SUMMARY
Biological roles for most long non-coding RNAs (lncRNAs) remain mysterious. Here, using forward genetics, we identify lep-5, a lncRNA acting in the C. elegans heterochronic (developmental timing) pathway. Loss of lep-5 delays hypodermal maturation and male tail tip morphogenesis (TTM), hallmarks of the juvenile-to-adult transition. We find that lep-5 is a ~600-nt cytoplasmic RNA that is conserved across Caenorhabditis and possesses three essential secondary structure motifs but no essential open reading frames. lep-5 expression is temporally controlled, peaking prior to TTM onset. Like the Makorin LEP-2, lep-5 facilitates the degradation of LIN-28, a conserved miRNA regulator specifying the juvenile state. Both LIN-28 and LEP-2 associate with lep-5 in vivo, suggesting that lep-5 directly regulates LIN-28 stability and may function as an RNA scaffold. These studies identify a key biological role for a lncRNA: by regulating protein stability, it provides a temporal cue to facilitate the juvenile-to-adult transition.
Keywords: C. elegans, lncRNA, lincRNA, heterochronic, developmental timing, male tail, morphogenesis
Graphical Abstract
eTOC BLURB
The functions of most long non-coding RNAs (lncRNAs) are unknown, despite their abundance in biological systems. Here, by characterizing C. elegans mutants with developmental delays, Kiontke et al. identify lep-5, a ~600-nt lncRNA. lep-5 regulates developmental timing by binding to and destabilizing LIN-28, a conserved regulator of miRNA biogenesis.
INTRODUCTION
Long non-coding RNAs (lncRNAs), once thought to be biological noise, are now appreciated as important components of many biological processes. These ≥200nt long non-coding transcripts are associated with remarkably diverse molecular functions, including regulation of chromatin topology and modification, transcriptional activation, control of miRNA availability, and scaffolding of proteins and RNAs. lncRNAs act in diverse developmental processes including pluripotency, patterning, and differentiation, and are also implicated in the pathogenesis of neurodegeneration and cancer (Cech and Steitz, 2014; Delas and Hannon, 2017; Fatica and Bozzoni, 2014; Geisler and Coller, 2013; Quinn and Chang, 2016; Ransohoff et al., 2018). While most lncRNAs function in the nucleus, some have cytoplasmic functions, e.g., TINCR brings together Staufen and mRNAs that promote epidermal differentiation (Kretz et al., 2013) and HOTAIR scaffolds the E3 ubiquitin ligases Dzip3 and Mex3b with their respective substrates Ataxin-1 and Snurportin-1 to prevent premature senescence (Yoon et al., 2013). However, biological roles and molecular functions remain unknown for the vast majority of lncRNAs, particularly for those that act in the cytoplasm.
In the nematode C. elegans, numerous lncRNAs have been detected by high-throughput approaches, but relatively little is known about their functions or roles in biological processes (Liu et al., 2017b; Nam and Bartel, 2012; Wei et al., 2019). For another important family of ncRNAs, the microRNAs (miRNAs), essential insights came from forward genetic approaches. The first two known miRNAs in any system, lin-4 and let-7, were identified in a series of classic studies on C. elegans developmental timing mutants (Ambros, 1989; Lee et al., 1993; Reinhart et al., 2000; Wightman et al., 1993). Both of these miRNAs function in the C. elegans heterochronic pathway, a mechanism that controls developmental progression through four larval stages into adulthood (Rougvie and Moss, 2013). In heterochronic mutants, certain stage-specific developmental events occur too early (“precocious” mutants) or too late (“retarded” or delayed mutants). Interestingly, regulation by non-coding RNAs figures prominently in the heterochronic pathway: in addition to lin-4 and let-7, the lin-4-related mir-237 and three other let-7-like miRNAs, mir-48, −84 and −241, also have key roles (Abbott et al., 2005; Tsialikas et al., 2017). Expression of these miRNAs in successive temporal waves during larval development keeps the activities of their targets in check until the appropriate time (Ambros, 2011).
Several key components of the heterochronic pathway, most notably the miRNA let-7 and its negative regulator LIN-28, are functionally conserved in animals (Faunes and Larrain, 2016). While lin-28 orthologs promote an immature state associated with stemness and multipotentiality, let-7 promotes differentiation and maturation. For example, Drosophila let-7 regulates the timing of neuromuscular remodeling during metamorphosis (Caygill and Johnston, 2008; Sokol et al., 2008) and the temporal patterning of neural cell fates in the brain (Wu et al., 2012). In mammals, LIN28 promotes stem cell pluripotency (Copley et al., 2013; Zhang et al., 2016) and is genetically linked to the timing of the juvenile-to-adult transition (Faunes et al., 2017; Ong et al., 2009; Perry et al., 2009; Zhu et al., 2010).
The lin-28-let-7 system also controls the juvenile-to-adult transition in C. elegans (Del Rio-Albrechtsen et al., 2006; Herrera et al., 2016; Rougvie and Moss, 2013; Vadla et al., 2012). A hallmark of this transition is male tail tip morphogenesis (TTM). TTM occurs during the fourth larval stage (L4) and involves the fusion and retraction of the four tail-tip hypodermal cells, hyp8-11, to generate the rounded tail tip characteristic of the adult male (Figs. 1A, B) (Nguyen et al., 1999). In retarded lin-41(gf), let-7(rf) and lep-2(lf) mutants, TTM is delayed or absent. This leads to the perseverance of a juvenile tail tip in adults, a phenotype called Lep (leptoderan). Conversely, TTM initiates early in lin-41(lf) and lin-28(lf) mutants, resulting in an over-retracted (Ore) phenotype in adults (Del Rio-Albrechtsen et al., 2006; Herrera et al., 2016; Vadla et al., 2012). The Lep and Ore phenotypes of these mutants result from alterations in the timing of the expression of dmd-3, a doublesex-family transcription factor that governs execution of the TTM program (Mason et al., 2008; Nelson et al., 2011). While the role of the heterochronic pathway has been intensively investigated in the division and differentiation of the lateral seam cells, little is known about this pathway in other cell types, or the extent to which it regulates other aspects of developmental timing. TTM provides an outstanding opportunity to address this gap.
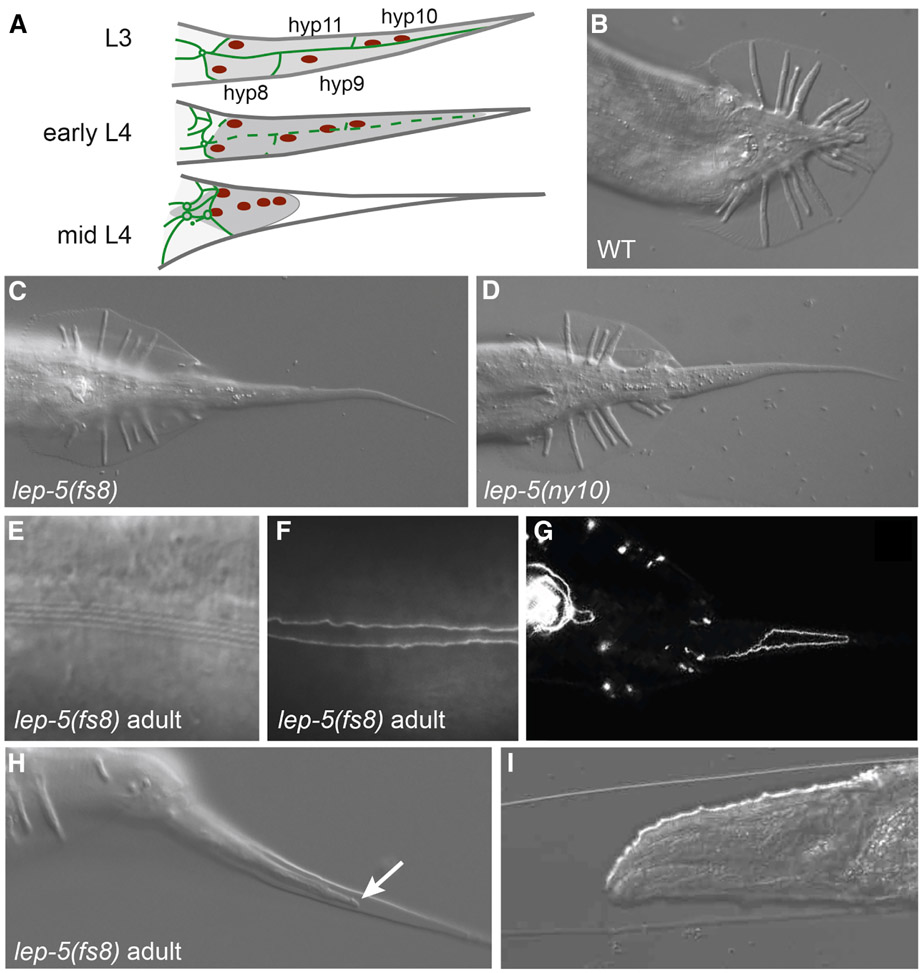
Figure 1. lep-5 is required for male tail tip morphogenesis.
(A) Schematic of TTM in lateral view. Tail tip cytoplasm shaded dark grey, adherens junctions green, nuclei red. The L3 tail tip is long and pointed. In early L4, TTM begins with fusion of the four tail tip cells (adherens junctions degrade) and retraction away from the cuticle in the posterior. By mid-L4, the tail tip syncytium has rounded and is migrating anteriorly. (B-D) Adult tails of WT, lep-5(fs8) and lep-5(ny10) mutant males in ventral view. (E) DIC image of normal alae in a lep-5(fs8) adult male. (F) ajm-1::GFP labeling of adherens junctions shows fully fused lateral seam in a lep-5(fs8) adult male. (G) Incompletely fused tail tip cells in an adult lep-5(fs8) male display persisting ajm- 1::GFP. (H) Adult TTM in a lep-5(fs8) male; arrow points to retracting tissue under the adult cuticle; lateral view. (I) Anterior end of a lep-5 adult trapped inside the cuticle from a supernumerary molt. See also Figure S1.
Here, we report the identification and characterization of lep-5, mutations in which disrupt the onset of TTM as well as other aspects of the larval-to-adult transition. We find that lep-5 expression is under temporal control and that it acts in the heterochronic pathway to promote the degradation of LIN-28. Surprisingly, lep-5 acts as a lncRNA that is conserved across the Caenorhabditis genus. These findings highlight the role of lncRNAs as mediators of protein stability and emphasize the importance of ncRNAs in developmental timing.
RESULTS
lep-5 mutant males fail to undergo normal tail tip morphogenesis
Using a forward genetic approach, we identified two X-linked mutants, ny10 and fs8, with defects in TTM. As adult males, both mutants exhibit long, pointed (Lep) tail tips that protrude far outside the cuticular fan (Fig. 1C, D). Other aspects of male tail anatomy appeared normal. In particular, the “anterior retraction” process, which generates the rays and fan and is mechanistically distinct from TTM (Nguyen et al., 1999; Sulston et al., 1980) was not disrupted in lep-5 males (Figs.1C, D). Complementation tests showed that ny10 and fs8 are recessive and allelic (see STAR Methods). We named the gene identified by these mutations lep-5. The Lep phenotype of ny10 was completely penetrant at 15°C and 25°C, whereas that of fs8 was temperature-sensitive (Table 1), suggesting that fs8 is a hypomorphic allele.
Table 1.
Tail tip phenotypes of adult males
| Genotype | % Lep | % WT | % Ore | n |
|---|---|---|---|---|
| lep-5(ny10), 25ºC | 99 | 1 | 0 | 94 |
| lep-5(fs8), 25°C | 100 | 0 | 0 | 86 |
| lep-5(fs8), 20°C | 98 | 2 | 0 | 52 |
| lep-5(fs8), 15°C | 67 | 33 | 0 | 51 |
| lep-5(ny10), post-dauer | 29 | 71 | 0 | 49 |
| lep-5(ny10), fed on HT115 | 97 | 3 | 0 | 73 |
| lep-5(ny28), 25°C | 100 | 0 | 0 | 30 |
| lep-5(ny28), 20°C | 100 | 0 | 0 | 37 |
| lep-5(ny28), 15°C | 98 | 2 | 0 | 46 |
| lep-5(fs18), 25°C | 0 | 100 | 0 | 31 |
| lep-5(fs18), 20°C | 0 | 100 | 0 | 50 |
| lep-5(fs18), 15°C | 0 | 100 | 0 | 47 |
| lep-5(fs19), 25°C | 100 | 0 | 0 | 53 |
| lep-5(fs19), 20°C | 100 | 0 | 0 | 37 |
| lep-5(fs19), 15°C | 100 | 0 | 0 | 43 |
| lep-5(fs21), 25°C | 100 | 0 | 0 | 33 |
| lep-5(fs21), 20°C | 100 | 0 | 0 | 39 |
| lep-5(fs21), 15°C | 98 | 2 | 0 | 50 |
| lep-5(fs22), 25°C | 100 | 0 | 0 | 31 |
| lep-5(fs22), 20°C | 100 | 0 | 0 | 43 |
| lep-5(fs22), 15°C | 100 | 0 | 0 | 43 |
| lep-5(fs21fs25), 25°C | 0 | 100 | 0 | 43 |
| lep-5(fs21fs25), 20°C | 0 | 100 | 0 | 51 |
| lep-5(fs21fs25), 15°C | 0 | 100 | 0 | 43 |
| lin-41(ma104) | 0 | 0 | 100 | 61 |
| lin-41(ma104); lep-5(fs8) | 0 | 0 | 100 | 48 |
| lin-41(RNAi) | 0 | 28 | 74 | 40 |
| lin-41(RNAi); lep-5(ny10) | 11 | 39 | 50 | 158 |
| zaIs3[let-7(+) + myo-3::GFP]a | 0 | 100 | 0 | 55 |
| lep-5(ny10); zaIs3 | 2 | 89 | 0 | 111 |
| lin-28(RNAi) | 0 | 46 | 54 | 24 |
| lin-28(RNAi); lep-5(ny10) | 43 | 57 | 0 | 28 |
| lin-14(RNAi) | 0 | 72 | 28 | 32 |
| lin-14(RNAi); lep-5(ny10) | 98 | 2 | 0 | 140 |
72% of males begin tail tip retraction during the L3 stage (n = 54)
Consistent with the Lep phenotype, we observed that tail tip cells in lep-5 L4 males failed to undergo normal migration and retraction. Using the adherens junction marker AJM- 1::GFP, we found that some hyp cells remained unfused even in adult lep-5 males (Fig. 1G). Thus, lep-5 influences both cell fusion and retraction, indicating that it regulates the execution of the entire TTM program (Nguyen et al., 1999).
In addition to TTM defects, we observed several mutant phenotypes that suggested a more general role for lep-5 in developmental timing. In some lep-5 males, delayed TTM occurred in adults (Fig. 1H). Furthermore, 42% of lep-5 males (n = 101) and 62% of lep-5 hermaphrodites (n = 101) molted again in adulthood, a characteristic of some other developmental timing mutants (Ambros and Horvitz, 1984) (Fig. 1I, Fig. S1). In males, these supernumerary molts were invariably lethal, while in hermaphrodites, they led to defects in vulva morphology (Fig. S1K). The adult alae, stage-specific specializations of the lateral hypodermis, appeared normal in males, but in hermaphrodites, alae were weak or partially absent in most young adults (Fig. S1). Despite this, adult seam cell numbers (n = 50 sides) and seam cell fusion were normal in lep-5 mutants of both sexes, and we found no evidence for additional seam cell divisions in adults. We conclude that lep-5 function is important for some but not all somatic features of the juvenile-to-adult transition in C. elegans.
lep-5 alleles identify a previously uncharacterized gene
Using standard methods, we mapped lep-5 to the uncharacterized predicted gene H36L18.2 (Fig. S2). Transcriptome sequencing indicates that H36L18.2 produces a mature polyadenylated RNA of ~600 nt after the removal of two introns and trans-splicing to the SL1 splice leader (Gerstein et al., 2010) (Fig. 2A). We found that ny10 was a large, ~80-kb deletion encompassing 32 predicted genes (Fig. S2), while fs8 was a point mutation (G23A) in the first nucleotide following the SL1 acceptor site (Fig. S2C). We also engineered a lep-5 null allele, ny28, by CRISPR. While most experiments described below were carried out using ny10 and fs8, we found that lep-5(ny28) null mutants phenocopied lep-5(ny10) with respect to all phenotypes described above (Table 1). Furthermore, the lep-5(ny10) phenotypes in males and hermaphrodites are rescued by a transgene covering a region from 3838 nt upstream to 248 nt downstream of the wild-type H36L18.2 locus (n > 50).
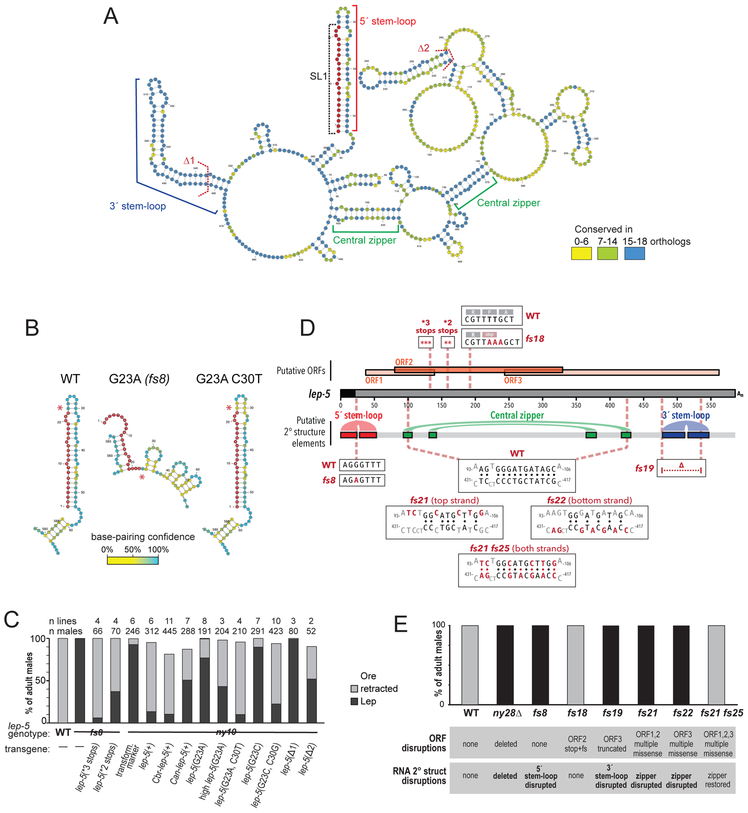
Figure 2. lep-5 is a long non-coding RNA.
(A) The secondary structure of the lep-5 lncRNA as predicted by Turbofold. The conserved 5’ stem-loop (red bracket) includes the SL1 trans-spliced leader (in red). The 3’ stem-loop (blue bracket) and two base-paired regions forming the “central zipper” (green brackets) are also conserved. Dashed lines: boundaries of two hairpins deleted in the rescue experiments (Δ1 and Δ2). The polyA tail is omitted for clarity. For alignments, see Fig. S3 and STAR Methods. The 5’ region from predicted full-length structures of wild-type lep-5, lep-5(fs8 = G23A) and lep-5(G23A, C30T). Asterisks indicate position 23. SL1 is in red. See also Figures S4, S5. (C) Rescue experiments with transgenes containing stop codons in the predicted coding region (*3 or *2 stops), the lep-5 orthologs of C. briggsae and C. angaria and constructs containing nucleotide substitutions that discrupt and restore the lep-5 secondary structure (high lep-5(G23A) = transgene at 10x concentration). (D) Schematic of the SL1-spliced lep-5 RNA with putative ORFs shown above and selected predicted secondary structure features shown below. Boxes show the sequence of various mutant alleles compared with the wild-type allele. (E) Male tail tip phenotype at 20ºC of lep-5 mutants in which the secondary structure and/or the predicted ORFs are disrupted.
The mature H36L18.2 transcript has three potentially translatable regions, encoding conceptual products of 34 (ORF1), 83 (ORF2), and 106 (ORF3) amino acids. One of these (starting at M12 of ORF2) was identified as predicted ORF in the WormBase genome annotation WS250. None of the three possible translation products have detectable domains or homology to any other known proteins. Remarkably, however, the primary nucleotide sequence of lep-5 was strongly conserved in the genomes of 18 other Caenorhabditis species in the Elegans group (Kiontke et al., 2011) and less well conserved in seven more distantly related species (Fig. S3). While most of these lep-5 orthologs contain potentially translatable regions, there is no detectable similarity in their potential protein products or in their positions in the predicted transcripts (Fig. S4).
These findings raised the possibility that lep-5 function depends on nucleotide sequence itself rather than coding potential. As an initial test of this idea, we asked if expression of lep-5 orthologs from C. briggsae (Cbr-lep-5) or the more highly divergent C. angaria (Can-lep-5) could rescue the tail-tip defects of C. elegans lep-5 (Cel-lep-5) mutants. Cbr-lep-5 and Can-lep-5 each have one potential ORF, but these share no coding potential with each other or with the potential ORFs in Cel-lep-5. Remarkably, we found that expression of Cbr-lep-5 completely rescued and Can-lep-5 partially rescued the Lep defect of C. elegans lep-5 mutants (Fig. 2C). These results very strongly suggest that the putative ORFs of lep-5 are not required for its function. Note that polyadenylation, as observed in lep-5, is a feature of many non-coding transcripts (Kopp and Mendell, 2018; Nam and Bartel, 2012).
lep-5 is a lncRNA with several prominent secondary structure motifs
To predict lep-5 secondary structure, we used Turbofold (Harmanci et al., 2011) for comparative analysis of lep-5 orthologs from 19 species in the Elegans group of Caenorhabditis. This revealed several notable features (Figs. 2A, S4, S5). First, lep-5 RNA is predicted to be highly structured, with multiple stem-loops, several prominent single-stranded regions, and a central “zipper” region. Most of the base-paired regions show high conservation; many predicted single-stranded positions are also strongly conserved, suggesting that these might serve as sites for interactions in trans (Fig. 2A). Second, the extensive base-pairing, particularly in the central zipper region, suggests that the mature RNA adopts a compact structure. Third, the very 5´ end of the lep-5 RNA is predicted to fold into a 53-nt stem-loop structure that includes the 22-nt trans-spliced leader SL1. A consensus structure derived from alignment of the 21 Elegans-group lep-5 genes, as well as individual Turbofold-predicted structures, indicated that three key features are conserved in all orthologs: the 5´ SL1-containing stem-loop, the central zipper region, and a stem-loop near the 3´ end (Fig. 2A, Fig. S4).
Interestingly, the position altered in the lep-5(fs8) mutant (G23A) lies near the tip of the 5´ stem-loop and is predicted to base-pair with C30. This mutation dramatically altered the predicted structure of the 5´ region, replacing the large stem-loop with several smaller double-stranded regions (Fig. 2B, S3B). To ask whether base-pairing between positions 23 and 30 is important for lep-5 function, we created rescue constructs containing the G23A mutation alone and in combination with a second mutation, C30T, a compensatory change predicted to restore the 5´ stem-loop (Fig. 2B). While the G23A mutant transgene had poor rescue activity, the G23A C30T double mutant transgene completely rescued the lep-5 tail tip phenotype (Fig. 2C). Introducing a different mutation at this position, G23C, alone and together with its corresponding compensatory change, C30G, yielded the same pattern of results (Fig. 2C). These experiments indicate that base-pairing between G23 and C30 is critical for lep-5 function, strongly supporting the existence of the 5´ stem-loop in vivo.
To probe additional regions that could be important for lep-5 function, we deleted two predicted internal stem-loops from the rescue construct. “Δ1” Δ482-G542) removes most of the large, well-conserved predicted stem-loop near the 3´ end of lep-5 (Fig. 2A). This deletion abolished rescue activity (Fig. 2C). Deletion of a less well-conserved smaller internal stem-loop (“Δ2”, ΔT216-A257) reduced, but did not eliminate, rescue (Figs. 2B, C). In contrast, introducing multiple stop codons into the putative ORF2, at codons 15, 16, and 17 (“*3 stops”) or at codons 25 and 26 (“*2 stops”), did not diminish the ability of these constructs to rescue lep-5 (Figs. 2C,D).
To further explore the structure of the lep-5 lncRNA and to confirm the dispensability of its putative ORFs for lep-5 function, we created several new lep-5 CRISPR alleles (Fig. 2D). lep-5(fs18) is a TT193AAA change that introduces a stop codon and frameshift into ORF2 but is not expected to significantly alter RNA secondary structure. These mutants were phenotypically wild-type (Figs. 2D, E; Table 1). lep-5(fs19) mutants replace 60 nt (G476-T535) with CA, eliminating the 3´ stem-loop and truncating ORF3, but leaving the other ORFs intact; these mutants phenocopy the lep-5(ny28) null allele (Figs. 2D, E; Table 1). Most tellingly, we created three mutants to disrupt the predicted central “zipper” and then restore it with predicted compensatory changes ~230 nucleotides away (Fig. 2D, E; Table 1). In lep-5(fs21), six point mutations were introduced into the top strand of the zipper, dramatically weakening its potential to form a double-stranded region. These mutations also cause missense changes to putative ORFs 1 and 2. Separately, lep-5(fs22) introduced seven point mutations into the lower strand of the zipper, similarly disrupting it; this causes five missense changes in ORF3. Both lep-5(fs21) and lep-5(fs22) mutants had completely penetrant TTM defects that phenocopy the lep-5 null allele (Fig. 2E). Thus, lep-5 function can be eliminated by two separate mutants that disrupt a central secondary-structure feature but cause no common lesion to putative coding sequence. Finally, we introduced mutations equivalent to fs22 into fs21 to create the “double mutant” fs21fs25, which is predicted to restore the secondary structure of the lep-5 RNA but leave extensive coding sequence changes in all putative ORFs (Fig. 2D). Strikingly, lep-5(fs21fs25) mutant males are phenotypically wild-type (Fig. 2E; Table 1). Thus, the integrity of the central “zipper” region of lep-5 RNA is essential for its function, and overwhelming evidence indicates that lep-5 activity is independent of the coding potential of its ORFs.
Consistent with an RNA-based function for lep-5, several previous studies have found the lep-5 RNA in ribonucleoprotein complexes in vivo. Two regions of lep-5, C335-G392 and T415-A467, were identified in CLIP-seq experiments with the Argonaute ALG-1 (Grosswendt et al., 2014). Another region, A531-T551, was suggested to interact with the Dicer DCR-1 (Rybak-Wolf et al., 2014). Note that Dicer can bind to many classes of RNA, including lncRNAs and mRNAs, and that this interaction does not necessarily imply cleavage (Rybak-Wolf et al., 2014). In HITS-CLIP experiments, lep-5 was found to interact with the heterochronic factor LIN-28, also through a region in its 3´ end (G434-G584) (Stefani et al., 2015).
Because of the extensive involvement of miRNAs in C. elegans developmental timing (Abbott et al., 2005; Lee et al., 1993; Reinhart et al., 2000), and because of lep-5’s association with ALG-1 and DCR-1, we considered the possibility that the full-length lep-5 transcript might serve as a precursor for one or more smaller RNAs. However, deep sequencing from multiple stages and both sexes in C. elegans has identified no small RNAs derived from this region (Gerstein et al., 2010; Kato et al., 2009), despite high abundance of the full-length lep-5 RNA. These results cannot exclude the possibility that very low-abundance, temporally regulated, or unstable miRNAs might be made from the lep-5 locus. However, the structure-function experiments above, together with results detailed below, are more consistent with a lncRNA-based function for lep-5.
lep-5 lncRNA is expressed in a temporal wave and localizes to the cytoplasm
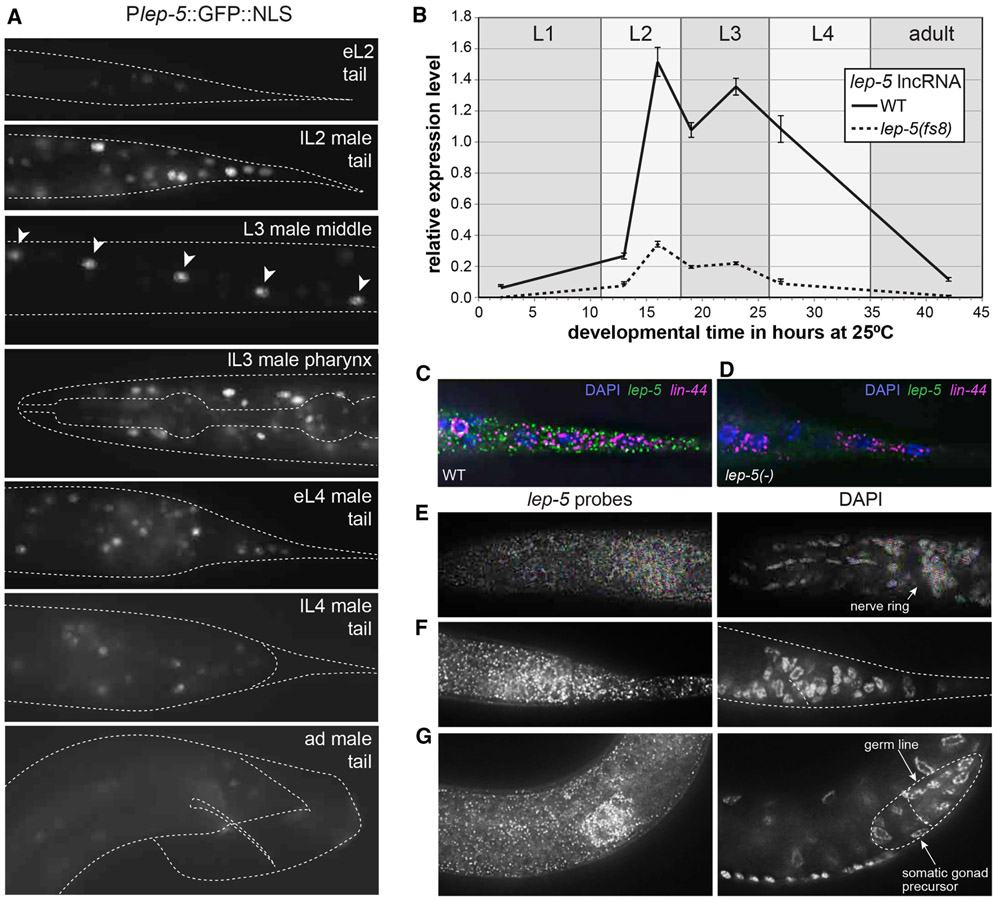
To examine the lep-5 expression pattern, we fused ~4 kb of upstream sequence to GFP. This Plep-5::GFP reporter was expressed in larvae of both sexes in several cell types including the tail tip, pharynx, nervous system, vulva, and seam cells (Fig. 3A, Fig. S6). Notably, expression of the reporter was temporally controlled: GFP was first weakly detectable in early L2, became stronger in late L2 and persisted until late L3. Expression was weak in L4 animals and nearly undetectable in adults. By qRT-PCR, lep-5 abundance showed a similar pattern, with expression low during L1 but rising dramatically by late L2 and remaining high through early L4 (Fig. 3B). lep-5 levels were markedly reduced in lep-5(fs8) mutants (Fig. 3B), suggesting that the 5´ stem-loop is important for RNA stability. SL1-trans-spliced lep-5 transcripts were still detectable in lep-5(fs8) mutants (not shown), indicating that the fs8 mutation does not abolish trans-splicing, though it is possible that such processing is impaired.
Figure 3. lep-5 lncRNA expression is temporally regulated.
(A) Temporal expression of Plep- 5::GFP::NLS in late L1 animals (top) through adults (bottom). Expression is observed in the tail epidermis including the tail tip cells, in pharynx muscles, in neurons in the head and cloacal region, and in seam cells (arrowheads). (B) Temporal expression of lep-5 lncRNA from L1 to adult as measured by qPCR. Error bars show SD. (C, D) lep-5 lncRNA visualized with smFISH probes in the tail tip of a wild-type and lep-5(ny10) L2 animal. lin-44 probes were used as positive control. DAPI staining indicates nuclei. (E-G) Late L2 male. lep-5 lncRNA visualized with smFISH probes (left) and nuclei stained with DAPI (right) showing areas of concentrated lep-5 lncRNA expression in the ganglia in the pharynx (E) and rectal (F) region and in the somatic portion of the developing gonad (G). See also Figure S6.
To determine the subcellular localization of the lep-5 lncRNA, we carried out single molecule fluorescent in situ hybridization (smFISH). lep-5 RNA was readily detectable in the tail tip of an L2 male (Fig. 3C, D). Co-staining with DAPI indicated that lep-5 RNA is predominantly, if not exclusively, cytoplasmic. Consistent with the broad expression of the transcriptional reporter, lep-5 RNA was also detectable throughout the body, including neuronal ganglia of the head and tail (Fig. 3E, F) and in the developing male somatic gonad (Fig. 3G).
lep-5 is necessary for timely activation of the master TTM regulator dmd-3
TTM is initiated by male-specific expression of the doublesex ortholog dmd-3 in the tail tip (Mason et al., 2008). dmd-3 expression follows a characteristic temporal pattern, with expression first detectable in the tail tip cells in early L4 (Fig. 4A) (Mason et al., 2008). By late L4, TTM is complete and tail tip expression of dmd-3 becomes undetectable. In lep-5(ny10) L4 males, dmd-3 expression was absent in the tail tip cells, while in lep-5(fs8), it was variably lost and delayed (Fig. 4A and data not shown). Moreover, many lep-5 males showed aberrant expression of dmd-3 in adulthood (Fig. 4A), likely accounting for the adult activation of TTM described above (Fig. 1H). Thus, lep-5 regulates the timing of TTM by controlling the temporal dynamics of dmd-3 expression in the tail tip.
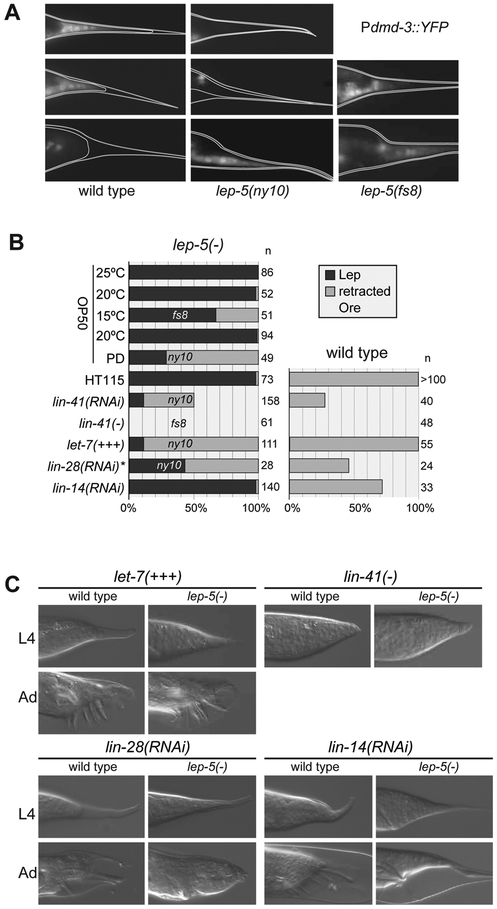
Figure 4. lep-5 functions upstream of lin-28 in the heterochronic pathway to control TTM.
(A) Expression of Pdmd-3::YFP in wild-type and mutant male tail tips at early (e), mid (m) and late (l) L4 stage. (B) Penetrance and expressivity of tail tip phenotypes in single mutants, double mutants and mutant/RNAi knockdown combinations. Left column shows results in a lep-5 mutant background (relevant allele indicated); right column shows corresponding result in a wild-type background. PD = post dauer, HT115 = RNAi control bacterial strain, asterisk: RNAi fed to L1 larvae. (C) DIC images of representative L4 and adult male tails from the experiments summarized in (B). A rounded tail tip in L4 results from precocious TTM in L3. Adults shown for lin-14(RNAi) have just molted and remain surrounded by the L4 cuticle. See also Figure S7.
lep-5 acts in the heterochronic pathway
The altered timing of TTM in lep-5 mutants, as well as the supernumerary molts and hermaphrodite alae defects, led us to consider whether lep-5 might act in the C. elegans heterochronic pathway (Rougvie and Moss, 2013). Consistent with this, we found that passage through the dauer stage, an L3 alternative used in times of stress, strongly suppressed the morphogenesis defect of lep-5 males (Table 1, Fig. 4B). Suppression by dauer is a characteristic feature of several heterochronic mutants (Liu and Ambros, 1991). Furthermore, the phenotype of lep-5(ny10) mutants was enhanced by RNAi knockdown of the RISC component ain-1, which by itself causes only weak heterochronic defects but modifies the phenotypes of many heterochronic mutants (Ding et al., 2005). While ain-1(RNAi) males had no male tail defects, all ain-1(RNAi); lep-5(ny10) males displayed severe defects in anterior tail retraction and TTM (n = 28, Fig. S1). ain-1(RNAi) also strongly enhanced the frequency of supernumerary molts of lep-5(ny10) adults to 100% in both sexes (n > 20). However, we observed no effect of ain-1 RNAi on seam cell development in lep-5 mutants, as all ain-1(RNAi); lep-5(ny10) adults had a normal number of seam cells (n = 27), which fused normally (n = 67), and males had normal alae (n = 38). Together, these findings indicate that lep-5 is a component of the heterochronic pathway.
We carried out several experiments to determine the regulatory relationships between lep-5 and other heterochronic genes. let-7 and its key target, the NHL/TRIM gene lin-41, regulate the timing of TTM (Del Rio-Albrechtsen et al., 2006; Mason et al., 2008). We found that lin-41(lf) and lin-41(RNAi) suppressed the lep-5 TTM phenotype (Table 1; Figs. 4B, C). Moreover, Plin-41::GFP::lin-41 3´UTR expression, normally downregulated during L4, persisted into adulthood in lep-5 mutants (Fig. S7). These observations indicate that lep-5 functions upstream of lin-41. Furthermore, five-fold overexpression of let-7 via the transgene zaIs3 (Bussing et al., 2010), while causing no tail tip phenotype in a wild-type background, was able to almost completely suppress the lep-5 mutant phenotype (Figs. 4B, C). Thus, lep-5 likely acts upstream of let-7.
The RNA-binding protein LIN-28 acts upstream of let-7 and is a central regulator of developmental progression across species. In both C. elegans and mammals, a key role of LIN-28 is to repress the biosynthesis of mature let-7 (Tsialikas and Romer-Seibert, 2015). Consistent with the precocious developmental phenotypes of lin-28 mutants (Ambros and Horvitz, 1984; Vadla et al., 2012), these animals exhibit premature TTM (leading to over-retraction; Ore) (Herrera et al., 2016). We observed an Ore phenotype in many lin-28(RNAi) males and found that lin-28(RNAi) also strongly suppressed the lep-5 TTM phenotype (Figs. 4B, C). Here, residual lin-28 activity may prevent complete suppression, as the penetrance of the lin-28(RNAi) TTM phenotype indicates incomplete knockdown (Herrera et al., 2016). These results suggest that lep-5 is a negative regulator of lin-28. Earlier in development, the transcription factor LIN-14 controls the progression between L1 and L2 stages by activating lin-28 (Seggerson et al., 2002). Consistent with previous findings (Herrera et al., 2016), lin-14(RNAi) males also precociously retracted their tail tips, but lin-14(RNAi) had no effect on TTM in lep-5(ny10) mutants (Figs. 4B, C). Together, these experiments support a model in which lep-5 functions in the heterochronic pathway downstream of lin-14 and upstream of lin-28 and let-7.
lep-5 is required for the timely decay of LIN-28
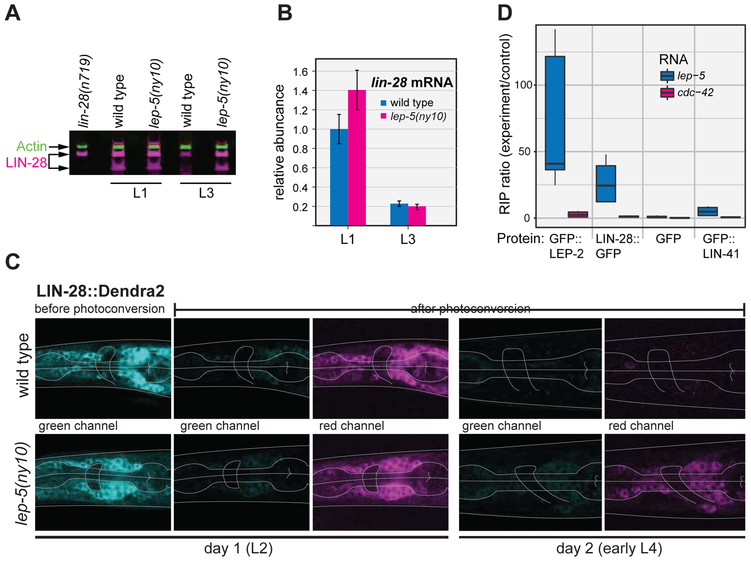
Normal progression through larval development requires the downregulation of LIN-28 by the L3 stage (Moss et al., 1997). This allows the production of mature let-7 miRNA and has other, let-7-independent, consequences (Vadla et al., 2012; Van Wynsberghe et al., 2011). Previous work has demonstrated that the control of lin-28 mRNA stability and translation, mediated through its 3´ UTR, is an important contributor to LIN-28 downregulation (Morita and Han, 2006). To ask if lep-5 has a role in this process, we examined LIN-28 protein levels by Western blot (Fig. 5A). As expected, LIN-28 abundance in wild-type larvae was high in L1 and decreased significantly by L3. In lep-5 mutants, LIN-28 levels were similar to those seen in wild-type during L1, but were markedly elevated in L3, indicating a defect in LIN-28 downregulation. By qRT-PCR, however, we found that the loss of lep-5 had no effect on lin-28 mRNA abundance in L3 (Fig. 5B). Thus, lep-5 is necessary for the developmental decline in LIN-28 protein, but it does not affect lin-28 mRNA levels. Consistent with this function, we found that overexpression of a LIN-28::GFP fusion protein in lep-5 mutants, but not in wild type, caused lethality due to highly penetrant supernumerary adult molts in both sexes (data not shown).
Figure 5. lep-5 lncRNA promotes LIN-28 degradation.
(A) Detection of endogenous LIN-28 protein by Western blot (actin used for a normalization control). (B) lin-28 mRNA levels determined by qPCR. Values are normalized to wild-type L1s. (C) Analysis of a LIN-28::Dendra2 fusion protein in WT (top) and lep-5(ny10) (bottom) males. Shown are neurons in the head region (pharynx and nerve ring indicated by grey lines). From left to right: green fluorescent signal before photoconversion, diminished green signal after photoconversion, red signal after photoconversion, green and red signals in the same animals 24 hours later. (D) Ratios of lep-5 lncRNA and control mRNA for cdc-42 in immune-precipitation of GFP-tagged proteins using anti-GFP relative to negative control IP measured by qPCR (see STAR Methods for details). For each set of triplicate experiments, the first to third quartile is represented as a box, the median as a black bar and the maximum and minimum values as whiskers.
Given these results, we considered two models for lep-5-mediated regulation of lin-28. First, lep-5 might be important for repressing translation of lin-28 mRNA. Such a mechanism is used by the heterochronic genes daf-12, lin-66, and sea-2, which repress lin-28 translation through its 3´ UTR (Huang et al., 2011; Morita and Han, 2006; Seggerson et al., 2002). Alternatively, lep-5 could promote the degradation of LIN-28 protein between L1 and L3. To distinguish between these possibilities, we used a photoconvertible LIN-28::Dendra2 fusion protein to monitor the stability of LIN-28 protein during development (Herrera et al., 2016). As expected, LIN-28::Dendra2 abundance was high in L2 animals, and UV illumination converted essentially all of this fusion protein from green to red (Fig. 5C). When these animals reached early L4, the pool of pre-existing (red) LIN-28::Dendra2 was nearly undetectable, and we observed no newly synthesized (green) LIN-28::Dendra2. In lep-5 mutants, LIN-28::Dendra2 levels were comparable to wild-type at the L2 stage, consistent with our Western blot results. Similarly, very little newly synthesized (green) LIN-28::Dendra2 was detectable in early L4 following photoconversion in L2. However, unlike wild-type larvae, lep-5 mutants exhibited significant amounts of pre-existing (red) LIN-28::Dendra2 protein in L4. Thus, the pool of LIN-28::Dendra2 synthesized before photoconversion was markedly more stable in lep-5 mutants than in wild type. This indicates that lep-5 regulates developmental timing by promoting the timely degradation of LIN-28 protein.
lep-5 RNA associates with LIN-28 and LEP-2 in vivo
lep-2, another recently identified heterochronic gene, shares many phenotypic similarities with lep-5 (Herrera et al., 2016). Both mutants have severe TTM defects, both genes act in the heterochronic pathway, and both are required for the timely degradation of LIN-28. lep-2 encodes the sole C. elegans Makorin, a conserved but poorly understood family of proteins with RNA-binding and E3 ubiquitin-ligase capacities (Cassar et al., 2015; Gray et al., 2000; Kim et al., 2005; Lee et al., 2012; Lee et al., 2009; Liu et al., 2017a; Salvatico et al., 2010). Correspondingly, Herrera et al. (2016) hypothesized that LEP-2 might act as the E3 ligase that tags LIN-28 for proteasomal degradation. Because both LIN-28 and LEP-2 are RNA-binding proteins, we considered the possibility that they might both bind to lep-5.
We therefore immunoprecipitated in vivo-crosslinked RNA-protein complexes to ask if LEP-2 and LIN-28 bound specifically to the lep-5 RNA. Using animals carrying a functional GFP::LEP-2 transgene, we carried out anti-GFP immunoprecipitation to recover GFP::LEP-2 along with any covalently-bound RNAs. After washing and crosslink reversal, we measured the recovery of lep-5 RNA and a control mRNA expressed at similar levels, cdc-42, by qRT-PCR and compared these to the recovery from negative-control (empty beads) immunoprecipitations. We robustly detected lep-5 RNA in the GFP::LEP-2 immunoprecipitate, while cdc-42 RNA was present only in trace amounts (Fig. 5D). Using a similar approach, we isolated RNAs associated in vivo with GFP::LIN-28 and with a negative control RNA-binding protein, GFP::LIN-41. Again, we found lep-5 RNA, but not significant amounts of cdc-42 RNA, in association with GFP::LIN-28. In contrast, negligible amounts of both RNAs were recovered in GFP::LIN-41 complexes (Fig. 5D). Together, these results demonstrate that LIN-28 and LEP-2 bind specifically to lep-5 RNA in vivo, suggesting that lep-5 might act as an RNA scaffold in a tripartite LEP-2—lep-5—LIN-28 complex that mediates the degradation of LIN-28.
DISCUSSION
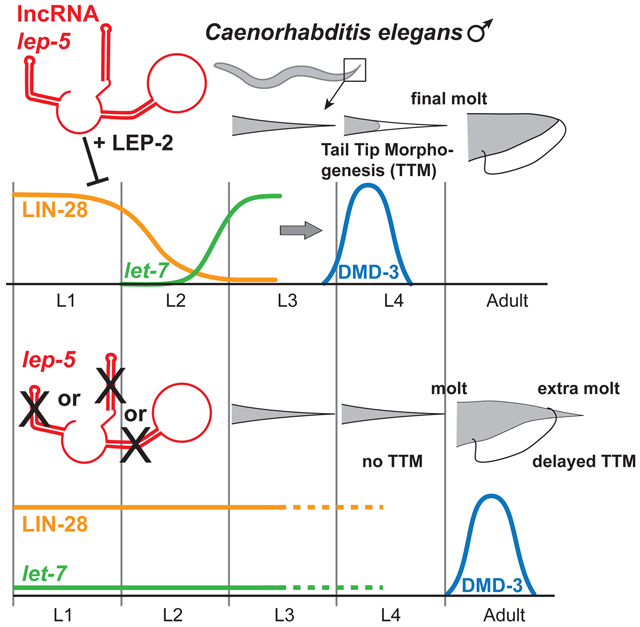
Here, we report the discovery of a C. elegans lncRNA, lep-5, and demonstrate that it regulates the timing of two events in the juvenile-to-adult transition: male tail morphogenesis and the final molt. lep-5 functions upstream of lin-28, a central regulator of developmental transitions—including the juvenile-to-adult transition—throughout the animal kingdom. lep-5 promotes the timely degradation of LIN-28, an essential step for properly coordinated larval development. This regulation is likely direct and might reflect a scaffolding ability of lep-5, through which it may promote LIN-28 proximity to an E3 ligase, LEP-2, whose mutant phenotype is nearly identical to that of lep-5. The lep-5 lncRNA is found in the cytoplasm, where LIN-28 and LEP-2 are localized and predicted to be active (Herrera et al., 2016), thus adding an important regulator to the small list of known cytoplasmic lncRNAs. To date, few lncRNAs have been characterized in C. elegans: rncs-1 is thought to play a role in the response to starvation (Hellwig and Bass, 2008), and tts-1 regulates ribosome levels to promote the extended lifespan of insulin receptor mutants (Essers et al., 2015). A recent study identified behavioral and developmental phenotypes for 23 putative C. elegans lncRNAs, finding that most appear to regulate transcription or bind to endogenous miRNAs (Wei et al., 2019). These studies did not include lep-5, as H36L18.2 was not originally annotated as a predicted lncRNA. Our findings that lep-5 acts cytoplasmically to regulate LIN-28 stability highlight the extensive use of non-coding RNA in the heterochronic pathway.
lep-5 is a lncRNA component of the C. elegans heterochronic pathway
The delay of TTM, the presence of supernumerary molts and partially defective alae, the suppression by passage through dauer, and the enhancement by ain-1(RNAi) suggest that lep-5 functions in the heterochronic pathway. The finding that lep-5´s key function is to promote the timely degradation of LIN-28 provides a straightforward mechanism to explain the Lep defect. Perdurance of LIN-28 function would block biogenesis of the mature let-7 miRNA (Tsialikas and Romer-Seibert, 2015), which would then be unable to downregulate its target lin-41. Because lin-41(gf) blocks dmd-3 activation and the onset of TTM (Del Rio-Albrechtsen et al., 2006; Mason et al., 2008), the persistence of lin-41 in lep-5 mutants accounts for these heterochronic defects. That lep-5 mutants do not completely phenocopy some other mutants that disrupt LIN- 28 degradation—particularly with respect to defects in seam cell lineages—strongly suggests cell-type-specificity in the control of LIN-28 (see below). We believe that lep-5 provides an instructive temporal cue for TTM, as overexpression of wild-type lep-5 is sometimes sufficient to cause premature morphogenesis (Ore) (Fig. 2C).
While multiple genes have been shown to be necessary for the proper temporal decline in lin-28 activity, many of these (e.g., lin-4, lin-66, daf-12, and sea-2) function through the 3´ UTR of lin-28 to repress its translation (Hochbaum et al., 2011; Huang et al., 2011; Morita and Han, 2006; Moss et al., 1997). In contrast, lep-5, like ced-3 and lep-2 (Herrera et al., 2016; Weaver et al., 2014), acts to promote LIN-28 protein degradation. The caspase CED-3, a protease best known for its role in programmed cell death, can act directly on LIN-28 (Weaver et al., 2017; Weaver et al., 2014). The Makorin LEP-2, as a putative E3 ligase, may promote LIN-28 degradation via ubiquitination (Herrera et al., 2016).
lep-5 may act as a molecular scaffold
How could the lep-5 lncRNA regulate LIN-28 stability? In other systems, many functional roles for lncRNAs have been described, especially the regulation of transcription or chromatin state of nearby genes (Fatica and Bozzoni, 2014; Geisler and Coller, 2013). We considered such a function for lep-5 but found no obvious nearby candidate target genes. Moreover, the cytoplasmic localization of lep-5 RNA makes this possibility unlikely. Many lncRNAs function by direct base-pairing to other RNAs (Cech and Steitz, 2014; Fatica and Bozzoni, 2014; Kretz et al., 2013), but lep-5 has no significant complementarity to any known transcribed regions of the C. elegans genome (BlastN e-values > 0.3) and it does not harbor a target site for a relevant miRNA (see STAR Methods).
IncRNAs can also function as molecular scaffolds by recruiting factors into a functional complex (Ransohoff et al., 2018; Wang and Chang, 2011). Our results favor this hypothesis for lep-5. By immunoprecipitation of intact ribonucleoprotein complexes, we found that both LIN-28 and LEP-2 bind to lep-5 in vivo. LIN-28 possesses two RNA binding domains (Tsialikas and Romer-Seibert, 2015) and can bind RNA directly (Van Wynsberghe et al., 2011). In previous HITS-CLIP experiments, lep-5 was found among the ~2000 RNA species that interact with LIN-28 in vivo (Stefani et al., 2015). LEP-2 is the sole C. elegans Makorin (Herrera et al., 2016), a family of conserved proteins that can bind nucleic acids and act as E3 ubiquitin ligases (Arumugam et al., 2007; Gray et al., 2000; Lee et al., 2009; Liu et al., 2017a; Salvatico et al., 2010). Because both lep-5 and lep-2 facilitate the degradation of LIN-28, we propose that the key function of lep-5 is to scaffold a tripartite LEP-2-lep-5-LIN-28 complex. As such, lep-5 would provide an instructive switch, allowing LEP-2 to act on LIN-28, causing it to be ubiquitinated and ultimately degraded by the proteasome (Fig. 6). Such a mechanism would be similar to that of HOTAIR, an lncRNA that unites RNA-binding E3 ligases with their substrates (Yoon et al., 2013). Once LIN-28 levels decline, pre-let-7 is able to be processed by Dicer, and the resulting increase in mature let-7 miRNA promotes the juvenile-to-adult transition. This model accounts for the phenotypic similarity between lep-2 and lep-5, and explains why LEP-2, despite being nearly ubiquitously present from embryo to adult, is apparently only active during the period when lep-5 is expressed (Herrera et al., 2016). Intriguingly, Argonaut binding is suggested to be involved in destabilizing HOTAIR RNA (Yoon et al., 2013), which, given the potential binding of ALG-1 to lep-5 (Grosswendt et al., 2014), could also be the case here. Such predictions will be the focus of future work.
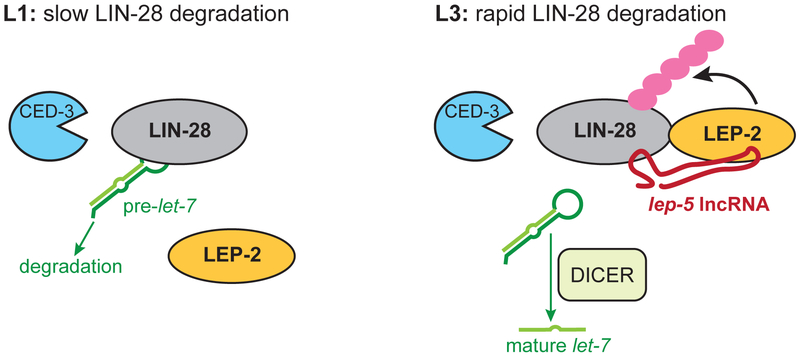
Figure 6. Model for lep-5 function as an instructive switch for LIN-28 degradation.
(Left) Early during C. elegans development, before the onset of lep-5 expression, slow degradation of LIN-28 is facilitated by CED-3 (Weaver et al., 2014). LEP-2, a putative E3 ligase, is present but does not efficiently ubiquitinate LIN-28. LIN-28 facilitates the degradation of pre-let-7. (Right) Later, e.g. in L3, lep-5 acts as a scaffold to bring LEP-2 and LIN-28 into close proximity, allowing efficient ubiquitination of LIN-28. Ubiquitinated LIN-28 would then be subject to rapid proteasomal degradation. When LIN-28 is absent, let-7 biogenesis can proceed, leading to adult-specific programs of TTM, production of adult cuticle and exit from the molting cycle.
Cell-type specificity in developmental timing mechanisms
In contrast to their roles in the timing of TTM and the cessation of molting, neither lep-5 nor lep-2 (Herrera et al., 2016) is required for stage-specific patterns of seam cell division or their terminal differentiation and fusion, both of which are canonical aspects of heterochronic control (Rougvie and Moss, 2013). This separation of phenotypes indicates that there may be cell-type specific characteristics of heterochronic regulation. This idea has precedent: lin-29, the terminal regulator of the larval-to-adult switch in seam cells, is not required for TTM (Ambros and Horvitz, 1984; Del Rio-Albrechtsen et al., 2006). Instead, dmd-3 fulfills this role (Mason et al., 2008). Nevertheless, given that lep-5 acts upstream of lin-28, it is surprising that lep-5 mutants display no strong defects in the seam. One possibility is that cell-type-specific mechanisms are important for regulating lin-28 activity. Additionally, different tissues may differ in their thresholds for lin-28 activity, such that the seam might be less sensitive and tail tip and body hypodermis (hyp7) more sensitive to increased lin-28 function. In any case, the identification of lep-5 and lep-2 indicate that the male tail tip provides an important and sensitive read-out for studies of the heterochronic pathway, key aspects of which could be missed by focusing exclusively on seam cells. Additionally, cell-type-specific variation in the heterochronic pathway could provide important developmental and evolutionary flexibility. Indeed, tissue-specific changes in developmental timing ("heterochrony") are thought to have important roles in morphological evolution (Gould, 1977).
Structural features of lep-5 RNA
The predicted secondary structure of lep-5 features extensive base-pairing, indicating that its higher-order structure is important for its activity. Disrupting the secondary structure of the central zipper as well as the 5´ stem-loop in vivo completely eliminated lep-5 function; restoring secondary structure with complementary mutations restored function. Interestingly, the predicted stem-loop at the 5´ end includes the trans-spliced leader SL1. While SL1 is speculated to promote translation initiation of C. elegans mRNAs (Blumenthal, 2005), our results indicate that SL1 can also have a role in RNA secondary structure and lncRNA function. The 5´ stem-loop might be an important binding site for lep-5 interactors; it is also important for RNA stability, as lep-5 levels were reduced ~5-fold in lep-5(fs8) mutants.
lep-5 lncRNA directly interacts with LIN-28 and LEP-2/Makorin. Both proteins—and even their roles in regulating developmental transitions, including the juvenile-to-adult transition — are highly conserved in animals (Abreu et al., 2013; Faunes et al., 2017; Faunes and Larrain, 2016; Gray et al., 2000; Herrera et al., 2016; Thornton and Gregory, 2012). Although one might expect this conservation to extend to lep-5, we did not find homologs outside of Caenorhabditis. It seems unlikely that Caenorhabditis evolved a special lncRNA-mediated mechanism to catalyze the LIN-28 degradation that must occur in all animals. Rather, we propose that the lep-5 primary sequence evolves rapidly and is therefore difficult to detect in more distantly related species by sequence similarity. Even identifying the C. angaria lep-5— which rescued C. elegans lep-5 mutants—required synteny and bioinformatic analyses in addition to BlastN (see STAR Methods). In general, lncRNAs are known to evolve rapidly, and orthologs of many functionally important lncRNAs are not easily found outside closely related species (Diederichs, 2014). One explanation is that functional conservation in some lncRNAs may depend more on 3D structure than primary sequence. Considering this and the conservation of the LIN28-let-7 regulatory module (Tsialikas and Romer-Seibert, 2015), we propose that lep-5 orthologs could indeed exist in other species but might not be identifiable based on sequence alone. Notably, recent work has shown that several lncRNAs are important for pluripotency and differentiation in mammalian stem cell systems (Flynn and Chang, 2014); the same biological processes often feature regulation by LIN28-let-7 (Shyh-Chang and Daley, 2013). Indeed, a recently identified rodent-specific lncRNA, Ephemeron, modulates the exit of embryonic stem cells from pluripotency by regulating Lin28a; however, unlike lep-5, Ephemeron promotes Lin28a expression (Li et al., 2017). Several lncRNAs have also been implicated in the regulation of LIN28 in cancer cells, in some cases through a feedback loop involving let-7-family miRNAs (Gao et al., 2017; He et al., 2018; Peng et al., 2017; Wang et al., 2016). Our results in C. elegans indicate that lncRNA regulation of lin-28 is an anciently conserved component of mechanisms that control cell state changes and developmental progression.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Douglas Portman (douglas.portman@rochester.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
These studies used the nematode C. elegans, which was cultured as described by Brenner (1974). Most experiments were carried out on larval and adult males, as these studies focus on a male-specific morphogenetic process. Most strains used here contained the mutation him-5(e1490), which increases the frequency of spontaneous XO males among the self-progeny of XX hermaphrodites.
METHOD DETAILS
Microscopy and phenotypic analysis
For microscopy, worms were placed in a drop of 20 mM sodium azide or 1 mM levamisole on 5% agar pads and studied at 400x or 1000x magnification with a Zeiss Axioskop with Nomarski (DIC) and epifluorescence. Images were recorded with a C4742-95 “Orca” Hamamatsu digital camera and Openlab software, ver. 3.0.9 (Improvision). Confocal images were obtained with a Leica TCS SP8 X microscope using the 63x objective. Images were taken as Z-stacks with 0.5 or 1 μm steps. Simple image editing was performed with ImageJ. To investigate cell-fusion phenotypes caused by lep-5 mutations and assess the number of seam cells in adult animals, we examined strain DF235 carrying the lep-5(ny10) mutation, the adherens junction reporter AJM-1::GFP (Koppen et al., 2001), and the seam cell reporter pF09D12.1::GFP.
Isolation and mapping of lep-5 mutants
The lep-5(ny10) deletion allele was isolated in a screen for Lep male phenotypes in which CB4088 hermaphrodites were mutagenized with trimethylpsoralen (TMP) and exposure to UV light. Using competitive genome hybridization (CGH), we identified two large deletions on X in lep-5(ny10) genomic DNA (see below; Fig. S2A). lep-5(fs8) was isolated in a mutagenesis screen in which bxIs14 him-5(e1490) hermaphrodites were mutagenized with ethylmethanesulfonate (EMS). In both screens, F1 hermaphrodites were isolated to produce the F2 generation, which were segregated. F3 male progeny of individual F2s were scored at 400x magnification for defects in ray development and tail tip morphology. Mutants were outcrossed several times with CB4088 to generate the strains used here. By SNP mapping, fs8 was found to lie in a 2.7-Mb region that was deleted in ny10.
A fosmid covering one end of the ny10 deletion rescued the lep-5 phenotype; subsequent truncations of this region indicated that a ~3 Kb fragment containing the predicted gene H36L18.2 was sufficient for rescue (Figs. S2B, C). By sequencing lep-5(fs8), we identified a single G-to-A change (G23A) in the first nucleotide following the SL1 trans-splicing acceptor site (Fig. S2C). Introducing this mutation into the 3-Kb rescuing fragment markedly reduced its activity (Fig. 2B). Together, these results indicate that lep-5 corresponds to H36L18.2.
Array comparative genomic hybridization to map deletions in lep-5(ny10)
The ny10 allele of lep-5 was mapped by comparative genomic hybridization by Nimblegen (Roche). Genomic DNA was isolated from the lep-5 mutant (strain DF70) and the wild type reference strain (CB4088) using the Gentra Puregene Tissue Kit (Qiagen). The tiling arrays contained 385,179 probes with a median spacing of 167 nt covering the whole C. elegans genome. Reference and mutant DNA were differentially labeled with Cy3 or Cy5 dye and hybridized to the tiling array. The fluorescence intensity for Cy3 and Cy5 was measured at each probe position and the expression values were log2-normalized. To visualize the tiling array data, the ratio of the normalized expression values for each probe was plotted according to its position within the C. elegans genome (Fig. 2A). We detected a ~24.6kb deletion on the left arm of ChrX (6,554,074 to 6,578,133 bp) and an ~86kb deletion on the right arm of ChrX (12,576,888 to 12,662,886 bp). To separate the deletions, we performed crosses with a strain that carries unc-18 and dpy-6 mutations between the deletions (EM122). Recombinant Dpy nonUnc or nonDpy Unc hermaphrodites were isolated and their male progeny screened for tail tip phenotypes. The Lep phenotype was only found in nonDpy Unc males, demonstrating that the lep-5 lesion was located on the right arm of ChrX. The lep-5(ny10) mutant was backcrossed to CB4088 multiple times, resulting in the homozygous strain DF135.
Complementation tests to show that fs8 and ny10 are allelic
Because lep-5 is located on the X chromosome and the Lep phenotype is only observed in males, we used XX pseudomales to test whether lep-5(ny10) and fs8 are dominant or recessive mutations and for genetic complementation of the two alleles. First, males from a tra-1(e1488)III; him-5(e1490)Vstrain were crossed with hermaphrodites from a him-5(e1490)V; unc-18(e81) lep-5(ny10)X strain. nonUnc F1 hermaphrodites were allowed to self and the nonUnc F2 pseudomales were scored for tail tip phenotypes. All of these animals had wild-type tails, demonstrating that lep-5(ny10) is recessive. The Unc F2 progeny from this cross were used to establish a tra-1; him-5; unc-18 lep-5(ny10) strain. Hermaphrodites with this genotype were then crossed with post-dauer lep-5(fs8) males. nonUnc F1 hermaphrodites were allowed to self. If fs8 and ny10 were non-allelic, only 1/3 of the of the Tra nonUnc F2 pseudomales should display the Lep phenotype. We examined 79 such animals and found that all were Lep, indicating that both alleles are mutations in the same gene. To exclude that the proportion of homozygous fs8 pseudomales was significantly distorted by the X chromosome non-disjunction in oocytes due to the him-5 mutation in the background of all strains, we followed the progeny of these hermaphrodites; 48 animals were sterile, 10 gave only nonUnc progeny (and were therefore homozygous for fs8). 21 yielded F3 progeny of which 25% were Unc, confirming that these animals were indeed heterozygous for fs8 and ny10. We concluded that ny10 and fs8 are alleles of lep-5.
Transgenesis
Transgenes for rescue experiments and expression constructs were made by overlap extension PCR (Nelson and Fitch, 2011) or by modification of plasmids using site directed mutagenesis. PCR products used for transgenesis were gel-purified with the Promega Wizard® SV Gel and PCR Clean-Up System. Unless otherwise noted, we used the pRF4[rol-6(d)] plasmid at a concentration of 100ng/μl as injection marker. PCR products were microinjected at a concentration of 0.5 or 1ng/μl into the gonads of young hermaphrodites. Sequences of oligos used to generate DNA constructs for transgenesis are listed in Table S1.
Fosmid and PCR rescue
Bacteria containing Geneservice fosmid clones WRM062bG06, WRM0629cE12, WRM0628aE08 and WRM0640cA10 were grown according to the company’s protocol. Fosmid clones were amplified using Epicentre CopyControl induction solution and isolated with the QIAprep Spin Miniprep kit (Qiagen). The fosmid identities were verified by sequencing (pCC1-forward). Fosmids were then linearized by digestion with SfiI, purified with QIAquick spin columns and injected into lep-5(ny10) hermaphrodites at a concentration of 4nμl. Fosmids were injected in groups of two or separately and transgenic males scored for rescue of the Lep phenotype. WRM0640cA10 was the only fosmid that rescued. Sections of its sequence, covering one or two predicted genes were generated by PCR and injected at a concentration of 1ng/μl. A 6506nt long PCR product obtained with primers H36L18.2_F and H36L18.2_R and covering the gene H36L18.2 rescued the phenotype of lep-5(ny10). To determine the minimal rescuing fragment for lep-5, subsequently smaller PCR products were injected into lep-5(fs8) mutants with Punc-122::GFP as injection marker.
Transgenes with secondary structure modifications of lep-5
Six different transgenes covering 3133 nt of the lep-5 transcribed and upstream sequence were made by PCR and injected into the gonads of lep-5(ny10) hermaphrodites at a concentration of 0.5 or 1 ng/μl At least 4 lines were scored for each experiment.
(1) lep-5(+) PCR fragment was amplified from N2 genomic DNA with primers KKlp5_expr-9 and KKlp5_expr-10.
(2) lep-5(G23A) PCR fragment was amplified from genomic DNA of lep-5(fs8) mutants with primers KKlp5_expr-9 and KKlp5_expr-10. Because the lep-5(fs8) mutation is temperature-sensitive and the construct lep-5(G23A) showed some degree of rescue when injected at 1ngμl, we also injected it at a ten-fold concentration.
Four transgenes were made by overlap extension PCR using N2 genomic DNA as template. The modifications noted were introduced into the reverse primer of the first PCR product (A piece) and the forward primer of the second PCR product (B piece). The final product was amplified with primers KKlp5_expr-9 and KKlp5_expr-10.
(3) lep-5(G23A, C30T). Primers for A piece (forward and reverse): KKlp5_expr-1 and KKlp5_23A+30T-R. Primers for B piece: KKlp5_23A+30T-F and KKOLp5-8a.
(4) lep-5(G23C). Primers for A piece: KKlp5_expr-1 and KKlp5_23C_R. Primers for B piece: KKlp5_23C_F and KKOLp5-8a.
(5) lep-5(G23C, C30G). Primers for A piece: KKlp5_expr-1 and KKlp5_23C+30G-R. Primers for B piece: KKlp5_23C+30G-F and KKOLp5-8a.
(6) PCel-lep-5::Cbr-lep-5(+). Primers for A piece (forward and reverse): KKlp5_expr-1 and KKlp5_Cbr-AR used on N2 DNA; Primers for B piece: KKlp5_Cbr-BF and KKOLp5-8a used on C. briggsae PB800 DNA. The final product was amplified with KKlp5_expr-9 and KKlp5_Cbr-nR.
(7) PCel-lep-5::Can-lep-5(+). Primers for A piece (forward and reverse): KKlp5_expr-1 and KKlp5_Can-AR used on N2 DNA; Primers for B piece: KKlp5_Can-BF and KKOLp5-8a used on C. angaria PS1010 DNA. The final product was amplified with KKlp5_expr-9 and KKlp5_Can-nR. All final products were sequenced with primer RHOLp5-2 to confirm the presence of the desired modification.
Transgenes with stop codons and deletions
Site-directed mutagenesis with QuikChangeXL (Agilent Technologies) was used to introduce stop codons into a plasmid containing the minimal rescuing fragment for lep-5 following the manufacturer’s instructions. This plasmid was generated by cloning a PCR product, made with attBlep-5F6 and attBlep-5R6, into the Gateway vector pDONR™P4-P1R (Life Technologies). Primers AA4,5,7F and AA4,5,7R were used to convert amino acids 4, 5 and 7 of the predicted ORF of H36L18.2 into stop codons. Amino acids 14 and 15 were converted into stop codons using primers AA14,15F and AA14,15R. The resulting plasmids were sequenced to confirm the changes and injected into lep-5(fs8) hermaphrodites with Punc-122::GFP as injection marker.
To create deletion constructs, two PCR products, covering the minimal rescuing fragment for lep-5 with a gap at the intended deletion were amplified with primers that introduced an AatII restriction site at the 3’ end of the first and the 5’ end of the second piece. The PCR products were digested with AatII, gel-purified and ligated with T4 DNA ligase. Replacement of the intended sequence by the 6-bp tag GACGTC was confirmed by sequencing. The constructs were injected into lep-5(ny10) hermaphrodites at a concentration of 10ng/μl with Punc-122::GFP as injection marker. The sequence cagtgaccataacaatgtatgcacaacctcttcggacttttctgcatctatggtggcgctg, containing an ALG-1 binding site (Zisoulis et al., 2010), was deleted with primer pairs lep-5F6 + Δ1R and Δ1F + lep-5R6. Sequence ttttccattattcattccaacttcttaaatgataatcgaaaa, forming a predicted stem-loop, was deleted with primer pairs ep-5F6 + Δ2R and Δ2F + lep-5R6.
Genome editing using CRISPR/Cas9
ny28:
To delete the endogenous lep-5 locus, genome editing with CRISPR/Cas9 with dpy-10 coCRISPR (Arribere et al., 2014) was performed as described in the protocol from the Dernburg lab published on the Integrated DNA Technologies (IDT) website. Two guide RNAs were designed with ChopChop (Labun et al., 2016; Montague et al., 2014). The crRNAs, tracrRNA and CAS9 were purchased from IDT. The RNAs were reconstituted to a concentration of 200μM in the IDT duplex buffer. 1μl of each lep-5-specific crRNA and 2μl of tracrRNA and 1μl of dpy-10 crRNA and 1μl of tracrRNA were mixed and incubated at 95°C for 5 minutes. RNA duplexes for lep-5 and dpy-10 were mixed 12:1 and diluted to 62μM. 0.5 μl RNA duplex mix was mixed with 0.5 μl of Cas9 enzyme and incubated at room temperature for 10 minutes. 1μl RNP complex was used for 10μl injection mix, supplemented with 0.2xTE buffer and 0.1μM single-stranded DNA oligo to edit the dpy-10 locus. Young him-5(−) hermaphrodites were injected into both gonads and their offspring screened for the Rol and Dpy phenotype. Pools of 5 F1 hermaphrodites from plates with the most Rol and Dpy worms were screened for edits at the lep-5 locus by PCR with primers KK_CR_lep-5_1 and and KK_CR_lep-5_2. Several lines with deletion of the lep-5 locus were obtained, one of which was retained (lep-5(ny28). This allele deletes 572 nucleotides of the lep-5 gene, from 104 nt upstream of the transcription start site through 468 nt of the transcript, replacing this region with 7 random nucleotides and leaving 59 nt at the 3′ end.
fs18, fs19, fs21 and fs22:
To modify the endogenous lep-5 locus, CRISPR/Cas9 genome editing using dpy-10 coCRISPR was performed as described (Arribere et al., 2014; Paix et al., 2015). One or two lep-5 guide RNAs were designed with CRISPOR (Haeussler et al., 2016) and/or ApE software using the algorithm of Doench et al. (2014). The tracrRNA and crRNAs were purchased from Dharmacon. ssODNs and primers were made by IDT. Recombinant Cas9 (25 μg/μl) was prepared as described (Paix et al., 2015). tracrRNA was reconstituted at 4 μg/μl with RNase-free Tris Buffer. Similarly, each crRNA was reconstituted at 8 μg/μl with Tris Buffer. Each ssODN was dissolved in RNase-free water at 500 ng/μl. For microinjection mixtures, we followed the published protocol (Paix et al., 2015) strictly, except that the total volume of each mixture was halved from 20 μl to 10 μl. Microinjection mixtures were incubated at 37ºC for 10 min before injection and used within 1h after preparation. Young him-5(−) hermaphrodites were injected and their F1s were screened for Rol, Dpy or DpyRol phenotype. Pools of 3 F1 hermaphrodites from plates with the most Rol and Dpy worms were screened for edits at the lep-5 locus by PCR and restriction enzymes digest. For each allele, one or two lines were obtained and kept. All strains were backcrossed to N2 or him-5(e1490) at least four times. Fragments containing the entire lep-5 gene, along with ~500 nt flanking regions, were confirmed by Sanger sequencing.
fs21 fs25:
Molecularly, fs25 and fs22 are identical alleles of lep-5. To generate fs21 fs25, the injection mix used for fs22 was injected into young him-5/+ V; lep-5(fs21)/oxTi1015 X hermaphrodites. Non-fluorescent offspring, which must be lep-5(fs21) homozygotes, were selected for future screening. Outcrossing and sequencing was done as for other fs alleles.
lep-5 transcriptional reporter
A GFP reporter construct driven by the lep-5 regulatory region was generated by OES-PCR. Primers KKOLp5-1 and KKkp5-GFPA-R were used to amplify the region upstream of the transcription start of lep-5 from N2 genomic DNA (piece A). This PCR product overlaps with piece B containing 4x NLS-GFP and the unregulated 3’ UTR of let-8583, generated with KKlp5_GFPB-f and MN-lin-44_8 from Plasmid pPD122.13 (Andrew Fire Vector Kit, Addgene). The final product was amplified with primers RHOLp5-7 and MN-lin-44_9, gel-purified and injected into CB4088 hermaphrodites; 8 lines were obtained.
lep-5 lncRNA expression via smFISH
For smFISH, we used custom Stellaris probes from Biosearch Technologies specific for lep-5 lncRNA (22 probes) labeled with Quasar570 (excitation 548nm, emission 566nm) and for lin-44 (39 probes) labeled with Quasar670 (excitation 647nm, emission 670nm). The probes were designed by Biosearch Technologies. Sample preparation and hybridization were performed in tubes using a modification of the protocols by Ji and van Oudenaarden (2012) and the protocol provided by Biosearch Technologies. Briefly: mixed stage or synchronized L2 worms from strains CB4088 (WT) and DF135 (lep-5(ny10)) were collected, washed in M9 buffer and fixed in 4% paraformaldehyde in 1x PBS for 40 minutes at room-temperature. Fixed worms were washed twice with 1x PBS and transferred to 70% ethanol and kept on a rotating shaker at 4ºC overnight. Worms were then washed with wash buffer (10% deionized formamide in 2x SSC) and hybridized in hybridization buffer (100mg/ml dextran sulfate and 10% formamide in 2x SSC) with 75nM lin-44 probes and 250nM lep-5 probes for 4 hours at 37ºC in a hybridizer. The hybridization solution was washed off with wash buffer and the samples incubated in wash buffer for 30 minutes at 30ºC. Worms were washed in 2x SSC and mounted in ProLong Gold with DAPI (Life Technologies) in imaging chambers as described by Ji and van Oudenaarden (2012). The slides were imaged on the DeltaVision Elite Imaging System using solid-state illumination with filters for DAPI, TRITC (with Quasar570 labeled probes) and Cy5 (with Quasar670 labeled probes). Images were recorded with an Evolve 512 EMCCD camera and processed using the softWoRx software package.
Sequences of the Stellaris RNA FISH probes
Probes for lin-44:
aatcaaaaggagctgctcgc gttgaaagagcagtcgattg ggccagggagcaaagaataa tgatcgtcgggatctcattc tttggcttaggtggttgaac tgggcaaccctgtttcaaaa gagcacgtgaatgcagaaga gcatgccagttgaattgatc attactgtagcaggatgtgt ttcctgaactccttcgaacg gattcgcacagttttgaagt tcccattgctgaaatctcaa aatatttccagcttccgaac tcaataacggcggatcatgc gagaagactctcggaaaccc ggcagatgcagatgacaacg caatcgtcaatccatccttg attcattttgacccatctgg tgagtacatccgccgaactc cgttattccgtgttgaacac ttgtgagcagctttcgacta aacaaagtattcaccgctcc cttcaaattgtgcttctcca tctttttgatggccaatctc ggcatttgcaggaagagatt tgacaagaaccggatactcc ttcgtttccagcaagttttc atcggtaatgtgctcaaggg cgtgcatacttttccactaa atcatccgtatagagcttgg aaatccgtcgtctttaccac atccggagacgcttctaaat caactgattttgccttgcaa cttgggtatgcgtctcattc caacacagccgatcacaatc tcgtgacgaatgctgaatcc tcacagtcacacttcacacg cagattgcaacaccacacga gcgatgttggatacaatcct tcccattacacgtggatatc aaaattaggcttttcggcgg
Probes for lep-5:
gcccatgtctttgggaaaac tggaagagccaggcctaaat gtcataagacaaatcgcgga gctatcatcccacttacgat tgttggtttagcattacacc catcttgtcacaacactggg tatttgatgggtgacatcgc acaccctaattgaactccaa gcataagcgaatttgacgat atatggtctaactttcgcgt tatgcgctaaacaagtacgg taataatgttgatggccacg accagtcgttgtgtgatttt taaaacggaccacctagttg gatagcgacatccaattccg caacaaggcctgaggaggga cagcttttgatcagttacaa tatggtcactgatccaccag aagaggttgtgcatacattg ccatagatgcagaaaagtcc caataaccagatacagcgcc cggtcccaacattctttaat
Time-course of lep-5 expression via qPCR
Staged animals were obtained with the hatch-off method (Pepper et al., 2003) and collected 0, 12, 15, 18, 22, 26 and 41 hours after hatching at 25ºC and flash-frozen. RNA was extracted using a modification of the tissue protocol for the RNeasy micro kit from Qiagen. Frozen worms were ground with a plastic pestle fitting into a 500μl tube. When the worms began to thaw, the tube was dipped into liquid nitrogen to re-freeze. This process was repeated 5 to 6 times before buffer was added and the sample disrupted by passing it through a syringe needle. A DNase treatment was performed on-column. Wash-steps were performed according to the protocol. RNA was eluted with water and concentrations checked by NanoDrop (Thermo Scientific). cDNA was generated from 250ng total RNA using the double-primed RNA to cDNA EcoDry Premix (Takara/Clontech). qPCR was performed using the iQ SYBR Green Supermix with a two-step PCR protocol (3 min 95ºC; 40× 10 sec 95ºC, 60 sec 60ºC; followed by melt curve analysis) on the CFX96 real time PCR detection system (BioRad). Y45F10D.4 served as reference gene (Hoogewijs et al., 2008) For primer sequences see Table S1. Unknowns were run in triplicates, standards in singles. Data were evaluated using the standard-curve method.
RNAi knockdown
RNA-interference was performed by feeding E. coli transformed with inducible RNAi vectors to mothers or larvae as previously described (Nelson et al., 2011). Bacterial strains for lin-14 and lin-41 RNAi were from the J. Ahringer library, and those for lin-28 from the M. Vidal library (Source Bioscience). For lin-14 RNAi, L4 hermaphrodites were treated and their male progeny scored as L3, L4 and adults for defects in tail tip morphogenesis. For lin-28 and lin-41 RNAi, L1 larvae were fed and scored as L4 and adults. Bacterial strain HT115 carrying the L4440 plasmid was used as negative control.
lin-28 mRNA levels
Staged WT (CB4088) and lep-5(ny10) animals were collected by 4h hatch-off. Some of the L1 were flash-frozen immediately, the rest was plated onto seeded plates and placed at 20ºC. L3 larvae were collected and flash frozen 27 hours later. RNA extraction and cDNA synthesis (using 440ng RNA) were performed as described above. qPCR was performed with primers lin-28_FW2 and lin-28_RV2 and Y45F10D.4 as reference gene as above.
Western blot
Western blot analysis by SDS-PAGE was performed according to standard procedure. For the L1 sample, arrested L1s were placed on food and collected a few hours later. L3 samples were collected 24 hours after plating arrested L1s at 20°C. Animals were washed twice with PBS, frozen in liquid nitrogen and ground in the presence of a protease inhibitor (Halt, Pierce), SDS buffer was added. Samples were heated to 95°C for 10 minutes and centrifuged to pellet the insoluble fraction. An aliquot was kept for Bradford assay. Approximately 10-20 μg of total protein was loaded onto a 10% Bis-Tris Bolt gel (Invitrogen). After electrophoresis and transfer to a nitrocellulose membrane, the blot was incubated overnight at 4°C with a rabbit anti-LIN-28 polyclonal antibody (gift from E. Moss; 1:5,000 dilution) and a mouse anti-actin monoclonal antibody (Sigma; 1:5,000 dilution) as a loading control. The blot was incubated 45 minutes in the dark with fluorescent secondary antibodies (Licor; 1:25,000) and scanned on a Licor infrared fluorescence scanner.
Analysis of LIN-28 degradation
To observe LIN-28 dynamics in lep-5 mutants, we used a Plin-28::Dendra2::lin-28_3'UTR reporter gene (Herrera et al., 2016). In the lep-5 genetic background, this transgene caused bursting of hermaphrodites, and no stable line could be established. Therefore, hemizygous male cross-progeny of him-5; nyEx56[Plin-28::Dendra2::lin-28_3'UTR] males and him-5; lep-5(ny10) hermaphrodites were investigated. The fluorescence signal of the reporter was visualized with a confocal microscope (Leica, SP8) as described in Herrera et al. (2016): L2 stage animals were examined and fluorescence in the pharynx region was captured for the green and red channels by sequential scans. Dendra2 photoconversion was performed by exposing a section of the pharynx to brief flashes of 405 nm light in 10-20 slices with ten successive scans of the z-plane with short rest periods inbetween. Sequential z-stacks of the exposed region were taken for the red and green channel to record the post-photoconversion fluorescent signal. The animal was recovered onto a plate with food and kept at 20°C for 24 h. The worms (now L3) were remounted, and another sequential z-stack was recorded. The image stacks were analyzed with ImageJ.
RNA co-IP with LEP-2, LIN-28 and LIN-41
Worms were synchronized by L1 arrest and grown for 17 hours at 25ºC to the L2 stage (DF237 carrying LIN-28::GFP and DF282 carrying GFP::LIN-41), or mixed stages were used (DF293 carrying GFP::LEP, DF302 carrying plep-5::GFP). Worms were washed twice in M9 buffer and irradiated in a BioRad crosslinker with 800 mJ/cm2 at 254nm. UV-treated worms were washed once in lysis buffer (20 mM Tris/Cl pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.5 % NP-40 Igepal), resuspended in an equal volume of lysis buffer supplemented with HALT protease inhibitor (Invitrogen), RNaseOUT (Thermo Fisher) and DTT and flash frozen in liquid nitrogen. At least three biological replicates were obtained for each genotype. The frozen worms were manually ground in Takara Biomasher tubes and lysed in 200μl lysis buffer supplemented with HALT protease inhibitor and RNaseOUT. IP was performed with Chromotek GFP-Trap MA and Chromotek binding control magnetic agarose beads according to the guidelines by the company: The beads were equilibrated in dilution buffer (20 mM Tris/Cl pH 7.4, 150 mM NaCl, 5 mM MgCl2) and resuspended in dilution buffer supplemented with RNase OUT, DTT and 400μM vanadyl ribonucleoside complexes. 40-90μl lysate was added to each kind of beads and incubated for 60 minutes at 4ºC while tumbling end over end. 20% of this volume (8 - 18μl) was set aside as input sample. Beads were washed twice with dilution buffer and twice with wash buffer (20 mM Tris/Cl pH 7.4, 250 mM NaCl, 1 mM MgCl2, 0.025% SDS, 0.05 % NP-40 Igepal). Beads and input samples were then treated for 20 minutes with Proteinase K (100μl PBS with 1μl 10% SDS and 2.5% Proteinase K (Invitrogen)) at 65ºC on a Thermomixer (Eppendorf) to reverse the crosslink and digest the proteins. RNA was extracted with Trizol and BCP, precipitated over night at −80ºC after adding Glycoblue™ (Ambion), sodium acetate and isopropanol. The pellet was washed once with 70% ethanol, air dried and resuspended in 44μl water. A DNase treatment with rDNase I (Ambion) was performed for 20 minutes at 37ºC. The RNA was re-purified immediately using the Qiagen RNeasy Micro kit following the manufacturer’s protocol. The RNA was eluted with 12μl water. 10μl RNA was used in a 20μl cDNA synthesis reaction with the Clontech/Takara double-primed EcoDry cDNA synthesis kit, 2pg luciferase control mRNA (Promega) was added to each reaction as a control for cDNA synthesis. The cDNA was diluted 1:7 for qPCR on a BioRad CFX instrument using the BioRad iQ SYBR Green Supermix and primers for lep-5, cdc-42 and luciferase. cdc-42 was chosen as a control gene because it was present at similar levels as lep-5 (i.e., in qPCR of the input sample, the Ct values for these genes differed by no more than 1.5 except in the LIN-41::GFP strain, which showed especially high levels of lep-5). All samples were run in triplicates. The results were evaluated using the Pfaffl Method (Pfaffl, 2001) with empirical efficiencies of 2 for luciferase, 1.98 for lep-5 and 1.91 for cdc-42, comparing Ct values for GFP-trap beads (experiment) to binding control. Results for lep-5 and cdc-42 were normalized against the spiked-in luciferase to account for differences in the efficiency of the RT reaction. A moderated T-test implemented in the Limma package was performed to test for significance of the results. This test is specifically designed for expression analyses with small sample sizes (Ritchie et al., 2015).
Identification of lep-5 orthologs from other Caenorhabditis species
Sequences of lep-5 transcripts for C. brenneri, C. elegans and C. japonica were confirmed by ESTs in WormBase. Genomic lep-5 sequences for 15 other Elegans supergroup species (Kiontke et al., 2011) were extracted from whole genomes (WormBase, and http://caenorhabditis.org) after BlastN search (Camacho et al., 2009). An unambiguous lep-5 homolog was not identified in the C. sp. 26 genome. All sequences contained two introns, which were edited out manually by assuming that their positions are homologous and that they bear the typical GT and AG motif at their 5’ and 3’ ends, respectively. No significant matches were found by BlastN alone in Caenorhabitis species outside of the Elegans supergroup. However, after multiple sequence alignment of a syntenic region in C. angaria and 7 Elegans supergroup species, we identified a sequence in the C. angaria genome with similarity to lep-5. A BlastN search subsequently yielded a partial C. angaria lep-5 cDNA sequence that had been previously assembled from RNA-seq reads, as part of the C. angaria genome project (Mortazavi et al., 2010). This sequence was used to design internal primers, which together with a primer complementary to SL1 and an anchored oligo(dT) primer amplified two overlapping fragments of lep-5 from C. angaria cDNA (made from total RNA with the double-primed RNA to cDNA EcoDry Premix by Takara/Clontech). The fragments were sequenced to identify the full C. angaria lep-5 transcript. This transcript was used for a BlastN search of draft genome assemblies for Caenorhabditis species outside of the Elegans supergroup (available from the laboratory of Mark Blaxter at http://download.caenorhabditis.org). This yielded lep-5 loci from C. castelli, C. sp. 38, C. plicata, and C. virilis, which we confirmed by BlastN with the C. virilis lep-5 genomic region. We used MAFFT v7.266 (Katoh and Standley, 2013) (to align the predicted lep-5 transcripts of 19 Elegans group species (Figure S1), our lep-5 cDNA and genomic sequences from C. angaria, and the four lep-5 genomic regions from non-Elegans group species. To maximize its accuracy and control the order of aligned sequences, MAFFT was run with the arguments '--localpair --maxiterate 16 --inputorder'. We manually edited the resulting alignment in Jalview 2.9.0b2 (Waterhouse et al., 2009) to remove trailing or poorly aligned flanking regions. Via the hmmbuild and hmmpress programs from HMMER 3.1b2 (Eddy, 2011), we converted the edited lep-5 alignment into a hidden Markov model (HMM). Searching other non-Elegans supergroup Caenorhabditis genomes with the 24-species HMM and nhmmer (Wheeler and Eddy, 2013) identified two more lep-5 loci from Caenorhabditis spp. 43 and 31. We also attempted HMM searches of non-Caenorhabditis species; although this gave various weak similarities, they were neither statistically significant nor similar to one another. We finally aligned the lep-5 sequences from all 26 Caenorhabditis species with MAFFT and visualized their alignment with Jalview (Figure S3).
Analysis of the secondary structure of the lep-5 transcript
The processed lep-5 RNA sequence, including the SL1 trans-spliced leader, along with similar information from lep-5 orthologs from twenty Elegans group Caenorhabditis species, was analyzed using TurboFold (Harmanci et al., 2011), part of the RNAstructure package (http://rna.urmc.rochester.edu/RNAstructure.html). To model lep-5(G23A) and lep-5(G23A C30T), corresponding changes were made to all twenty orthologous sequences. The folding temperature was set at 293.15°K (20°C); default parameters were used for all other settings. Structures were drawn using VARNA (Darty et al., 2009). Nucleotide positions were color-coded for confidence level and primary sequence conservation as described in the text. VARNA linear-format representations of Turbofold structures for all species were arranged according to an unpublished phylogenetic tree for Caenorhabditis inferred by K. Kiontke using molecular data from 17 loci (RhabditinaDB, rhabditina.org), as shown in Figure S4).
Using an independent method of alignment and structure analysis produced very similar results. Specifically, we used CARNA (ver. 1.3.3, linking LocARNA 1.9.1, Gecode 5.0.0, Vienna RNA package 2.3.2; http://rna.informatik.uni-freiburg.de) (Raden et al., 2018; Sorescu et al., 2012) and RNAalifold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAalifold.cgi). CARNA generated a dot-plot of probabilities of basepairing which was used to generate a consensus sequence and structure, used in an iterative manner to optimize an alignment based on secondary structure conservation. This alignment was very slightly modified by hand to allow a couple additional gaps to further optimize the alignment to the average structure. This alignment was used as input to RNAalifold to generate a consensus RNA structure in which alignment positions were highlighted with regard to conservation and basepair co-variance. Although the resulting structure differed in several details from that shown in Fig. 2A of this paper, there were three regions that were essentially identical: the 5' stem-loop involving SL1, the 3' stem-loop, and the central “zipper” region. Thus, very different structure-alignment methods yield essentially similar results with regard to these three important regions (highlighted red, blue and green, respectively, in Figs. 2 and S4).
Search for miRNA binding sites in the lep-5 transcript
The lep-5 sequence was scanned for miRNA binding sites using algorithms available on www.microrna.org,www.targetscan.org and the website of the Segal lab https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html (Kertesz et al., 2007). High scoring miRNAs found by all three algorithms are miR-76, miR-265, miR-1830 and miR-2220. None of these miRNAs has been implicated in the heterochronic pathway.
QUANTIFICATION AND STATISTICAL ANALYSIS
For statistical analysis of the RNA co-IP experiments, we used a moderated T-test as described above in the Method Details, “RNA co-IP with LEP-2, LIN-28 and LIN-41” section.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-Actin Monoclonal Antibody, Unconjugated, Clone AC-40 | Sigma-Aldrich | Sigma-Aldrich Cat# A4700, RRID:AB_476730 |
| Rabbit anti-LIN-28 polyclonal antibody | Eric Moss labSeggerson et al. 2002 | n/a |
| IRDye 800CW Goat anti-Mouse | LI-COR | LI-COR Biosciences Cat# 827-08364, RRID:AB_10793856 |
| IRDye 680RD Goat anti-Rabbit | LI-COR | LI-COR Biosciences Cat# 926-68171, RRID:AB_10956389 |
| Bacterial and Virus Strains | ||
| OP50-1 | RRID:WB-STRAIN:OP50-1 | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Alt-R® S.p. Cas9 Nuclease | Integrated DNA Technologies (IDT) | Cat#1081058 |
| Restriction enzyme SfiI | New England Biolabs | Cat#R0123S |
| Restriction enzyme AatII | New England Biolabs | Cat#R0117S |
| Critical Commercial Assays | ||
| QuikChangeXL site-directed mutagenesis kit | Agilent Technologies | Cat#200516 |
| Custom Stellaris RNA-FISH probes for lep-5 labeled with Quasar570 (sequences in STAR methods) | Biosearch Technologies | https://www.biosearchtech.com/products/rna-fish |
| Custom Stellaris RNA-Fish probes for lin-44 labeled with Quasar670 (sequences in STAR methods) | Biosearch Technologies | https://www.biosearchtech.com/products/rna-fish |
| RNeasy micro kit | Qiagen | Cat#74004 |
| double-primed RNA to cDNA EcoDry Premix | Takara/Clontech | Cat #639547 |
| iQ SYBR Green Supermix | BioRad | Cat#1708882 |
| GFP-Trap MA and | Chromotek | Cat#gtma-20 |
| Binding control for GFP-Trap MA | Chromotek | Cat#bmab-20 |
| Experimental Models: Organisms/Strains | ||
| C. elegans CB4088 = him-5(e1490)V | CGC | https://cgc.umn.edu/strain/CB4088 |
| C. elegans DF135 = him-5(e1490) V; lep-5(ny10) X | This paper | n/a |
| C. elegans DF137 = him-5(e1490) V; lep-5(ny10) X; fsIs2[Pdmd-3::yfp + CC::GFP] | This paper | n/a |
| C. elegans DF174 = him-5(e1490) V; lep-5(ny10) X ; nyEx23[Plin-41::NLS::GFP + rol-6] | This paper | n/a |
| C. elegans DF213 = him-5(e1490) V; zaIs3[let-7(+) + myo-3::GFP] made by crossing CT19 with CB4088 | This paper | n/a |
| C. elegans DF223 = him-5(e1490) V; lep-5(ny10)X; zaIs3[let-7(+) + myo-3::GFP] | This paper | n/a |
| C. elegans DF235 = him-5(e1490) V; lep-5(ny10) X; wIs78[unc-119(+) + ajm-1::GFP + scm::GFP + F58E10(+)] | This paper | n/a |
| C. elegans DF237 = him-5(e1490) V; maIs108[lin-28::gfp + rol-6] | This paper | n/a |
| C. elegans DF293 = lep-2(ny23) IV; him-5(e1490) V; nyEx53[lep-2>GFP::lep-2b + pRF4] | This paper | n/a |
| C. elegans DF282 = lin-41(tn1541[GFP::tev::s::lin-41]) I; him-5(e1490) V | This paper | n/a |
| C. elegans DF302 = him-5(e1490) V; nyEx61[lep-5>GFP + rol-6] | This paper | n/a |
| C. elegans DF271 = him-5(e1490) V ; nyEx56[lin-28::Dendra2::lin-28_3'UTR + rol-6] | Herrera et al. 2016 | n/a |
| C. elegans EM122 = him-5(e1490) V ; unc-18(e81) dpy6(e14) X | CGC | https://cgc.umn.edu/strain/EM122 |
| C. elegans UR290 = him-5(e1490) V; lep-5(fs8) X | This paper | n/a |
| C. elegans UR1140 = him-5(e1490) V; lep-5(fs18) X | This paper | n/a |
| UR157 = fsIs2[dmd-3promxln2::YFP + CCGFP] I; him-5(e1490) V | This paper | n/a |
| C. elegans UR1141 = him-5(e1490) V; lep-5(fs19) X | This paper | n/a |
| C. elegans UR1207 = him-5(e1490) V; lep-5(fs21) X | This paper | n/a |
| C. elegans UR1208 = him-5(e1490) V; lep-5(fs22) X | This paper | n/a |
| C. elegans UR1211 = him-5(e1490) V; lep-5(fs21 fs25) X | This paper | n/a |
| C. elegans UR1226 = lin-41(ma104) I ; him-5(e1490)V; lep-5(fs8) X | This paper | n/a |
| Oligonucleotides | ||
| See Table S1 for list of oligonucleotides | ||
| Recombinant DNA | ||
| Gateway vector pDONR™P4-P1R | Life Technologies | Not available (replaced by new product) |
| RNAi feeding clone for C. elegans lin-14: Ahringer library sjj_T25C12.1 | Source BioScience | sjj_T25C12.1 |
| RNAi feeding clone for C. elegans lin-41: Ahringer library sjj_C12C8.3 | Source BioScience | sjj_C12C8.3 |
| RNAi feeding clone for C. elegans lin-28: Vidal library GHR-11014@G03 | Source BioScience | GHR-11014@G03 |
| L4440 control plasmid for RNAi feeding experiments | Addgene | RRID:Addgene_1654 |
| pRF4[rol-6(d)] | ||
| Punc-122::GFP C. elegans expression vector | Addgene | RRID:Addgene_19325 |
| GFP::NLS C. elegans expression vector | Addgene | RRID:Addgene_1622 |
| Dendra2 C. elegans expression vector | Addgene | RRID:Addgene_40077 |
| Fosmid clones WRM062bG06, WRM0629cE12, WRM0628aE08 and WRM0640cA10 | Source BioScience | https://www.sourcebioscience.com/ |
| Software and Algorithms | ||
| imageJ | https://imagej.nih.gov | |
| ChopChop | Labun et al., 2016; Montague et al., 2014 | http://chopchop.cbu.uib.no |
| CRISPOR | Haeussler et al., 2016 | http://crispor.tefor.net |
| ApE | Doench et al., 2014 | http://jorgensen.biology.utah.edu/wayned/ape/ |
| LIMMA | https://bioconductor.org/packages/release/bioc/html/limma.html | |
| HMMER 3.1b2 | Eddy, 2011 | http://hmmer.org |
| MAFFT v7.266 | Katoh and Standley, 2013 | https://mafft.cbrc.jp/alignment/software/ |
| Jalview 2.9.0b2 | Waterhouse et al., 2009 | http://www.jalview.org |
| TurboFold | Harmanci et al., 2011 | http://rna.urmc.rochester.edu/RNAstructure.html |
| VARNA | Darty et al., 2009 | http://varna.lri.fr |
| CARNA | Raden et al., 2018; Sorescu et al., 2012 | http://rna.informatik.uni-freiburg.de |
| RNAalifold | http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAalifold.cgi | |
| TargetScan | http://www.targetscan.org/worm_52/ | |
| Online MicroRNA prediction tool | Kertesz et al. 2007 | https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html |
| microRNA.org | http://www.microrna.org/microrna/home.do | |
| Other | ||
| Genome sequence of Caenorhabditis afra | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. brenneri | https://wormbase.org/species/c_brenneri#4--10 | |
| Genome sequence of C. briggsae | https://wormbase.org/species/c_briggsae#0--10 | |
| Genome sequence of C. doughertyi | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. japonica | https://wormbase.org/species/c_japonica#0--10 | |
| Genome sequence of C. kamaaina | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. latens | https://parasite.wormbase.org/Caenorhabditis_latens_prjna248912/Info/Index | |
| Genome sequence of C. macrosperma | nematode.org | |
| Genome sequence of C. nigoni | https://parasite.wormbase.org/Caenorhabditis_nigoni_prjna384657/Info/Index | |
| Genome sequence of C. nouraguensis | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. remanei | https://wormbase.org/species/c_remanei#0--10 | |
| Genome sequence of C. sinica | https://parasite.wormbase.org/Caenorhabditis_sinica_prjna194557/Info/Index | |
| Genome sequence of C. sp. 28 | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. sp. 29 | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. sp. 32 | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. sp. 40 | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. tropicalis | https://parasite.wormbase.org/Caenorhabditis_tropicalis_prjna53597/Info/Index | |
| Genome sequence of C. wallacei | nematode.org | |
| Genome sequence of C. angaria | https://wormbase.org/species/all#0123--10 | |
| Genome sequence of C. castelli | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. sp. 38 | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. pilcata | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. virilis | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. sp. 43 | http://ensembl.caenorhabditis.org/index.html | |
| Genome sequence of C. sp. 31 | http://ensembl.caenorhabditis.org/index.html | |
HIGHLIGHTS.
lep-5 acts in the heterochronic pathway to promote the larval-to-adult transition
lep-5 is a ~600-nt, highly structured lncRNA that is conserved across Caenorhabditis
Like the Makorin LEP-2, lep-5 promotes the degradation of LIN-28 protein
lep-5 may act as a scaffold to bring LEP-2 into close proximity with LIN-28
ACKNOWLEDGEMENTS
We thank K. Nguyen for technical help in the screen yielding ny10 and A. Woronik for help with statistical analyses. We thank V. Ambros, G. Ruvkun, T. Montgomery, D. Mathews, S. Bellaousov, A. Mason, A. Samuelson, L. Maquat, D. Moerman, C. Hammel, S. West and WormBase for constructive discussions and information, F. Slack, V. Ambros, and A. Rougvie for strains, and E. Moss for generously providing the LIN-28 antibody. Some strains were provided by the Caenorhabditis Genetics Center (University of Minnesota), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This research was supported by NIH R01 GM100140, NSF DEB 0922012, NYU RCF 002456, and research funds from NYU Shanghai (D.H.A.F.); and NIH R01 GM108885 and NSF IOS 1353075 (D.S.P.). E.V. was supported by the University of Rochester Medical Scientist Training Program, NIH T32 GM007356, and R.A.H. was supported by the New York Consortium for the Advancement of Postdoctoral Scholars, NIH K12 GM102778.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, and Ambros V (2005). The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 9, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, Cukier P, Thompson IR, Navarro VM, Gagliardi PC, et al. (2013). Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med 368, 2467–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V (1989). A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57, 49–57. [DOI] [PubMed] [Google Scholar]
- Ambros V (2011). MicroRNAs and developmental timing. Curr Opin Genet Dev 21, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, and Horvitz HR (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam TU, Davies E, Morita EH, and Abe S (2007). Sequence, expression and tissue localization of a gene encoding a makorin RING zinc-finger protein in germinating rice (Oryza sativa L. ssp. Japonica) seeds. Plant Physiol Biochem 45, 767–780. [DOI] [PubMed] [Google Scholar]
- Blumenthal T (2005). Trans-splicing and operons WormBook : the online review of C elegans biology, 1–9. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussing I, Yang JS, Lai EC, and Grosshans H (2010). The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J 29, 1830–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: architecture and applications. BMC Bioinf 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassar PA, Carpenedo RL, Samavarchi-Tehrani P, Olsen JB, Park CJ, Chang WY, Chen Z, Choey C, Delaney S, Guo H et al. (2015). Integrative genomics positions MKRN1 as a novel ribonucleoprotein within the embryonic stem cell gene regulatory network. EMBO Rep 16, 1334–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, and Johnston LA (2008). Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol 18, 943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, and Steitz JA (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94. [DOI] [PubMed] [Google Scholar]
- Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K , et al. (2013). The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol 15, 916–925. [DOI] [PubMed] [Google Scholar]
- Darty K, Denise A, and Ponty Y (2009). VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio-Albrechtsen T, Kiontke K, Chiou S-Y, and Fitch DH (2006). Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis. Dev Biol 297, 74–86. [DOI] [PubMed] [Google Scholar]
- Delas MJ, and Hannon GJ (2017). lncRNAs in development and disease: from functions to mechanisms. Open Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S (2014). The four dimensions of noncoding RNA conservation. Trends Genet 30, 121–123. [DOI] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, and Han M (2005). The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in elegans. Mol Cell 19, 437–447. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, and Root DE (2014). Rational design of highly active sgRNAs for CRISPR-Cas9- mediated gene inactivation. Nat Biotechnol 32, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR (2011). Accelerated Profile HMM Searches. PLoS Comput Biol 7, e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers PB, Nonnekens J, Goos YJ, Betist MC, Viester MD, Mossink B, Lansu N, Korswagen HC, Jelier R, Brenkman AB, et al. (2015). A Long Noncoding RNA on the Ribosome Is Required for Lifespan Extension. Cell Rep. [DOI] [PubMed] [Google Scholar]
- Fatica A, and Bozzoni I (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15, 7–21. [DOI] [PubMed] [Google Scholar]
- Faunes F, Gundermann DG, Munoz R, Bruno R, and Larrain J (2017). The heterochronic gene Lin28 regulates amphibian metamorphosis through disturbance of thyroid hormone function. Dev Biol 425, 142–151. [DOI] [PubMed] [Google Scholar]
- Faunes F, and Larrain J (2016). Conservation in the involvement of heterochronic genes and hormones during developmental transitions. Dev Biol 416, 3–17. [DOI] [PubMed] [Google Scholar]
- Flynn RA, and Chang HY (2014). Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14, 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang Z, Li K, Gong L, Yang Q, Huang X, Hong C, Ding M, and Yang H (2017). Linc-DYNC2H1–4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death Dis 8, e2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, and Coller J (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K , et al. (2010). Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ (1977). Ontogeny and phylogeny (Cambridge, Mass: Belknap Press of Harvard University Press; ). [Google Scholar]
- Gray TA, Hernandez L, Carey AH, Schaldach MA, Smithwick MJ, Rus K, Marshall Graves JA, Stewart CL, and Nicholls RD (2000). The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics 66, 76–86. [DOI] [PubMed] [Google Scholar]
- Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, and Rajewsky N (2014). Unambiguous identification of miRNA: target site interactions by different types of ligation reactions. Mol Cell 54, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on- target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmanci AO, Sharma G, and Mathews DH (2011). TurboFold: iterative probabilistic estimation of secondary structures for multiple RNA sequences. BMC Bioinf 12, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Song Z, Chen H, Chen Z, Yang P, Li W, Yang Z, Zhang T, Wang F, Wei J, et al. (2018). Long noncoding RNA PVT1–214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S, and Bass BL (2008). A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci U S A 105, 12897–12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera RA, Kiontke K, and Fitch DH (2016). Makorin ortholog LEP-2 regulates LIN-28 stability to promote the juvenile-to-adult transition in Caenorhabditis elegans. Development 143, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum D, Zhang Y, Stuckenholz C, Labhart P, Alexiadis V, Martin R, Knolker HJ, and Fisher AL (2011). DAF-12 regulates a connected network of genes to ensure robust developmental decisions. PLoS Genet 7, e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, and Vanfleteren JR (2008). Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang H, and Zhang H (2011). The zinc-finger protein SEA-2 regulates larval developmental timing and adult lifespan in C. elegans. Development 138, 2059–2068. [DOI] [PubMed] [Google Scholar]
- Ji N, and van Oudenaarden A (2012). Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos WormBook : the online review of C elegans biology, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, and Slack FJ (2009). Dynamic expression of small noncoding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, and Segal E (2007). The role of site accessibility in microRNA target recognition. Nat Genet 39, 1278–1284. [DOI] [PubMed] [Google Scholar]
- Kim JH, Park SM, Kang MR, Oh SY, Lee TH, Muller MT, and Chung IK (2005). Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev 19, 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke KC, Felix MA, Ailion M, Rockman MV, Braendle C, Penigault JB, and Fitch DH (2011). A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol 11, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, and Mendell JT (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, and Hardin JD (2001). Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol 3, 983–991. [DOI] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J , et al. (2013). Control of somatic tissue differentiation by the long noncoding RNA TINCR. Nature 493, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, and Valen E (2016). CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44, W272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EW, Kim JH, Ahn YH, Seo J, Ko A, Jeong M, Kim SJ, Ro JY, Park KM, Lee HW, et al. (2012). Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat Commun 3, 978. [DOI] [PubMed] [Google Scholar]
- Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, Ha NC, Lane DP, and Song J (2009). Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J 28, 2100–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, and Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Li MA, Amaral PP, Cheung P, Bergmann JH, Kinoshita M, Kalkan T, Ralser M, Robson S, von Meyenn F, Paramor M , et al. (2017). A lncRNA fine tunes the dynamics of a cell state transition involving Lin28, let-7 and de novo DNA methylation. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kong X, and Chen F (2017a). Mkrn3 functions as a novel ubiquitin E3 ligase to inhibit Nptx1 during puberty initiation. Oncotarget 8, 85102–85109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Yu E, Chen S, Ma X, Lu Y, and Liu X (2017b). Spatiotemporal expression profiling of long intervening noncoding RNAs in Caenorhabditis elegans. Sci Rep 7, 5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Ambros V (1991). Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature 350, 162–165. [DOI] [PubMed] [Google Scholar]
- Mason D, Rabinowitz J, and Portman D (2008). dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135, 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, and Valen E (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42, W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, and Han M (2006). Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J 25, 5794–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Schwarz EM, Williams B, Schaeffer L, Antoshechkin I, Wold BJ, and Sternberg PW (2010). Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res 20, 1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, and Ambros V (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646. [DOI] [PubMed] [Google Scholar]
- Nam JW, and Bartel DP (2012). Long noncoding RNAs in C. elegans. Genome Res 22, 25292540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MD, and Fitch DH (2011). Overlap extension PCR: an efficient method for transgene construction. Methods Mol Biol 772, 459–470. [DOI] [PubMed] [Google Scholar]
- Nelson MD, Zhou E, Kiontke K, Fradin H, Maldonado G, Martin D, Shah K, and Fitch DH (2011). A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans. PLoS Genet 7, e1002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CQ, Hall DH, Yang Y, and Fitch DH (1999). Morphogenesis of the Caenorhabditis elegans male tail tip. Dev Biol 207, 86–106. [DOI] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U et al. (2009). Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet 41, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, and Seydoux G (2015). High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M , et al. (2017). H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis 8, e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AS, Killian DJ, and Hubbard EJ (2003). Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163, 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E , et al. (2009). Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet 41, 648–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, and Chang HY (2016). Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17, 47–62. [DOI] [PubMed] [Google Scholar]
- Raden M, Ali SM, Alkhnbashi OS, Busch A, Costa F, Davis JA, Eggenhofer F, Gelhausen R, Georg J, Heyne S , et al. (2018). Freiburg RNA tools: a central online resource for RNA-focused research and teaching. Nucleic Acids Res 46, W25–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, and Khavari PA (2018). The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, and Ruvkun G (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, and Moss EG (2013). Developmental transitions in C. elegans larval stages. Curr Top Dev Biol 105, 153–180. [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, and Rajewsky N (2014). A variety of dicer substrates in human and C. elegans. Cell 159, 1153–1167. [DOI] [PubMed] [Google Scholar]
- Salvatico J, Kim JH, Chung IK, and Muller MT (2010). Differentiation linked regulation of telomerase activity by Makorin-1. Mol Cell Biochem 342, 241–250. [DOI] [PubMed] [Google Scholar]
- Seggerson K, Tang L, and Moss EG (2002). Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol 243, 215–225. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, and Daley GQ (2013). Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, and Ambros V (2008). Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev 22, 1591–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorescu DA, Mohl M, Mann M, Backofen R, and Will S (2012). CARNA--alignment of RNA structure ensembles. Nucleic Acids Res 40, W49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Chen X, Zhao H, and Slack FJ (2015). A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans. RNA 21, 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, and Thomson JN (1980). The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol 78, 542–576. [DOI] [PubMed] [Google Scholar]
- Thornton JE, and Gregory RI (2012). How does Lin28 let-7 control development and disease? Trends Cell Biol 22, 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsialikas J, Romens MA, Abbott A, and Moss EG (2017). Stage-Specific Timing of the microRNA Regulation of lin-28 by the Heterochronic Gene lin-14 in Caenorhabditis elegans. Genetics 205, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsialikas J, and Romer-Seibert J (2015). LIN28: roles and regulation in development and beyond. Development 142, 2397–2404. [DOI] [PubMed] [Google Scholar]
- Vadla B, Kemper K, Alaimo J, Heine C, and Moss EG (2012). lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet 8, e1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, and Pasquinelli AE (2011). LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol 18, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, and Chang HY (2011). Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhu Y, Li S, Chen X, Jiang G, Shen Z, Qiao Y, Wang L, Zheng P, and Zhang Y (2016). Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting beta-catenin via Ezh2. Oncotarget 7, 25668–25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BP, Weaver YM, Mitani S, and Han M (2017). Coupled Caspase and N-End Rule Ligase Activities Allow Recognition and Degradation of Pluripotency Factor LIN-28 during Non- Apoptotic Development. Dev Cell 41, 665–673 e666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BP, Zabinsky R, Weaver YM, Lee ES, Xue D, and Han M (2014). CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. Elife 3, e04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Chen H, Dzakah EE, Yu B, Wang X, Fu T, Li J, Liu L, Fang S, Liu W , et al. (2019). Systematic evaluation of C. elegans lincRNAs with CRISPR knockout mutants. Genome Biol 20, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, and Eddy SR (2013). nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, and Ruvkun G (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Wu YC, Chen CH, Mercer A, and Sokol NS (2012). let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell 23, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al. (2013). Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 4, 2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M , et al. (2016). LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 19, 66–80. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. (2010). Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet 42, 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, and Yeo GW (2010). Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.