Abstract
Background:
Methicillin-resistant coagulase-negative staphylococci (MR-CoNS) are multidrug-resistant bacteria that are difficult to treat because of their ability to form biofilms.
Objectives:
In the present study, we evaluated the antibiotic-resistant phenotypes, biofilm-forming ability, and biofilm associated genes of 55 clinical MR-CoNS isolates obtained from two hospitals in Thailand.
Materials and Methods:
MALDI-TOF-MS and tuf gene sequencing were performed to determine the species of all isolates. Biofilm production was determined using Congo red agar (CRA) and the microtiter plate (MTP) assay. Biofilm-associated genes were characterized using polymerase chain reaction (PCR).
Results:
Among the 55 MR-CoNS isolates, five species were identified as Staphylococcus haemolyticus (34.5%), Staphylococcus epidermidis (32.7%), Staphylococcus capitis (18.2%), Staphylococcus cohnii (9.1%), and Staphylococcus hominis (5.5%). The antimicrobial susceptibility pattern of MR-CoNS isolates indicated high resistance to cefoxitin (100%), penicillin (98.2%), erythromycin (96.4%), ciprofloxacin (67.3%), sulfamethoxazole/trimethoprim (67.3%), gentamicin (67.3%), and clindamycin (63.6%). All the isolates were susceptible to vancomycin and linezolid. The biofilm production was detected in 87.3% isolates through the CRA method and in 38.1% isolates through the MTP assay. The prevalence rates of icaAD, bap, fnbA, and cna were 18.2%, 12.7%, 47.3%, and 27.3%, respectively. There were significant differences in the presence of these biofilm-associated genes among the MR-CoNS isolates. Moreover, quantitative biofilm formation was significantly different among MR-CoNS species.
Conclusion:
The present study revealed that biofilm-associated genes are important for biofilm biomass in MR-CoNS isolates, and the findings of this study are essential for finding new strategies to control biofilm formation and prevent the spread of MR-CoNS infectious diseases.
Keywords: Biofilm, biofilm-associated protein, intracellular adhesion AD, methicillin-resistant coagulase-negative staphylococcus
INTRODUCTION
Methicillin-resistant coagulase-negative staphylococci (MR-CoNS) are Gram-positive bacteria considered to be opportunistic pathogens that cause nosocomial and community-acquired infections, including skin and tissue infections, pneumonia, endocarditis, and septicemia.[1] To date, the prevalence of MR-CoNS infections worldwide ranges from 20% to 30% and can be isolated from various clinical samples, such as blood, sputum, urine, and pus.[2] Among MR-CoNS, Staphylococcus epidermidis, and Staphylococcus haemolyticus have become the two major causes of nosocomial infections, which are difficult to treat with antimicrobial agents owing to their capacity to form biofilms on implant medical devices.[3] Staphylococcal infections can be treated with antimicrobial agents, but most bacteria strains have developed resistance to methicillin and most beta-lactam antibiotics.[4] The resistance to methicillin and other beta-lactam antibiotics in both MR Staphylococcus aureus and MR-CoNS is primarily caused by the acquisition of the mec A gene, which encodes a modified penicillin-binding protein 2a that has a low binding affinity for all beta-lactam antibiotics.[5,6]
Slime or biofilm formation permits microorganisms to adhere to different materials, such as prostheses and intravenous devices.[7] Cells within the biofilm are highly resistant to sanitation procedures. The interaction of the host immune system with antimicrobial agents and the development of the biofilm begins with the bacteria adhering to a biotic or an abiotic surface mediated by microbial surface components that recognize adhesive matrix molecules.[8] The bacteria then multiply to form a multilayered biofilm, and this is associated with the production of the polysaccharide intracellular adhesion (PIA) protein, which mediates cell-to-cell adhesion.[1] Three major proteins that play an important role in biofilm development are PIA, biofilm-associated protein (Bap), and fibronectin binding protein (FnbA). PIA is encoded by icaABCD genes located within the intracellular adhesion (ica) operon, and Bap is encoded by bap gene involved in initial attachment and ica.[9,10,11] In addition, fnbA plays an important role in the accumulation phase of biofilm formation either through homophilic interactions or through binding of the proteins to surface receptors of adjacent cells.[12] The distinct nature of species clusters was suspected to be the cause of this difference.[13] Therefore, the process of biofilm formation of MR-CoNS isolated from different sources of clinical specimens should be studied. In the present study, we detected an association between antimicrobial resistance and biofilm formation in different phenotypes of MR-CoNS isolated from various clinical specimens. Understanding the virulence of pathogen biofilms is essential for finding new strategies to lower the severity and prevalence of infectious diseases.
MATERIALS AND METHODS
Samples
A total of 55 clinical isolates of MR-CoNS were obtained. Of which 31 clinical isolates were collected from patients who were admitted to Chiangrai Prachanukroh hospital, and 24 isolates were obtained from patients hospitalized in Naresuan University Hospital. The Chiangrai Prachanukroh Hospital is located in the upper northern region of Thailand, and the Naresuan University Hospital is located in the lower northern region of Thailand. The clinical samples were collected between November 2014 and October 2015. All MR-CoNS isolates were collected from blood (80%), pus (7.3%), and other body fluid (12.7%) samples.
Species identification of methicillin-resistant staphylococci
The bacteria were initially identified by colony morphology, mannitol fermentation, Gram characteristics, catalase, coagulase test, and DNase activity. All isolates were subsequently confirmed as staphylococci by PCR using 16S RNA primers specific to staphylococci.[14] Cefoxitin disk (30 μg) on Mueller-Hinton agar and detection of mecA gene by PCR method was performed to confirm the methicillin resistance. The direct MALDI-TOF-MS was carried out to distinguish the species level of MR-CoNS. Briefly, several colonies were harvested from Mueller-Hinton agar and suspended in 100 μl of sterile water. 1 μl of this mixture was then deposited on a target plate (Bruker Daltonics, Germany) in two replicates and allowed to dry at room temperature. One microliter of absolute ethanol (Merck, Darmstadt, Germany) was then added to each well and dried at room temperature. Subsequently, 1 μl of matrix, α-cyano-4-hydroxycinnamic acid (Bruker Daltonics, Germany) dissolved in a solution of 50% acetonitrile, 2.5% trifluoroacetic acid, and 47.5% water (Sigma-Aldrich, Fluka, MO, USA). MALDI-TOF-MS Spectrometer Autoflex speed (Bruker Daltonics, Germany) and FlexControl software (version 3.4.135, Bruker Daltonics, Germany) were processed to detect the protein and identified the difference between species. A score of 2.000–3.000 indicated species-level identification. The score from 1.700–1.999 indicated genus-level identification and a score of <1.700 was an unreliable identification.[15] The sequencing of tuf genes was used to confirm the species level of isolates that could not be identified directly through MALDI-TOF MS.[16]
Determination of antimicrobial susceptibility pattern
Disc diffusion tests were performed with the following 15 antibiotics (Oxoid, Basingstoke, England): Penicillin (P, 10 units), clindamycin (DA; 2 μg), chloramphenicol (C; 30 μg), gentamicin (CN; 10 μg), erythromycin (E; 15 μg), cefoxitin (FOX; 30 μg), oxacillin (OX; 1 μg), sulfamethoxazole/trimethoprim (SXT; 1.25/23.75 μg), vancomycin (VA; 30 μg), rifampicin (RD; 5 μg), linezolid (LZD; 30 μg), mupirocin (MUP; 5 μg), ciprofloxacin (CIP; 5 μg), fusidic acid (FD; 10 μg), and novobiocin (NV; 5 μg). The results were interpreted according to Clinical and Laboratory Standards Institute guidelines.[17]
Phenotypic biofilm assay
Congo red agar (CRA) test was performed to determine slime production. All MR-CoNS isolates were inoculated on the CRA plate and incubated at 35°C under aerobic conditions for 24–48 h. The slime production was characterized by the color change of the colonies from red to black color. The colonies that remain red were classified as nonslime producers.[18] To determine the quantitative biofilm formation, microtiter plate (MTP) assay test was performed by culturing MR-CoNS isolates overnight in 96-well polystyrene tissue culture MTPs at 37°C, with Trypticase soy broth and 0.25% glucose as the growth medium. The culture medium was then removed and fixed with 95% ethanol, then stained with 1% crystal violet. Each isolate was tested in triplicate and absorbance at 570 nm was determined. Biofilm formation was interpreted as follows: highly positive (OD570 ≥ 1), low-grade positive (0.1 ≤ OD570 < 1), or negative (OD570 < 0.1).[19]
Molecular detection of biofilm-associated gene
The presence of icaAD, fnbA, and bap genes was detected through PCR using primers designed for MR-CoNS gene sequences from previous studies.[13] Primers specific to the cna gene of S. aureus were used for amplification.[20] The primer sets are shown in Table 1. The amplified PCR products were analyzed through electrophoresis on a 1% agarose gel.
Table 1.
List of primers used in this study
| Target gene | Primer | Size (bp) | Tm (p) | Reference |
|---|---|---|---|---|
| 16S rRNA | F: CGAAAGCCTGACGGAGCAAC | 528 | 52 | [14] |
| R: AACCTTGCGGTCGTACTCCC | ||||
| tuf | F: CCAATGCCACAAACTCGTGA | 480 | 62 | [16] |
| R: CAGCTTCAGCGTAGTCTAATAATTTACG | ||||
| mecA | F: TGGCTATCGTGTCACAATCG | 310 | 58 | [21] |
| R: CTGGAACTTGTTGAGCAGAG | ||||
| icaAD | F: GACAGTCGCTACGAAAAG | 211 | 55 | [13] |
| R: AATAAGCTCTCCCTAACTA | ||||
| fnbA | F: CCCTCTTCGTTATTCAGCC | 422 | 58 | [13] |
| R: CAGGAGGCAAGTCACCTTG | ||||
| bap | F: GGCGCAAGCAGCAGAATTA | 901 | 63 | [13] |
| R: CATAGTTCTTTGTGGTGTTGC | ||||
| cna | F: AAAGCGTTGCCTAGTGGAGA | 192 | 55 | [20] |
| R: AGTGCCTTCCCAAACCTTTT |
Statistical analysis
All data were analyzed in Excel and Stata 12.0 software (StataCorporation, College Station, TX, USA). Biofilm biomass formation by all clinical isolates was monitored based on OD570 values. The values were transformed to be normal distribution by a taking log. These log-transformed data were statistically analyzed using parametric statistics. The comparison of biofilm biomass production between different groups was analyzed using one-way ANOVA test. In the case of analysis among two different species group, Student's t-test was performed.
RESULTS
Species distribution of methicillin-resistant coagulase-negative staphylococci
Species-level characterization of the clinical isolates was performed by biochemical tests, PCR, MALDI-TOF-MS, and tuf gene sequencing. The five different species identified were S. haemolyticus (34.5%), S. epidermidis (32.7%), Staphylococcus capitis (18.2%), Staphylococcus cohnii (9.1%), and Staphylococcus hominis (5.5%) [Figure 1]. The prevalence of S. haemolyticus was high in the clinical isolates from Chiangrai Prachanukroh Hospital, while S. epidermidis was predominant among MR-CoNS isolated from Naresuan University Hospital.
Figure 1.
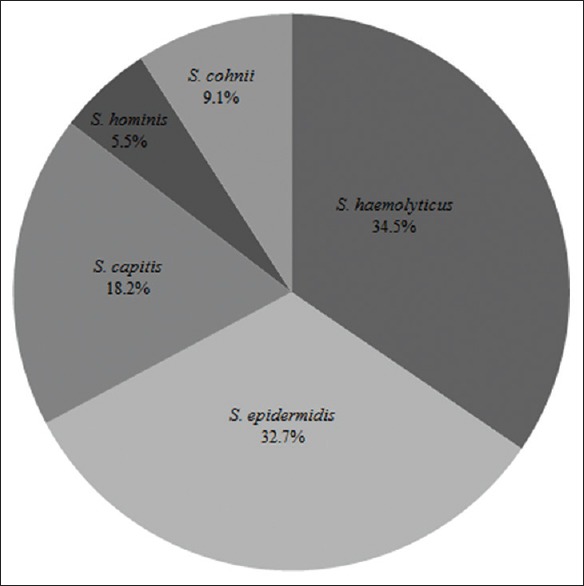
Different species of methicillin-resistant coagulase-negative staphylococci isolated from hospitals in Northern Thailand
Antimicrobial susceptibility testing
All MR-CoNS isolates were tested for their susceptibility against 15 commonly used antibiotics. All isolates showed the resistance to at least one antibiotic class. The isolates were resistant to oxacillin (98.2%), cefoxitin (100%), ciprofloxacin (67.3%), penicillin (98.2%), erythromycin (96.4%), sulfamethoxazole/trimethoprim (67.3%), chloramphenicol (12.7%), rifampicin (20.0%), gentamicin (67.3%), fusidic acid (23.6%), clindamycin (63.6%), mupirocin (38.2%), and novobiocin (9.1%); however, all isolates were sensitive to linezolid and vancomycin [Figure 2].
Figure 2.
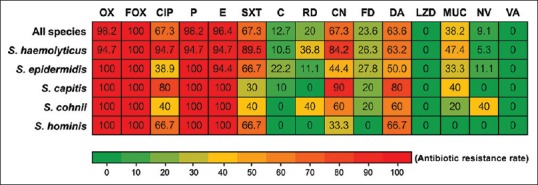
Drug resistance of methicillin-resistant coagulase-negative staphylococci isolated from hospitals in Northern Thailand
Determination of biofilm formation
Isolates showing biofilm formation ability detected through CRA and MTP methods are indicated in Table 2. Using the CRA method, it was observed that seven out of 55 (12.7%) of MR-CoNS isolates formed red colonies, 27 isolates (49.1%) formed black colonies, and 21 isolates (38.2%) formed intensely black colonies. MTP assay demonstrated that 21 out of the 55 MR-CoNS isolates were biofilm producers, out of which 19 (34.5%) isolates showed low-grade positivity, 2 (3.6%) isolates showed high-grade positivity, and 34 (61.8%) isolates were nonbiofilm producers.
Table 2.
Presence of biofilm formation and adhesion genes in methicillin-resistant coagulase-negative staphylococci
| Biofilm formation | Clinical samples | |||||
|---|---|---|---|---|---|---|
| S. haemolyticus (n=19) (%) | S. epidermidis (n=18) (%) | S. capitis (n=10) (%) | S. cohnii (n=5) (%) | S. hominis (n=3) (%) | Total (n=55) (%) | |
| CRA | ||||||
| Red (%) | 0 | 2 (11.1) | 2 (20) | 3 (60) | 0 | 7 (12.7) |
| Black (%) | 12 (63.2) | 8 (44.4) | 4 (40) | 1 (20) | 2 (66.7) | 27 (49.1) |
| Very black (%) | 7 (36.8) | 8 (44.4) | 4 (40) | 1 (20) | 1 (33.3) | 21 (38.2) |
| MTP | ||||||
| Negative (%) | 18 (94.7) | 9 (50) | 1 (10) | 4 (80) | 2 (66.7) | 34 (61.8) |
| Low-grade positive (%) | 1 (5.3) | 7 (38.9) | 9 (90) | 1 (20) | 1 (33.3) | 19 (34.5) |
| Highly positive (%) | 0 | 2 (11.1) | 0 | 0 | 0 | 2 (3.6) |
| Adhesion genes | ||||||
| icaAD (%) | 0 | 4 (22.2) | 6 (60) | 0 | 0 | 10 (18.2) |
| bap (%) | 0 | 1 (5.6) | 5 (50) | 1 (20) | 0 | 7 (12.7) |
| fnbA (%) | 8 (42.1) | 15 (83.3) | 0 | 3 (60) | 0 | 26 (47.3) |
| cna (%) | 4 (21.1) | 10 (55.6) | 0 | 1 (20) | 0 | 15 (27.3) |
| mecA genes | 19 (100) | 18 (100) | 10 (100) | 5 (100) | 3 (100) | 55 (100) |
S. haemolyticus: Staphylococcus haemolyticus, S. epidermidis: Staphylococcus epidermidis, S. capitis: Staphylococcus capitis, S. cohnii: Staphylococcus cohnii, S. hominis: Staphylococcus hominis, CRA: Congo red agar, MTP: Microtiter plate
Detection of the intracellular adhesion AD, biofilm-associated protein, fibronectin binding protein and cna genes
The results given in Table 2 indicates that the icaAD gene was detected in 18.2% of MR-CoNS isolates, belonging to S. epidermidis (22.2%) and S. capitis (60.0%) species, respectively. Clinical isolates of S. haemolyticus, S. hominis, and S. cohnii did not possess this gene. The presence of the bap gene was found in 12.7% of MR-CoNS isolates that belonged to S. capitis (50.0%), S. cohnii (20%), and S. epidermidis (5.6%), respectively. However, the bap gene was not detected in S. haemolyticus and S. hominis. The fnbA gene was present in 47.3% of MR-CoNS isolates and belonged to S. epidermidis (83.3%), S. cohnii (60%), and S. haemolyticus (42.1%), respectively. The cna gene was present in 27.3% of the MR-CoNS isolates, harbored by S. haemolyticus (21.1%), S. epidermidis (55.6%), and S. cohnii (20%), respectively.
Association of biofilm genotypes and biofilm biomass in Methicillin-resistant coagulase-negative staphylococci isolates
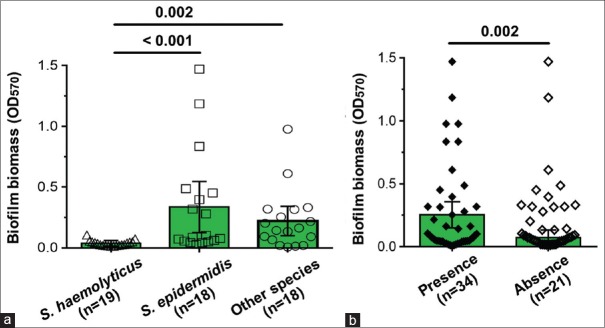
The biofilm biomass production (median OD570) of MR-CoNS isolates was investigated, and the biofilm-forming ability of each species was compared, as shown in [Figure 3a]. The biofilm biomass of S. epidermidis and other species (S. capitis, S. cohnii, and S. hominis) were significantly higher than that of S. haemolyticus (P < 0.05). The correlation between the presence of biofilm-associated genes and the biofilm phenotype of MR-CoNS isolates was statistically evaluated [Figure 3b]. We found that the presence of biofilm-associated genes in MR-CoNS significantly made more biofilm biomass than strains without these biofilm-associated genes.
Figure 3.
Biofilm producing ability of methicillin-resistant coagulase-negative staphylococci obtained from clinical samples. (a) The comparison of OD570 among Staphylococcus haemolyticus, Staphylococcus epidermidis, and other staphylococcal species (Staphylococcus capitis, Staphylococcus cohnii and Staphylococcus hominis). (b) Comparisons of biofilm forming ability between the present and absent of biofilm-associated genes
DISCUSSION
MR-CoNS are the predominant cause of nosocomial infections, which greatly limit therapeutic options for opportunistic infections. It is caused by their ability to adhere the surface of biomaterials and form biofilms.[22] The principal aim of the current study was to determine the prevalence of MR-CoNS species and the biofilm production of MR-CoNS isolated from hospitalized patients in Thailand. In our study, among the 55 clinically significant isolates of MR-CoNS belonging to 5 different species, S. haemolyticus was observed to be the most commonly distributed species. The second-most common isolate was S. epidermidis, followed by S. capitis, S. cohnii, and S. hominis, respectively. The findings agree with those of Teeraputon et al., who reported the prevalence MR-CoNS at Maesot Hospital, Tak province, western Thailand, and documented S. haemolyticus as the most prevalent species (37.55%), followed by S. epidermidis (21.83%), S. saprophyticus (11.79%), and S. hominis (11.35%), respectively.[23] The prevalence of S. haemolyticus was found predominantly in bloodstream infections, and S. epidermidis was found to be one of the most prevalent pathogens implicated in catheter-related bloodstream infections. In addition, treating infections caused by MR-CoNS can be difficult due to their high level of drug resistance.[1] Regarding antimicrobial susceptibility, the MR-CoNS isolates in this study showed high antibiotic resistance, especially to cefoxitin (100%), penicillin (98.2%), and erythromycin (96.4%), which is similar to MR-CoNS isolates in Thailand.[23] All the 55 MR-CoNS isolates were susceptible to vancomycin and linezolid, and this result is consistent with that of the MR-CoNS isolated from clinical samples by Shrestha et al.[24] However, some studies have reported the existence of vancomycin resistance. Mashaly and El-Mahdy (2017) observed that all the clinical isolates were susceptible to vancomycin. However, 15.5% isolates could grow on BHI agar containing 4 μg/mL vancomycin.[25]
Biofilm formation remains the most important mechanism of pathogenicity among staphylococci, especially MR-CoNS. We found that 87.3% of the MR-CoNS isolated in this study were biofilm producers based on the CRA method. These results were similar to findings reported by Shrestha et al., which evidenced that 85% of all MR-CoNS isolated from clinical specimens were biofilm producers.[24] Oliveira and Cunha Mde reported that 75% of the clinical staphylococci isolates were biofilm positive as determined by the CRA method.[26] Using the MTP method to determine biofilm production, we found that 3.6% of the isolates were highly positive and 34.5% were low-grade positive. This prevalence was lower than that in the biofilm-producing MR-CoNS isolated from hospital environments, in which 66 (26.3%) isolates were highly positive and 166 (66.1%) were low-grade positive.[13] However, we found that the biofilm-producing ability of MR-CoNS obtained from various species was different. The biofilm-producing ability of S. epidermidis and other species was significantly higher than that of S. haemolyticus (P < 0.05). This finding correlated with the study conducted by Thilakavathy et al., which reported that out of the 96 MR-CoNS isolated from clinical samples, biofilm production was highest in S. epidermidis (38.54%) followed by S. saprophyticus (1.04%).[27]
Comparing the qualitative and the quantitative methods, it was observed that CRA method is better for biofilm detection than the MTP method. All the S. haemolyticus isolates were identified as biofilm producers by CRA method, while 5.3% were indicated to be low-grade positive by the MTP method. A low correspondence between both methods was also demonstrated by Mathur et al., (2006) who showed that the screening on CRA did not correlate with the MTP screening of staphylococcal isolates.[28] Several studies have evidenced that nosocomial infections caused by staphylococci is associated with the presence of biofilm-associated genes.[10,26] In the present study, the icaAD and bap genes were detected in 18.2% and 12.7% of MR-CoNS isolates, respectively. On the other hand, S. haemolyticus and S. hominis did not possess icaAD and bap genes. These findings correlated with the studies conducted by Seng et al., which demonstrated that S. haemolyticus and other species of staphylococci isolated from community environments did not possess icaAD and bap genes.[13] All of S. capitis isolates that possessed icaAD genes formed biofilms as detected by the MTP and CRA methods, while S. haemolyticus, S. hominis, and S. cohnii that have the ability to produce biofilm when assessed by the CRA method lack the icaAD gene. The biofilm-forming ability of some isolates in absence of icaAD gene, as detected by the CRA method, indicates that they form biofilm through icaAD-independent mechanisms.[29]
Regarding the important role of genes associated with biofilm biomass production, the icaAD gene was found to be involved in biofilm formation, while the bap, fnbA, and cna genes were found to play a role in attachment to biotic or abiotic surfaces, which represents the first step of the process of biofilm formation.[8] In this study, 61.82% of the isolates that harbored icaAD, bap, fnbA, and cna genes alone or in combination were found to produce biofilm significantly via the MTP method. These results were in accordance with the study conducted by Oliveira and Cunha Mde, who demonstrated that 81% of the isolates were positive in the Tissue culture plate test and showed the presence of the icaAD genes.[26] Similarly, the study conducted by Nasr et al. demonstrated that 50% of the icaAD-positive isolates were found to be positive for biofilm formation through the MTP method.[30] In addition, this study indicated that the fnbA gene is present in 47.3% of MR-CoNS isolates. These findings correlated with studies conducted by Giormezis et al., who detected the fnbA gene in 41.4% of biofilm-producing isolates from patients exhibiting bloodstream infections.[31] However, this study concluded that the CRA method might be less precise for the identification of biofilm-forming isolates when compared to PCR, which is used for the detection of the genes involved in biofilm production.
CONCLUSION
All MR-CoNS were belonged to different species, and all the isolates were observed to be multidrug-resistant bacteria. The presence of biofilm associated-genes was detected in the MR-CoNS isolates, and this study has demonstrated an association between biofilm-associated genes and the biofilm phenotype of MR-CoNS isolates. These results represent an important area for further research, and the regulation of biofilm expression is in need to play a central role in the disease prevention.
Financial support and sponsorship
This study was supported by a grant from Office of the higher Education Thailand (HERP 2559) (R2559A006) to SS.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We would like to thank microbiology staff from Chiangrai Prachanukroh Hospital and Naresuan University hospital for specimen collection.
REFERENCES
- 1.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saber H, Jasni AS, Jamaluddin TZ, Ibrahim R. A review of staphylococcal cassette chromosome mec (SCCmec) types in coagulase-negative staphylococci (CoNS) species. Malays J Med Sci. 2017;24:7–18. doi: 10.21315/mjms2017.24.5.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barros EM, Ceotto H, Bastos MC, Dos Santos KR, Giambiagi-Demarval M. Staphylococcus haemolyticus as an important hospital pathogen and carrier of methicillin resistance genes. J Clin Microbiol. 2012;50:166–8. doi: 10.1128/JCM.05563-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma XX, Wang EH, Liu Y, Luo EJ. Antibiotic susceptibility of coagulase-negative staphylococci (CoNS): Emergence of teicoplanin-non-susceptible coNS strains with inducible resistance to vancomycin. J Med Microbiol. 2011;60:1661–8. doi: 10.1099/jmm.0.034066-0. [DOI] [PubMed] [Google Scholar]
- 5.García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tulinski P, Fluit AC, Wagenaar JA, Mevius D, van de Vijver L, Duim B, et al. Methicillin-resistant coagulase-negative staphylococci on pig farms as a reservoir of heterogeneous staphylococcal cassette chromosome mec elements. Appl Environ Microbiol. 2012;78:299–304. doi: 10.1128/AEM.05594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinheiro L, Brito CI, Pereira VC, Oliveira Ad, Camargo CH, Cunha Mde L, et al. Reduced susceptibility to vancomycin and biofilm formation in methicillin-resistant Staphylococcus epidermidis isolated from blood cultures. Mem Inst Oswaldo Cruz. 2014;109:871–8. doi: 10.1590/0074-0276140120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–28. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maira-Litr T, 8, Kropec A, Abeygunawardana C, Joyce J, Mark G, 3rd, Goldmann DA, et al. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70:4433–40. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cucarella C, Tormo MA, Ubeda C, Trotonda MP, Monz M M, Peris C, et al. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun. 2004;72:2177–85. doi: 10.1128/IAI.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tormo MA, Knecht E, GE F, Lasa I, Penaden JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: Evidence of horizontal gene transfer? Microbiology. 2005;151:2465–75. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 12.Herman-Bausier P, El-Kirat-Chatel S, Foster TJ, Geoghegan JA, Dufrfrf YF. Staphylococcus aureus fibronectin-binding protein A mediates cell-cell adhesion through low-affinity homophilic bonds. MBio. 2015;6:e00413–15. doi: 10.1128/mBio.00413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seng R, Kitti T, Thummeepak R, Kongthai P, Leungtongkam U, Wannalerdsakun S, et al. Biofilm formation of methicillin-resistant coagulase negative staphylococci (MR-coNS) isolated from community and hospital environments. PLoS One. 2017;12:e0184172. doi: 10.1371/journal.pone.0184172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohner P, Uhl J, Kolbert C, Persing D, Cockerill F., 3rd Comparison of susceptibility testing methods with mecA gene analysis for determining oxacillin (methicillin) resistance in clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus spp. J Clin Microbiol. 1999;37:2952–61. doi: 10.1128/jcm.37.9.2952-2961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al Laham NA. Species identification of clinical coagulase-negative staphylococci isolated in Al-Shifa hospital Gaza using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Curr Res Bacteriol. 2017;10:1–8. [Google Scholar]
- 16.Loonen AJ, Jansz AR, Bergland JN, Valkenburg M, Wolffs PF, van den Brule AJ, et al. Comparative study using phenotypic, genotypic, and proteomics methods for identification of coagulase-negative staphylococci. J Clin Microbiol. 2012;50:1437–9. doi: 10.1128/JCM.06746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. [Google Scholar]
- 18.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42:872–4. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bekir K, Ben Abdallah F, Ellafi A, Bakhrouf A. Adherence assays and slime production of Staphylococcus aureus strains after their incubation in seawater microcosms. Ann Microbiol. 2011;61:819–23. [Google Scholar]
- 20.Smith K, Gould KA, Ramage G, Gemmell CG, Hinds J, Lang S, et al. Influence of tigecycline on expression of virulence factors in biofilm-associated cells of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:380–7. doi: 10.1128/AAC.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryffel C, Tesch W, Birch-Machin I, Reynolds PE, Barberis-Maino L, Kayser FH, et al. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene. 1990;94:137–8. doi: 10.1016/0378-1119(90)90481-6. [DOI] [PubMed] [Google Scholar]
- 22.Talebi M, Shafiee M, Sadeghi J, Moghadam NA, Saifi M, Pourshafie MR, et al. Genotypic diversity of methicillin-resistant coagulase-negative staphylococci isolated from inpatients and outpatients. Microb Drug Resist. 2016;22:147–54. doi: 10.1089/mdr.2014.0195. [DOI] [PubMed] [Google Scholar]
- 23.Teeraputon S, Santanirand P, Wongchai T, Songjang W, Lapsomthob N, Jaikrasun D, et al. Prevalence of methicillin resistance and macrolide-lincosamide-streptogramin B resistance in Staphylococcus haemolyticus among clinical strains at a tertiary-care hospital in Thailand. New Microbes New Infect. 2017;19:28–33. doi: 10.1016/j.nmni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrestha LB, Bhattarai NR, Khanal B. Antibiotic resistance and biofilm formation among coagulase-negative staphylococci isolated from clinical samples at a tertiary care hospital of Eastern Nepal. Antimicrob Resist Infect Control. 2017;6:89. doi: 10.1186/s13756-017-0251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mashaly GE, El-Mahdy RH. Vancomycin heteroresistance in coagulase negative Staphylococcus blood stream infections from patients of intensive care units in Mansoura university hospitals, Egypt. Ann Clin Microbiol Antimicrob. 2017;16:63. doi: 10.1186/s12941-017-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira A, Cunha Mde L. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res Notes. 2010;3:260. doi: 10.1186/1756-0500-3-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thilakavathy P, Priyan RM, Jagatheeswari PA, Charles J, Dhanalakshmi V, Lallitha S, et al. Evaluation of ica gene in comparison with phenotypic methods for detection of biofilm production by coagulase negative staphylococci in a tertiary care hospital. J Clin Diagn Res. 2015;9:DC16–9. doi: 10.7860/JCDR/2015/11725.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A, et al. Detection of biofilm formation among the clinical isolates of staphylococci: An evaluation of three different screening methods. Indian J Med Microbiol. 2006;24:25–9. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 29.Qin Z, Yang X, Yang L, Jiang J, Ou Y, Molin S, et al. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J Med Microbiol. 2007;56:83–93. doi: 10.1099/jmm.0.46799-0. [DOI] [PubMed] [Google Scholar]
- 30.Nasr RA, Abu Shadady HM, Hussein SH. Biofilm formation and presence of icaAD gene in clinical isolates of staphylococci. Egypt J Med Hum Genet. 2012;13:269–74. [Google Scholar]
- 31.Giormezis N, Kolonitsiou F, Foka A, Drougka E, Liakopoulos A, Makri A, et al. Coagulase-negative staphylococcal bloodstream and prosthetic-device-associated infections: The role of biofilm formation and distribution of adhesin and toxin genes. J Med Microbiol. 2014;63:1500–8. doi: 10.1099/jmm.0.075259-0. [DOI] [PubMed] [Google Scholar]