to adjust lysis timing would be modulating the supply of the SAR endolysin. This would mean changes in translational efficiency, since almost invariably phage endolysins are co-transcribed with essential morphogenesis cistrons. Translational signals are tiny genetic targets, and the alternative, changing the activity of the catalytic site of the muralytic enzyme, is even more difficult. The next step would be the acquisition of a pinholin gene, possibly derived from pre-existing regulated pore proteins like McsS. This would allow active regulation of the SAR endolysin and create the potential for either regulation of holin activity by an antiholin or rapid evolution to new triggering times by mutations in the TMDs of the holin. The final step, the evolution of the canonical holins, would be favored because of the inherent weakness of pinholin-SAR endolysin systems, in that the SAR endolysins “leak” out of the membrane at a certain gene-specific rate, so that lysis eventually occurs even in the absence of the pinholin (Oliveira et al., 2013). Thus pinholins have a limited dynamic range in terms regulation of lysis time. The canonical endolysins are trapped within the cytoplasm and have no effect until released by the hole-forming ability of the holin. An interesting evolutionary question might be worth addressing experimentally: can a pinholin be evolved into a canonical holin?
Supplementary Material
A culture of MG1655 cells carrying a thermally inducible lambda prophage was induced for lysis. Approximately 45 min after induction, an aliquot of cells was taken and gently covered with a coverslip on a glass slide. Lysis was monitored using an Zeiss Axio Observer inverted microscope and a 100x/1.46 NA phase-contrast objective.
A culture of MG1655 cells carrying a thermally inducible lambda prophage with a nonsense mutation in Rz was induced for lysis. Approximately 45 min after induction, an aliquot of cells was taken and gently covered with a coverslip on a glass slide. The shape conversion was monitored using a Zeiss Axio Observer inverted microscope and a 100x/1.46 NA phase contrast objective.
ACKNOWLEDGMENT
This study was supported by Public Health Service grant GM27099 and the Center for Phage Technology at Texas A&M University, jointly sponsored by Texas AgriLife.
ABBREVIATIONS
- CWB
cell wall binding
- IM
inner membrane
- LIN
lysis inhibition
- OM
outer membrane
- PG
peptidoglycan
- PMF
proton motive force
- PRR
proline-rich region
- SAR
signal anchor release
- Sgl
single gene lysis protein
- TCD
Transporter Classification Database
- TMD
transmembrane domain
REFERENCES
- Abedon ST, 1989. Selection for bacteriophage latent period length by bacterial density: a theoretical examination. Microb. Ecol. 18, 79. [DOI] [PubMed] [Google Scholar]
- Abedon ST, Herschler TD, Stopar D, 2001. Bacteriophage latent-period evolution as a response to resource availability. Appl. Environ. Microbiol. 67, 4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenboim M, Chang CY, dib Hajj F, Young R, 1999. Characterization of the dual start motif of a class II holin gene. Mol. Microbiol. 32, 715. [DOI] [PubMed] [Google Scholar]
- Bass RB, Strop P, Barclay M, Rees DC, 2002. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298, 1582. [DOI] [PubMed] [Google Scholar]
- Beremand MN, Blumenthal T, 1979. Overlapping genes in RNA phage: a new protein implicated in lysis. Cell 18, 257. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Wang IN, Struck DK, Young R, 2001. A protein antibiotic in the phage Qβ virion: diversity in lysis targets. Science 292, 2326. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R, 2002. The Escherichia coli FKBP-type PPIase SlyD is required for the stabilization of the E lysis protein of bacteriophage ϕX174. Mol. Microbiol. 45, 99. [DOI] [PubMed] [Google Scholar]
- Berry J, Summer EJ, Struck DK, Young R, 2008. The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Mol. Microbiol. 70, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Savva C, Holzenburg A, Young R, 2010. The lambda spanin components Rz and Rz1 undergo tertiary and quaternary rearrangements upon complex formation. Protein Sci. 19, 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Rajaure M, Pang T, Young R, 2012. The spanin complex is essential for lambda lysis. J. Bacteriol. 194, 5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JD, Rajaure M, Young R, 2013. Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol. Microbiol. 88, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Szewczyk K, Taylor A, 1980. Murein transglycosylase from phage λ lysate: purification and properties. Biochim. Biophys. Acta 615, 489. [DOI] [PubMed] [Google Scholar]
- Bläsi U, Nam K, Hartz D, Gold L, Young R, 1989. Dual translational initiation sites control function of the λS gene. EMBO J. 8, 3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsi U, Chang CY, Zagotta MT, Nam K, Young R, 1990. The lethallambda S gene encodes its own inhibitor. EMBO J. 9, 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonovich MT, Young R, 1991. Dual start motif in two lambdoid S genes unrelated to lambda S. J. Bacteriol. 173, 2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger P, Letellier L, 1992. Ion channels are likely to be involved in the two steps of phage T5 DNA penetration into Escherichia coli cells. J. Biol. Chem. 267, 3168. [PubMed] [Google Scholar]
- Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R, 2007. Muralytic activity and modular structure of the endolysins of Pseu-domonas aeruginosa bacteriophages phiKZ and EL. Mol. Microbiol. 65, 1334. [DOI] [PubMed] [Google Scholar]
- Cahill J, Rajaure M, Holt A, Moreland R, O’Leary C, Kulkarni A, Sloan J, Young R, 2017a. Suppressor analysis of the fusogenic lambda spanins. J. Virol. 91 (14). pii: e00413–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill J, Rajaure M, O’Leary C, Sloan J, Marrufo A, Holt A, Kulkarni A, Hernandez O, Young R, 2017b. Genetic analysis of the lambda spanins Rz and Rz1: identification of functional domains. Genes Genomes Genet. 7 (2), 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalão MJ, Gil F, Moniz-Pereira J, Pimentel M, 2010. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidogly-can. Mol. Microbiol. 77, 672. [DOI] [PubMed] [Google Scholar]
- Catalao MJ, Gil F, Moniz-Pereira J, Pimentel M, 2011. Functional analysis of the holin-like proteins of mycobacteriophage Ms6. J. Bacteriol. 193, 2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalao MJ, Gil F, Moniz-Pereira J, Sao-Jose C, Pimentel M, 2013. Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol. Rev. 37, 554. [DOI] [PubMed] [Google Scholar]
- Chamakura KR, Sham L-T, Davis RM, Min L, Cho H, Ruiz N, Bernhardt TG, Young R, 2017. A viral protein antibiotic inhibits lipid II flippase activity. Nat. Microbiol. 2, 1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Young R, 2016. The last r locus unveiled: T4 RIII is a cytoplasmic co-antiholin. J. Bacteriol. 198 (18), 2448–2457. (JB-00294). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumby N, Reimer K, Mengin-Lecreulx D, Davidson AR, Maxwell KL, 2015. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK 97. Mol. Microbiol. 96, 437. [DOI] [PubMed] [Google Scholar]
- Deaton J, Savva CG, Sun J, Holzenburg A, Berry J, Young R, 2004. Solubilization and delivery by GroEL of megadalton complexes of the lambda holin. Protein Sci. 13, 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi S, Shimoshige H, Tsudome M, Mukai S. a., Corkery RW, Ito S, Horikoshi K, 2011. Microbial growth at hyperaccelerations up to 403,627 × g. Proc. Natl. Acad. Sci. 108, 7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchick P, Koch AL, 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey JS, Savva CG, White RL, Vitha S, Holzenburg A, Young R, 2010. Micron-scale holes terminate the phage infection cycle. Proc. Natl. Acad. Sci. U. S. A. 107, 2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doermann AH, 1948. Intracellular growth of bacteriophage. In: Carnegie Institution of Washington Yearbook. vol. 47 p. 176. [Google Scholar]
- Fernandes S, São-José C, 2017. Probing the function of the two holin-like proteins of bacteriophage SPP1. Virology 500, 184. [DOI] [PubMed] [Google Scholar]
- Fischetti VA, 2005. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13, 491. [DOI] [PubMed] [Google Scholar]
- Fischetti VA, 2010. Bacteriophage endolysins: a novel anti-infective to control Grampositive pathogens. Int. J. Med. Microbiol. 300, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett JM, Young R, 1982. Lethal action of bacteriophage lambda S gene. J. Virol. 44, 886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B, Holtje JV, Schwarz U, 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263, 10088. [PubMed] [Google Scholar]
- Graschopf A, Blasi U, 1999. Molecular function of the dual-start motif in the lambda S holin. Mol. Microbiol. 33, 569. [DOI] [PubMed] [Google Scholar]
- Gründling A, Blasi U, Young R, 2000. Genetic and Biochemical analysis of dimer and oligomer interactions of the λ S Holin. J. Bacteriol. 182 (21), 6082–6090. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC94742/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A, Manson MD, Young R, 2001. Holins kill without warning. Proc. Natl. Acad. Sci. U. S. A. 98, 9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Xin Y, Zhang C, Kong J, 2018. A cytoplasmic antiholin is embedded in-frame with the holin in a Lactobacillus fermentum bacteriophage. Appl. Environ. Microbiol. 84 (6). pii: e02518–17 (AEM-02518). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadas H, Einav M, Fishov I, Zaritsky A, 1997. Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology 143, 179. [DOI] [PubMed] [Google Scholar]
- Hernandez-Morales A, Lessor L, Wood T, Migl D, Mijalis E, Cahill J, Russell W, Young R, Gill J, 2018. Genomic and biochemical characterization of acinetobacter podophage petty reveals a novel lysis mechanism and tail-associated depolymerase activity. J. Virol. 92, e01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey AD, 1946. Spontaneous mutations in bacterial viruses. Cold Spring Harb. Symp. Quant. Biol. 11, 67. [Google Scholar]
- Holtje JV, 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA III, Sinsheimer RL, 1966. The process of infection with bacteriophage ϕX174. X. Mutations in a f X lysis gene. J. Mol. Biol. 18, 429. [DOI] [PubMed] [Google Scholar]
- Jacob F, Fuerst CR, 1958. The mechanism of lysis by phage studied with defective lysogenic bacteria. J. Gen. Microbiol. 18, 518. [DOI] [PubMed] [Google Scholar]
- Jacob F, Fuerst C, Wollman E, 1957. Study of defective lysogenic bacteria. II.-physiological types resulting from prophage mutations. Ann. Inst. Pasteur 93, 724. [PubMed] [Google Scholar]
- Josslin R, 1970. The lysis mechanism of phage T4: mutants affectinglysis. Virology 40, 719. [DOI] [PubMed] [Google Scholar]
- Josslin R, 1971. Physiological studies of the t gene defect in T4-infected Escherichia coli. Virology 44, 101. [DOI] [PubMed] [Google Scholar]
- Karnik S, Billeter M, 1983. The lysis function of RNA bacteriophage Qβ is mediated by the maturation (A2) protein. EMBO J. 2, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska S, Wawrzynów A, Taylor A, 1996. The Rz1 gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene 168, 1. [DOI] [PubMed] [Google Scholar]
- Kemp P, Gupta M, Molineux IJ, 2004. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol. Microbiol. 53, 1251. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Prokhorov NS, Shashkov AS, Ovchinnikova OG, Zdorovenko EL, Liu B, Kostryukova ES, Larin AK, Golomidova AK, Letarov AV, 2015. Variations in O-antigen biosynthesis and O-acetylation associated with altered phage sensitivity in Escherichia coli 4s. J. Bacteriol. 197, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongari R, Rajaure M, Cahill J, Rasche E, Mijalis E, Berry J, Young R, 2018a. Phage spanins: diversity, topological dynamics and gene convergence. BMC Bioinf. 19, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongari R, Snowden J, Berry JD, Young R, 2018b. Localization and regulation of the T1 unimolecular spanin. J. Virol. (JVI-00380). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MP, Isenbarger TA, 2000. Structural determinants of purple membrane assembly. Biochim. Biophys. Acta Bioenerg. 1460, 15. [DOI] [PubMed] [Google Scholar]
- Krogh S, Jorgensen ST, Devine KM, 1998. Lysis genes of the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 180, 2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovič M, Bamford DH, 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6, 941. [DOI] [PubMed] [Google Scholar]
- Kuty GF, Xu M, Struck DK, Summer EJ, Young R, 2010. Regulation of a phage endolysin by disulfide caging. J. Bacteriol. 192, 5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labedan B, Letellier L, 1981. Membrane potential changes during the first steps of coli-phage infection. Proc. Natl. Acad. Sci. 78, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letellier L, Labedan B, 1985. Release of respiratory control in Escherichia coli after bacteriophage adsorption: process independent of DNA injection. J. Bacteriol. 161, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Besanceney C, Song Y, Kielian M, 2010. Pseudorevertants of asemliki forest virus fusion-blocking mutation reveal a critical interchain interaction in the core trimer. J. Virol. 84, 11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner MJ, 2005. Bacteriophage endolysins—current state of research and applications. Curr. Opin. Microbiol. 8, 480. [DOI] [PubMed] [Google Scholar]
- Martens S, McMahon HT, 2008. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9, 543. [DOI] [PubMed] [Google Scholar]
- Mukai F, Streisinger G, Miller B, 1967. The mechanisms of lysis in phage T4-infected cells. Virology 33, 398. [DOI] [PubMed] [Google Scholar]
- Oliveira H, Melo LD, Santos SB, Nóbrega FL, Ferreira EC, Cerca N, Azeredo J, Kluskens LD, 2013. Molecular aspects and comparative genomics of bacteriophage endolysins.J. Virol. 87 (8), 4558–4570. (JVI-03277). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Munson R, 1974. Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 31, 642. [DOI] [PubMed] [Google Scholar]
- Pang T, Savva CG, Fleming KG, Struck DK, Young R, 2009. Structure ofthelethal phage pinhole. Proc. Natl. Acad. Sci. U. S. A. 106, 18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang T, Park T,Young R, 2010. Mutational analysis of the S21 pinholin. Mol. Microbiol. 76, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang T, Fleming TC, Pogliano K, Young R, 2013. Visualization of pinholin lesions in vivo. Proc. Natl. Acad. Sci. U. S. A. 110, E2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Struck DK, Deaton JF, Young R, 2006. Topological dynamics ofholins in programmed bacterial lysis. Proc. Natl. Acad. Sci. U. S. A. 103, 19713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Struck DK, Dankenbring CA, Young R, 2007. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J. Bacteriol. 189, 9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, 2013a. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinformatics 42 (1). 3–1, Unit 3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, 2013b. Selecting the right similarity-scoring matrix. Curr. Protoc. Bioinformatics 43, 3.5. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab R, Neal G, Garrett J, Grimaila R, Fusselman R, Young R, 1986. Mutational analysis of bacteriophage lambda lysis gene S. J. Bacteriol. 167, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaure M, Berry J, Kongari R, Cahill J, Young R, 2015. Membrane fusion during phage lysis. Proc. Natl. Acad. Sci. U. S. A. 112, 5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanculov ER, Young R,2001. Genetic analysis of the T4 holin: timing and topology. Gene 265, 25. [DOI] [PubMed] [Google Scholar]
- Reader RW, Siminovitch L, 1971. Lysis defective mutants of bacteriophage lambda: on the role of the S function in lysis. Virology 43, 623. [DOI] [PubMed] [Google Scholar]
- Reddy BL, Saier MH Jr., 2013. Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim. Biophys. Acta Biomembr. 1828, 2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed CA, Langlais C, Wang IN, Young R, 2013. A2 expression and assembly regulates lysis in Qß infections. Microbiology 159, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennell D, Poteete AR, 1985. Phage P22 lysis genes: nucleotide sequences and functional relationships with T4 and X genes. Virology 143, 280. [DOI] [PubMed] [Google Scholar]
- Rost B, 1999. Twilight zone of protein sequence alignments. Protein Eng. 12, 85. [DOI] [PubMed] [Google Scholar]
- Ryan GL, Rutenberg AD, 2007. Clocking out: modeling phage-induced lysis of Escherichia coli. J. Bacteriol. 189, 4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr., Tran CV, Barabote RD, 2006. TCDB: the transporterclassification database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr., Reddy BL, 2015. Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J. Bacteriol. 197, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva CG, Dewey JS, Deaton JF, White RL, Struck DK, Holzenburg A, Young R, 2008. The holin of bacteriophage lambda forms rings with large diameter. Mol. Microbiol. 69, 784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva CG, Dewey JS, Moussa SH, To KH, Holzenburg A, Young R, 2014. Stable micron-scale holes are a general feature of canonical holins. Mol. Microbiol. 91, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Velleman M, Arber W, 1996. Three functions of bacteriophage P1 involved in cell lysis. J. Bacteriol. 178, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Struck DK, Scholtz JM, Young R, 1998. Purification and biochemical characterization of the lambda holin. J. Bacteriol. 180, 2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summer EJ,Berry J, Tran TAT, Niu L, Struck DK,Young R, 2007. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 373, 1098. [DOI] [PubMed] [Google Scholar]
- Sun Q, Kuty GF, Arockiasamy A, Xu M, Young R, Sacchettini JC, 2009. Regulation of a muralytic enzyme by dynamic membrane topology. Nat. Struct. Mol. Biol. 16, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA, 2005. Viruses in the sea. Nature 437, 356. [DOI] [PubMed] [Google Scholar]
- Taylor A, 1971. Endopeptidase activity of phage lamba endolysin. Nat. New Biol. 234, 144. [DOI] [PubMed] [Google Scholar]
- Taylor A, Kedzierska S, WawrzynÓW A, 1996. Bacteriophage λ lysis gene product modified and inserted into Escherichia coli outer membrane: Rz1 lipoprotein. Microb. Drug Resist. 2, 147. [DOI] [PubMed] [Google Scholar]
- Tedin K, Resch A, Steiner M, Bläsi U, 1995. Dual translational start motif evolutionarily conserved in the holin gene of Bacillus subtilis phage φ29. Virology 206, 479. [DOI] [PubMed] [Google Scholar]
- Top D, Read JA, Dawe SJ, Syvitski RT, Duncan R, 2012. Cell-cell membrane fusion induced by p15 fusion-associated small transmembrane (FAST) protein requires a novel fusion peptide motif containing a myristoylated polyproline type II helix. J. Biol. Chem. 287, 3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TA, Struck DK, Young R, 2007. The T4 RI antiholin has an N-terminal signal anchor release domain that targets it for degradation by DegP. J. Bacteriol. 189, 7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN, Dykhuizen DE, Slobodkin LB, 1996. The evolution ofphage lysis timing. Evol. Ecol. 10, 545. [Google Scholar]
- Wang IN, Smith DL, Young R, 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54, 799. [DOI] [PubMed] [Google Scholar]
- Watson JD, Hopkins NH, Roberts JW, Steitz JA, Weiner AM, 1987. Molecular Biology of the Gene. Benjamin/Cummings Publishing Co., Inc., Menlo Park, CA [Google Scholar]
- Watts NR, Jones LN, Cheng N, Wall JS, Parry DA, Steven AC, 2002. Cryo-electron microscopy of trichocyte (hard α-keratin) intermediate filaments reveals a low-density core. J. Struct. Biol. 137, 109. [DOI] [PubMed] [Google Scholar]
- Weaver LH, Rennell D, Poteete AR, Mathews BW, 1985. Structure of phage P22 gene 19 lysozyme inferred from its homology with phage T4 lysozyme. Implications for lysozyme evolution. J. Mol. Biol. 184, 739. [DOI] [PubMed] [Google Scholar]
- Wilson DB, 1982. Effect of the lambda S gene product on properties of the Escherichia coli inner membrane. J. Bacteriol. 151, 1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter RB, Gold L, 1983. Overproduction of bacteriophage Qβ maturation (A2) protein leads to cell lysis. Cell 33, 877. [DOI] [PubMed] [Google Scholar]
- Xu M, Struck DK, Deaton J, Wang IN, Young R, 2004. The signal arrest-release (SAR) sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. U. S. A. 101, 6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R, 2005. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307, 113. [DOI] [PubMed] [Google Scholar]
- Yao Z, Kahne D, Kishony R, 2012. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol. Cell 48, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R, 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Ry., 2013. Phage lysis: do we have the hole story yet? Curr. Opin. Microbiol. 16(6), 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R, 2014. Phage lysis: three steps, three choices, one outcome. J. Microbiol. 52, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R,Way S, Yin J, Syvanen M, 1979. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J. Mol. Biol. 132, 307. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Struck DK, Dankenbring CA, Young R, 2008. Evolutionary dominance of holin lysis systems derives from superior genetic malleability. Microbiology 154, 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A culture of MG1655 cells carrying a thermally inducible lambda prophage was induced for lysis. Approximately 45 min after induction, an aliquot of cells was taken and gently covered with a coverslip on a glass slide. Lysis was monitored using an Zeiss Axio Observer inverted microscope and a 100x/1.46 NA phase-contrast objective.
A culture of MG1655 cells carrying a thermally inducible lambda prophage with a nonsense mutation in Rz was induced for lysis. Approximately 45 min after induction, an aliquot of cells was taken and gently covered with a coverslip on a glass slide. The shape conversion was monitored using a Zeiss Axio Observer inverted microscope and a 100x/1.46 NA phase contrast objective.


