Abstract
Recently, numerous organizations, including governmental regulatory agencies in the U.S. and abroad, have proposed using data from New Approach Methodologies (NAMs) for augmenting and increasing the pace of chemical assessments. NAMs are broadly defined as any technology, methodology, approach or combination thereof that can be used to provide information on chemical hazard and risk assessment that avoids the use of intact animals. High-throughput transcriptomics (HTTr) is a type of NAM that uses gene expression profiling as an endpoint for rapidly evaluating the effects of large numbers of chemicals on in vitro cell culture systems. As compared to targeted high-throughput screening (HTS) approaches that measure the effect of chemical X on target Y, HTTr is a non-targeted approach that allows researchers to more broadly characterize the integrated response of an intact biological system to chemicals that may affect a specific biological target or many biological targets under a defined set of treatment conditions (time, concentration, etc.). HTTr screening performed in concentration-response mode can provide potency estimates for the concentrations of chemicals that produce perturbations in cellular response pathways. Here, we discuss study design considerations for HTTr concentration-response screening and present a framework for the use of HTTr-based biological pathway-altering concentrations (BPACs) in a screening-level, risk-based chemical prioritization approach. The framework involves concentration-response modeling of HTTr data, mapping gene level responses to biological pathways, determination of BPACs, in vitro-to-in vivo extrapolation (IVIVE) and comparison to human exposure predictions.
Introduction
High-throughput screening (HTS) is a general term that refers to large-scale experiments where combinations of robotic automation, liquid handling devices, instruments for detecting assay-specific outputs and data processing and analysis pipelines are used to evaluate the biological effects of hundreds to thousands of chemicals tested in parallel, typically in in vitro test systems [1, 2]. HTS studies are a mainstay of the pharmaceutical and agrochemical industries and are used extensively for screening of chemical libraries and identifying biologically active chemicals [3–5]. High-throughput transcriptomics (HTTr) is a type of HTS that uses gene expression profiling as a highly-multiplexed endpoint for rapidly evaluating the biological effects of large numbers of chemicals. A variety of high-throughput transcriptomic technologies can be categorized by the breadth of genes measured, ranging from dozens of genes using qRT-PCR arrays [6, 7], hundreds or thousands of genes using targeted transcriptomic panels (i.e. L1000, S1500+) [8, 9], or the whole transcriptome using microarrays, RNA-Seq or targeted RNA-Seq [10, 11]. The selection of a particular transcriptomics technology or gene panel will vary by use case. For instance, if a researcher is concerned with a specific type of toxicity (i.e. genotoxicity, phospholipidosis, etc.), then they may choose to develop a transcriptomic panel with genes of known relevance to the toxicological responses of interest [7, 12]. If less is known regarding the potential mechanism of chemical toxicity, or what targets or signaling pathways may be affected, a whole transcriptome approach may be more appropriate. Increasing efficiency and declining cost of generating transcriptomic profiles, including whole transcriptome profiles, has made HTTr a feasible approach for chemical screening and assessment.
Transcriptomic technologies have been used in toxicology, and other scientific fields, for many years. These technologies provide broad insight into the molecular signaling networks that are perturbed following chemical exposures and that underlie functional and pathological changes in tissues upon which traditional chemical risk assessments are based [13]. To date, data from transcriptomics studies have not been routinely used in chemical risk assessment dossiers submitted to regulatory agencies [14, 15]. However, transcriptomics data has been used to a limited extent in risk assessment approaches employed by private industry to support internal decision-making processes. These approaches have primarily focused on hazard identification, specifically elucidation, establishment and categorization of mode(s)-of-action in the context of a product development pipeline [15].
Depositing transcriptomics datasets in publicly-accessible databases is a near universal prerequisite for publication of such data in scientific journals. As a result, transcriptomics datasets from low-throughput in vivo and in vitro toxicology studies have been accumulating in public databases for many years [16, 17]. The combination of proprietary transcriptomics data available in some industry settings, as well as publicly-available transcriptomic databases represent a rich and largely-untapped resource for supporting chemical risk assessment. Further, in vitro chemical screening using HTTr has the potential to greatly increase the amount of transcriptomics data available for potential use in risk assessment applications. However, like low-throughout transcriptomics data, HTTr data will encounter similar hurdles to regulatory use until data analysis frameworks demonstrate technological reproducibility, data interpretability and concrete applications.
Recently, numerous organizations, including some with regulatory authority, have proposed the use of data from New Approach Methodologies (NAMs) to modernize and increase the pace of chemical assessments [18–20]. NAMs are defined as any technology, methodology, approach or combination thereof that can be used to provide information on chemical hazard and risk assessment that avoids the use of intact animals [18]. Therefore, HTTr assays performed in in vitro test systems can be considered NAMs. Specific use-cases for NAM data in risk assessment include screening-level evaluations of “data-poor” chemicals (i.e. those lacking an in vivo toxicity knowledgebase) or use in filling mechanistic knowledge gaps for “data-rich” chemicals (i.e. those where some in vivo toxicity data are available).
Also in recent years, a variety of research groups have explored how low-throughput in vitro and in vivo transcriptomics data as well as HTS data, respectively, may be used in the context of chemical assessment [21–26]. The utility of transcriptomics for risk assessment has been evaluated in a number of areas including: computational approaches for concentration-response modeling of transcriptomics datasets (often containing information on the expression of thousands of genes) to identify transcriptional points-of-departure (PODT) [21, 27, 28], comparing the concordance between PODT and PODs derived from traditional toxicity testing [13, 22, 29], and comparisons of cross-species sensitivity to chemicals [30–32]. With respect to HTS and risk assessment, a general framework for in vitro-to-in vivo extrapolation (IVIVE) has emerged that involves concentration-response modeling of HTS data to identify biological pathway altering concentrations (BPAC), and conversion to administered dose equivalents (ADEs) using high-throughput toxicokinetic (HTTK) modeling and reverse dosimetry [23, 25]. The resulting ADEs can then be used with measured or predicted exposures to perform bioactivity to exposure ratio (BER) analyses as a means of prioritizing or deprioritizing chemicals for further scrutiny [24, 33, 34]. The sections below examine these concepts in greater detail and proposes a framework where methodologies from the transcriptomics and HTS research fields are combined for analysis of HTTr data and potential application to chemical risk assessment.
Study Design Considerations for High Throughput Transcriptomics
The variety of in vitro models appropriate for use in HTTr screening is vast but could include cancer or immortalized cell lines, primary cell culture and/or mixed cell cultures grown in two- or three-dimensional formats. It is advisable that the domain of applicability (DOA) of an in vitro test system in terms of biological target expression and suitability for evaluating chemical collections of interest be characterized prior to undertaking large-scale HTTr screening studies. An in-depth discussion regarding the choice of appropriate cell models, media formulations, exposure conditions, test chemical compatibility, dose spacing, etc., is beyond the scope of this article. Instead the section below focuses on operational and quality control aspects commonly associated with HTS with regards to their application to HTTr screening.
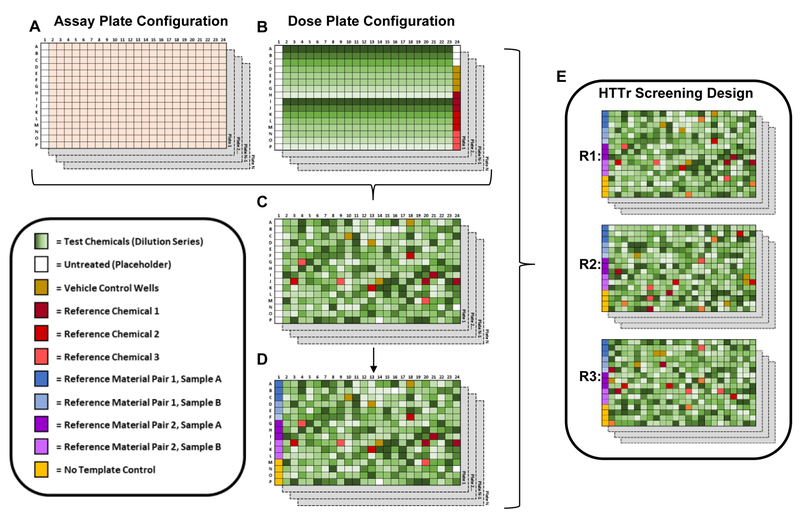
The experimental design for a HTTr chemical screening assay should incorporate features commonly employed in HTS, such as the use of parallel cytotoxicity assays, reference chemicals, as well as unique types of quality control (QC) samples for assessing performance of a transcriptomic assay. Figure 1 illustrates a generic experimental design for an HTTr concentration-response screen involving hundreds of chemicals. This example assumes that cultures will be prepared in 384-well format, that test chemicals will be applied using some type of high-throughput microfluidics technology and that automated (or partially automated) workflows will be used to process the biological samples contained in each well to generate gene expression profiles. The term “dose plate” refers to a multi-well plate containing solubilized chemicals, at various concentrations, to be applied to an “assay plate” that contains cells in culture. In this example, chemicals are tested in eight-point concentration-response with an n=1 technical replicate per plate and three culture (i.e. biological) replicates. A randomized dosing pattern is used. However, principles illustrated in this example would also be applicable to designs using a non-randomized (i.e. invariable) dose patterning with respect to treatment well positioning. In addition, while this example uses a 384-well format and single technical replicates, the principles illustrated in this example are also applicable to lower-density microtiter plate formats and designs that use multiple technical replicates within an assay plate.
Figure 1. Generic experimental design for HTTr chemical-concentration response screening.
A) Example of a configuration for a series of assay plates in an HTTr screen. Columns 2–24 contain cells in culture. Column 1 is empty. B) Example of a configuration for a series of dose plates in an HTTr screen. A total of 44 DMSO-solubilized chemicals in eight-point dilution series are arranged from high to low concentration (varying shades of green), vertically in a column, in the top and bottom halves of the dose plate, respectively. In Column 24, vehicle wells (i.e. DMSO only) are shown in brown and triplicate wells of three different reference compounds are shown in various shades of red. Wells A24, B24 and C24 are empty and are used as placeholders for untreated culture wells. C) Test chemicals housed on the dose plates are applied to the assay plates in a randomized pattern using microfluidics. The first column of the plates is empty of cultures and test substances to facilitate dispensing of QC samples as illustrated in panel D. In this design each test chemical x dose combination has a single technical replicate per plate while DMSO controls, untreated controls and reference chemicals have 3–4 technical replicates per plate D) Shades of blue in the first column represents triplicates of two commercially-available reference materials (e.g. purified RNAs). Shades of purple in the first column represents triplicates of two reference materials derived from the matrix under study. For example, bulk lysates or purified RNAs of DMSO-treated cells and cell treated with a pharmacological agent, such as an HDAC inhibitor. The purpose of paired reference materials (A & B; C & D) to ensure that the same biological samples, from the same lot or stock, are tested on every assay plate and for derivation of plate-level DEG profiles for evaluating reproducibility of assay performance. E) Chemicals in the dose plate series are tested in three replicate cultures.
Demonstration of assay reproducibility has been identified as a criterion for the incorporation of NAMs into risk-based decision-making processes [35]. Unlike HTS assays where one (or a few) reference chemicals are used to consistently evaluate reproducibility of a single endpoint or functional outcome (i.e. chemical X activating receptor Y), the multiplexed nature of HTTr data makes performance evaluation a more complex problem. In HTS, statistical indices of assay dynamic range and endpoint variability, such as Z-factors and/or signal windows, are used to judge assay performance [36, 37]. While these measures of assay performance may potentially be useful when measuring small numbers of genes, the breadth of biological responses covered using large panels of genes or the whole transcriptome limit its application. For example, the dynamic range of responses for one pathway or target (e.g., activating receptor Y) may be different than another (e.g., inhibiting enzyme Z). In addition, it is well known that identification of significantly-affected genes is bound to include some level of type I error, even when stringent false discovery rate (FDR) corrections are used [38–40]. The practical implication is that researchers should not expect the exact same set of transcripts to be identified as significant or responsive in repeated, independent comparisons of like samples or treatments using a simple statistical test. Therefore, Z-factor or signal window approaches using sentinel genes would likely not provide a comprehensive assessment of HTTr assay performance; nor should quantifying the number of differentially expressed genes (DEGs) from a pairwise statistical comparison, and comparing the percentage of overlapping genes (POG) between like comparisons, be considered a reliable metric of assay performance [40].
A more appropriate approach for evaluating performance includes multiple measures that capture the reproducibility and signal-to-noise characteristics of the HTTr assay using a combination of reference materials and reference chemicals. The term “reference materials” refers to standardized biological samples that are either manufactured commercially or generated in bulk at a research laboratory. Reference materials could be purified RNAs or a biological sample type (i.e. biological matrix) that contains RNA. Aliquots of reference materials are incorporated into each assay plate and processed in parallel with test samples.
In the Figure 1 example, two reference material pairs are incorporated into the experimental design. Reference material pair 1 (blue) represents a pair of commercially available purified-RNA products with highly divergent gene expression patterns, such as purified human RNAs from visceral and nervous system tissues used in the Microarray Quality Control Consortium (MAQC) studies [39]. Reference material pair 2 (purple) represents a pair of bulk-generated reference materials with divergent gene expression patterns that are of the same biological matrix used in the HTTr study. For example, cell lysates from DMSO-treated cells and cells treated with a reference chemical, such as a histone deacetylase (HDAC) inhibitor. In this example, reference materials are located in a static position across all assay plates, but could also be randomized across the plate. Paired reference materials with divergent gene expression patterns are compared to generate DEG profiles with a large range of fold-change (FC) values across genes. Since the same reference materials are evaluated on every plate, large plate-to-plate inconsistencies in the DEG profile, resulting pathway profile or similarity to a reference profile may be indicative of a poor performing HTTr assay. In contrast, instances where a reference material DEG profile, pathway profile or similarity score for a specific plate is inconsistent with other like comparisons within an HTTr screen (or historical data from past screens) may be indicative of a sporadic plate or sample failure. Use of matrix-specific bulk reference material provides an additional means to evaluate assay performance in terms of consistency of DEG profiles and provides information on whether the study-specific biological matrix influences HTTr assay performance using purified reference materials as the benchmark. In addition, “no template controls” (NTCs) are also included in the experimental design in Figure 1 (yellow wells). NTCs contain no RNA or other biological material and are used to identify potential contamination of assay reagents with exogenous genetic material that would interfere with the performance on an HTTr assay.
Reference chemicals are defined as treatments that produce a robust change in the gene expression profile of the cell type being studied. On a plate, the reference chemical treatments can then be paired with DMSO control wells to generate DEG profiles and evaluate reproducibility of an HTTr assay performance across plates, screening blocks and experiments; a similar approach to that described for reference materials. Unlike reference materials, reference chemical treatments can be used to identify potential malfunctions or poor performance related to automated chemical transfer from dose plates to assay plates (i.e. dosing). In analyzing the reference chemical treatments, a simple profile comparison approach involves mapping of DEGs to biological signaling pathways and determining the consistency in the identity of affected pathways from plate-to-plate. Previous studies have demonstrated that pathway-centric analyses are able to identify hallmark effects of model toxicants across different in vitro experiments even when the POG identified via statistical and fold-change cut-offs is low [27, 41, 42]. Alternatively, signature-based approaches, such as Connectivity Mapping [43], compares the pattern of gene expression from reference chemical treatment with a database of reference profiles enabling the evaluation of assay performance based on the strength of association. The Figure 1 example illustrates the use of three reference chemical treatments in triplicate (Figure 1, red wells of varying shades) for evaluating HTTr assay performance across plates and screening blocks or studies performed over many weeks/months/years of experimentation.
The use of commercially-available paired reference materials, assay matrix-specific paired bulk reference materials, reference chemical treatments and NTCs would allow a researcher to evaluate many different aspects of an HTTr experimental workflow. Results from these QC samples can be used in a combinatorial fashion for plate-based QC evaluations. For instance, failure of one of the QC comparisons to meet established performance criteria may trigger more in-depth evaluation of data from an assay plate, but may not necessarily indicate that all the samples on that plate performed poorly. However, failure of many of the QC comparisons on a plate to meet established performance criteria would provide stronger evidence that data on that plate is of poor quality and may need to be repeated. Review of QC could be considered as part of the decision making process for inclusion of HTTr data in a chemical risk assessment. In addition, the use of reference materials and reference treatments also allows comparisons of results across technology platforms (e.g. microarrays, RNA-seq, qRT-PCR, etc.) for evaluating comparative performance of a particular HTTr assay or integrating results from multiple transcriptomic data types.
High-Throughput Transcriptomics and Potential Applications in Risk Assessment
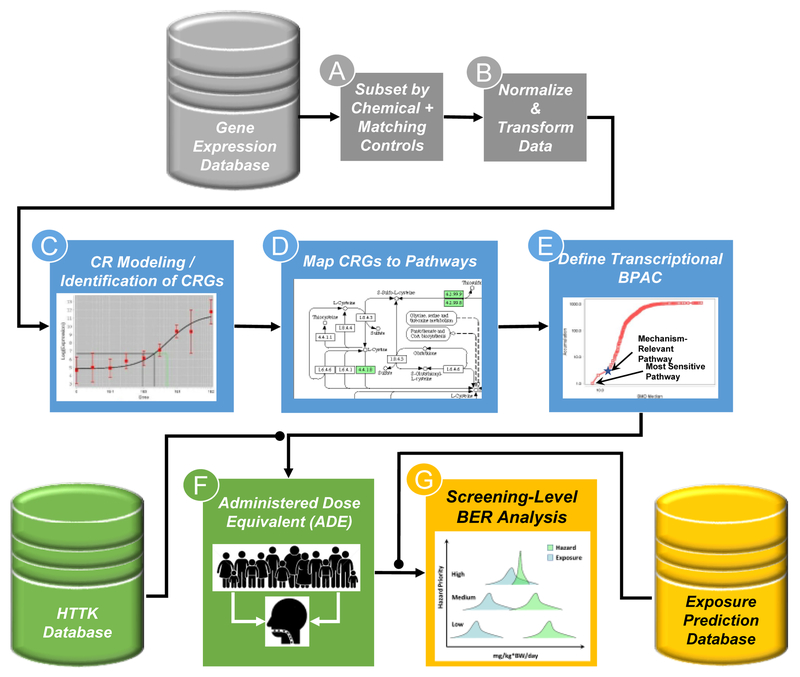
Figure 2 illustrates a generalized workflow for a potential use of HTTr data in chemical risk assessment, namely screening-level bioactivity-to-exposure ratio (BER) analysis. Importantly, in addition to a database of transcriptomic profiles obtained from an HTTr screen (Figure 2, gray cylinder) downstream steps of this workflow require drawing information from other databases; namely those containing experimentally-derived or predicted human toxicokinetic data (Figure 2, green cylinder) and chemical exposure predictions for a population of interest (Figure 2, yellow cylinder).
Figure 2. Framework for the use of HTTr data in screening level risk assessment.
HTTr screening data is generated and housed in a database (gray cylinder). For analysis, data are subset by chemical of interest and corresponding vehicle controls (A) and the data is normalized and transformed (B) in a manner appropriate for the HTTr assay being used. The normalized and transformed data is then used for gene-level concentration response (CR) modeling, identification of concentration-responsive genes (CRGs) and calculation of potency estimates for biological effects (C). Typically, a pre-filter (i.e. Trend test or ANOVA) is applied to filter out probes that do not show any indication of concentration-dependent changes in expression prior to modeling. CRGs are then mapped to pathways using existing knowledgebases (D). Pathway-level aggregate potency values are calculated and a biological pathway altering concentration (BPAC) is identified corresponding to the most sensitive mechanistically-relevant pathway (star) or the most sensitive overall pathway (E). BPACs are converted to ADE using experimentally-derived or predicted estimates of hepatic clearance and plasma protein binding (green cylinder) and reverse dosimetry using high-throughput toxicokinetic (httk) modeling of a heterogeneous human population (F). The ADEs are then compared to known or predicted human exposure levels (yellow cylinder), including estimates of uncertainty, to determine if an overlap dose of a chemical expected to produce perturbations in human biology and the dose of a chemical that may result from human exposure in the environment (G). Chemicals with a BPAC ADE that overlaps with predicted exposure estimates may be of more concern than those that do not.
Concentration-response modeling for BPAC determination
The first step in the proposed workflow is selection of a subset of data from the HTTr database corresponding to the chemical(s) of interest, along with corresponding controls (Figure 2A). The data is then normalized and transformed as appropriate for the transcriptomic assay used in the HTTr screen (Figure 2B). For HTTr studies that screen multiple concentrations of test chemicals (as illustrated in Figure 1), a combination of statistical tests (e.g. trend tests or ANOVA) and curve-fitting is used to identify concentration-responsive genes (CRGs); to determine concentration-response functions (curve shapes) that best describes the transcript-level data; and use those functions to calculate transcript or gene level potency estimates for a given level of biological response (Figure 2C). Potency estimates for CRGs are then mapped to a pre-defined gene set or pathway (Figure 2D) to calculate “pathway-level” potency estimates. Pathway-level potency estimates are then ranked to identify the most sensitive pathways or to determine at what concentration pathways known or suspected to be associated with the toxicity of a chemical are perturbed (Figure 2E). Either approach lends itself to the identification of a biological pathway altering concentration (BPAC) that can be used as an input for screening-level BER analysis.
A popular approach for concentration-response analysis of transcriptomics data that follows this scheme is benchmark dose modeling using the BMDExpress software package [44]. BMD modeling was originally developed by the USEPA as a data-driven approach for identifying PODs from in vivo toxicity studies for use in chemical risk assessment and is described in detail elsewhere [45, 46]. The concept of adapting BMD modeling for transcriptomics data originated as a proposed means of incorporating transcriptomics data into a chemical risk assessment framework [21]. Thomas et al. leveraged the statistical procedures contained in the USEPA Benchmark Dose Modeling Software (BMDS) package [47], applied them to gene-level curve fitting of normalized, log2-transformed Affymetrix microarray data and introduced the innovative approach of aggregating gene-level BMDs into Gene Ontology (GO) or pathway-based potency estimates [21, 22, 29]. In brief, expression data is treated as a continuous variable and a benchmark response (BMR) corresponding to a quantitative change in expression (i.e. % change or number of standard deviations relative to the vehicle control) is selected. Then, concentration-response curves of various shapes (i.e. Hill, polynomials, exponentials, etc.) are fit to the data and the curve that best describes the data according to an objective criterion (i.e. Akaike’s Information Criteria, AIC) is selected as the best-fitting model. The BMD is defined as the point along the best-fitting curve that intersects with the BMR. An upper and lower confidence limit on the BMD may also be calculated (known as the BMDU and BMDL, respectively). Dose-responsive transcripts are then categorized in terms of membership within a GO category or pathway and a composite BMD or BMDL (such as the mean or median BMD or BMDL across all affected transcripts within a category) is reported as an estimate of chemical potency for pathway perturbation. The BMD analysis approach has been used to analyze data from a variety of transcriptomic assay types including microarray, RNA-Seq and qPCR arrays [27, 48]. A software application known as BMDExpress was developed to facilitate high-throughput curve fitting of transcriptomics data and reporting of results [49]. The program has since been modified to facilitate different types of pre-filtering options based on analysis of variance (ANOVA) or trend testing, pathway enrichment tests and now includes a user-friendly graphical user interface for interactive analysis and a command-line mode for batch execution suitable for large-scale studies (https://github.com/auerbachs/BMDExpress-2/wiki) [44]. BMD modeling of transcriptomics data has seen a surge in popularity in recent years and has recently been adopted by the National Toxicology Program (NTP) as the preferred methods for dose-response modeling of transcriptomics data, with an emphasis on estimating the biological potency of test substances, promoting consistency in reporting of transcriptomic dose-response data and facilitating uptake by the regulatory community [50]. In addition, other concentration-response modeling approaches and/or packages such as PROAST [51], tcpl [52] and non-parametric methods have been either suggested or implemented for analyzing transcriptomics data [50, 53].
Transcriptomic dose-response information from in vivo studies modeled and aggregated as described above has shown excellent correlation between the composite BMD for the most sensitive pathway and the BMDs for apical cancer and non-cancer effects [13, 22, 27, 29, 54]. Importantly, for the chemicals studied, the ratio of apical and transcriptional BMDs for a target tissue was small, indicating that transcriptional effects were not occurring at doses that were substantially lower than those associated with the most sensitive apical endpoints [13, 22, 27, 29, 54, 55]. Therefore, transcriptomic BMDs were either slightly more conservative or approximately equal to BMDs derived from traditional toxicity endpoints. Furthermore, transcriptional BMD values from in vivo studies were generally concordant across exposure durations indicating that shorter duration studies could provide comparable potency values than longer duration (sub-chronic or chronic) studies [13]. A previous study has also explored the impact of data pre-filtering and different methods for pathway-based and non-pathway-based aggregation of gene level BMD results. Overall, pre-filtering of transcriptomics data to remove probes with no evidence of dose-responsiveness prior to BMD modeling tended to produce more conservative (i.e. lower) pathway-level BMD estimates [27]. Different approaches for aggregating gene level BMDs to produce an overall POD tended to vary no more than one order of magnitude [28], demonstrating that different ways of summarizing BMD modeling results has comparatively small impact with regards to determining the threshold where significant perturbations in biology occur. In addition to POD determination, transcriptomic BMD modeling and pathway aggregation has been applied to more complex research questions that are applicable to chemical risk assessment such as investigating cross-species sensitivities to toxicants [30, 31, 54], characterizing the relative potency of structurally-related chemicals [32] and exploring dose-dependent transitions in toxicological responses [56, 57].
In in vivo transcriptomics studies, biological pathways relevant to the apical tissue-level effects of a chemical can often be identified within the list of pathways affected using the BMD modeling approach [13, 21, 55, 58, 59]. However, in vivo transcriptomic BMD modeling studies have also demonstrated that while the quantitative relationship between the most sensitive pathway and most sensitive apical adverse response is strong, the most sensitive pathway does not always have a clearly discernable mechanistic linkage with the most sensitive apical effect. Likewise, in in vitro transcriptomics studies, the list of perturbed pathways often contains pathways with known or strongly suspected mechanistic linkage with the chemical or agent under study [31, 56, 60]. Like the in vivo situation, the most sensitive pathway does not always have a clearly discernable mechanistic linkage to an apical cellular or tissue level effect or a chemical. In terms of HTTr screening studies, it is anticipated that most of the chemicals being studied will fall into the “data-poor” category, meaning that often mechanistic linkage between perturbed pathways and an apical biological effect cannot be inferred, chiefly due to lack of existing toxicity data. These observations have led to two potential routes for incorporation of HTTr data into a risk assessment framework. In instances where there is evidence for a causal relationship between an affected pathway and an apical effect, the BMD of the putative causal pathway could be used as a candidate BPAC. In instances involving “data poor” chemicals, the BMD of the most sensitive pathway could serve as a protective, if not predictive, BPAC until gaps in the existing mechanistic knowledge-base are filled [22, 61]. BPACs derived using the latter approach may have limited predictive value for specific toxicities, but are hypothesized to provide a reliable potency estimate for the threshold concentration where biological signaling is perturbed by a chemical in an in vitro system.
In vitro-to-in vivo extrapolation (IVIVE) and bioactivity to exposure ratio (BER) analysis
A BPAC is a concentration of a chemical, expressed as a nominal media concentration, that produces a significant perturbation of a cellular process in an in vitro test system. To perform a human-relevant, bioactivity-to-exposure ratio (BER) analysis based on in vitro data, three steps are required: 1) conversion of the BPAC to an administered dose equivalent (ADE) that human populations may encounter (Figure 2F) using experimentally-derived human plasma binding and hepatic clearance data (Figure 2, green cylinder) and reverse dosimetry, 2) obtaining an estimate of exposure in the human populations of interest (Figure 2, yellow cylinder) and 3) comparison of ADEs and exposure predictions to determine if an overlap exists between the dose of a chemical expected to produce perturbations in human biology and the dose of a chemical that humans may receive from the environment (Figure 2G). In the case of HTTr data, the transcriptomic BPAC is assumed to be equivalent to the concentration in the blood that would produce perturbation of a biological pathway in vivo. The process of converting in vitro potency estimates, such as BPACs, to human-relevant ADEs with accompanying estimates of human population variability has been described extensively elsewhere [23–26, 33]. In addition, the processes and challenges associated with generating high-throughput human exposure predictions with uncertainty bounds have also been described elsewhere [25, 26, 33, 62, 63] and will not be discussed further here.
In the final step of an IVIVE-based BER analysis, the distribution of ADEs corresponding to the transcriptional BPAC are compared to the distribution of the high-throughput exposure predictions. A chemical for which the predicted exposure does not overlap with the predicted ADE would be of less concern than a chemical where there is an overlap. To prioritize chemicals in this manner, they are ranked by the separation between the lower bound of the ADE distribution and the upper-bound of the exposure prediction. Previous analyses of HTS data in this manner have demonstrated that a substantial gap exists between ADEs calculated from in vitro BPACs and the corresponding high-throughput human exposure prediction for most environmental chemicals [24, 26, 33]. Likewise, occasions where the BPAC-derived ADEs overlap with the high-throughput human exposure predictions are rare [24, 33]. It is yet to be determined whether results from HTTr screening of environmental chemical libraries, such as the ToxCast and Tox21 libraries used in the aforementioned studies, will yield similar results.
Putative mechanism of action prediction using HTTr data
In addition to providing quantitative estimates for potential threshold(s) for chemical bioactivity, HTTr data also has potential use for predicting putative mechanisms of action for chemical toxicity [20]. There are many approaches that may be used for putative mechanism of action prediction, including (but not limited to): 1) pathway mapping of CRGs to collections of gene sets (as described in Figure 2), 2) DEG signature querying with transcriptional biomarkers and 3) connectivity mapping [43] of transcriptomic signatures to databases of annotated transcriptomic reference profiles. Each of these can be considered hypothesis-generating approaches for identification of putative molecular mechanisms of action that may be confirmed through testing in orthogonal functional assays or compared to supporting information from the scientific literature in a weight-of-evidence framework. In depth discussion of each of these approaches is beyond the scope of this article; however a general, conceptual overview of potential applications for mechanism of action prediction is presented below.
Pathway mapping of CRGs to collections of gene sets is a straightforward approach for discerning putative mechanisms of action from concentration-response modeling data. For example, if an estrogenic chemical is tested in an estrogen-responsive cell line and the CRGs then mapped to a gene set collection containing an estrogen signaling pathway, one would expect that this pathway would be included in the list of perturbed pathways and reasonably hypothesize that the chemical in question targets the estrogen receptor. As detailed above, there are numerous examples where mapping CRGs to gene set collections yielded biological pathways with plausible mechanistic linkage to the chemical of interest [21, 30, 31, 55–57]. Approaches such as Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA) are designed to assess the coordinated responses of functionally related genes contained within a defined pathway/gene set/ontology structure using rank order statistical methods [64, 65]. These types of approaches are sensitive to small magnitude changes in coordinated sets of genes, a phenomenon that may be expected to occur near the threshold for chemical bioactivity in the context of a tested concentration range. Gene set enrichment approaches have typically been used in “case-control” (i.e. two sample) comparison but have potential utility for concentration-response modeling of enrichment scores for identification of BPACs. As with the CRG mapping approach, a set of pathways enriched in response to chemical treatment could be used to generate hypotheses regarding the putative mechanisms of action of a chemical of interest.
Transcriptional biomarker-based approaches have also been proposed as a means for identification of putative mechanisms of action for chemicals using HTTr data. “Toxicity-centric” approaches use gene expression patterns produced by model toxicants with well-characterized mechanisms to identify panels of genes whose expression levels change with the onset of cellular stress or injury. The toxicity-centric approach has been used to develop a robust transcriptional biomarker for distinguishing genotoxic from non-genotoxic chemicals (i.e. the TGx-DDI biomarker) [66–68]. This biomarker was developed to augment and increase the classification accuracy of existing in vitro test batteries for genotoxicity that often produce “positive” results for non-genotoxic chemicals [66]. “Target-centric” approaches use gene expression patterns produced by pharmacological modulation or genetic knockdown of a discrete molecular target to identify panels of genes whose expression levels are affected by the functional status of the target under study. Examples of target-centric transcriptional biomarkers include those developed for estrogen receptor alpha [69], androgen receptor [70], constitutive androstane receptor [71, 72], aryl hydrocarbon receptor [73] and peroxisome proliferator-activated receptor alpha [74]. In both approaches transcriptional biomarkers are constructed by comparing the transcriptomic response profiles of multiple perturbagens that affect the same molecular target, act through the same mechanism or produce the same apical cellular effect. Genes that are consistently upor down-regulated across the profiles are identified and incorporated in a transcriptional biomarker profile. The biomarker profiles can then be queried against the transcriptomic profiles of unknown chemicals to determine whether there is evidence for a particular molecular target/mechanism of action being affected or activated. Clustering-based approaches have been used to demonstrate similarity in transcriptional biomarker response profiles across a chemical set and illustrate predictive value [66, 67]. In addition, rank-order statistical tests have been used to quantify the strength of association between transcriptional biomarkers and transcriptomic profiles of chemicals under investigation [69, 70]. Overall, cross-validation testing using chemical training sets has demonstrated that transcriptional biomarker-based approaches like those described above have high balanced accuracy for classification of chemicals as “positive” or “negative” for affecting a particular molecular target or acting through a particular mechanism of action. However, it is worth noting that development of transcriptional biomarkers with high predictive value depends upon the availability of transcriptomic data from multiple reference treatments that is not confounded by off-target effects that may arise with accumulating tissue doses above those needed to activate the target/toxicity of interest or as a result of off-target effects of genetic manipulations (i.e. promiscuous knockdown). Incorporation of treatments that produce off-target effects in the input set used to construct a transcriptional biomarker will likely have a negative impact on the predictive value of said biomarker. Likewise, the predictive value of a transcriptional biomarker may be limited to a particular domain of applicability such as cell types that express a particular functional target or the cellular machinery necessary for manifestation of a particular toxicity. For example, using a biomarker approach to identify PPAR active chemicals in an HTTr data set derived from a cell type that does not express PPAR may lead to spurious or inaccurate mechanistic predictions.
Connectivity mapping is yet another approach that can be used to identify putative mechanisms of action for a chemical using HTTr data [43]. In this approach, a gene expression profile for a treatment of interest is compared to a matching control and all genes are rank-ordered according to their differential expression (i.e. up- or down-regulated). The differential expression signature is then queried against a large reference database containing rank-ordered differential expression signatures from a large variety of treatments. If up- and down-regulated genes in the query signature appear at the upper or lower extremes, respectively, of a reference signature, then the two conditions have strong positive connectivity (i.e. are correlated). Conversely, if up- and down-regulated genes in the query signature appear at the opposite extremes, respectively, of a reference signature, then the two conditions have strong negative connectivity (i.e. are anti-correlated). The connectivity scores across the entire reference database can then be ranked to identify treatments that are the most similar (or opposite) to said query. Strong positive connectivity may indicate that the query and reference treatment affect a molecular target or biological pathway in the same manner. Strong negative connectivity may indicate that the query and reference treatment have opposite effects on a biological process, such as agonism or antagonism of a receptor. The Connectivity Mapping approach has been used for putative identification of novel therapeutics and drug repurposing based on similarity in gene expression signatures [75–77] and has also shown promise for predictive toxicology [78, 79]. In order for Connectivity Mapping to be useful for putative mechanism of action prediction in an assessment context, the reference profile database must be populated with data from treatments (pharmacological or genetic) that are unequivocally associated with a specific effect on a molecular target. A putative mechanism of action for a treatment of interest could then be inferred via strong negative or positive connectivity with target-annotated reference profile(s). Similar to transcriptional biomarker-based approaches, Connectivity Mapping for mechanism of action identification should be used cautiously, taking into consideration the applicability domains of the in vitro model used to generate the HTTr data and the model used to generate the reference profiles. For example, it is anticipated that the predictive power of a Connectivity Mapping approach for a particular target would be negatively affected if either the cell type used to generate the query signature, or the cell type used to generate the reference signature, lacked functional expression of the target of interest.
Recently, machine learning approaches have been applied to transcriptomics data for prediction of drug-target interactions, grouping of chemicals into mechanistic and therapeutic classes and deriving transcriptional biomarkers for various chemical classes [80–82]. These studies primarily used drugs and well-studied, well-annotated model toxicants to train prediction models. Machine learning approaches have potential application for HTTr-based MOA prediction for environmental chemicals, including data poor chemicals, but has yet to be widely applied for this purpose. Development of large datasets of transcriptomic reference profiles specific to the environmental chemical space would aid in evaluating the utility of machine learning approaches for mechanism of action prediction and chemical grouping.
Summary and Conclusions
Technological advancements in transcriptomics assays, as well as increasing efficiency and declining cost, has made HTTr feasible for chemical hazard screening. In addition, there is an interest in many governmental and non-governmental organizations for incorporation of data from NAMs, such as HTTr, into chemical assessment practices. Demonstrating HTTr assay performance and reproducibility is a key prerequisite for including such data in an assessment context. Incorporating standardized reference materials and reference chemical treatments can demonstrate the performance and reproducibility of HTTr assay technology and laboratory workflows to support their utilization in a chemical assessment dossier. Furthermore, the BER analysis framework presented here is one of the ways HTTr data may be used in regulatory decisions, particularly in the context of prioritization and screening level chemical assessments. Additional work is required to establish HTTr performance and reporting standards that would support use of HTTr data in regulatory decision making and for development of chemical of chemical case studies that illustrate application of HTTr data in a variety of regulatory decision contexts.
Acknowledgements
The authors declare no conflicts of interest regarding this manuscript. The U.S. Environmental Protection Agency has provided administrative review and has approved this paper for publication. The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Reference to commercial products or services does not constitute endorsement. The authors thank Drs. Keith Houck and Nisha Sipes for their helpful technical reviews of the manuscript.
Funding
The United States Environmental Protection Agency through its Office of Research and Development provided funding for the development of this article.
References
- 1.Jones E, Michael S, and Sittampalam GS, Basics of Assay Equipment and Instrumentation for High Throughput Screening, in Assay Guidance Manual, Sittampalam GS, et al. , Editors. 2004: Bethesda (MD). [PubMed] [Google Scholar]
- 2.Inglese J and Auld DS, High Throughput Screening (HTS) Techniques: Applications in Chemical Biology, in Wiley Encyclopedia of Chemical Biology, Begley TP, Editor. 2008, Wiley & Sons, Inc: Hoboken, NJ: p. 260–274. [Google Scholar]
- 3.Janzen WP, Screening technologies for small molecule discovery: the state of the art. Chem Biol, 2014. 21(9): p. 1162–70. [DOI] [PubMed] [Google Scholar]
- 4.Tietjen K, Drewes M, and Stenzel K, High throughput screening in agrochemical research. Comb Chem High Throughput Screen, 2005. 8(7): p. 589–94. [DOI] [PubMed] [Google Scholar]
- 5.Hughes JP, et al. , Principles of early drug discovery. Br J Pharmacol, 2011. 162(6): p. 1239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ates G, et al. , A novel genotoxin-specific qPCR array based on the metabolically competent human HepaRG() cell line as a rapid and reliable tool for improved in vitro hazard assessment. Arch Toxicol, 2018. 92(4): p. 1593–1608. [DOI] [PubMed] [Google Scholar]
- 7.Sawada H, Taniguchi K, and Takami K, Improved toxicogenomic screening for drug-induced phospholipidosis using a multiplexed quantitative gene expression ArrayPlate assay. Toxicol In Vitro, 2006. 20(8): p. 1506–13. [DOI] [PubMed] [Google Scholar]
- *8.Mav D, et al. , A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLoS One, 2018. 13(2): p. e0191105. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe the development and application of a targeted gene panel for high-throughput transcriptomics screening. The targeted gene panel was created through a combination of data-driven and knowledge-driven gene selection and may serve as a cost-effective substitute for whole transcriptome profiling.
- 9.Subramanian A, et al. , A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell, 2017. 171(6): p. 1437–1452 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Yeakley JM, et al. , A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS One, 2017. 12(5): p. e0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe a novel ligation-based targeted whole transcriptome gene expression profiling assay suiteable for high-throughput transcriptomics (HTT) screening, demonstrate utility for detecting changes in gene expression in response to chemical exposure and consistency in gene expression signatures for a model toxicant across cell types and transcriptomics platforms.
- 11.Bush EC, et al. , PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens. Nat Commun, 2017. 8(1): p. 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yauk CL, et al. , Application of the TGx-28.65 transcriptomic biomarker to classify genotoxic and non-genotoxic chemicals in human TK6 cells in the presence of rat liver S9. Environ Mol Mutagen, 2016. 57(4): p. 243–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas RS, et al. , Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci, 2013. 134(1): p. 180–94. [DOI] [PubMed] [Google Scholar]
- 14.Buesen R, et al. , Applying ‘omics technologies in chemicals risk assessment: Report of an ECETOC workshop. Regul Toxicol Pharmacol, 2017. 91 Suppl 1: p. S3–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauer UG, et al. , The challenge of the application of ‘omics technologies in chemicals risk assessment: Background and outlook. Regul Toxicol Pharmacol, 2017. 91 Suppl 1: p. S14–S26. [DOI] [PubMed] [Google Scholar]
- 16.Kolesnikov N, et al. , ArrayExpress update--simplifying data submissions. Nucleic Acids Res, 2015. 43(Database issue): p. D1113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, et al. , NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res, 2013. 41(Database issue): p. D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.USEPA, Strategic Plan to Promote the Development and Implementation of Alternative Test Methods Within the TSCA Program 2018, U.S. Environmental Protection Agency, Office of Chemical Safety and Pollution Prevention: Washington, DC. [Google Scholar]
- 19.ECHA, New Approach Methodologies in Regulatory Science 2016, European Chemicals Agency: Helsinki, Finland. [Google Scholar]
- **20.Thomas RS, et al. , The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency. Toxicol Sci, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript describes the strategic and operational approaches that will by used by the US Environmental Protection Agency over the next five years to foster broader acceptance of computational toxicology for application to higher teir regulatory decisions, such as chemical assessments.
- 21.Thomas RS, et al. , A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol Sci, 2007. 98(1): p. 240–8. [DOI] [PubMed] [Google Scholar]
- 22.Thomas RS, et al. , Integrating pathway-based transcriptomic data into quantitative chemical risk assessment: a five chemical case study. Mutat Res, 2012. 746(2): p. 135–43. [DOI] [PubMed] [Google Scholar]
- 23.Judson RS, et al. , Estimating toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem Res Toxicol, 2011. 24(4): p. 451–62. [DOI] [PubMed] [Google Scholar]
- **24.Sipes NS, et al. , An Intuitive Approach for Predicting Potential Human Health Risk with the Tox21 10k Library. Environ Sci Technol, 2017. 51(18): p. 10786–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe an bioactivity exposure ration (BER) approach that uses in vitro-to-in vivo extrapolation (IVIVE) to convert potency estimates from high-throughput screening (HTS) data to administered equivalent doses (AEDs). This was followed by comparison to computational exposure predictions and chemical grouping based on overlap or lack of overlap between biological activity and exposure.
- 25.Rotroff DM, et al. , Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol Sci, 2010. 117(2): p. 348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wetmore BA, et al. , Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol Sci, 2012. 125(1): p. 157–74. [DOI] [PubMed] [Google Scholar]
- 27.Webster AF, et al. , Impact of Genomics Platform and Statistical Filtering on Transcriptional Benchmark Doses (BMD) and Multiple Approaches for Selection of Chemical Point of Departure (PoD). PLoS One, 2015. 10(8): p. e0136764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Farmahin R, et al. , Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch Toxicol, 2017. 91(5): p. 2045–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe and compare different approaches for determining a transcriptional point-of-departure from transcriptomics dose-response studies and demonstrate marked concordance with traditional points-of-departure based on tissue level effects.
- 29.Thomas RS, et al. , Application of transcriptional benchmark dose values in quantitative cancer and noncancer risk assessment. Toxicol Sci, 2011. 120(1): p. 194–205. [DOI] [PubMed] [Google Scholar]
- 30.Thomas RS, et al. , Cross-species transcriptomic analysis of mouse and rat lung exposed to chloroprene. Toxicol Sci, 2013. 131(2): p. 629–40. [DOI] [PubMed] [Google Scholar]
- 31.Black MB, et al. , Cross-species comparisons of transcriptomic alterations in human and rat primary hepatocytes exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci, 2012. 127(1): p. 199–215. [DOI] [PubMed] [Google Scholar]
- 32.Rowlands JC, et al. , A genomics-based analysis of relative potencies of dioxin-like compounds in primary rat hepatocytes. Toxicol Sci, 2013. 136(2): p. 595–604. [DOI] [PubMed] [Google Scholar]
- 33.Wetmore BA, et al. , Incorporating High-Throughput Exposure Predictions With Dosimetry-Adjusted In Vitro Bioactivity to Inform Chemical Toxicity Testing. Toxicol Sci, 2015. 148(1): p. 121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin HM, et al. , Risk-Based High-Throughput Chemical Screening and Prioritization using Exposure Models and in Vitro Bioactivity Assays. Environ Sci Technol, 2015. 49(11): p. 6760–71. [DOI] [PubMed] [Google Scholar]
- 35.Bal-Price A, et al. , Recommendation on test readiness criteria for new approach methods in toxicology: Exemplified for developmental neurotoxicity. ALTEX, 2018. 35(3): p. 306–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang JH, Chung TD, and Oldenburg KR, A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen, 1999. 4(2): p. 67–73. [DOI] [PubMed] [Google Scholar]
- 37.Iversen PW, et al. , A comparison of assay performance measures in screening assays: signal window, Z’ factor, and assay variability ratio. J Biomol Screen, 2006. 11(3): p. 247–52. [DOI] [PubMed] [Google Scholar]
- 38.Fan X, et al. , Investigation of reproducibility of differentially expressed genes in DNA microarrays through statistical simulation. BMC Proc, 2009. 3 Suppl 2: p. S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium M, et al. , The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol, 2006. 24(9): p. 1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JJ, et al. , Reproducibility of microarray data: a further analysis of microarray quality control (MAQC) data. BMC Bioinformatics, 2007. 8: p. 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakhshandehroo M, et al. , Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PLoS One, 2009. 4(8): p. e6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, et al. , The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol, 2014. 32(9): p. 926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamb J, et al. , The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science, 2006. 313(5795): p. 1929–35. [DOI] [PubMed] [Google Scholar]
- **44.Phillips JR, et al. , BMDExpress 2: Enhanced transcriptomic dose-response analysis workflow. Bioinformatics, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript introduces BMDExpress 2, an open-access software package for concentration-response modeling of transcriptomics data.
- 45.Filipsson AF, et al. , The benchmark dose method--review of available models, and recommendations for application in health risk assessment. Crit Rev Toxicol, 2003. 33(5): p. 505–42. [PubMed] [Google Scholar]
- 46.Crump KS, A new method for determining allowable daily intakes. Fundam Appl Toxicol, 1984. 4(5): p. 854–71. [DOI] [PubMed] [Google Scholar]
- 47.Davis JA, Gift JS, and Zhao QJ, Introduction to benchmark dose methods and U.S. EPA’s benchmark dose software (BMDS) version 2.1.1. Toxicol Appl Pharmacol, 2011. 254(2): p. 181–91. [DOI] [PubMed] [Google Scholar]
- 48.Black MB, et al. , Comparison of microarrays and RNA-seq for gene expression analyses of dose-response experiments. Toxicol Sci, 2014. 137(2): p. 385–403. [DOI] [PubMed] [Google Scholar]
- 49.Yang L, Allen BC, and Thomas RS, BMDExpress: a software tool for the benchmark dose analyses of genomic data. BMC Genomics, 2007. 8: p. 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **50.NTP, NTP Research Report on National Toxicology Program Approach to Genomic Dose-Response Modeling 2018, National Institute of Environmental Health Sciences, Division of the National Toxicology Program: Research Triangle Park, NC. [PubMed] [Google Scholar]; This document describes the National Toxicology Program’s proposed approach to transcriptomics concentration-response modeling with a focus on identifying the biological potency of test substances.
- 51.Slob W, Dose-response modeling of continuous endpoints. Toxicol Sci, 2002. 66(2): p. 298–312. [DOI] [PubMed] [Google Scholar]
- 52.Filer DL, et al. , tcpl: the ToxCast pipeline for high-throughput screening data. Bioinformatics, 2017. 33(4): p. 618–620. [DOI] [PubMed] [Google Scholar]
- 53.House JS, et al. , A Pipeline for High-Throughput Concentration Response Modeling of Gene Expression for Toxicogenomics. Front Genet, 2017. 8: p. 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moffat I, et al. , Comparison of toxicogenomics and traditional approaches to inform mode of action and points of departure in human health risk assessment of benzo[a]pyrene in drinking water. Crit Rev Toxicol, 2015. 45(1): p. 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bercu JP, et al. , Toxicogenomics and cancer risk assessment: a framework for key event analysis and dose-response assessment for nongenotoxic carcinogens. Regul Toxicol Pharmacol, 2010. 58(3): p. 369–81. [DOI] [PubMed] [Google Scholar]
- 56.Woods CG, et al. , Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol Appl Pharmacol, 2009. 238(1): p. 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qutob SS, et al. , The application of transcriptional benchmark dose modeling for deriving thresholds of effects associated with solar-simulated ultraviolet radiation exposure. Environ Mol Mutagen, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dodd DE, et al. , Subchronic hepatotoxicity evaluation of 1,2,4-tribromobenzene in Sprague-Dawley rats. Int J Toxicol, 2012. 31(3): p. 250–6. [DOI] [PubMed] [Google Scholar]
- 59.Recio L, et al. , Impact of Acrylamide on Calcium Signaling and Cytoskeletal Filaments in Testes From F344 Rat. Int J Toxicol, 2017. 36(2): p. 124–132. [DOI] [PubMed] [Google Scholar]
- 60.Chauhan V, et al. , Transcriptional benchmark dose modeling: Exploring how advances in chemical risk assessment may be applied to the radiation field. Environ Mol Mutagen, 2016. 57(8): p. 589–604. [DOI] [PubMed] [Google Scholar]
- *61.Kavlock RJ, et al. , Accelerating the Pace of Chemical Risk Assessment. Chem Res Toxicol, 2018. 31(5): p. 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe a framework for advancing the applications of NAM data for evaluating the safety of data poor chemicals and filling information gaps relating to both chemical hazard and exposure.
- 62.Wambaugh JF, et al. , High throughput heuristics for prioritizing human exposure to environmental chemicals. Environ Sci Technol, 2014. 48(21): p. 12760–7. [DOI] [PubMed] [Google Scholar]
- 63.Isaacs KK, et al. , SHEDS-HT: an integrated probabilistic exposure model for prioritizing exposures to chemicals with near-field and dietary sources. Environ Sci Technol, 2014. 48(21): p. 12750–9. [DOI] [PubMed] [Google Scholar]
- 64.Subramanian A, et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 2005. 102(43): p. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanzelmann S, Castelo R, and Guinney J, GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics, 2013. 14: p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li HH, et al. , Development and validation of a high-throughput transcriptomic biomarker to address 21st century genetic toxicology needs. Proc Natl Acad Sci U S A, 2017. 114(51): p. E10881–E10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li HH, et al. , Development of a toxicogenomics signature for genotoxicity using a dose-optimization and informatics strategy in human cells. Environ Mol Mutagen, 2015. 56(6): p. 505–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *68.Corton JC, Williams A, and Yauk CL, Using a gene expression biomarker to identify DNA damage-inducing agents in microarray profiles. Environ Mol Mutagen, 2018. 59(9): p. 772–784. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe the use of the TGx-DDI transcriptional biomarker for differentiating DNA damage inducing agents and those that do not produce DNA damage. This biomarker approach may be broadly applicable for predicting putative mechanisms of action for chemicals across a vareity of molecular targets and target classes.
- 69.Ryan N, et al. , Moving Toward Integrating Gene Expression Profiling Into High-Throughput Testing: A Gene Expression Biomarker Accurately Predicts Estrogen Receptor alpha Modulation in a Microarray Compendium. Toxicol Sci, 2016. 151(1): p. 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rooney JP, et al. , Identification of Androgen Receptor Modulators in a Prostate Cancer Cell Line Microarray Compendium. Toxicol Sci, 2018. 166(1): p. 146–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rooney JP, et al. , Chemical Activation of the Constitutive Androstane Receptor (CAR) Leads to Activation of Oxidant-Induced Nrf2. Toxicol Sci, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oshida K, et al. , Identification of chemical modulators of the constitutive activated receptor (CAR) in a gene expression compendium. Nucl Recept Signal, 2015. 13: p. e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oshida K, et al. , Screening a mouse liver gene expression compendium identifies modulators of the aryl hydrocarbon receptor (AhR). Toxicology, 2015. 336: p. 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oshida K, et al. , Identification of modulators of the nuclear receptor peroxisome proliferator-activated receptor alpha (PPARalpha) in a mouse liver gene expression compendium. PLoS One, 2015. 10(2): p. e0112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smalley JL, et al. , Connectivity mapping uncovers small molecules that modulate neurodegeneration in Huntington’s disease models. J Mol Med (Berl), 2016. 94(2): p. 235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stumpel DJ, et al. , Connectivity mapping identifies HDAC inhibitors for the treatment of t(4;11)-positive infant acute lymphoblastic leukemia. Leukemia, 2012. 26(4): p. 682–92. [DOI] [PubMed] [Google Scholar]
- 77.Wen Q, et al. , A gene-signature progression approach to identifying candidate small-molecule cancer therapeutics with connectivity mapping. BMC Bioinformatics, 2016. 17(1): p. 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smalley JL, Gant TW, and Zhang SD, Application of connectivity mapping in predictive toxicology based on gene-expression similarity. Toxicology, 2010. 268(3): p. 143–6. [DOI] [PubMed] [Google Scholar]
- 79.De Abrew KN, et al. , Grouping 34 Chemicals Based on Mode of Action Using Connectivity Mapping. Toxicol Sci, 2016. 151(2): p. 447–61. [DOI] [PubMed] [Google Scholar]
- 80.Wei X, et al. , Identification of biomarkers that distinguish chemical contaminants based on gene expression profiles. BMC Genomics, 2014. 15: p. 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pabon NA, et al. , Predicting protein targets for drug-like compounds using transcriptomics. PLoS Comput Biol, 2018. 14(12): p. e1006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aliper A, et al. , Deep Learning Applications for Predicting Pharmacological Properties of Drugs and Drug Repurposing Using Transcriptomic Data. Mol Pharm, 2016. 13(7): p. 2524–30. [DOI] [PMC free article] [PubMed] [Google Scholar]