Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) carriage and infection are well documented in the human and veterinary literature; however only limited information is available regarding MRSA carriage and infection in laboratory NHP populations. The objective of this study was to characterize MRSA carriage in a representative research colony of rhesus and cynomolgus macaques through a cross-sectional analysis of 300 animals. MRSA carriage was determined by using nasal culture. Demographic characteristics of carriers and noncarriers were compared to determine factors linked to increased risk of carriage, and MRSA isolates were analyzed to determine antimicrobial susceptibility patterns, staphylococcal chromosome cassette mec (SCCmec) type, and multilocus sequence type (ST). Culture results demonstrated MRSA carriage in 6.3% of the study population. Animals with greater numbers of veterinary or experimental interventions including antibiotic administration, steroid administration, dental procedures, and surgery were more likely to carry MRSA. Susceptibility results indicated that MRSA isolates were resistant to β-lactams, and all isolates were resistant to between 1 and 4 nonβ-lactam antibiotics. In addition, 73.7% of MRSA isolates were identified as ST188-SCCmec IV, an isolate previously observed in an unrelated population of macaques and 15.8% were ST3268-SCCmec V, which has only been described in macaques. A single isolate had a novel sequence type, ST3478, and carried SCCmec V. These results suggest that NHP-adapted strains of MRSA exist and highlight the emergence of antimicrobial resistance in laboratory NHP populations.
Abbreviations: ACME, arginine catabolic mobile element; CA-MRSA, community-associated MRSA; CoNS, coagulase-negative staphylococcal species; MLST, multilocus sequence typing; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; PVL, Panton–Valentine leucocidin; SCCmec, staphylococcal chromosome cassette mec; TMS, trimethoprim–sulfamethoxazole
Laboratory NHP have many of the same risk factors that predispose the hospitalized human population to infection with methicillin-resistant Staphylococcus aureus (MRSA), including a high density of individuals, use of indwelling devices, frequent antibiotic use, and immunosuppression.8,34 MRSA carriage substantially raises the risk of MRSA infection in humans,19,38 and knowledge of carrier status may prompt steps to minimize the risk of transmission and infection. However, little information regarding the prevalence, risk factors, and strains associated with MRSA carriage in populations of laboratory macaques is available to guide clinical decision making.
The emergence of MRSA has been widely documented in human and animal populations. S. aureus is part of the normal flora present on the skin and mucosal surfaces of humans, NHP, and livestock species but has the potential to cause a variety of infections, including skin and soft tissue infections, endocarditis, pneumonia, and sepsis.41 Distinct lineages of S. aureus are often differentiated by multilocus sequence types (ST) that are determined according to the sequences of 7 core genes in the genome. MRSA strains arise through acquisition of mecA or, less commonly, mecC, which is carried on the mobile genetic element SCCmec that integrates site-specifically into the genome. mecA and mecC encode an acquired penicillin-binding protein, PBP2a, with low affinity for all β-lactam antibiotics, thereby conferring antimicrobial resistance. Frequently MRSA strains, especially those associated with the healthcare setting, also have resistance to nonβ-lactam classes of antibiotics, including lincosamides, macrolides, aminoglycosides, and fluoroquinolones. Because SCCmec is horizontally acquired, the same ST can be found among both MRSA and methicillin-susceptible S. aureus (MSSA) strains. However, SCCmec has entered into a limited number of ST, suggesting that restriction to uptake or integration exists among different lineages of S. aureus. MRSA strains are not necessarily more virulent than MSSA, given that each strain carries a unique set of virulence traits and resistance genes that define its potential for transmission, disease severity, and options for therapy.
MRSA was initially described as a nosocomial infection, becoming pervasive in the hospitalized human population during the late 1970s and early 1980s, with the most recent large-scale analysis of US hospitals indicating a total carriage rate of 6.64% among inpatients.14 However, this rate varies considerably between the populations studied, ranging from 3.8% of inpatients in a children's hospital in 201032 to 24.4% of ICU patients in a 2008 survey.27 This carrier status is important because, although MRSA can survive in the environment for limited periods of time, it is susceptible to most commonly used disinfectants. Transmission is most frequently through direct contact with infected or colonized persons, with colonized patients serving as the main reservoir of S. aureus in healthcare facilities.38
NHP share many predisposing factors with humans, including natural nasal carriage of S. aureus.37 In a previous study of 731 laboratory rhesus macaques, 39% had S. aureus isolated from the nasal passages, comparable to the approximately 35% of the human population that carries the bacteria commensally in the nares.19 In a study of 596 laboratory macaques, MRSA was isolated from 17.6%.31 Multiple case reports have presented the effects of MRSA infection in previously healthy macaques, including pneumonia26 and necrotizing stomatitis,20 and in association with risk factors that predispose NHP to opportunistic infections, including immunosuppression,3,17,19 cranial implants,15 and chronic, exteriorized catheters.35 A population- based study in laboratory chimpanzees revealed a MRSA carriage rate of 69%, with isolates identified as primarily human community-associated (CA) MRSA strains.11
MRSA carriage in laboratory NHP is problematic from both the perspective of preventing research interference and from the risk of zoonotic and anthropozoonotic transmission between animals and their human caretakers. Animals in research colonies may have varied origins and travel histories, allowing for the introduction of bacterial strains from multiple sources. The current study evaluates MRSA carriage in a population of laboratory macaques, showing prevalence rates lower than those observed in previous studies of NHP but close to that of the hospitalized human population, with increased rates of carriage associated with greater degrees of veterinary intervention. However, the isolates in the studied population do not belong to prevalent local (Chicago, IL) human sequence types, with some seen sporadically in human populations in Asia and Australia, some currently undescribed in humans, and others that were identified during the course of this work. Our data suggest these are NHP-adapted MRSA strains that can be harbored in laboratory colonies of NHP.
Materials and Methods
Animals.
Samples obtained from 148 rhesus macaques (Macaca mulatta) and 152 cynomolgus macaques (Macaca fascicularis) were used in a cross-sectional analysis. The study population comprised all macaques present in the institution's NHP colony at the initiation of the study, as well as all newly arrived animals until a total study population of 300 animals was achieved. All animals were housed in accordance with the Guide for the Care and Use of Laboratory Animals,12 Public Health Service Policy,24 and the Animal Welfare Act1 and Regulations2 in an AAALAC-accredited institution. All animal procedures described were determined, in consultation with the institution's IACUC, to be part of standard veterinary care and that a separate IACUC approval was unnecessary. Animals were maintained in an indoor facility kept at 22 ± 2 °C, 30% to 70% humidity, with 100% conditioned air and 12 to 15 air-changes hourly and lighting on a 12:12-h light:dark cycle (lights on, 0600). NHP were kept in visual and auditory contact with congenerics and cohoused in pairs whenever possible. Animals had access to toys and manipulanda in the cage, and speakers in the room provided auditory enrichment through music and natural sounds. Animals were fed 15% Monkey Diet (8714, Harlan-Teklad, Madison, WI) in weight-appropriate amounts once daily and had free access to municipal tap water. Foraging material or fresh produce was offered once daily. All NHP tested negative for STLV and SIV by serology and negative for SRV types 1 through 5 by serology and PCR analysis. All animals were determined to be free of tuberculosis through twice-yearly skin testing and were deemed healthy by annual physical examination, including evaluation of hematology, serum chemistry, and fecal flotation.
Sample collection.
Swabs were taken from the nares while NHP were sedated under ketamine (10 mg/kg IM; Henry Schein, Dublin, OH) or ketamine (10 mg/kg IM)–xylazine (1 to 2 mg/kg IM; Lloyd Laboratories, Shenandoah, IA) for routine healthcare or study-related purposes. Nasal cultures were collected over a period of 9 mo, with a single culture swab collected from each animal. A swab (Liquid Stuart's Minitip Swab, Becton–Dickinson, Sparks, MD) was inserted into one nare until resistance was met or to 2 cm. The swab was then slowly rotated against the nasal mucosa while being retracted from the first nare; this process was repeated by using the same swab in the second nare. The swab was immediately placed in the sterile collection tube, stored at 4 °C, and transported to the University of Illinois Veterinary Diagnostic Lab within 24 to 48 h.
Isolation of methicillin-resistant and -sensitive S. aureus strains and identification of carriers.
Swabs were immersed in 1 mL trypticase soy broth to remove and suspend bacteria from the nasal secretions. A fresh swab was used to transfer the suspension and inoculate plates of Columbia blood agar, Columbia blood agar containing colistin and naladixic acid, mannitol salt agar, and oxacillin (6 μg/mL) in agar (Remel Microbiology Labs, ThermoFisher Scientific, Lenexa, KS). After overnight incubation, staphylococcal isolates (a minimum of 3 colonies per plate for 4 media) demonstrating hemolysis or acid production from mannitol were subcultured to oxacillin-containing medium. Another 3 colonies were selected from the oxacillin medium and replated on Columbia blood agar to confirm their identification as staphylococci. Catalase and coagulase tests were performed to screen putative S. aureus isolates. Trek Sensititire GPID (ThermoFisher Scientific) or GP2 panels (Biolog, Hayward, CA) were used to confirm the identity of any atypical isolates. Representative isolates identified as S. aureus that grew on agar containing 6 μg/mL oxacillin were characterized regarding the minimal inhibitory concentration of oxacillin by using E-strips (bioMérieux, Lombard, IL). Oxacillin-resistant (≥4.0 μg/mL) isolates were confirmed phenotypically as S. aureus by means of MALDI–TOF mass spectrometry using the Bruker Biotyper (Bruker Daltonics, Billerica, MA) according to the manufacturer's tube extraction methods. A Main Spectrum library was created for further comparison of the isolates.
Isolates were genetically confirmed as MRSA through mecA PCR testing. A single colony of S. aureus was inoculated into 1 mL trypticase soy broth and incubated overnight. Broth cultures were centrifuged for 10 min at 6000 × g to pellet the cells. The cell pellets were resuspended in 180 µl of lysostaphin (200 μg/mL; 20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1.2% Triton; Sigma-Aldrich, St Louis, MO) and incubated at 37 °C for 30 min. After the addition of 20 µL proteinase K (stock, 20 mg/mL) and 200 µL of buffer AL, the tubes were incubated at 55 °C for 30 min followed by 10 min at 95 °C. Genomic DNA was extracted from these lysates by using QiaAmp DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. DNA was eluted in 100 µL of buffer AE. All buffer components used were part of the QiAmp – BioSprint 96 One-for-All Vet Kit (Qiagen, Leipzig, Germany). PCR analysis for mecA was performed as previously described.30 Isolates were identified as MRSA when S. aureus was isolated and the mecA gene was present. Isolates identified as S. aureus that lacked mecA were classified as MSSA. In addition, the subset of randomly selected MSSA isolates used for antimicrobial susceptibility testing was confirmed to lack the mecA gene. Other staphylococci were classified on the basis of coagulase testing and hemolysis pattern on blood agar. NHP were classified as MRSA carriers when at least one MRSA isolate was obtained on nasal culture; MSSA carriers were identified in light of isolation of at least one S. aureus isolate on nasal culture but without MRSA isolates. Animals were determined to be noncarriers only when no S. aureus organisms were identified.
Antimicrobial sensitivity.
Antimicrobial sensitivity was performed by using GPALL 3F (Sensititre, West Lake, OH) and enrofloxacin E-strips (bioMérieux, Marcy-l'Étoile, France) for all MRSA carriers (n = 19) and a subset of MSSA carriers (n = 15). MSSA carriers for analysis were selected by using the random-number generator function in Excel (Microsoft, Redmond, WA). Interpretation was based on Clinical and Laboratory Standards Institute criteria for companion animals (category A of document Vet A-04 S-1) or humans (category B of document M-100 S-23) for included antibiotics.4 Drugs included in the analysis were selected on the basis of their usage in veterinary medicine, the presence of interpretative criteria, and the ability to obtain representative drugs from commonly used classes of antibiotics. Antibiotic-specific minimal inhibitory concentrations were determined by using TREK Sensititre Panel GPALL (ThermoFisher Scientific) for ampicillin, ceftriaxone, chloramphenicol, clindamycin, erythromycin, gentamicin, oxacillin, penicillin, rifampin, tetracycline, and trimethoprim–sulfamethoxazole (TMS). Minimal inhibitory concentrations of enrofloxacin and vancomycin were determined by using E-strips (bioMérieux).
blaZ PCR analysis.
MRSA isolates and the majority of MSSA isolates that were evaluated for antimicrobial susceptibility underwent PCR for the blaZ gene, which encodes penicillin resistance. PCR analysis was performed as described previously with slight modification.10 The PCR mixture consisted of 32 pmol of each primer (stau-blaz-fwd, 5′ CAA AGA TGA TAT AGT TGC TTA TTC TCC 3′; stau-blaz-rev, 5′ TGC TTG ACC ACT TTT ATC AGC 3′), 100 ng of genomic DNA, and an Illustra PuReTaq Ready-To-GoPCR Analysis Bead (GE Healthcare Life Sciences, Pittsburg, PA) in a total volume of 25 µL. The following thermal cycling conditions were applied: initial denaturation at 95 °C for 5 min; 35 amplification cycles each consisting of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 30 s; and a final extension at 72 °C for 7 min. Amplification of a 421-bp product confirmed the presence of the blaZ β-lactamase gene and penicillin resistance. PCR products were electrophoresed at 150 V for 50 min on a 2% agarose gel containing 0.5 µg/mL ethidium bromide.
Acquisition of demographic information.
Complete medical records from time of colony entry and, when appropriate, importation into the United States (USFW 3-177), were obtained for all study participants (n = 300), and information on species, sex, age, weight, length of time in colony, country of origin, length of time in the United States, investigator, IACUC protocol assignment, history of surgical and dental procedures, history of illness or injury, history of antibiotic treatment, and history of steroid treatment was extracted (Table 1).
Table 1.
Significance of demographic factors relative to MRSA carrier status
| General | Factor | Category | MRSA+ | MRSA– | Analysis method | P |
| Animal | Species | M. fasicularis | 11 | 137 | ||
| M. mulatta | 8 | 144 | χ2 | 0.590 | ||
| Sex | Male | 14 | 188 | |||
| Female | 5 | 90 | χ2 | 0.769 | ||
| Age | <4 y | 4 | 85 | |||
| 5–15 y | 12 | 172 | ||||
| >15 y | 3 | 24 | Fisher exact test | 0.442 | ||
| Weight | <5 kg | 9 | 152 | |||
| 5–10 kg | 9 | 100 | ||||
| >10 kg | 1 | 24 | Fisher exact test | 0.323 | ||
| Geographic history | Time in US | <1 y | 4 | 91 | ||
| 1–5 y | 10 | 75 | ||||
| >5 y | 3 | 39 | Fisher exact test | 0.168 | ||
| Time in colony | 0–42 d | 3 | 72 | |||
| 43 d–1 y | 7 | 112 | ||||
| 1–3 y | 7 | 64 | ||||
| >3 y | 2 | 33 | Fisher exact test | 0.559 | ||
| Vendor | 16 categories | na | na | Fisher exact test | 0.599 | |
| Origin type | Academic | 0 | 5 | |||
| Import | 17 | 218 | ||||
| National Primate Center | 2 | 31 | ||||
| Pharmaceutical | 0 | 20 | ||||
| Unknown | 0 | 3 | Fisher exact test | 0.795 | ||
| Research use | Protocol | 24 protocols | na | na | Logistic regression | None significant |
| Investigator | A | 0 | 16 | |||
| B | 1 | 36 | ||||
| C | 0 | 30 | ||||
| D | 11 | 78 | ||||
| E | 5 | 74 | ||||
| F | 2 | 47 | Fisher exact test | 0.149 | ||
| History of surgery | Yes | 9 | 46 | |||
| No | 10 | 231 | Fisher exact test | 0.003 | ||
| No. of surgeries | 1 | 7 | 27 | |||
| 2 | 2 | 5 | ||||
| 3 | 0 | 10 | ||||
| 4 | 0 | 1 | Fisher exact test | 0.534 | ||
| History of dental procedure | Yes | 8 | 38 | |||
| No | 11 | 239 | Fisher exact test | 2.91 × 10−3 | ||
| No. of dental procedures | 1 | 7 | 26 | |||
| 2 | 1 | 8 | ||||
| 3 | 0 | 2 | ||||
| 4 | 0 | 1 | ||||
| 6 | 0 | 1 | Fisher exact test | 0.847 | ||
| History of clinical incident | Yes | 9 | 69 | |||
| No | 10 | 207 | Fisher exact test | 0.055 | ||
| Implant | Yes | 1 | 23 | |||
| No | 18 | 242 | Fisher exact test | 1 | ||
| History of steroid administration | Yes | 5 | 9 | |||
| No | 14 | 272 | Fisher exact test | 0.824 × 10−3 | ||
| History of antibiotic administration | Yes | 13 | 91 | |||
| No | 6 | 183 | Fisher exact test | 4.32 × 10−3 | ||
| Specific antibiotic administered | 7 categories | na | na | Fisher exact test | None significant |
Statistical analysis of demographic information.
The association between the demographic factors with the response subgroups (MRSA carrier status) was examined. For categorical factors (for example, sex, species), the χ2 test and the Fisher exact test were used as appropriate. The Fisher exact test was applied when there were 5 observations or fewer in one or more subcategories. For continuous factors (for example, age, body weight), a logistic regression model was used for the binary response variable (MRSA carrier and noncarrier). The analyses were implemented by using R 3.2.2.25 A P value of less than 0.05 was considered significant.
Molecular typing.
One or more isolates from 18 of 19 MRSA-positive animals underwent identity confirmation by using the Staphaurex Plus Test (Remel) followed by molecular typing involving SCCmec typing, multilocus sequence typing (MLST), and PCR amplification for the detection of the arcA gene within the arginine catabolic mobile element (ACME arcA) and Panton–Valentine leucocidin (PVL) as described.7
MLST was performed by DNA sequencing of the PCR amplicons of 7 S. aureus-specific housekeeping genes by using primer sets described on the S. aureus organism page of the MLST Database.16 Sequence types were assigned through submission of sequences to the MLST web portal, which compares the alleles deposited on the S. aureus organism page.
Results
Carriage rate.
In the 300 macaques studied, S. aureus was isolated from the nares of 58.7% (n = 176), representing 50.7% (n = 75) of the rhesus macaques and 66.4% (n = 101) of the cynomolgus macaques. Of the total study population, 6.3% (n = 19; 8 rhesus, 11 cynomolgus) yielded S. aureus that supported amplification of the mecA gene and were classified as MRSA carriers (Figure 1). The remaining S. aureus carriers, accounting for 52.3% (n = 157) of the total population, did not have phenotypic resistance to methicillin or support amplification of the mecA gene and were classified as MSSA carriers. Of the total 176 animals from which S. aureus was isolated, 10.8% were classified as MRSA carriers, with the remaining 89.2% classified as MSSA carriers.
Figure 1.
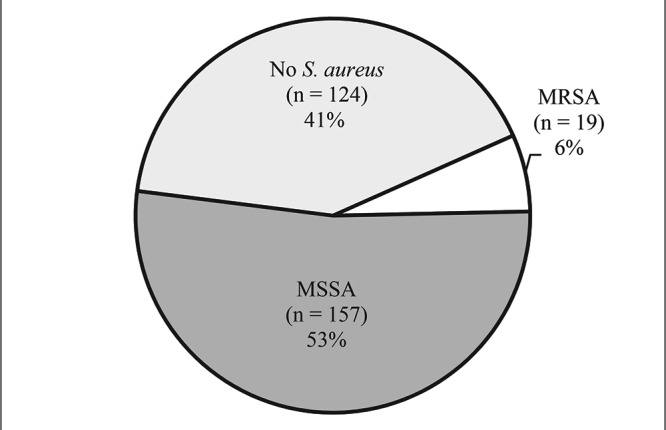
Carriage of MSSA and MRSA in relation to total study population (n = 300).
Demographic analysis.
Statistical analysis of information from medical records revealed no significant association between MRSA carrier status and species, sex, age, body weight, length of time since entry into the University of Illinois at Chicago primate colony, country of origin, length of time in the US, assigned investigator, IACUC protocol assignment, presence of an implanted device, or history of illness or injury.
A history of at least one surgical procedure was significantly (P = 2.45 × 10−3) linked to MRSA carriage. Macaques that underwent surgical procedures had histories documenting between 1 and 4 surgical procedures, but there was no trend toward increased rates of carriage according to the number of surgical procedures. A history of at least one dental procedure was significantly (P = 2.91 × 10−3) linked to MRSA carriage. Dental procedures included endodontic procedures and surgical extractions. Macaques that underwent dental procedures had histories documenting between 1 and 6 dental procedures, but there was no trend toward increased rates of MRSA carriage according to the number of dental procedures. A history of antibiotic treatment was significantly (P = 4.32 × 10−3) linked to MRSA carriage rate; 34.7% (n = 104) of the study population had a documented history of receiving at least one antibiotic during their residence in the colony. The majority of these animals (n = 92) received cefazolin, 40 received clindamycin, and few (less than 10 each) had histories that included administration of ampicillin–sulbactam, amoxicillin–clavulanic acid, ceftiofur, enrofloxacin, erythromycin, or TMS. Within the population of animals that had a history of antibiotic administration, no single drug was significantly linked to MRSA carriage rate. A history of steroid treatment had a statistically significant link to MRSA carriage, with the smallest P value (P = 8.24 × 10−4). All animals in the study population with a history of steroid use received oral prednisone as part of a single experimental protocol (Table 1).
Molecular characterization of isolates.
MRSA isolates had DNA extracted and were analyzed for the presence of virulence genes, multilocus sequence type, and SCCmec type (Table 2). All isolates were negative for PVL, which is commonly carried by community-associated (CA) MRSA strains in the United States. In addition, all isolates were negative for the ACME arcA gene, which is associated with the most common human CA-MRSA clone in the United States, USA300/ ST8. However, 94.4% of the macaque MRSA isolates were positive for blaZ, which codes for penicillin resistance.
Table 2.
Presence or absence of mecA, PVL, ACME arcA, and blaZ genes grouped according to methicillin resistance and sequence type.
|
mecA |
PVL |
ACME arcA |
blaZ |
||||||
| + | – | + | – | + | – | + | – | ||
| MRSA | ST188 | 14 | 0 | 0 | 14 | 0 | 14 | 13 | 1 |
| ST3268 | 3 | 0 | 0 | 3 | 0 | 3 | 3 | 0 | |
| ST3478 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | |
| not typed | 1 | 0 | Not done | Not done | 1 | 0 | |||
| MSSA | ST3479 | 0 | 1 | 0 | 1 | Not done | 1 | 0 | |
| ST3480 | 0 | 1 | 0 | 1 | Not done | 0 | 1 | ||
| not typed | 0 | 13 | Not done | Not done | 2 | 11 | |||
MRSA ST188 (n = 14), MRSA ST3268 (n = 3), MRSA ST3478 (n = 1), MRSA-not typed (n = 1), MSSA ST3479 (n = 1), MSSA ST3480 (n = 1), MSSA-not typed (n = 13).
According to MLST and SCCmec typing, 73.7% of MRSA carriers (n = 14) harbored ST188-SCCmec IV isolates; 15.8% (n = 3) of isolates, from 3 individuals, were identified as ST3268-SCCmec V; and a single MRSA isolate was identified as a novel sequence type, ST3478, carrying SCCmec V. No macaque had more than one MRSA sequence type isolated. However, on repeat analysis, one sample was determined to contain heterogeneous strain types, consisting of 1 MRSA isolate and 2 MSSA isolates, which also underwent MLST. In addition to a ST188-SCCmec IV MRSA, this sample included 2 previously undescribed MSSA isolates, ST3479 and ST3480. These isolates likely grew on oxacillin medium due to overproduction of β-lactamase, and 1 of the 2 MSSA was positive for blaZ. Alternately, the 2 MSSA could have been cocultured on the oxacillin-containing agar, where they benefited from the methacillinase produced by the MRSA strain.
Antibiotic susceptibility.
Isolates underwent antibiotic susceptibility testing by using gram-positive sensitivity panels and enrofloxacin test strips to identify antibiotic resistance patterns for antibiotics in veterinary use. This analysis revealed remarkable differences between MSSA and MRSA isolates (Tables 3 and 4). The prevalence of antibiotic resistance among MSSA isolates tested was low: 87% of nontyped MSSA isolates were susceptible to enrofloxacin. However, 1 of the 2 novel strains of MSSA, ST3480, was resistant to enrofloxacin. All nontyped MSSA were susceptible to all other nonβ-lactam antibiotics tested, with the exception of a single MSSA isolate that was intermediately resistant to chloramphenicol. Compared with other MSSA strains, ST3479 and ST3480 had higher levels of resistance, with both showing resistance to clindamycin and erythromycin and intermediate resistance to vancomycin (Table 5). The high number of isolates exhibiting phenotypic susceptibility to penicillin among the MSSA isolates was surprising given the nearly universal presence of blaZ, which encodes the penicillin-hydrolyzing enzyme β-lactamase, among S. aureus isolates from human and animal populations.22 Accordingly, 80% of MSSA isolates analyzed were blaZ-negative, confirming penicillin susceptibility.
Table 3.
Percentage (%) of nontyped MSSA isolates (n = 15) susceptible to antimicrobials
| Susceptible | Intermediate | Resistant | |
| Enrofloxacin | 87 | 0 | 13 |
| Ampicillin | 100 | 0 | 0 |
| Ceftriaxone | 100 | 0 | 0 |
| Chloramphenicol | 93 | 7 | 0 |
| Clindamycin | 100 | 0 | 0 |
| Erythromycin | 100 | 0 | 0 |
| Gentamicin | 100 | 0 | 0 |
| Oxacillin | 100 | 0 | 0 |
| Penicillin | 100 | 0 | 0 |
| Rifampin | 100 | 0 | 0 |
| Tetracycline | 100 | 0 | 0 |
| TMS | 100 | 0 | 0 |
| Vancomycin | 100 | 0 | 0 |
Table 4.
Antimicrobial susceptibility of MRSA isolates, grouped according to sequence type; data in each column are given as percentage of total isolates
| ST188 (total n = 14) |
ST3268 (total n = 3) |
|||||||
| S | I | R | S | I | R | ST3478 (n = 1) | Not typed (n = 1) | |
| Enrofloxacin | 0% | 0% | 100% | 0% | 0% | 100% | S | S |
| Ampicillin | 7% | 0% | 93% | 0% | 0% | 100% | R | R |
| Ceftriaxone | 0% | 14% | 86% | 0% | 33% | 67% | I | R |
| Chloramphenicol | 7% | 93% | 0% | 33% | 67% | 0% | I | I |
| Clindamycin | 14% | 0% | 86% | 0% | 67% | 33% | S | R |
| Erythromycin | 14% | 0% | 86% | 0% | 67% | 33% | S | R |
| Gentamicin | 7% | 0% | 93% | 0% | 33% | 67% | R | R |
| Oxacillin | 0% | 0% | 100% | 0% | 0% | 100% | R | R |
| Penicillin | 7% | 0% | 93% | 0% | 0% | 100% | R | R |
| Rifampin | 100% | 0% | 0% | 100% | 0% | 0% | S | S |
| Tetracycline | 86% | 0% | 14% | 33% | 0% | 67% | R | S |
| TMS | 43% | 0% | 57% | 67% | 0% | 33% | S | R |
| Vancomycin | 100% | 0% | 0% | 100% | 0% | 0% | S | S |
I, intermediate; R, resistant; S, susceptible
Table 5.
Antimicrobial susceptibility of newly described MSSA isolates ST3479 and ST3480
| ST3479 | ST3480 | |
| Enrofloxacin | Sensitive | Resistant |
| Ampicillin | Resistant | Sensitive |
| Ceftriaxone | Sensitive | Sensitive |
| Chloramphenicol | Sensitive | Sensitive |
| Clindamycin | Resistant | Resistant |
| Erythromycin | Resistant | Resistant |
| Gentamicin | Sensitive | Sensitive |
| Oxacillin | Sensitive | Sensitive |
| Penicillin | Resistant | Sensitive |
| Rifampin | Sensitive | Sensitive |
| Tetracycline | Sensitive | Sensitive |
| TMS | Sensitive | Sensitive |
| Vancomycin | Intermediate | Intermediate |
As expected for MRSA, isolates were uniformly resistant to β-lactam antibiotics. A single MRSA isolate belonging to ST188 showed phenotypic susceptibility to penicillin and ampicillin, testing negative for the blaZ gene. The remaining MRSA isolates were blaZ positive (Table 2). MRSA isolates had a greater tendency to be resistant to nonβ-lactam antibiotics than the MSSA in our study population. Among the 9 nonβ-lactam drugs tested, all MRSA isolates were resistant to at least 1 drug, with the majority resistant to 3 or 4 additional drugs. However, none of the MRSA isolates in our study population was resistant to rifampin or vancomycin. No clear patterns regarding antibiotic resistance were present when isolates were stratified according to ST (Table 5).
Discussion
We characterized the prevalence of MRSA colonization in a representative academic research colony of NHP, with the goals of determining factors linked to increased rates of carriage and gaining insight into the origin of strains present within our study population. It is noteworthy that the MRSA carriage rate of 6.3% in our NHP is close to that reported in hospitalized human populations but much lower than carriage rates in human ICU and long-term care settings33 and in other populations of laboratory macaques and chimpanzees.11,31 This difference may reflect that 25% of the study population had been in the colony for less than 42 d, the minimal length of the quarantine period, during which no study-related activities were conducted, or the variable degree to which animals had undergone study-related or veterinary manipulation. However, we did find a surprisingly high rate of nasal S. aureus carriage, with the bacteria isolated from 58.7% of the study population; this rate is almost twice that in many studies of human CA-MRSA and previous data from laboratory macaques.18,22,38 It is unknown whether this difference was due to culture methods that were more sensitive for detection of S. aureus or whether this high carriage rate is a true reflection of the carriage rate in the population studied.
Our analysis of risk factors showed a significant association between MRSA carriage and veterinary or experimental interventions that included dental procedures, surgical procedures, antibiotic administration, and steroid administration. Although the factors identified do appear to be linked to MRSA carriage, determining the individual effect of each factor is challenging. At our institution, perioperative antibiotics are regularly given, and antibiotics are frequently administered in conjunction with dental procedures where infection is a component of the presenting pathology. In addition, the protocol under which animals received oral prednisone included a surgical procedure, for which perioperative antibiotics were given. Furthermore, due to advanced age, many animals under this protocol had dental pathology, requiring dental procedures prior to experimental use. Although it is impossible to ascertain the influence of each individual factor within the current study population, the current data support increased veterinary intervention affecting MRSA carriage status. These risks correlate with many factors thought to predispose the hospitalized human population to MRSA infection. However, in the human literature, although antibiotic administration does predispose to MRSA carriage,18 there is no evidence that the remaining factors are linked to increased risk of MRSA carriage compared with carriage of MSSA.
No associations between MRSA carriage rate and time in the colony, animal origin, or the protocol or investigator to which an animal was assigned were identified, suggesting that MRSA does not enter the colony from a single origin or transfer between animals that share human handlers or equipment. The lack of significant associations for these factors may be due to the fact that our study population was extremely heterogeneous and, with only a few positive animals, it is difficult to show significant statistical association. Although our methods grouped together animals that shared an experimental protocol or investigator, these signifiers did not distinguish between those animals that had undergone protocol-related manipulation and those awaiting use. This situation may explain the statistically significant link between veterinary or experimental intervention and carriage rate, whereas no association occurred between either investigator or protocol and carriage rate, despite the fact that many interventions linked to increased carriage rate were protocol-based.
Antibiotic susceptibility patterns between MRSA and MSSA isolates differed widely. MSSA in our study population had low levels of resistance to most antibiotics tested, including penicillin, thus suggesting that isolates in our population differ from those in the human population, where penicillin resistance is currently greater than 90% in human clinical isolates22 and 78% in commensal nasal flora.36 Penicillin resistance is also prevalent and well documented in groups of animals with extensive human contact or exposure to antibiotics, including companion animals, livestock, and NHP. A study in captive lion tamarins demonstrated penicillin resistance in all S. aureus isolates,21 and studies of S. aureus carried by sanctuary chimpanzees showed as many as 75% of isolates to be resistant to penicillin.28,29 In the chimpanzee study,28,29 a high degree of concordance with sequence types found in the local human population was observed, supporting anthropozoonotic spread of penicillin-resistant isolates. In contrast, the high degree of antibiotic susceptibility among MSSA isolates in the current study indicates that these strains differ from those circulating in the human population.
MRSA isolates in our population showed very broad antibiotic-resistance patterns, which is more characteristic of healthcare-associated MRSA than CA-MRSA. Although most isolates were resistant to 3 or 4 nonβ-lactam drugs, there were no consistent patterns of susceptibility when isolates were grouped according to sequence type. All of the MRSA isolates from our study population carried SCCmec type IV or V, which are present among CA-MRSA. These mobile genetic elements are smaller than other types and generally associated with more virulent strains. However, none of these isolates carried the PVL gene, which codes for a pore-forming toxin associated with increased virulence that is commonly identified in CA-MRSA isolates.9
Evidence supports the frequent zoonotic and anthropozoonotic transmission of methicillin-resistant staphylococcal species. For example, MRSA strains identified in pets frequently mirror those seen in the local human population,40 and pork producers are frequently carriers of ST398, a strain common in pigs.5,6 Our data, in contrast to observations in captive laboratory chimpanzees,11 show a lack of local human strains and suggest the presence of NHP-associated MRSA strains.
ST188-SCCmec IV accounted for almost 75% of the MRSA isolates that we found in our study population. Notably, ST188-SCCmec-IVa was identified in nasal samples from NHP at a National Primate Center, as well as in the local environment and in a nasal sample from a member of the research staff.31 This ST has been identified in the human population in the Asia–Pacific region, primarily as MSSA, but has not been documented in the local human population of Chicago, where the current study was conducted. ST188 is most closely related to MRSA of clonal complex 1, which contains USA400, a common CA-MRSA. However, genetic differences between the 2 groups are dispersed across the genome in a manner that suggests multiple recombination events over a complex natural history.16 Reports of ST188 in humans vary, including association with both hospital and community-acquired infections and in both PVL-positive and -negative forms.13,23,42 In addition, ST188 MRSA has been found in raw chicken in China.39 The isolates of ST188 in NHP at the earlier-mentioned National Primate Center as both MRSA and MSSA were closely related to each other but were easily differentiated from the human isolate of ST188 from Hong Kong used to generate the draft genome sequence.13,31 The ST188 isolates in our study showed variable phenotypic resistance to all antibiotics tested, except for oxacillin, to which all were resistant, and rifampin and vancomycin, to which all were susceptible. These findings suggest that despite a shared sequence type, these isolates comprise a heterogeneous population.
The second genotype identified, ST3268-SCCmec V, was present in 3 of the 19 MRSA carriers in our study population. These animals consisted of 2 rhesus and 1 cynomolgus macaque. The rhesus macaques were imported by 2 separate suppliers from China in 2013 and 2014, whereas the cynomolgus macaque was imported from Cambodia in 2008. All 3 NHP arrived in separate shipments within a 1-mo span of time and were sampled for carriage between 19 and 113 d after colony entry. ST3268 has not been reported in humans, but ST3268-SCCmec V was identified in macaques at the earlier-mentioned National Primate Center.31 The most closely related strain is ST2817, a human clinical MRSA isolate identified in Singapore.16 Isolates of ST3268 varied in resistance to all nonβ-lactam antibiotics tested, except for enrofloxacin, to which all were resistant, and rifampin and vancomycin, to which all were susceptible. The third sequence type identified in our colony was ST3478. This previously undescribed ST was identified from a single animal and, of the nonβ-lactam drugs tested, was resistant to gentamicin and tetracycline, with intermediate resistance to chloramphenicol. This novel MRSA background has multiple alleles (glpF, pta) that do not correspond to previously described sequences, but it is most closely related to ST2096, an MSSA isolated from a macaque nasal swab in the Netherlands that does not fit within previously described clonal complexes.37 A previous study investigating S. aureus nasal carriage in rhesus macaques found that 59% of isolates were previously undescribed and unrelated to identified human strains, thus supporting NHP-specific MSSA lineages.37 In our current study, the identification of isolates that are unidentified in humans yet that have been identified in unrelated populations of laboratory macaques is consistent with this previous finding and suggests the presence of NHP-adapted MRSA with the potential to pass between macaques and humans.
Our findings provide no evidence of MRSA entering the study population from the local human population in Chicago; however, the potential for zoonotic transmission bears further investigation. ST188, the most prevalent isolate type identified in the study population, is most frequently observed in Asia, where the majority of new arrivals to the colony originated. This strain may enter from the human population in Asia or may be an NHP-specific strain transmitting into humans. Further investigation of MRSA carriage in both humans and animals associated with macaque breeding facilities and analysis of macaque populations from multiple origins may shed light on this dynamic.
Our current study describes the nasal carriage of MRSA in a population of laboratory macaques. The carriage rate in the study population was comparable to that of many studied hospitalized human populations. But, much as variation exists between human populations, the carriage rate may differ widely between facilities, with our results dependent on characteristics specific to the population studied. However, the current findings place laboratory NHP in the context of broader patterns of emergence of antimicrobial resistant bacteria in humans and animals. Taken as a whole, our data strongly support the potential for NHP-associated MRSA strains and lay the groundwork for further investigations of MRSA in NHP.
Acknowledgments
We thank Alex Castenada and Heather Charles (UIC) for assistance with sample collection and processing, Kat Coda (UIC) for assistance collecting demographic information, and Dr Saraswathi Lanka (UIUC-VDL) for mecA and blaZ PCR analysis. This article was prepared while Susan (Daum) Boyle-Vavra was employed at the University of Chicago. The opinions expressed in this article are the authors’ own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States Government.
References
- 1.Animal Welfare Act as Amended. 2008. 7 USC §2131–2156.
- 2.Animal Welfare Regulations. 2008. 9 CFR § 3.129.
- 3.Baxter JR, Jasmin BH, Hankenson F, Behrman A, Rankin S. 2008. Epidemiologic investigation of a multidrug-resistant Staphylococcus aureus outbreak in 2 macaque colonies. Abstract presented at the AALAS 59th National Meeting, Indianapolis, Indiana, 9–13 November 2008. J Am Assoc Lab Anim Sci 47:89. [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: second informational supplement. CLSI document VET01-S2 (ISBN 1-56238-879-7). Wayne (PA): Clinical and Standards Institute. [Google Scholar]
- 5.Cohn LA, Middleton JR. 2010. A veterinary perspective on methicillin-resistant staphylococci. J Vet Emerg Crit Care (San Antonio) 20:31–45. 10.1111/j.1476-4431.2009.00497.x. [DOI] [PubMed] [Google Scholar]
- 6.Cuny C, Freidrich A, Kozytska S, Layer F, Nubel U, Ohlsen K, Strommenger B, Walther B, Wieler L, Witte W. 2010. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int J Med Microbiol 300:109–117. 10.1016/j.ijmm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 7.David MZ, Acree ME, Sieth JJ, Boxrud DJ, Dobbins G, Lynfield R, Boyle-Vavra S, Daum RS. 2015. Pediatric Staphylococcus aureus isolate genotypes and infections from the dawn of the community-associated methicillin-resistant S. aureus epidemic era in Chicago, 1994 to 1997. J Clin Microbiol 53:2486–2491. 10.1128/JCM.00096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2012. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spatTyping, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton–Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a US medical center. J Clin Microbiol 51:814–819. 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Feghaly RE, Stamm JE, Fritz SA, Burnham CA. 2012. Presence of the bla(Z) β-lactamase gene in isolates of Staphylococcus aureus that appear penicillin susceptible by conventional phenotypic methods. Diagn Microbiol Infect Dis 74:388–393. 10.1016/j.diagmicrobio.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Hanley PW, Barnhart KF, Abee CR, Lambeth SP, Weese JS. 2015. Methicillin-resistant Staphylococcus aureus prevalence among captive chimpanzees, Texas, USA, 2012. Emerg Infect Dis 21:2158–2160. 10.3201/eid2112.142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Institute for Laboratory Animal Research. 2011. Guide for the care and use of laboratory animals, 8th ed Washington (DC): National Academies Press. [Google Scholar]
- 13.Ip M, Wang Z, Lam WY, Zhou H, Tsui S. 2014. Draft genome sequence of methicillin-resistant Staphylococcus aureus CUHK_188, a health care-associated bacteremic isolate from Hong Kong. Genome Announc 2:1–2. 10.1128/genomeA.00255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarvis WR, Jarvis AA, Chin RY. 2012. National prevalence of methicillin-resistant Staphylococcus aureus in inpatients at United States healthcare facilities, 2010. Am J Infect Control 40:194–200. 10.1016/j.ajic.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Jasmin BH, Gold JI, Vite CH, Rankin SC, Baxter J, Hankenson F. 2008. Methicillin-resistant Staphylococcus aureus infection and cranial abscessation in a rhesus macaque (Macaca mulatta). Abstract presented at the 59th AALAS National Meeting, Indianapolis, Indiana, 9–13 November 2008. J Am Assoc Lab Anim Sci 47:87. [Google Scholar]
- 16.Jolley KA, Maiden MCJ. [Internet]. 2019. Staphylococcus aureus MLST website. [Cited 30 July 2019]. Available at: https://pubmlst.org/saureus/
- 17.Kim TM, Park H, Lee KW, Choi EW, Moon SH, Lee YS, Cho K, Park WJ, Park JB, Kim SJ. 2017. A simple way to eradicate methicillin-resistant Staphylococcus aureus in cynomolgus macaques (Macaca fascicularis). Comp Med 67:356–359. [PMC free article] [PubMed] [Google Scholar]
- 18.Kluytmans J, van Belkum A, Verbrugh H. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10:505–520. 10.1128/CMR.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluytmans JA, Wertheim H. 2005. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection 33:3–8. 10.1007/s15010-005-4012-9. [DOI] [PubMed] [Google Scholar]
- 20.Lee JI, Kim KS, Oh BC, Kim NA, Kim IH, Park CG, Kim SJ. 2011. Acute necrotic stomatitis (noma) associated with methicillin-resistant Staphylococcus aureus infection in a newly acquired rhesus macaque (Macacca mulatta). J Med Primatol 40:188–193. 10.1111/j.1600-0684.2011.00470.x. [DOI] [PubMed] [Google Scholar]
- 21.Lilenbaum W, Moraes IS, Cardoso VS, Varges RG, Ferreira AMR, Pissinatti AL. 2006. Antibiotic resistance in Staphylococci isolated from the vaginas of captive female Leontopithecus (Callitrichidae Primates). Am J Primatol 68:825–831. 10.1002/ajp.20282. [DOI] [PubMed] [Google Scholar]
- 22.Lowy FD. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111:1265–1273. 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic, and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:1–24. 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Office of Laboratory Animal Welfare. [Internet]. 2015. PHS policy on humane care and use of laboratory animals. [Cited 30 July 2019]. Available at: https://olaw.nih.gov/policies-laws/phs-policy.htm.
- 25.R Project. [Internet]. 2015. R-3.2.2. for Windows (32/64 bit) [Cited 30 July 2019]. Available: https://cran.r-project.org/bin/windows/base/old/3.2.2/
- 26.Rivas K, Garcia A, Morales PR, Matchett CA, Saucedo A, Wagner JL. 2007. Methicillin-resistant Staphylococcus aureus (MRSA) pneumonia in a rhesus macaque. Abstract presented at the AALAS National Meeting, Charlotte, North Carolina, 14–18 October 2007. J Am Assoc Lab Anim Sci 46:102. [Google Scholar]
- 27.Sarikonda KV, Micek ST, Doherty JA, Reichley RM, Warren D, Kollef MH. 2010. Methicillin-resistant Staphylococcus aureus nasal colonization is a poor predictor of intensive care unit-acquired methicillin-resistant Staphylococcus aureus infections requiring antibiotic treatment. Crit Care Med 38:1991–1995. 10.1097/CCM.0b013e3181eeda3f. [DOI] [PubMed] [Google Scholar]
- 28.Schaumburg F, Mugisha L, Peck B, Becker K, Gillespie TR, Peters G, Leendertz FH. 2012. Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am J Primatol 74:1071–1075. 10.1002/ajp.22067. [DOI] [PubMed] [Google Scholar]
- 29.Schaumburg F, Mugisha L, Kappeler P, Fichtel C, Köck R, Köndgen S, Becker K, Boesch C, Peters G, Leendertz F. 2013. Evaluation of noninvasive biologic samples to monitor Staphylococcus aureus colonization in great apes and lemurs. PLoS One 81–6. 10.1371/journal.pone.0078046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schissler JR, Hillier A, Daniels JB, Cole LK, Gebreyes WA. 2009. Evaluation of clinical laboratory standards institute interpretive criteria for methicillin-resistant Staphylococcus pseudintermedius isolated from dogs. J Vet Diagn Invest 21:684–688. 10.1177/104063870902100514. [DOI] [PubMed] [Google Scholar]
- 31.Soge OO, No D, Michael KE, Dankoff JD, Lane J, Vogel K, Smedley J, Roberts MC. 2016. Transmission of MDR MRSA between primates, their environment, and personnel at a United States primate centre. J Antimicrob Chemother 71:2798–2803. 10.1093/jac/dkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song X, Cogen J, Singh N. 2013. Incidence of methicillin-resistant Staphylococcus aureus infection in a children's hospital in the Washington metropolitan area of the United States, 2003–2010. Emerg Microbes Infect 2:1–5. 10.1038/emi.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone ND, Lewis DR, Johnson TM, 2nd, Hartney T, Chandler D, Byrd-Sellers J, McGowan JE, Jr, Tenover FC, Jernigan JA, Gaynes RP; Southeast Veterans Affairs Long-Term Care Methicillin-Resistant Staphylococcus aureus Cooperative. 2012. Methicillin-resistant Staphylococcus aureus (MRSA) nasal carriage in residents of Veterans Affairs long-term care facilities: role of antimicrobial exposure and MRSA acquisition. Infect Control Hosp Epidemiol 33:551–557. 10.1086/665711. [DOI] [PubMed] [Google Scholar]
- 34.Taylor AR. 2013. Methicillin-resistant Staphylococcus aureus infection. Prim Care 40:637–654. 10.1016/j.pop.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Taylor WM, Grady AW. 1998. Catheter-tract infections in rhesus macaques (Macaca mulatta) with indwelling intravenous catheters. Lab Anim Sci 48:448–454. [PubMed] [Google Scholar]
- 36.van Bijnen EM, Paget J, Lange-de Klerk ES, den Heijer CD, Versporten A, Stobberingh EE, Goossens H, Schellevis FG, collaboration with the APRES Study Team. 2015. Antibiotic exposure and other risk factors for antimicrobial resistance in nasal commensal Staphylococcus aureus: an ecological study in 8 European countries. PLoS One 10:1–11. 10.1371/journal.pone.0135094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Berg S, van Wamel WJ, Snijders SV, Ouwerling B, de Vogel CP, Boelens HA, Willems RJ, Huijsdens XW, Verreck FA, Kondova I, Heidt PJ, Verbrugh HA, van Belkum A. 2011. Rhesus macaques (Macaca mulatta) are natural hosts of specific Staphylococcus aureus lineages. PLoS One 6:1–14. 10.1371/journal.pone.0026170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Eiff C, Becker K, Macheka K, Stammer H, Peters G. 2001. Nasal carriage of Staphylococcus aureus bacteremia. N Engl J Med 344:11–16. 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Li G, Xia X, Yang B, Xi M, Meng J. 2014. Antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog Dis 11:281–286. 10.1089/fpd.2013.1643. [DOI] [PubMed] [Google Scholar]
- 40.Weese JS, van Duijkeren EV. 2010. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet Microbiol 140:418–429. 10.1016/j.vetmic.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 41.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–762. 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 42.Yu F, Li T, Huang X, Xie J, Xu Y, Tu J, Qin Z, Parsons C, Wang J, Hu L, Wang L. 2012. Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn Microbiol Infect Dis 74:363–368. 10.1016/j.diagmicrobio.2012.08.015. [DOI] [PubMed] [Google Scholar]


