Summary
Clinical isolates of Acinetobacter baumannii have a tendency to develop antimicrobial resistance against commonly prescribed antimicrobial agents, including aminoglycoside agents, particularly in hospitalized patients worldwide. Resistance mechanisms of the bacterium to aminoglycosides are diverse and commonly involve production of aminoglycoside-modifying enzymes and efflux systems. The aim of this study was to investigate the frequency of gene encoding aminoglycoside-modifying enzymes and expression level of adeB efflux gene in A. baumannii isolates recovered from burn wound colonization. A total of 47 clinical isolates of A. baumannii were obtained from burned patients admitted to the Burns Teaching Hospital, Tehran, in 2018. Standard antimicrobial susceptibility screening was performed to determine resistance pattern. A polymerase chain reaction (PCR) assay was performed to determine aminoglycoside-modifying genes ACC(6’), aph(3’)-Via, aph(3’)-IIb, aadA1, aphA1 and aph6. Semi-quantitative RT-PCR was also carried out to quantify the expression level of the adeB gene. According to the results of the present study, the acc(6’) was the predominant aminoglycoside-modifying enzyme gene (80.9%), followed by aph(3’)-via, aph6, aph(3’)-IIb and aphA1, which was detected in 59.6%, 42.6%, 14.9% and 14.9% of isolates, respectively. None of the A. baumannii isolates harboured the aadA1 gene. The up regulation of adeB gene expression was observed in 63.8% of strains. Moreover, we indicated that there is a relationship between adeB expression and high resistance to gentamicin. Our results revealed that aminoglycoside resistance could be explained by the production of one or a combination of known aminoglycoside-modifying enzymes rather than overexpression of adeB.
Keywords: Acinetobacter baumannii, aminoglycoside, burned patients, adeB gene, efflux pump
Abstract
Acinetobacter baumannii (AB) est de plus en plus fréquemment isolé de prélèvements cliniques de par le monde. Il est très susceptible de développer des résistances aux antibiotiques, parmi lesquels les aminosides, en particulier dans les hôpitaux. Les mécanismes sont variables, le plus souvent enzymatiques ou par efflux. Le but de cette étude était d’évaluer les fréquences des gènes codant pour des enzymes modifiant les aminosides et le niveau d’expression du gène de pompe d’efflux adeB chez 47 AB isolés de zones brûlées dans le CTB du CHU de Téhéran. Les gènes codant pour AAC(6’), aph(3’)-Via, aph(3’) IIb, aadA1, aphA1 et aph6 ont été recherchés par PCR. Le niveau d’expression du gène adeB a été étudié par PCR semi-quantitative : aac(6’) était le gène le plus fréquemment retrouvé (80,9%), suivi par aph(3’)-Via (59,6%), aph6 (42,6%), aph(3’) IIb (14,9%) et aphA1 (14,9%). Nous n’avons pas mis en évidence aadA1. Une surexpression de adeB a été observée chez 63,8 % des souches, reliée à une résistance élevée à la gentamicine. Ces résultats montrent que la résistance de AB aux aminosides est plus d’origine enzymatique que liée à un efflux.
Introduction
Acinetobacter spp., especially Acinetobacter baumannii, have indeed emerged as opportunistic pathogens responsible for causing a variety of severe life-threatening nosocomial infections in immunocompromised individuals, particularly in burn patients. 1,2 The bacteria are capable of surviving in dehydrated and harsh environmental conditions for prolonged periods of time and might colonize in hospitalized patients, which could cause delayed wound healing, sepsis, and even death. A. baumannii can cause complicated infections, such as wound infections, hospital-acquired pneumonia, osteomyelitis and bacteremia among patients with burns.3,4
In recent decades, clinical isolates of A. baumannii have gained antimicrobial resistance against commonly prescribed antimicrobial agents, exclusively in hospitalized patients worldwide. It has accurately been described that one of the most important factors contributing to the high mortality of patients with nosocomial infections caused by A. baumannii is the ability for the acquisition of a wide variety of antibiotic resistance genes and rapid development of multidrug- resistant (MDR), extensively drug-resistant (XDR) and even pan-drug resistant (PDR) strains.5,6 The emergence and spread of drug resistant A. baumannii strains has significantly limited the choice of available therapeutic options for treatment of infections caused by the microorganisms and their worse clinical outcome.7,8 Numerous mechanisms are responsible for the development of multidrug resistance in A. baumannii, including decreased membrane permeability due to loss of porins, acquisition of extended-spectrum β - lactamase, and multidrug efflux systems.9
The antibiotic class of aminoglycosides is widely used to treat hospital-acquired infections caused by gram-negative bacilli, including A. baumannii strains. However, high resistance to traditional aminoglycoside agents such as gentamicin and kanamycin is common among clinical isolates of A. baumannii. In addition, strains of A. baumannii resistant to newer semisynthetic aminoglycosides such as amikacin, tobramycin, isepamicin and sisomicin are increasingly being reported in many countries worldwide.10,11 The resistance mechanisms of A. baumannii to aminoglycoside agents are diverse and commonly involve production of aminoglycosidemodifying enzymes, which can be classified into aminoglycoside acetyltransferases (AAC), aminoglycoside phosphotransferases (APH), and/or aminoglycoside nucleotidyltransferases (ANT or AAD). The genes for this group of enzymes are commonly present on mobile elements, such as plasmids and transposons, and are transferred among the A. baumannii population.12 The synthesis of AAC(3)-I, APH(3’)-VI and ANT(3’’)-I has been found to be predominant by several investigations on A. baumannii isolates, but there are considerable regional variances in their genotypes.13,14
Moreover, aminoglycoside resistance mediated by efflux systems has been reported in A. baumannii strains. Among these efflux systems, AdeABC in the resistance-nodulation-division (RND) superfamily has been well demonstrated to be associated with high resistance to aminoglycoside agents.15
In Iran, although the prevalence of aminoglycoside resistance has been estimated to be high, especially among MDR A. baumannii isolates,16 the overall prevalence of aminoglycoside resistance genes and the mechanisms of resistance among A. baumannii clinical isolates from burn infections have not been well elucidated experimentally. Therefore, the aim of the present study was to provide an insight into the frequency of gene encoding aminoglycoside-modifying enzymes and expression level of AdeB efflux gene in A. baumannii isolates recovered from burn infections.
Materials and methods
Samples and bacterial isolation
The present study was performed on 47 clinical isolates of A. baumannii recovered from burn patients admitted to the Burns Teaching Hospital, Tehran, during June 2018 and August 2018. The study protocol was approved by the Ethics Committee of the National Institute for Medical Research Development (NIMAD). Bacterial identification was carried out using conventional biochemical tests such as growth on MacConkey agar (Merck co., Germany), Gram-staining, oxidase test, oxidative fermentative (OF) test, growth in 44°C and motility. In addition, to confirm A. baumannii identification, amplification and sequencing of intrinsic blaOXA-51-like genes were carried out using specific primers, as previously described.17 All strains were preserved in Tryptic Soy Broth (TSB; Merck, Germany) containing 20% glycerol at -70°C for further analysis.
Antibiotic susceptibility testing
In vitro susceptibility testing was performed using a panel of three representative aminoglycosides via Kirby–Bauer disc diffusion method according to the Clinical and Laboratory Standards Institute (CLSI 2018) guidelines. The antimicrobial agents tested included, gentamicin, amikacin and tobramycin. Escherichia coli ATCC 25922 was used as a quality control strain in every test run.
Detection of aminoglycoside resistance genes
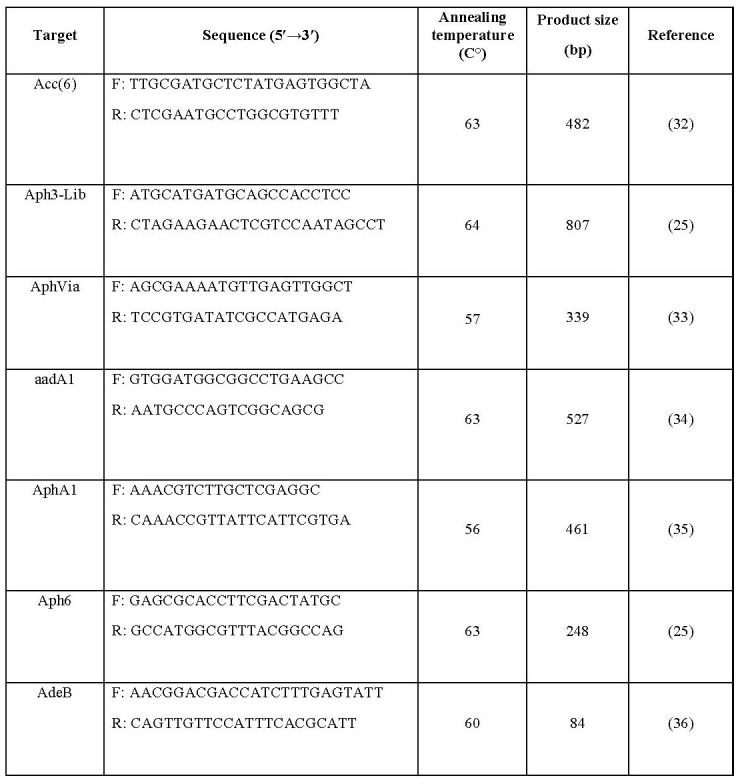
Genomic DNA of the isolates was extracted using the boiling method as described previously.18 The existence of aminoglycoside resistance genes ACC(6’), aph(3’)-Via, aph(3’)-IIb, aadA1, aphA1 and aph6 was evaluated using PCR via specific primers presented in Table I. The PCR products were detected by agarose gel electrophoresis (1.5%) then they were stained with ethidium bromide and visualized under (UV) light (UVItec, Cambridge, UK).
Table I. Oligonucleotide primers used in this study.
Semi-quantitative Real-Time RT-PCR
Semi-quantitative RT-PCR was performed to quantify the expression rates of the adeB gene among A. baumannii isolates. Accordingly, RNA extraction was carried out using the RNeasy Mini Kit (Roche Co., Germany) according to the manufacturer’s instructions with the addition of an extra DNase treatment (CinnaGen Co., Iran) following RNA purification. The absence of DNA contamination was verified by PCR amplification of the housekeeping 16srRNA gene. The relative expression was compared to the value of the standard strain ATCC19606. Reverse transcription was carried out using the cDNA synthesis kit (Wizbio Co., South Korea) according to the manufacturer’s instructions. Real-time PCR assays were carried out using the SYBR green PCR master mix (Amplicon Co., Denmark) via specific primers (Table I). The level of gene expression of the efflux pump was normalized with the 16S ribosomal RNA gene as an internal gene control. Amplification was designed including 10 of SYBR Green qPCR MasterMix, 0.5 μL of primers, 8 μL water, and 1 μL cDNA in a 20 μL total volume. The reaction conditions were 95°C for 5 minutes for the first denaturation; 95°C for 20 seconds for denaturation, 60°C for one minute for annealing for 40 cycles, and melt curve at 72 to 95°C.
Statistical analysis
The data were analyzed with SPSS version 22.0 (IBM Corp., USA).
Results
The results of antimicrobial susceptibility testing indicated that 97.9%, 89.5% and 87.5% of the tested isolates were resistant to amikacin, gentamicin and tobramycin, respectively. Our data also showed that 79.1% of isolates were non-susceptible to all of the tested aminoglycoside agents.
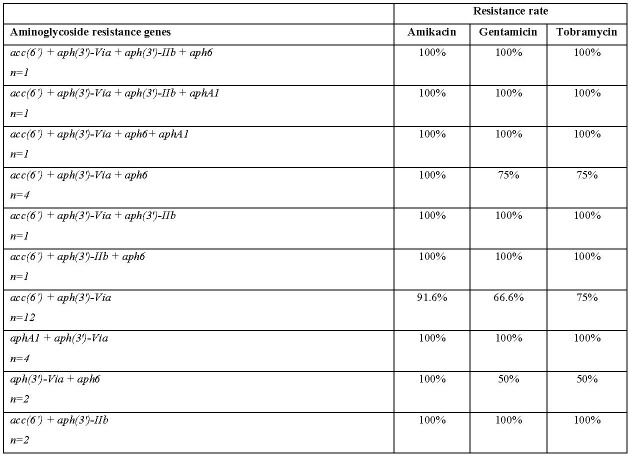
According to the results of the present study, the acc(6’) was the predominant aminoglycosidemodifying enzyme gene (80.9%), followed by aph(3’)-via, aph6, aph(3’)-IIb, and aphA1, which was detected in 59.6%, 42.6%, 14.9% and 14.9% of isolates, respectively. None of the A. baumannii isolates harboured the aadA1 gene. The distribution of aminoglycoside resistance genes among A. baumannii isolates is shown in Table II. Furthermore, the co-existence of aminoglycoside resistance genes is also indicated in Table III.
Table II. The distribution of aminoglycoside resistance genes in 48 A. baumannii isolated from burn infections.
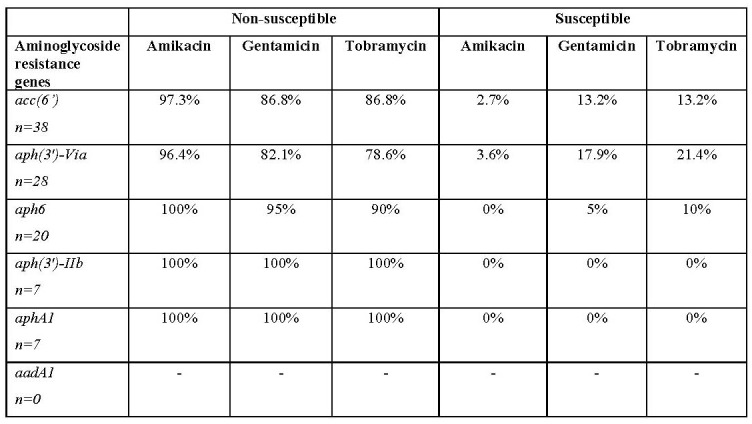
Table III. Aminoglycoside resistance gene profiles of the Acinetobacter baumannii isolates from burn infections.
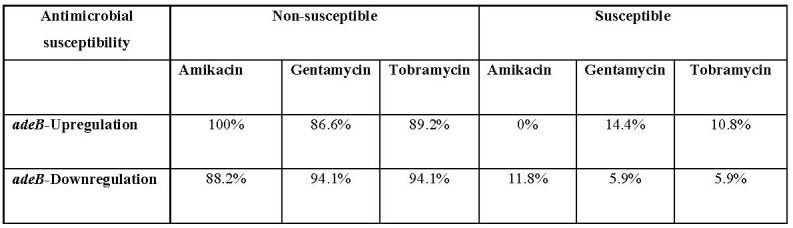
To understand whether the expression of adeB efflux pump gene effects aminoglycoside resistance, the adeB expression was analyzed by qRTPCR in the isolates. The expression levels of adeB upregulated in 63.8% of isolates. Accordingly, our analysis showed that there is no statistically significant correlation between adeB expression level and resistance rate to amikacin, gentamicin and tobramycin (p value 0.5) (Table IV).
Table IV. The correlation between the expression level of adeB efflux pump gene and aminoglycoside resistance.
Discussion
Although aminoglycosides present nephrotoxicity risks and other side effects, they are considered to be important antimicrobial agents and are used to treat nosocomial infections. The high rates of aminoglycoside resistance could cause a serious issue for combination therapy of aminoglycoside with broad-spectrum β-lactams including cephalosporins and carbapenems against A. baumannii infections.19 Aminoglycosidemodifying enzymes and efflux pumps are the most important sources of aminoglycoside resistance among A. baumannii isolates. The genes encoding these aminoglycoside resistance mechanisms can be distributed mobile elements.20 Previously, a high rate of resistance to aminoglycosides was reported by several investigators in Iran.21-24
Antimicrobial susceptibility screening revealed that 98.9% (46/47) of isolates were fully resistant to at least one of the tested aminoglycosides and at least one aminoglycoside-modifying enzyme gene was detected in these isolates. Furthermore, our findings indicated that resistance against amikacin is more related to the existence of aminoglycoside-modifying enzyme genes among A. baumannii isolates. Our results showed that the acc(6’) gene was the predominant aminoglycoside-modifying gene among the A. baumannii isolates from burn patients. Nie and colleagues suggested that the acc(6’) is related to high level aminoglycoside resistance in A. baumannii strains.25 To the best of our knowledge, there are insufficient data on the frequency of acc(6’) in clinical isolates of A. baumannii in Iran. In this study approximately half of the investigated clinical isolates of A. baumannii contained aph6 gene. This finding is in accordance with the results reported by Asadollahi and colleagues in Tehran.26 Our findings also indicated that the presence of the phosphotransferases genes aphA1 and aph(3’)-IIb was correlated with high resistance against amikacin, gentamicin and tobramycin. The prevalence of aphA1 and aph(3’)-IIb genes in this study was in accordance with the findings reported by Aliakbarzade et al. in Tabriz.22 However, Farsiani et al. reported a high prevalence of aphA1 (75%) among nosocomial A. baumannii isolates in the north-east of Iran.27 Among the aminoglycosidemodifying enzyme genes, aadA1 was not detected in the examined A. baumannii isolates. On the contrary, more recently, Salimizand et al. reported that aadA1 is the most frequent aminoglycoside-modifying enzyme among nosocomial A. baumannii isolates from Iranian patients.28 Considering these differences, it appeared that the aminoglycoside resistance genes were distributed among various genotypically distinct groups of A. baumannii strains. Thus, these data could reflect a widespread occurrence and clonal variation in nosocomial A. baumannii isolates in various hospitals in Iran. It has been clearly established that AdeABC efflux pumps are associated with high resistance against aminoglycoside agents.29 Magnet et al. first described that the adeB gene encoded an RND-type efflux pump involved in aminoglycoside resistance in A. baumannii strain BM4454.30 However, they stated that among the aminoglycoside agents, amikacin and kanamycin appeared to be less efficiently transported than other agents by AdeB efflux pump, because these compounds contain the highest density of hydroxyl groups and, consequently, are the most hydrophilic.
Therefore, we then evaluated the potential association between the expression level of the adeB and aminoglycoside resistance among clinical isolates of A. baumannii. Our findings revealed that all A. baumannii isolates with adeB overexpression were highly resistant to gentamicin (MIC90≥32). Previously, Rumbo et al. described that the overexpression of AdeABC system is associated with high-level resistance with a MIC above 8.0 μg/ml against gentamicin in MDR- A. baumannii isolates. However, they reported that these systems are not significantly associated with resistance to netilmicin or tobramycin.31 Accordingly, the resulting data obtained in this study indicated that there is no clear relationship between adeB expression and resistance to tobramycin among A. baumannii isolates.
Conclusion
A. baumannii clinical isolates collected from burn patients in Tehran showed high levels of aminoglycoside resistance. Several resistant aminoglycoside-modifying enzyme genes were also detected in the isolates, and coexistence of resistance genes was found in most strains. We also characterized the adeB expression level in A. baumannii clinical isolates, which exhibited that aminoglycoside resistance could be explained by production of one or a combination of known aminoglycoside-modifying enzymes rather than overexpression of adeB. Furthermore, exploration of other resistance mechanisms to aminoglycoside compounds, such as alteration of the ribosome-binding site and reduced uptake, can help determine the correct correlation between genotype and phenotype pattern.
Acknowledgments
Acknowledgement.The research reported in this publication was supported by the Elite Researcher Grant Committee under award number IR NIMAD REC 1396 223 from the National Institutes for Medical Research Development (NIMAD), Tehran, Iran.
Conflict of interest.There is no conflict of interest.
References
- 1.Trottier V, Segura PG, Namias N, King D. Outcomes of Acinetobacter baumannii infection in critically ill burned patients. J Burn Care Res. 2007;28(2):248–254. doi: 10.1097/BCR.0B013E318031A20F. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht MC, Griffith ME, Murray CK, Chung KK. Impact of Acinetobacter infection on the mortality of burn patients. J Am Coll Surg. 2006;203(4):546–550. doi: 10.1016/j.jamcollsurg.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Tekin R, Dal T, Bozkurt F, Deveci O. Risk factors for nosocomial burn wound infection caused by multidrug resistant Acinetobacter baumannii. J Burn Care Res. 2014;35(1):e73–e80. doi: 10.1097/BCR.0b013e31828a493f. [DOI] [PubMed] [Google Scholar]
- 4.Leseva M, Arguirova M, Nashev D, Zamfirova E, Hadzhyiski O. Nosocomial infections in burn patients: etiology, antimicrobial resistance, means to control. Ann Burns Fire Disasters. 2013;26(1):5–11. [PMC free article] [PubMed] [Google Scholar]
- 5.Salehi B, Goudarzi H, Nikmanesh B, Houri H. Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran, Iran. J Infect Chemother. 2018;24(7):515–523. doi: 10.1016/j.jiac.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Cheng VCC, Wong SC, Chen JHK, So SYC. Control of multidrug-resistant Acinetobacter baumannii in Hong Kong: role of environmental surveillance in communal areas after a hospital outbreak. Am J Infect Control. 2018;46(1):60–66. doi: 10.1016/j.ajic.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Manchanda V, Sanchaita S, Singh N. Multidrug resistant acinetobacter. J Glob Infect Dis. 2010;2(3):291–304. doi: 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asif M, Alvi IA, Rehman SU. Insight into Acinetobacter baumannii: pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect Drug Resist. 2018;11:1249–1260. doi: 10.2147/IDR.S166750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YJ, Kim SI, Kim YR, Hong KW. Carbapenem-resistant Acinetobacter baumannii: diversity of resistant mechanisms and risk factors for infection. Epidemiology and infection. 2012;140(1):137–145. doi: 10.1017/S0950268811000744. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Wang C, Wu J, Jiang R. A novel aminoglycoside-modifying enzyme gene aac(6’)-Ib in a pandrug-resistant Acinetobacter baumannii strain. J Hosp Infect. 2009;73(2):184–185. doi: 10.1016/j.jhin.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Upadhyay S, Khyriem AB, Bhattacharya P, Bhattacharjee A, Joshi SR. High-level aminoglycoside resistance in Acinetobacter baumannii recovered from Intensive Care Unit patients in Northeastern India. Indian J Med Microbiol. 2018;36(1):43–48. doi: 10.4103/ijmm.IJMM_17_225. [DOI] [PubMed] [Google Scholar]
- 12.Lin MF, Liou ML, Tu CC, Yeh HW, Lan CY. Molecular epidemiology of integron-associated antimicrobial gene cassettes in the clinical isolates of Acinetobacter baumannii from northern Taiwan. Indian J Med Microbiol. 2013;33(4):242–247. doi: 10.3343/alm.2013.33.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garneau-Tsodikova S, Labby KJ. Mechanisms of resistance to Aminoglycoside antibiotics: overview and perspectives. Med Chem Comm. 2016;7(1):11–27. doi: 10.1039/C5MD00344J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akers KS, Chaney C, Barsoumian A, Beckius M. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J Clin Microbiol. 2010;48(4):1132–1138. doi: 10.1128/JCM.02006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45(12):3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasani A, Sheikhalizadeh V, Ahangarzadeh Rezaee M, Rahmati-Yamchi M. Frequency of aminoglycoside-modifying enzymes and ArmA among different sequence groups of Acinetobacter baumannii in Iran. Microbiol Drug Resist. 2016;22(5):347–353. doi: 10.1089/mdr.2015.0254. [DOI] [PubMed] [Google Scholar]
- 17.Woodford N, Ellington MJ, Coelho JM, Turton JF. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Pitcher D, Saunders N, Owen R. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Applied Microbiol. 1989;8(4):151–156. [Google Scholar]
- 19.Krause KM, Serio AW, Kane TR, Connolly LE. Aminoglycosides: an overview. Cold Spring Harb Perspect Med. 6(6):a027029. doi: 10.1101/cshperspect.a027029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C-R, Lee JH, Park M, Park KS. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;(55) doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheikhalizadeh V, Hasani A, Ahangarzadeh Rezaee R, Rahmati-Yamchi M. Comprehensive study to investigate the role of various aminoglycoside resistance mechanisms in clinical isolates of Acinetobacter baumannii. J Infect Chemother. 2017;23(2):74–79. doi: 10.1016/j.jiac.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Aliakbarzade K, Farajnia S, Karimi Nik A, Zarei F, Tanomand A. Prevalence of aminoglycoside resistance genes in Acinetobacter baumannii isolates. Jundishapur J Microbiol. 2014;7(10):e11924. doi: 10.5812/jjm.11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.aponi-Nejad A, Farshad S, van Belkum A, Ghaznavi-Rad E. Novel cassette array in a class 1 integron in clinical isolates of Acinetobacter baumannii from central Iran. Int J Med Microbiol. 2013;303(8):645–650. doi: 10.1016/j.ijmm.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Sistanizad M, Kouchek M, Miri M, Goharani R. Carbapenem restriction and its effect on bacterial resistance in an intensive care unit of a Teaching Hospital. Iran J Pharm Re. 2013;12(3):503–509. [PMC free article] [PubMed] [Google Scholar]
- 25.Nie L, Lv Y, Yuan M, Hu X. Genetic basis of high level aminoglycoside resistance in Acinetobacter baumannii from Beijing, China. Acta Pharm Sin B. 2014;4(4):295–300. doi: 10.1016/j.apsb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asadollahi K, Taherikalani M, Maleki A, Alizadeh E. Diversity of aminoglycoside modifying enzyme genes among multidrug resistant Acinetobacter baumannii genotypes isolated from nosocomial infections in Tehran hospitals and their association with class 1 integrons. Acta Microbiol Immunol. 2011;58(4):359–570. doi: 10.1556/AMicr.58.2011.4.11. [DOI] [PubMed] [Google Scholar]
- 27.Farsiani H, Mosavat A, Soleimanpour S, Nasab MN. Limited genetic diversity and extensive antimicrobial resistance in clinical isolates of Acinetobacter baumannii in north-east Iran. J Med Microbiol. 2015;64(7):767–773. doi: 10.1099/jmm.0.000090. [DOI] [PubMed] [Google Scholar]
- 28.Salimizand H, Zomorodi AR, Mansury D, Khakshoor M. Diversity of aminoglycoside modifying enzymes and 16S rRNA methylases in Acinetobacter baumannii and Acinetobacter nosocomialis species in Iran; wide distribution of aadA1 and armA. Infect Genet Evol. 2018;66:195–199. doi: 10.1016/j.meegid.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 29.Verma P, Maurya P, Tiwari M, Tiwari V. In-silico interaction studies suggest RND efflux pump mediates polymyxin resistance in Acinetobacterbaumannii. J Biomol Struct Dyn. 2019;37(1):95–103. doi: 10.1080/07391102.2017.1418680. [DOI] [PubMed] [Google Scholar]
- 30.Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45(12):3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rumbo C, Gato E, López M, Ruiz de Alegría C. Contribution of efflux pumps, porins, and b - lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2013;57(11):5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6’)- Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006;50(11):3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khoshnood S, Eslami G, Hashemi A, Bahramian A. Distribution of aminoglycoside resistance genes among acinetobacter baumannii strains isolated from burn patients in Tehran, Iran. Arch Pediatr Infect Dis. 2017;5(3):e57263. [Google Scholar]
- 34.Zishiri OT, Mkhize N, Mukaratirwa S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J Vet Res. 2016;83(1):a1067. doi: 10.4102/ojvr.v83i1.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebreyes WA, Thakur S. Multidrug-resistant Salmonella enterica serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrob Agents Chemother. 2005;49(2):503–511. doi: 10.1128/AAC.49.2.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peleg AY, Adams J, Paterson DL. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(6):2065–209. doi: 10.1128/AAC.01198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]