Abstract
Tissue macrophages rapidly recognize and engulf apoptotic cells. These events require the display of ‘eat-me’ signals on the apoptotic cell surface, the most fundamental of which is phosphatidylserine (PtdSer). Externalization of this phospholipid is catalyzed by scramblase enzymes, several of which are activated by caspase cleavage. PtdSer is detected both by macrophage receptors that bind this phospholipid directly, and by receptors that bind a soluble ‘bridging protein’ that is independently bound to PtdSer. Prominent among the latter receptors are the Mer and Axl receptor tyrosine kinases. Eat-me signals also trigger macrophages to engulf virus-infected or metabolically traumatized, but still living cells; and this ‘murder by phagocytosis’ may be a common phenomenon. Finally, the localized presentation of PtdSer and other eat-me signals on delimited cell surface domains may enable the phagocytic pruning of these ‘locally dead’ domains by macrophages, most notably by microglia of the central nervous system.
Introduction
In long-lived organisms, abundant cell types are often short lived. In the human body for example, the lifespan of many white blood cells — including neutrophils, eosinophils, and platelets — is less than two weeks. For normal healthy humans, a direct consequence of this turnover is the routine generation of more than 100 billion dead cells each and every day of life1,2. This macroscopic mass of cell corpses, which is largely the product of apoptosis, must be recognized and cleared. These quotidian functions are carried out continuously, in a ‘silent’ non-inflammatory fashion, by tissue-resident macrophages, the dedicated undertakers of the immune system3. These often highly specialized cells mediate tissue homeostasis in all organs, and include marginal zone macrophages of the spleen, Kupffer cells of the liver, alveolar macrophages of the lungs, Langerhans cells of the skin, and microglia of the central nervous system4. In settings of fulminant infection or severe tissue trauma where cells may also die by immediate necrosis, the dead cell burden reaches even higher levels, but tissue-resident macrophages are again mobilized to eat these cells. Although apoptosis and necrosis are morphologically and physiologically distinct death processes — apoptotic cells shrink and their plasma membranes bleb but remain intact, whereas necrotic cells swell and their plasma membranes rupture5 — the principal phagocytes that deal with both dead cell types are macrophages.
As apoptosis accounts for the bulk of the everyday dead cell burden, this Review focuses on recent findings with respect to the phagocytosis of apoptotic cells by tissue-resident macrophages — a process termed ‘efferocytosis’, from the Latin efferre, to carry to the grave6 — and on the molecules that mediate this process. Efferocytosis is a remarkably efficient business: macrophages can engulf apoptotic cells in less than 10 minutes, and it is therefore difficult experimentally to detect free apoptotic cells in vivo, even in tissues where large numbers are generated7. Many of the molecules that macrophages and other phagocytes use to recognize dead cells are themselves the products of apoptosis, and are often generated via the action of the cysteine-aspartic acid proteases, most notably caspase 3 and caspase 7, that are the ultimate executioners of apoptotic signalling cascades8. Since defects in efferocytosis have significant consequences for tissue integrity, homeostasis and function, and can lead to the development of autoimmunity9,10, identifying these molecules and elucidating their mechanisms of action is of genuine importance to understanding and treating human disease.
Finding dead cells
Find-me signals.
Many macrophages are embedded within tissues in which apoptosis occurs continuously (for example, the neurogenic regions of the adult brain)11,12 or in which dead cells are delivered directly to phagocytes by the circulation (for example, the marginal zones of the spleen)13. In other settings, however, tissue-resident and blood-borne macrophages, or the processes of these cells, appear to be recruited and to migrate to apoptotic cell loci in response to cues. This may also be the case for tissues in which the number of resident macrophages is low relative to that of the cells surrounding them. In these settings, macrophages in vitro, and to a much more limited extent in vivo, have been shown to respond to a set of so-called ‘find-me’ signals14,15, released by injured and dying cells, and to use these signals to migrate toward sites of cell death. The most-studied find-me signals have been, firstly, the lysophospholipids lysophosphatidylcholine (LPC)16,17 and sphingosine-1-phosphate (S1P)18, and secondly, the nucleotides ATP, AMP, and UTP19. LPC production is triggered by caspase 3 activation of phospholipase A2 (PLA2)16. PLA2 generates LPC via partial hydrolysis (removal of one of the fatty acid chains) of phosphatidylcholine, and so its activation by caspase-mediated cleavage may be an intrinsic component of the apoptotic cascade. LPC release from cells is dependent on the ATP-binding cassette transporter 1 (ABCA1)20. This is consistent with the fact that ABCA1 is a homologue of ced-7, which was among the first gene products identified in genetic screens for mutations that perturb the engulfment of dead cells in Caenorhabditis elegans21.
ATP, AMP, and UTP are released from apoptotic but still intact cells via a mechanism that may also be caspase-dependent. This release involves export of the nucleotides through connexin-like Pannexin-1 channels in the plasma membrane, the gating of which is controlled by cleavage of the C-termini of oligomeric Pannexin-1 subunits by caspase 3 and caspase 722. These auto-inhibitory C-termini normally occlude the channel, and their proteolytic removal in mid-to-late apoptosis therefore opens Pannexin-1 for nucleotide transport23–25.
In addition to lysophospholipids and nucleotides, several other find-me signals have been investigated, although most analyses have been carried out in cells in culture, and the case for the in vivo biological relevance of many of these additional signals is therefore less clear. Among the strongest candidates is the chemokine CX3CL1 (also known as fractalkine), which has been implicated as a find-me signal that mediates macrophage chemotaxis towards apoptotic B cells in germinal centres26.
Find-me receptors.
Macrophages are thought to detect and respond to find-me signals using an array of receptor systems that have been reviewed previously15. For lysophospholipids, the G protein coupled receptor (GPCR) G2A appears to play a role in the chemotactic response of macrophages to LPC27, although the extent to which LPC binds to G2A is unclear, and the precise pathway that transduces LPC signalling in macrophages remains to be elucidated. There are five S1P receptors in the mouse, but only limited data as to which of these might control macrophage chemotaxis in vivo28. For nucleotides, tissue macrophage populations express several different P2Y purinoceptors, which are GPCRs that bind ATP, ADP, UTP, and UDP29,30. These include the P2Y2, P2Y6, P2Y12, P2Y13, and P2Y14 receptors, some of which show expression that is highly restricted to specific macrophage subsets31,32. The Gi-coupled P2Y12 receptor, for example, is very abundantly expressed in CNS microglia, where it plays important roles in the earliest stages of microglial chemotaxis and process extension30,33, but is not expressed by most other tissue macrophages. As such, P2Y12 is now used as a specific marker of ‘homeostatic’ as opposed to activated microglia33,34. How various receptors might mediate a response to gradients of find-me signals such that macrophages move in the right direction; that is, toward increasingly higher levels of a find-me signal, remains controversial and unresolved35; and in vivo gradients of these signals (in tissues where apoptotic cells are cleared) have not been quantified.
Distinguishing live cells from dead
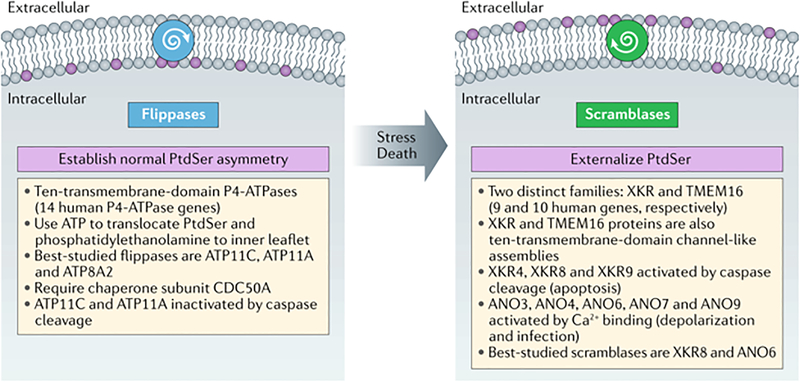
Phosphatidylserine, flippases, and scramblases.
Once macrophages are close enough to actually touch apoptotic cells, they rely on the expression of a set of cell surface molecules that tag these cells as dead. These tags are the so-called ‘eat-me’ signals for phagocytosis. Multiple eat-me candidates have been advanced (discussed below), but the most ubiquitous, efficacious, pleiotropic, and important of these is, without a doubt, phosphatidylserine (PtdSer)1,36,37. This humble glycerophospholipid is a component (at varying levels) of many different membranes – including those of the endoplasmic reticulum, the mitochondria, the Golgi apparatus, and the plasma membrane – within each and every cell of the body. Given this, it is difficult, a priori, to imagine how PtdSer might function as a signal for efferocytosis – or anything else. It can do so only because it is normally highly polarized with respect to its membrane bilayer localization (Box 1). In the plasma membrane of healthy cells, for example, nearly 100% of PtdSer is confined to the inner, cytoplasm-facing leaflet of the bilayer. (Phosphatidylinositol and phosphatidylethanolamine are also highly enriched in the inner leaflet.) Except in the special circumstances discussed below, PtdSer is never seen by the extracellular world38,39. When it is externalized on the plasma membrane surface, PtdSer serves as a signal — most generally to indicate that the cell has died by apoptosis or is en route to this end. As discussed below, externalized PtdSer is recognized by several different ligand/receptor systems on macrophages, but all of these systems function based on their detection of this everyday phospholipid.
Box 1. Stringent regulation of phosphatidylserine localization in the plasma membrane.
Mammalian genomes encode many 10-transmembrane-domain, channel-like enzymes that are devoted to the intra-bilayer translocation and localization of phosphatidylserine (PtdSer) and other phospholipids1,38–40,42,50,56. Although some members of the indicated families display other activities — the TMEM16A and TMEM16B proteins, for example, are Ca2+-activated chloride channels — many are catalytically active phospholipid translocases. A subset of ATP8 and ATP11 proteins, notably ATP11C, use ATP hydrolysis to establish the pronounced asymmetric distribution of PtdSer and other phospholipids seen at the plasma membrane (and internal membranes) of all healthy cells. Importantly, nearly 100% of PtdSer is normally confined to the cytoplasmic leaflet of the plasma membrane bilayer. Scramblases are ATP-independent enzymes that move PtdSer to the extracellular leaflet, where it is displayed to the external world. Several XKR scramblases, notably XKR8, are activated by caspase cleavage and are thought to function during apoptosis. Several TMEM16 scramblases, notably TMEM16F, are activated by Ca2+ binding subsequent to membrane depolarization and other signaling events.
The remarkable PtdSer membrane asymmetry of healthy cells is established by the action of a family of ion-channel-like, 10-transmembrane-domain phospholipid translocases commonly referred to as ‘flippases’40,41 (Box 1). These flippases are P4-type ATPases. There are 14 such intramembrane ATPases in human cells, distributed among different tissues and membrane compartments40,42; and several of these proteins — including ATP8A1, ATP8A2, ATP10A, ATP11A, and ATP11C — have been found to catalyze the translocation of phospholipids41,43,44. In general, flippases display substrate specificity either for aminophospholipids, such as PtdSer and phosphatidylethanolamine, or alternatively, for phosphatidylcholine41. At the plasma membrane, ATP11A and ATP11C flip essentially all of the PtdSer to the inner leaflet of the plasma membrane bilayer45,46. In some settings, flippase expression has been shown to be essential for the normal development of cell lineages. Mice that lack ATP11C, for example, present with a severe B cell deficiency that results from the fact that ATP11C-deficient B cell precursors display high levels of surface PtdSer, and are therefore aberrantly recognized and eaten by macrophages47. Flippases require a chaperone, generally CDC50A, in order to take up residence at their appropriate membrane locations41, and cells that lack CDC50A have been shown to lose all plasma membrane flippase activity and to constitutively expose PtdSer on the cell surface46,48.
If flippases set up PtdSer bilayer asymmetry, how is this asymmetry ever disrupted? Although a subset of flippases, notably ATP11A and ATP11C, are inactivated by caspase cleavage during the course of apoptosis45,46, this inactivation alone is insufficient for the exposure of PtdSer on the cell surface, as the inner-to-outer transmembrane exchange of any phospholipid has a high energy barrier (15–50 kcal/mol) and thus does not occur spontaneously49,50. Instead, the movement of PtdSer from the inner to the outer leaflet of the plasma membrane requires yet another set of phospholipid translocases – so-called ‘scramblases’ – which catalyze this reverse translocation (Box 1). It is these ATP-independent enzymes that allow PtdSer to act as a signal. There are now thought to be two major classes of scramblases — those of the TMEM16 and XKR families51–53. The latter has at least three members, XKR4, XKR8, and XKR9, that are cleaved and activated by caspase-3 and/or caspase-7 during apoptosis54,55, and correspondingly, caspase inhibitors antagonize PtdSer externalization by these XKR scramblases. XKR8 is thought to be especially important for the externalization of PtdSer on the plasma membrane of apoptotic cells52. A large second set of scramblases, those of the transmembrane protein 16 (TMEM16) family, are Ca2+-activated56–58. Also referred to as Anoctamins (they were originally thought to be anion channels with 8 transmembrane helices), these 10-transmembrane domain proteins are particularly interesting. Although two family members — anoctamin 1 (ANO1) and (ANO2) — appear to function exclusively as Ca2+-activated ion (chloride) channels, there is now good evidence that ANO3, ANO4, ANO5, ANO7, ANO9, and especially ANO6 (also known as TMEM16F) function as Ca2+-dependent phospholipid scramblases50,51,53,58. TMEM16F plays a key role in PtdSer externalization on activated platelets during blood coagulation59. As discussed below, the localized activation of XKR and/or TMEM16 family scramblases may allow for the localized externalization of PtdSer on only a small segment of the plasma membrane, as opposed to across the entire surface of the cell.
Receptors for apoptotic cell uptake
TIM4.
As PtdSer is the only universal apoptotic cell-intrinsic eat-me signal, the most intensively studied of the macrophage cell surface proteins that mediate apoptotic cell recognition are those that bind, either directly or indirectly, to this phospholipid. Prominent among these are the transmembrane receptors of the T cell immunoglobulin- and mucin-domain-containing molecule (TIM)60 and Tyro3, Axl and Mer (TAM)61 families (Fig. 1). The TIM family has three members in the human genome and eight in the mouse, but the best-studied of these with respect to macrophage efferocytosis is TIM4 (Fig. 1). TIM4 is a heavily glycosylated single transmembrane protein with a short (42-residue) cytoplasmic tail62. It binds PtdSer — but not phosphatidylcholine, phosphatidylinositol, or phosphatidylethanolamine — tightly and directly, with low nanomolar affinity, in a Ca2+-dependent reaction. When transduced into heterologous cells, such as NIH3T3 cells and mouse embryonic fibroblasts, TIM4 strongly supports efferocytosis of apoptotic cells62. This activity requires a secondary intracellular signalling transducer, as the short cytoplasmic domain of TIM4 is dispensable for signalling in response to PtdSer binding. TIM4 is presumed to use the TAM tyrosine kinase activities for signal transduction, since when it is introduced into fibroblasts that lack a TAM receptor (see below) it is unable to support efferocytosis63. Similarly, phagocytosis studies using PtdSer-coated beads and AD293 cells that heterologously expressed TIM4 suggested that TIM4 may also use β1, β3, and β5 integrins as co-receptors, and in turn, the cytoplasmic signalling proteins that are associated with these integrins (for example, Src-family kinases), as downstream intracellular effectors64. As discussed below, the β3 and β5 subunits also play important roles in other PtdSer-dependent phagocytosis pathways.
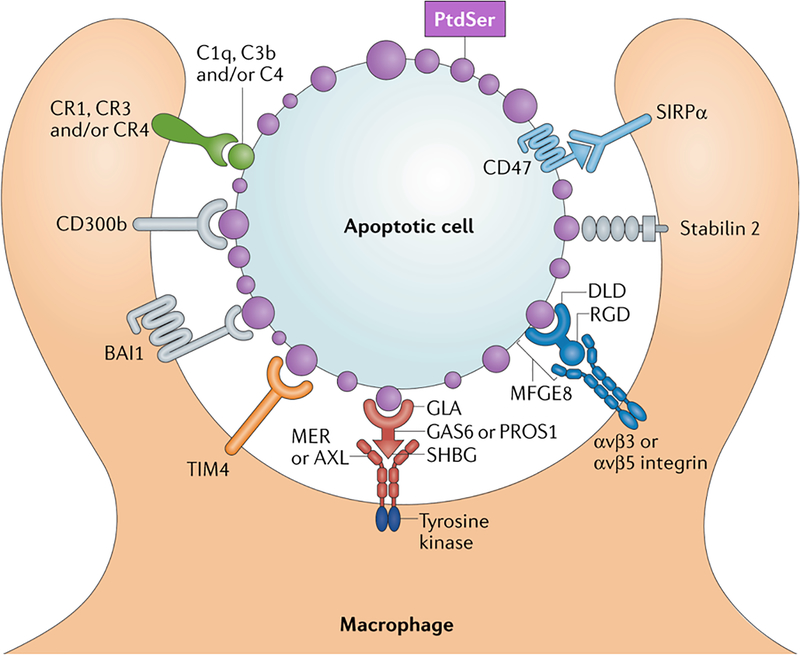
Figure 1. Receptors and ligands mediating apoptotic cell (AC) recognition and phagocytosis by macrophages.
Apoptotic cell surfaces are marked by a profusion of membrane blebs that universally display externalized phosphatidylserine (PtdSer, purple). PtdSer – the most potent ‘eat-me’ signal for apoptotic cell phagocytosis – is recognized by macrophage receptors that directly bind this phospholipid – including CD300b, BAI1, TIM4 (green), and Stabilin-2. PtdSer is also recognized by soluble, bi-functional ‘bridging’ proteins, including Gas6/Pros1 (red) and MFG-E8 (blue). These proteins carry one domain that binds PtdSer – the ‘Gla’ domains of Gas6/Pros1 and the Discoidin-like domain (Dld) of MFG-E8 – and a second domain that binds to phagocytic receptors expressed by macrophages – the SHBG domain of Gas6/Pros1 and an RGD motif within the second EGF-like domain of MFG-E8. The macrophage receptors for Gas6/Pros1 are the TAM receptor tyrosine kinases Mer and Axl (red), while those for MFG-E8 are αvβ3 and αvβ5 integrin dimers (blue). Complement proteins, including C1q, C3b, and C4 (closed gray circle), also decorate the surface of apoptotic cells by mechanisms that remain under study, but may involve the recognition of PtdSer. These complement factors are recognized by the complement receptors CR1, CR3, and CR4. Some apoptotic cells also express CD47 (light blue), which negatively regulates phagocytosis by acting as a ‘don’t-eat-me’ signal. Its receptor is SIRPα. All phagocytic receptors must engage signal transduction networks that ultimately activate Rho family GTPases, but the only receptors that carry intrinsic signaling activity are the TAM receptors, which are robust, ligand/apoptotic cell-activated tyrosine kinases (TKs).
AlthoughTIM4 is expressed in large peritoneal macrophages, which are frequently studied due to their ease of isolation, it is not detectably expressed by several other highly phagocytic macrophage populations, including alveolar macrophages and microglia32, and so its stimulation of efferocytosis is not common to all macrophages. Mice deficient in TIM4 do not display detectable accumulation of apoptotic cells in the spleen or other tissues, and do not present with splenomegaly or lymphadenopathy, although modestly elevated circulating levels of anti-dsDNA autoantibodies have been reported65,66. The related TIM1 and TIM3 proteins, which are prominently expressed in T helper (Th2) cells, plasmacytoid DCs, and Th1 cells, have also been reported to bind PtdSer62,67–69, but these proteins are not generally expressed by macrophages.
TAM receptors and their ligands.
The TAM proteins Tyro3, Axl, and Mer (gene name Mertk) are cell surface receptor tyrosine kinases (RTKs)61,70. These receptors do not bind PtdSer directly, but instead rely on their activating ligands — growth arrest specific Gas6 and Protein S (Pros1) – for this activity (Fig. 1)61,71–74. As such, Gas6 and Pros1 may be viewed as co-receptors that act in concert with the TAM RTKs to mediate PtdSer recognition and signalling. The C-terminal sex hormone-binding globulin (SHBG) domains of the ligands bind to the extracellular domains of the TAMs, while their N-terminal γ-carboxyglutamic acid (GLA) domains bind to PtdSer61,70,71,75,76. Gas6 binds and activates all three TAMs, whereas Pros1 binds and activates only Tyro3 and Mer71. In a very unusual arrangement, Gas6 is constitutively bound to Axl in tissues in vivo, and moreover, is entirely dependent on Axl for its stable expression in these tissues74,77. Both Gas6 and Pros1 only function as effective activators of TAM tyrosine kinase activity and drivers of phagocytosis when, first, they are simultaneously bound to PtdSer via their GLA domains and to a TAM receptor via their SHBG domains; second, the γ carbons of glutamic acid residues within their GLA domains are carboxylated in a vitamin K-dependent reaction; and third, 6–7 divalent cations (probably Ca2+) are also bound to the GLA domains61,71,73,78. In this way, the TAM ligands ‘bridge’ a TAM receptor-expressing phagocyte (for example, a macrophage) to a PtdSer-expressing phagocytic target (for example, an apoptotic cell).
The TAM receptors and their ligands are the most broadly expressed PtdSer recognition system in macrophages. Unlike TIM4, Mer appears to be universally expressed on all phagocytic macrophages at steady state31,32, and antibodies to Mer and the high-affinity Fcγ receptor are now routinely used as a marker pair to define and sort these cells. Multiple studies have shown that Mer is a critical mediator of efferocytosis by tissue macrophages throughout the body11,79–84. Axl is more restricted in its macrophage expression under basal conditions – to red pulp macrophages, Kuppfer cells, and alveolar macrophages, among others74. However, Axl expression is markedly up-regulated in all macrophages by most inflammatory stimuli, including type I interferons and interferon-γ (IFNγ), lipopolysaccharide (LPS), and poly I:C74,80. Dramatic Axl up-regulation is also seen when tissue macrophages in vivo are activated by trauma, disease, or viral infection11,80. Similarly to other RTKs, including MET, ERBB4 and EPHB2, Axl exhibits ‘shedding’ of a soluble version of its extracellular domain, which is generated via the action of ADAM family metalloproteases, subsequent to tyrosine kinase activation86,87. As such, soluble Axl in the blood, generated by proteolytic cleavage of the Axl ectodomain subsequent to receptor activation, is now commonly used as an inflammation indicator85.
Thus, macrophage Mer is thought to handle the bulk of steady state apoptotic cell phagocytosis that is associated with continuous tissue turnover and homeostasis, whereas Axl participates in apoptotic cell phagocytosis during the resolution of inflammation subsequent to infection and tissue trauma74,80. Correspondingly, the two receptors frequently — though not always — display a ‘yin and yang’ relationship with respect to their regulation in macrophages: when one goes up, the other tends to go down74. This is also true with respect to their expression in most DCs, where Axl is more prominent than Mer74,88. Tyro3 expression in macrophages is very limited, but this RTK is expressed in select DC populations, where it plays a role in the inhibition of type 2 immunity74,89. Importantly, mice with mutations in TAM receptor genes, most notably loss-of-function mutants in the Mertk gene, show prominent accumulation of apoptotic cells in multiple tissues — and develop splenomegaly, lymphadenopathy, autoantibodies, glomerular nephritis, rheumatoid arthritis and broad-spectrum autoimmune disease — in the absence of any other overt perturbation or challenge11,70,71,76,90–92.
Also unlike TIM4, the TAM RTKs do not need an accessory intracellular signal transducer to promote phagocytosis, as they carry very strong tyrosine kinases74,88. Indeed, the kinase activities of Mer and Axl are absolutely required for their stimulation of apoptotic cell phagocytosis by macrophages74. The TAMs are not mere signal transducers, however, since their kinase activity alone is not sufficient to stimulate efferocytosis. The TAM kinases can be strongly activated artificially using extracellular domain antibodies that cross-link the receptors in the absence of Gas6 or Pros1, but this artificial activation has no stimulatory effect on the phagocytosis of apoptotic cells by cultured bone marrow-derived macrophages in the absence of a TAM ligand74. It appears that in order for the TAM system to function in efferocytosis, the entire assembly of PtdSer exposure on apoptotic cells, TAM ligand bridging between apoptotic cells and macrophages, and TAM receptor kinase activation within macrophages must operate (Fig. 1). As noted at the outset, in most settings, routine efferocytosis is immunologically silent, in that it is not associated with the secretion of inflammatory cytokines. The TAM receptors also appear to play an essential role in this effect, as the activation of Mer and Axl in macrophages and DCs has repeatedly been found to be potently immunosuppressive83,88,91,93–95.
MFG-E8 and integrins.
In a manner that is very much analogous to the TAM ligands Gas6 and Pros1, the soluble extracellular matrix glycoprotein MFG-E8 (milk fat globule-EGF factor 8, also named lactadherin)96 functions during efferocytosis to bridge PtdSer on the surface of apoptotic cells to receptors that are expressed by phagocytic macrophages97,98 (Fig.1). In this case, these receptors are the αvβ3 and αvβ5 integrins, which bind to MFG-E8 via an Arg-Gly-Asp (RGD) motif located within its second EGF-like repeat. MFG-E8 ‘bridging’ of these macrophage-expressed integrins to PtdSer on the surface of apoptotic cells is mediated by a C-terminal discoidin-like domain rather than a Gla domain (which is not present in the protein)99,100 (Fig.1). Both MFG-E8 and the integrin proteins to which it binds are widely expressed in many cell types in addition to macrophages and their phagocytic targets, but introduction of these proteins into heterologous cells often strongly potentiates efferocytosis97. Consistent with its activity and expression, mice that lack MFG-E8 can display apoptotic cell accumulation in the spleen and lymph nodes, and often go on to develop splenomegaly, nephritis, autoantibodies and a systemic lupus erythematosus (SLE)-like autoimmune disease98,101. Recent studies suggest that glucocorticoid-mediated amelioration of autoimmunity in mice may be dependent on the ability of glucocorticoids to up-regulate MFG-E8 expression102.
Additional ‘eat-me’ signals and receptors.
There are additional receptor systems that have been found to play a role in direct PtdSer binding and recognition, and consequently, in the efferocytosis of apoptotic cells by select subsets of tissue macrophages (Fig. 1). These include the single Ig-domain type I transmembrane protein CD300b103, a member of the large CD300 receptor family104, which is expressed by peritoneal macrophages and neutrophils, but not microglia. This protein also binds PtdSer directly, is localized to the phagocytic cup of engulfing macrophages, and uses the adaptor protein DAP12 to nucleate intracellular signal transduction events that mobilize F-actin for phagocytosis103. Similarly, brain-specific angiogenesis inhibitor 1 (BAI1), an adhesion-related GPCR with a relatively large extracellular domain that carries an RGD motif and multiple thrombospondin type I repeats, has been reported to bind PtdSer, cardiolipin, phosphatidic acid, sulfatide and LPS, and to promote the phagocytosis of apoptotic cells (and Gram-negative bacteria) by coupling to downstream effectors105,106. While BAI1 is highly expressed in the brain, its expression in many macrophage populations is very low; and although mice deficient in BAI1 exhibit deficits in synaptic plasticity107, they do not, except upon challenge, display significant apoptotic cell accumulation in most tissues where clearance is mediated by macrophages108. An additional direct PtdSer binder implicated in apoptotic cell clearance, notably that of spent erythrocytes, is the large, 19-EGF-domain-containing, transmembrane protein Stabilin-2109,110. While highly expressed in red pulp macrophages and present in Kuppfer cells, it is not generally expressed by tissue macrophages.
Finally, there are several sets of plasma proteins that have been shown to decorate the surface of apoptotic cells and have been tied to efferocytosis. Among the most intriguing of these are proteins of the complement cascade, notably C1q and C3b111–113. C1q is a very large, 18-chain hexameric plasma glycoprotein assembly, structurally similar to mannose binding lectin and ficolins, which is an essential upstream activator of the classical complement cascade114. As might be expected given its size and abundance in serum, C1q has been reported to associate with many molecules (for example, β amyloid, calreticulin, fibronectin, DNA) in addition to its well-described binding to IgM and IgG immune complexes114. There are conflicting reports as to its ability to bind to PtdSer, and the macrophage/phagocyte receptors for C1q remain the subject of investigation114,115. C1q has nonetheless received considerable attention, as the majority of human patients with C1q deficiency eventually develop lupus116, and C1q binding to macrophages in culture is immunosuppressive117. C3b has also been found to decorate the surface of apoptotic cells, and to account for much of the ability of serum to potentiate apoptotic cell phagocytosis118,119. The phagocyte receptors for C3b engagement are thought to be CR3 (CD11b/CD18; αMβ2 integrin) and CR4 (CD11c/CD18; αxβ2 integrin)114. Interestingly, reduced plasma levels of the TAM RTK ligand Pros1 are observed in patients with SLE that have serositis, haematological, neurological and immune disorders, and these reduced Pros1 levels are in turn correlated with reductions in plasma levels of C3, which is consistent with complement consumption, in these patients120.
Don’t eat me.
Although most attention has been focused on signals that promote efferocytosis, a countervailing, phagocytosis-inhibiting activity, or ‘don’t eat-me’ signal, has been proposed to function as a negative regulator of engulfment. This activity is exhibited by the transmembrane, immunoglobulin-related cell surface protein CD47121 (Fig. 1). In addition to interacting with integrins and thrombospondin-1 to regulate neutrophil migration, neuronal axon growth and T cell co-stimulation, CD47 has been found to bind to signal regulatory protein alpha (SIRPα) on macrophages to inhibit phagocytosis122,123. This inhibition appears to require phosphorylation of immunoreceptor tyrosine-based inhibition motifs (ITIMs) in the SIRPα cytoplasmic domain, leading to the activation of the SHP1 and SHP2 phosphatases124, although the precise mechanism of inhibition remains the subject of investigation125. Tumour cells have frequently been found to up-regulate CD47 expression as a potential mechanism of immune evasion126, and antibodies that target the CD47–SIRPα interaction have shown efficacy as therapeutics in some mouse pre-clinical cancer models127,128. Correspondingly, four of these antibodies are now in clinical trials as therapeutics for haematological malgnancies129. The relative power of CD47 as a don’t eat-me signal versus PtdSer as an eat-me signal has not been measured, but it should be noted that B cell precursors deficient in the ATP11C flippase (mentioned above), which strongly express both signals, are readily engulfed47. Apart from the efferocytosis of apoptotic cells, the CD47–SIRPα axis has also recently been implicated in the negative regulation of more local membrane excision events carried out by microglia during ‘synaptic pruning’ in the postnatal brain130, as discussed below.
Phagocytic attack and engulfment
Efferocytosis requires: one, the engagement of signal transduction pathways that activate RAC1–CDC42 GTPases and dramatically re-organize the macrophage actin cytoskeleton; two, the extension of a phagocytic cup around the target cell; three, the internalization of this doomed cell into the macrophage; four, the routing of the internalized cargo to endosomes and lysosomes; and five, its eventual enzymatic degradation and turnover131–134. The genetic and biochemical pathways underlying these intracellular engulfment events are beyond the scope of this Review, but have recently been discussed135–137. The remainder of this Review will focus on our growing understanding of how it is not just dead and dying cells that are targeted by macrophages, but also, in some settings, the living.
Eaten alive.
Do eat-me signals only trigger macrophage phagocytosis of dead cells? Indeed, they do not. Recent findings indicate that macrophages do not always wait for cells to die before initiating phagocytic attack, and may kill living cells by eating them, if these cells express sufficient levels of externalized PtdSer. These findings harken back to much earlier observations in C. elegans, in which mutation of engulfment genes, including ced-6 and ced-7, whose products are required for the phagocytosis of dead and dying cells, led to the long-term survival of ‘half-dead’ embryonic cell populations (expressing a hypomorphic ced-3 allele) that would normally have been eliminated138,139. The recent findings support the hypothesis that scramblase activation and the externalization of PtdSer may result in a ‘murder by phagocytosis’ phenomenon that is both PtdSer- and TAM-dependent (Fig. 2). For example, the precursor B cells that are aberrantly cleared, by macrophages, in mice deficient for the ATP11C flippase, are alive at the time of their engulfment; and genetic and tyrosine kinase inhibitor analyses indicate that this fatal engulfment is dependent on the action of Axl and Mer47. In the adult brain, when dividing progenitor cells in the subventricular zone (SVZ) are pulse-labelled with BrdU during cell division, many more healthy, functional, BrdU-labelled neurons are observed in the olfactory bulb, to which newborn SVZ cells migrate, one month after labeling in Axl−/−Mertk−/− mice than in wild-type11. Conditional inactivation of the Mertk gene only in microglia again indicates the cells responsible for this effect are the brain’s macrophages11. Similarly, adenovirus-transduced astrocytes have been shown to transiently externalize PtdSer after infection. Over the course of the next several days, many of these PtdSer-displaying transduced cells are phagocytically cleared by activated microglia140. This phagocytosis is not observed in Axl−/−Mertk−/− mutants or when PtdSer externalization is inhibited140. Importantly, these cleared cells are unambiguously alive at the time of phagocytosis, since the surviving transduced cells seen in the Axl−/−Mertk−/− mutants persist as healthy functioning astrocytes for many months after infection – if they are not eaten140 (Fig. 2).
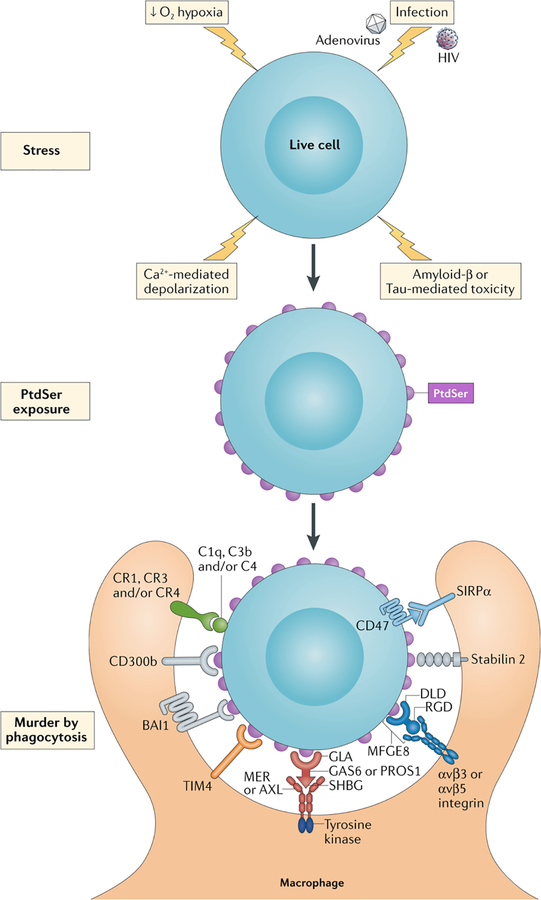
Figure 2. PtdSer-triggered phagocytic engulfment of living cells.
Recent findings indicate that a variety of stressors - including hypoxia precipitated by stroke, calcium influx triggered by excitatory and/or exitotoxic stimulation (depolarization), infection by HIV and adenoviruses (AdV), and the toxic effects of Amyloid β (Aβ) and phospho-Tau deposition (top) – can lead to the activation of XKR and TMEM16 scramblases and the exposure of PtdSer on the surface of living cells (middle). In some settings, this leads to the engulfment of stressed but still living cells – or murder by phagocytosis (bottom).
These findings of microglial murder by phagocytosis are also consistent with analyses of tissue recovery following focal brain ischemia, in which areas of neuronal atrophy were found to be markedly reduced in size, and functional recovery to be enhanced and accelerated, in Mertk−/− mice relative to wild-type animals141 (Fig. 2). They may also be relevant to neurodegenerative disease, since neurons in the P301S-tau mouse model of frontotemporal dementia were recently shown to externalize PtdSer and, as a consequence, to be phagocytosed by microglia while still alive142. In this instance, MFG-E8 appears to play an important role in microglial recognition of the externalized PtdSer142 (Fig. 2). This may also be the case for microglial phagocytosis of viable neurons during brain inflammation, since neuronal loss subsequent to in vivo LPS injections is also reduced in Mfge8−/− mice relative to wild-type143. Finally, recent work suggests that macrophage engulfment of HIV-1-infected but still living CD4+ T cells can occur, and that this engulfment of live cells is TAM-dependent, since it is triggered by; one, PtdSer externalization after infection; two, Pros1 or Gas6 binding to this externalized PtdSer; and three, engagement of macrophage Mer by these bound TAM ligands144. It is possible that the phagocytic killing of PtdSer-expressing but nonetheless living cells by microglia and other tissue macrophages, a process that is sometimes referred to as ‘phagoptosis’145,146, may be a common and numerically substantial phenomenon whose general significance has not heretofore been widely appreciated.
Phagocytosis of membrane segments.
There are select settings in which only small membrane segments of cells, as opposed to entire cells, are phagocytically engulfed. The best-understood of these selective ‘pruning’ phenomena occurs in the retina. For a few hours each morning around subjective dawn, the retinal pigment epithelial (RPE) cells of the eye, which are not macrophages but rather neuroectoderm-derived epithelia, nibble off the distal ends of photoreceptor outer segments147,148 (Fig. 3). This localized phagocytosis disposes of toxic oxidized proteins and lipids that are produced by reactive oxygen species during phototransduction; these oxidized products normally accumulate in the outer segments and would kill the photoreceptors if not removed. Students used to be taught that photoreceptors ‘shed’ the distal tips of their outer segments on a daily basis, but this is not entirely correct: these tips are instead actively eaten by RPE cells, the most vigorous phagocytes in the body148.
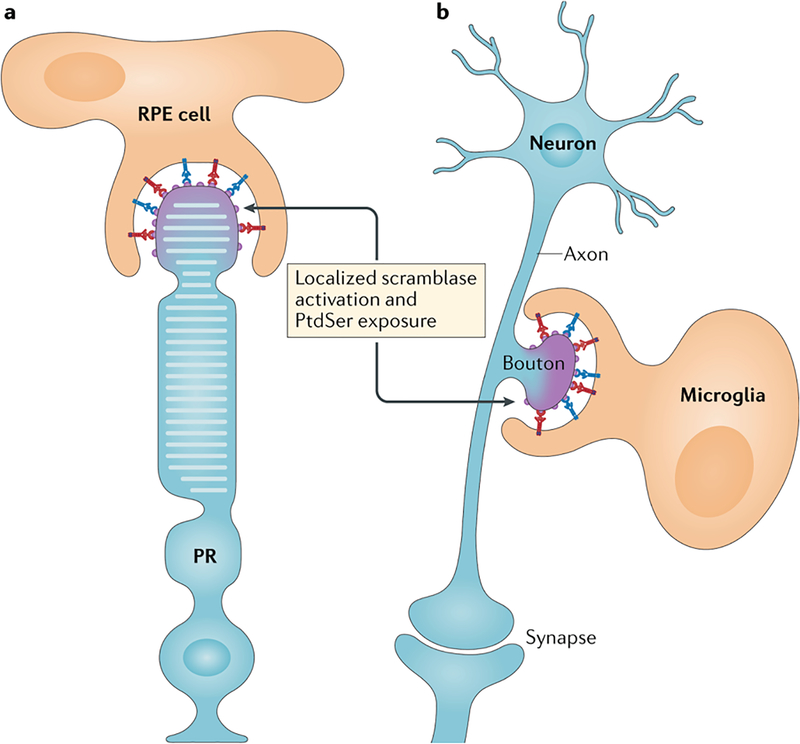
Figure 3. PtdSer-delimited phagocytosis of only small parts of cells.
a. The phagocytic retinal pigment epithelial (RPE) cells of the eye pinch of and engulf the distal ends of the rhodopsin-containing outer segments of photoreceptors (PR) each morning. This localized phagocytosis is entirely Mer-dependent (red) and is assisted by the MFG-E8 integrin system (blue). Patients with complete loss-of-function MERTK mutations have an inherited form of retinitis pigmentosa due to a failure in RPE phagocytosis. b. Microglia, the tissue macrophages of the brain, phagocytose unpaired pre-synaptic axonal boutons of the postnatal brain (those that are not part of an anatomically complete, functional synapse), in a process termed ‘synaptic pruning’. These boutons are decorated by several of the eat-me signals highlighted in Fig. 1, including complement proteins (gray). Importantly, the extent of the bouton to be pruned may again be delimited by the local externalization of PtdSer on its surface. The localized activation of scramblase activity for the events depicted in a and b is at this point a speculation.
This RPE phagocytosis displays an absolute requirement for Mer: RPE cells express Mer (and Tyro3), and mice, rats and humans with complete loss-of-function mutations in MERTK are blind due to a failure in RPE phagocytosis and the consequent death of most photoreceptors149–152. Many different human MERTK mutations account for inherited forms of retinitis pigmentosa151, and clinical trials involving subretinal injection of Mertk-expressing adeno-associated virus vectors have been initiated in patients carrying these mutations153. Gas6 and Pros1 function in concert as Mer ligands for this RPE-mediated phagocytosis154. The MFG-E8–αvβ5 integrin system highlighted above also plays a role, albeit a secondary one, in RPE phagocytosis of these photoreceptor outer segments155. Very importantly, the membrane segment that is phagocytosed by an RPE cell appears to be delimited by the precisely localized exposure of PtdSer at only the tips of photoreceptor outer segments around subjective dawn156. This externalized PtdSer provides a binding site for both the GLA domains of Gas6 and Pros1 (which bridge to RPE-expressed Mer) and the discoidin-like domain of MGF-E8 (which bridges to RPE-expressed αvβ5 integrin). That is, externalized PtdSer tells RPE phagocytes exactly how much of the outer segment membrane to eat, and is thus the essential signal for this localized phagocytosis.
Macrophages also prune. Microglia selectively phagocytose pre-synaptic elements (boutons) of axonal membrane during postnatal brain development, in a process termed ‘synaptic pruning’157–159 (Fig. 3). This very selective phagocytosis is known to be driven by neuronal electrical activity and is thought to be mediated at least in part by the deposition of complement proteins C1q, C3, and C4 on pre-synaptic elements that are to be engulfed by microglia159,160. Indeed, defects in synaptic pruning associated with complement deposition may contribute to neurodegenerative and neuropsychiatric disease161.
Complement proteins are not intrinsic to the neuron, however, and so how they might specifically decorate pre-synaptic boutons of the axon that are to be pruned, while not decorating others, has remained unclear. One recently advanced hypothesis is that PtdSer might again be an important signal75 (Fig. 3). Neuronal activity is associated with Ca2+ influx into boutons, which might in principle locally activate Ca2+-dependent scramblases that would in turn result in the local externalization of PtdSer — to which C1q has been reported to bind75,115. Interesting experimental support for this hypothesis has very recently been provided by the demonstration of preferential association of C1q with pre-synaptic membrane vesicles that are both positive for Annexin V binding; that is, have externalized PtdSer, and cleaved (that is, activated) caspase 3162. As highlighted above, PtdSer scramblases of the XKR family are activated by caspase 3 proteolysis. Together, these observations suggest that highly localized externalization of PtdSer on pre-synaptic axonal boutons that are not paired with a post-synaptic element (to form a complete synapse) may serve as a platform for the binding of Gas6 and/or Pros1, MFG-E8 or complement proteins to trigger microglial engulfment of these boutons (Fig. 3). How PtdSer externalization might be confined to small membrane domains of cells — unpaired pre-synaptic boutons or the tips of photoreceptor outer segments — remains the subject of speculation. Possibilities include the localized clustering of either plasma membrane or intracellular Ca2+ channels, which would locally activate Ca2+-dependent scramblases, and/or the localized expression of the scramblases themselves, and/or the localized cleavage (activation) of caspase 3 and caspase 7. Locally externalized PtdSer would in theory be prevented from diffusing within the plane of the plasma membrane by its immediate binding to the panoply of extracellular agents — for example, Gas6/Pros1-TAM, MFG-E8-integrin, TIM4 – described above, all of which are physically connected to another cell.
PtdSer-dependent phagocytosis of membrane segments highlights the vast scale over which PtdSer and TAM receptor signalling operates: most of the apoptotic and live cells illustrated in Fig. 1 and Fig. 2 have volumes ranging from 1,000 to 100,000 μm3, whereas the tips of photoreceptor outer segments that are engulfed by RPE cells have volumes of roughly 10 μm3, and the engulfed pre-synaptic boutons of neuronal axons measure in the range of 0.2 to 1μm3. PtdSer- and TAM-dependent phagocytosis of enveloped virus particles also occurs — in a process termed ‘apoptotic mimicry’163,164 — and these viruses have volumes on the order of 0.005 μm3. Together, these diverse phagocytic events, all of which are initiated by PtdSer externalization and most of which are known to be TAM-dependent, can capture engulfment targets whose sizes span over seven orders of magnitude.
Conclusion: what could go wrong?
The consequences of defective efferocytosis by macrophages, which leads to secondary necrosis and the presentation of autoantigens, are commonly assumed to be dire. However, the evidence for this with respect to disease in humans, as opposed to mouse models, is largely circumstantial. Deficient efferocytosis has been observed in advanced human atherosclerotic plaques, leading to the formation of highly inflammatory necrotic lesions165,166. In human inflammatory lung diseases, including chronic obstructive pulmonary disease, inadequate efferocytosis has also been strongly implicated in the exacerbation of disease pathology167. And defects in the clearance of apoptotic cells from the germinal centers of the lymph nodes of patients with SLE have been a consistently reported feature of the disease9,168. Nonetheless, these observations of apoptotic cell accumulation can only be said to correlate with disease, as human beings are not, except in the context of clinical trials, experimental animals.
As noted above, mice in which the TAM system, the MFG-E8/integrin system, or the enzymes that externalize PtdSer are either partially or fully disabled do indeed display very substantial accumulations of apoptotic cells and often develop severe autoimmune disease. Tyro3−/−Axl−/−Mertk−/− triple mutant mice, for example, are a mess. They present with prominent apoptotic cell accumulation and activated lymphocytes in many tissues (both lymphoid and non-lymphoid), are plagued by massive splenomegaly, rheumatoid arthritis, psoriasis, and nephritis, are infertile as males due to severely impaired phagocytosis of apoptotic germ cells by Sertoli cells in the testes, and are blind due a failure in RPE phagocytosis of photoreceptor outer segments in the retina90,91,154. In humans, there are now descriptions of a plethora of MERTK gene mutations that result in inherited retinal diseases152, but patients carrying these mutations have not been evaluated for immune dysfunction. Particularly for those patients with complete loss-of-function mutations in MERTK, such evaluations — for example, tests for splenomegaly and/or the presence of anti-dsDNA, anti-nuclear antigen, or anti-phospholipid autoantibodies in the circulation — are clearly warranted. Finally, the TAM RTKs play very important roles in the initiation, growth, metastasis and resistance of many different cancers, and several small molecule kinase inhibitors of these receptors are now in development or in clinical trials as cancer therapeutics169. Given the macrophage biology highlighted above, evaluation of the effects of these inhibitors on the development of autoimmune disease should also be a priority.
Acknowledgements
I thank the members of my laboratory for comments and discussions. Supported by grants from the NIH (AI101400, NS085296, AG060748), the Cure Alzheimer’s Fund, and the Leona M. and Harry B. Helmsley Charitable Trust.
Glossary terms:
- NIH3T3 cells
A cell line derived from mouse embryonic fibroblasts
- AD293 cells
A cell line derived from the HEK293 human embryonic kidney cell line
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Competing interests
The author declares no competing interests.
Reviewer information
Nature Reviews Immunology thanks S. Grinstein and the other anonymous reviewers for their contribution to the peer review of this work.
References
- 1.Nagata S Apoptosis and Clearance of Apoptotic Cells. Annu Rev Immunol 36, 489–517 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Elliott MR & Ravichandran KS The Dynamics of Apoptotic Cell Clearance. Dev Cell 38, 147–160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon S & Pluddemann A Macrophage Clearance of Apoptotic Cells: A Critical Assessment. Front Immunol 9, 127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epelman S, Lavine KJ & Randolph GJ Origin and functions of tissue macrophages. Immunity 41, 21–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krysko DV, Vanden Berghe T, D’Herde K & Vandenabeele P Apoptosis and necrosis: detection, discrimination and phagocytosis. Methods 44, 205–221 (2008). [DOI] [PubMed] [Google Scholar]
- 6.deCathelineau AM & Henson PM The final step in programmed cell death: phagocytes carry apoptotic cells to the grave. Essays Biochem 39, 105–117 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Surh CD & Sprent J T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature 372, 100–103 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Earnshaw WC, Martins LM & Kaufmann SH Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 68, 383–424 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Mahajan A, Herrmann M & Munoz LE Clearance Deficiency and Cell Death Pathways: A Model for the Pathogenesis of SLE. Front Immunol 7, 35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdolmaleki F et al. The Role of Efferocytosis in Autoimmune Diseases. Front Immunol 9, 1645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fourgeaud L et al. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper highlighted a role for microglial Mer in the steady state phagocytosis of apoptotic cells in the neurogenic regions of the adult brain.
- 12.Sierra A et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell stem cell 7, 483–495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGaha TL & Karlsson MC Apoptotic cell responses in the splenic marginal zone: a paradigm for immunologic reactions to apoptotic antigens with implications for autoimmunity. Immunol Rev 269, 26–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peter C, Wesselborg S, Herrmann M & Lauber K Dangerous attraction: phagocyte recruitment and danger signals of apoptotic and necrotic cells. Apoptosis 15, 1007–1028 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Medina CB & Ravichandran KS Do not let death do us part: ‘find-me’ signals in communication between dying cells and the phagocytes. Cell Death Differ 23, 979–989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauber K et al. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell 113, 717–730 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Peter C et al. Migration to apoptotic “find-me” signals is mediated via the phagocyte receptor G2A. J Biol Chem 283, 5296–5305 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Gude DR et al. Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a “come-and-get-me” signal. FASEB J 22, 2629–2638 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott MR et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461, 282–286 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peter C et al. Release of lysophospholipid ‘find-me’ signals during apoptosis requires the ATP-binding cassette transporter A1. Autoimmunity 45, 568–573 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Wu YC & Horvitz HR The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93, 951–960 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Chekeni FB et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandilos JK et al. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J Biol Chem 287, 11303–11311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandilos JK & Bayliss DA Physiological mechanisms for the modulation of pannexin 1 channel activity. J Physiol 590, 6257–6266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu YH et al. A quantized mechanism for activation of pannexin channels. Nature communications 8, 14324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truman LA et al. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood 112, 5026–5036 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Yang LV, Radu CG, Wang L, Riedinger M & Witte ON Gi-independent macrophage chemotaxis to lysophosphatidylcholine via the immunoregulatory GPCR G2A. Blood 105, 1127–1134 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Luo B et al. Erythropoeitin Signaling in Macrophages Promotes Dying Cell Clearance and Immune Tolerance. Immunity 44, 287–302 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Idzko M, Ferrari D & Eltzschig HK Nucleotide signalling during inflammation. Nature 509, 310–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haynes SE et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci 9, 1512–1519 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Gautier EL et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13, 1118–1128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ImmGen C Open-source ImmGen: mononuclear phagocytes. Nat Immunol 17, 741 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Ohsawa K et al. P2Y12 receptor-mediated integrin-beta1 activation regulates microglial process extension induced by ATP. Glia 58, 790–801 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Zrzavy T et al. Loss of ‘homeostatic’ microglia and patterns of their activation in active multiple sclerosis. Brain 140, 1900–1913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kronlage M et al. Autocrine purinergic receptor signaling is essential for macrophage chemotaxis. Sci Signal 3, ra55 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Fadok VA et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 148, 2207–2216 (1992). [PubMed] [Google Scholar]; This paper identified externalized PtdSer as an eat-me signal for phagocytosis of apoptotic cells.
- 37.Birge RB et al. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ 23, 962–978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leventis PA & Grinstein S The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 39, 407–427 (2010). [DOI] [PubMed] [Google Scholar]
- 39.van Meer G Dynamic transbilayer lipid asymmetry. Cold Spring Harb Perspect Biol 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen JP et al. P4-ATPases as Phospholipid Flippases-Structure, Function, and Enigmas. Front Physiol 7, 275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin HW & Takatsu H Substrates of P4-ATPases: beyond aminophospholipids (phosphatidylserine and phosphatidylethanolamine). FASEB J, fj201801873R (2018). [DOI] [PubMed] [Google Scholar]
- 42.Nagata S, Suzuki J, Segawa K & Fujii T Exposure of phosphatidylserine on the cell surface. Cell Death Differ 23, 952–961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pomorski TG & Menon AK Lipid somersaults: Uncovering the mechanisms of protein-mediated lipid flipping. Prog Lipid Res 64, 69–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang X, Halleck MS, Schlegel RA & Williamson P A subfamily of P-type ATPases with aminophospholipid transporting activity. Science 272, 1495–1497 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Segawa K, Kurata S & Nagata S Human Type IV P-type ATPases That Work as Plasma Membrane Phospholipid Flippases and Their Regulation by Caspase and Calcium. J Biol Chem 291, 762–772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segawa K et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168 (2014). [DOI] [PubMed] [Google Scholar]; This paper identified ATP11C as a PtdSer flippase whose enzymatic activity is lost upon caspase cleavage.
- 47.Segawa K et al. Phospholipid flippases enable precursor B cells to flee engulfment by macrophages. Proc Natl Acad Sci U S A (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coleman JA & Molday RS Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J Biol Chem 286, 17205–17216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kornberg RD & McConnell HM Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry 10, 1111–1120 (1971). [DOI] [PubMed] [Google Scholar]
- 50.Whitlock JM & Hartzell HC Anoctamins/TMEM16 Proteins: Chloride Channels Flirting with Lipids and Extracellular Vesicles. Annu Rev Physiol 79, 119–143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki J et al. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem 288, 13305–13316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki J, Imanishi E & Nagata S Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc Natl Acad Sci U S A 113, 9509–9514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki J, Umeda M, Sims PJ & Nagata S Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 (2010). [DOI] [PubMed] [Google Scholar]; This paper identified a widely-expressed PtdSer scramblase that is activated by Ca2+ binding.
- 54.Suzuki J, Denning DP, Imanishi E, Horvitz HR & Nagata S Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406 (2013). [DOI] [PubMed] [Google Scholar]; This paper identified a widely-expressed PtdSer scramblase that is activated by caspase cleavage.
- 55.Suzuki J, Imanishi E & Nagata S Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J Biol Chem 289, 30257–30267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bevers EM & Williamson PL Getting to the Outer Leaflet: Physiology of Phosphatidylserine Exposure at the Plasma Membrane. Physiol Rev 96, 605–645 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Yu K et al. Identification of a lipid scrambling domain in ANO6/TMEM16F. Elife 4, e06901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe R, Sakuragi T, Noji H & Nagata S Single-molecule analysis of phospholipid scrambling by TMEM16F. Proc Natl Acad Sci U S A 115, 3066–3071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujii T, Sakata A, Nishimura S, Eto K & Nagata S TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc Natl Acad Sci U S A 112, 12800–12805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Freeman GJ, Casasnovas JM, Umetsu DT & DeKruyff RH TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol Rev 235, 172–189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemke G Biology of the TAM receptors. Cold Spring Harbor Perspectives 5(11), doi: 10.1101/cshperspect.a009076. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyanishi M et al. Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439 (2007). [DOI] [PubMed] [Google Scholar]; This paper described TIM4 as a direct PtdSer binder.
- 63.Yanagihashi Y, Segawa K, Maeda R, Nabeshima YI & Nagata S Mouse macrophages show different requirements for phosphatidylserine receptor Tim4 in efferocytosis. Proc Natl Acad Sci U S A 114, 8800–8805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flannagan RS, Canton J, Furuya W, Glogauer M & Grinstein S The phosphatidylserine receptor TIM4 utilizes integrins as coreceptors to effect phagocytosis. Mol Biol Cell 25, 1511–1522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong K et al. Phosphatidylserine receptor Tim-4 is essential for the maintenance of the homeostatic state of resident peritoneal macrophages. Proc Natl Acad Sci U S A 107, 8712–8717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez-Manzanet R et al. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proc Natl Acad Sci U S A 107, 8706–8711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santiago C et al. Structures of T cell immunoglobulin mucin protein 4 show a metal-Ion-dependent ligand binding site where phosphatidylserine binds. Immunity 27, 941–951 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeKruyff RH et al. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J Immunol 184, 1918–1930 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tietjen GT et al. Molecular mechanism for differential recognition of membrane phosphatidylserine by the immune regulatory receptor Tim4. Proc Natl Acad Sci U S A 111, E1463–1472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lemke G & Rothlin CV Immunobiology of the TAM receptors. Nat Rev Immunol 8, 327–336; PMC2856445 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lew ED et al. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. eLife 3:e03385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper established the rules of engagement for TAM receptors and their ligands.
- 72.Stitt TN et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell 80, 661–670 (1995). [DOI] [PubMed] [Google Scholar]
- 73.Dransfield I, Zagorska A, Lew ED, Michail K & Lemke G Mer receptor tyrosine kinase mediates both tethering and phagocytosis of apoptotic cells. Cell death & disease 6, e1646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zagórska A, Través PG, Lew ED, Dransfield I & Lemke G Diversification of TAM receptor tyrosine kinase function. Nat Immunol 15, 920–928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identified Mer and Axl as mediators of efferocytosis under homeostatic and inflammatory conditions, respectively.
- 75.Lemke G Phosphatidylserine Is the Signal for TAM Receptors and Their Ligands. Trends Biochem Sci 42, 738–748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rothlin CV, Carrera-Silva EA, Bosurgi L & Ghosh S TAM receptor signaling in immune homeostasis. Annu Rev Immunol 33, 355–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujimori T et al. The Axl receptor tyrosine kinase is a discriminator of macrophage function in the inflamed lung. Mucosal Immunol 8, 1021–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsou WI et al. Receptor Tyrosine Kinases, TYRO3, AXL and MER, Demonstrate Distinct Patterns and Complex Regulation of Ligand-Induced Activation. J Biol Chem 289, 25750–25763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scott RS et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411, 207–211 (2001). [DOI] [PubMed] [Google Scholar]; This paper provided the first evidence for Mer as a mediator of efferocytosis.
- 80.Grabiec AM, Goenka A, Fife ME, Fujimori T & Hussell T Axl and MerTK receptor tyrosine kinases maintain human macrophage efferocytic capacity in the presence of viral triggers. Eur J Immunol 48, 855–860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cohen PL et al. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J Exp Med 196, 135–140 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thorp E, Cui D, Schrijvers DM, Kuriakose G & Tabas I Mertk receptor mutation reduces efferocytosis efficiency and promotes apoptotic cell accumulation and plaque necrosis in atherosclerotic lesions of apoe−/− mice. Arterioscler Thromb Vasc Biol 28, 1421–1428 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.A-Gonzalez N et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 31, 245–258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toda S, Segawa K & Nagata S MerTK-mediated engulfment of pyrenocytes by central macrophages in erythroblastic islands. Blood 123, 3963–3971 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Dengler M et al. Accurate Determination of Soluble Axl by Enzyme-Linked Immunosorbent Assay. Assay Drug Dev Technol 14, 543–550 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Ancot F, Foveau B, Lefebvre J, Leroy C & Tulasne D Proteolytic cleavages give receptor tyrosine kinases the gift of ubiquity. Oncogene 28, 2185–2195 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Orme JJ et al. Heightened cleavage of Axl receptor tyrosine kinase by ADAM metalloproteases may contribute to disease pathogenesis in SLE. Clin Immunol 169, 58–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB & Lemke G TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131, 1124–1136 (2007). [DOI] [PubMed] [Google Scholar]; This paper delineated a mechanism whereby Axl functions as an intrinsic negative feedback inhibitor of the innate immune response in DCs.
- 89.Chan PY et al. The TAM family receptor tyrosine kinase TYRO3 is a negative regulator of type 2 immunity. Science 352, 99–103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu Q et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728 (1999). [DOI] [PubMed] [Google Scholar]
- 91.Lu Q & Lemke G Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 293, 306–311 (2001). [DOI] [PubMed] [Google Scholar]; This paper described TAM receptor expression in macrophages, and the autoimmune phenotypes that appear in mice with mutations in TAM receptor genes.
- 92.Khan TN, Wong EB, Soni C & Rahman ZS Prolonged apoptotic cell accumulation in germinal centers of Mer-deficient mice causes elevated B cell and CD4+ Th cell responses leading to autoantibody production. J Immunol 190, 1433–1446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lemke G & Lu Q Macrophage regulation by Tyro 3 family receptors. Curr Opin Immunol 15, 31–36 (2003). [DOI] [PubMed] [Google Scholar]
- 94.Schell SL et al. Mer Receptor Tyrosine Kinase Signaling Prevents Self-Ligand Sensing and Aberrant Selection in Germinal Centers. J Immunol 199, 4001–4015 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Wallet MA et al. MerTK is required for apoptotic cell-induced T cell tolerance. J Exp Med 205, 219–232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaminska A, Enguita FJ & Stepien EL Lactadherin: An unappreciated haemostasis regulator and potential therapeutic agent. Vascul Pharmacol 101, 21–28 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Hanayama R et al. Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 (2002). [DOI] [PubMed] [Google Scholar]; This paper identified MFG-E8 as a soluble bridging protein that binds PtdSer.
- 98.Hanayama R et al. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 304, 1147–1150 (2004). [DOI] [PubMed] [Google Scholar]
- 99.Akakura S et al. The opsonin MFG-E8 is a ligand for the alphavbeta5 integrin and triggers DOCK180-dependent Rac1 activation for the phagocytosis of apoptotic cells. Exp Cell Res 292, 403–416 (2004). [DOI] [PubMed] [Google Scholar]
- 100.Raymond A, Ensslin MA & Shur BD SED1/MFG-E8: a bi-motif protein that orchestrates diverse cellular interactions. J Cell Biochem 106, 957–966 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng Y & Elkon KB Autoimmunity in MFG-E8-deficient mice is associated with altered trafficking and enhanced cross-presentation of apoptotic cell antigens. J Clin Invest 121, 2221–2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lauber K et al. Milk fat globule-EGF factor 8 mediates the enhancement of apoptotic cell clearance by glucocorticoids. Cell Death Differ 20, 1230–1240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murakami Y et al. CD300b regulates the phagocytosis of apoptotic cells via phosphatidylserine recognition. Cell Death Differ 21, 1746–1757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Borrego F The CD300 molecules: an emerging family of regulators of the immune system. Blood 121, 1951–1960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park D et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434 (2007). [DOI] [PubMed] [Google Scholar]
- 106.Das S et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc Natl Acad Sci U S A 108, 2136–2141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhu D et al. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J Clin Invest 125, 1497–1508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee CS et al. Boosting Apoptotic Cell Clearance by Colonic Epithelial Cells Attenuates Inflammation In Vivo. Immunity 44, 807–820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park SY et al. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ 15, 192–201 (2008). [DOI] [PubMed] [Google Scholar]
- 110.Kim S et al. Cross talk between engulfment receptors stabilin-2 and integrin alphavbeta5 orchestrates engulfment of phosphatidylserine-exposed erythrocytes. Mol Cell Biol 32, 2698–2708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Taylor PR et al. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J Exp Med 192, 359–366 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramirez-Ortiz ZG et al. The scavenger receptor SCARF1 mediates the clearance of apoptotic cells and prevents autoimmunity. Nat Immunol 14, 917–926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Galvan MD, Greenlee-Wacker MC & Bohlson SS C1q and phagocytosis: the perfect complement to a good meal. J Leukoc Biol 92, 489–497 (2012). [DOI] [PubMed] [Google Scholar]
- 114.Martin M & Blom AM Complement in removal of the dead - balancing inflammation. Immunol Rev 274, 218–232 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Paidassi H et al. C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J Immunol 180, 2329–2338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stegert M, Bock M & Trendelenburg M Clinical presentation of human C1q deficiency: How much of a lupus? Mol Immunol 67, 3–11 (2015). [DOI] [PubMed] [Google Scholar]
- 117.Clarke EV, Weist BM, Walsh CM & Tenner AJ Complement protein C1q bound to apoptotic cells suppresses human macrophage and dendritic cell-mediated Th17 and Th1 T cell subset proliferation. J Leukoc Biol 97, 147–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kenyon KD et al. IgG autoantibodies against deposited C3 inhibit macrophage-mediated apoptotic cell engulfment in systemic autoimmunity. J Immunol 187, 2101–2111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mevorach D, Mascarenhas JO, Gershov D & Elkon KB Complement-dependent clearance of apoptotic cells by human macrophages. J Exp Med 188, 2313–2320 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suh CH, Hilliard B, Li S, Merrill JT & Cohen PL TAM receptor ligands in lupus: protein S but not Gas6 levels reflect disease activity in systemic lupus erythematosus. Arthritis Res Ther 12, R146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao AG et al. Integrin-associated protein is a receptor for the C-terminal domain of thrombospondin. J Biol Chem 271, 21–24 (1996). [DOI] [PubMed] [Google Scholar]
- 122.Brown EJ & Frazier WA Integrin-associated protein (CD47) and its ligands. Trends Cell Biol 11, 130–135 (2001). [DOI] [PubMed] [Google Scholar]
- 123.Oldenborg PA et al. Role of CD47 as a marker of self on red blood cells. Science 288, 2051–2054 (2000). [DOI] [PubMed] [Google Scholar]
- 124.Barclay AN & Van den Berg TK The interaction between signal regulatory protein alpha (SIRPalpha) and CD47: structure, function, and therapeutic target. Annu Rev Immunol 32, 25–50 (2014). [DOI] [PubMed] [Google Scholar]
- 125.Bian Z et al. Cd47-Sirpalpha interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc Natl Acad Sci U S A 113, E5434–5443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jaiswal S et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138, 271–285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chao MP et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 142, 699–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Horrigan SK & Reproducibility Project: Cancer, B. Replication Study: The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Russ A et al. Blocking “don’t eat me” signal of CD47-SIRPalpha in hematological malignancies, an in-depth review. Blood Rev (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lehrman EK et al. CD47 Protects Synapses from Excess Microglia-Mediated Pruning during Development. Neuron 100, 120–134 e126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gumienny TL et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 (2001). [DOI] [PubMed] [Google Scholar]
- 132.Tosello-Trampont AC, Brugnera E & Ravichandran KS Evidence for a conserved role for CRKII and Rac in engulfment of apoptotic cells. J Biol Chem 276, 13797–13802 (2001). [DOI] [PubMed] [Google Scholar]
- 133.Leverrier Y & Ridley AJ Requirement for Rho GTPases and PI 3-kinases during apoptotic cell phagocytosis by macrophages. Curr Biol 11, 195–199 (2001). [DOI] [PubMed] [Google Scholar]
- 134.Kitano M, Nakaya M, Nakamura T, Nagata S & Matsuda M Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature 453, 241–245 (2008). [DOI] [PubMed] [Google Scholar]
- 135.Fond AM & Ravichandran KS Clearance of Dying Cells by Phagocytes: Mechanisms and Implications for Disease Pathogenesis. Adv Exp Med Biol 930, 25–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Park SY & Kim IS Engulfment signals and the phagocytic machinery for apoptotic cell clearance. Exp Mol Med 49, e331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hochreiter-Hufford A & Ravichandran KS Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb Perspect Biol 5, a008748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hoeppner DJ, Hengartner MO & Schnabel R Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412, 202–206 (2001). [DOI] [PubMed] [Google Scholar]
- 139.Reddien PW, Cameron S & Horvitz HR Phagocytosis promotes programmed cell death in C. elegans. Nature 412, 198–202 (2001). [DOI] [PubMed] [Google Scholar]
- 140.Tufail Y et al. Phosphatidylserine Exposure Controls Viral Innate Immune Responses by Microglia. Neuron 93, 574–586 e578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Neher JJ et al. Phagocytosis executes delayed neuronal death after focal brain ischemia. Proc Natl Acad Sci U S A 110, E4098–4107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provided in vivo evidence that live neurons may be phagocytosed by microglia in a Mer-dependent process.
- 142.Brelstaff J, Tolkovsky AM, Ghetti B, Goedert M & Spillantini MG Living Neurons with Tau Filaments Aberrantly Expose Phosphatidylserine and Are Phagocytosed by Microglia. Cell Rep 24, 1939–1948 e1934 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fricker M et al. MFG-E8 mediates primary phagocytosis of viable neurons during neuroinflammation. J Neurosci 32, 2657–2666 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chua BA et al. Protein S and Gas6 induce efferocytosis of HIV-1-infected cells. Virology 515, 176–190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brown GC & Neher JJ Microglial phagocytosis of live neurons. Nature reviews. Neuroscience 15, 209–216 (2014). [DOI] [PubMed] [Google Scholar]
- 146.Fricker M, Tolkovsky AM, Borutaite V, Coleman M & Brown GC Neuronal Cell Death. Physiol Rev 98, 813–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Strauss O The retinal pigment epithelium in visual function. Physiol Rev 85, 845–881 (2005). [DOI] [PubMed] [Google Scholar]
- 148.Sparrow JR, Hicks D & Hamel CP The retinal pigment epithelium in health and disease. Curr Mol Med 10, 802–823 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.D’Cruz PM et al. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet 9, 645–651 (2000). [DOI] [PubMed] [Google Scholar]
- 150.Duncan JL et al. An RCS-like retinal dystrophy phenotype in mer knockout mice. Invest Ophthalmol Vis Sci 44, 826–838 (2003). [DOI] [PubMed] [Google Scholar]
- 151.Gal A et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet 26, 270–271 (2000). [DOI] [PubMed] [Google Scholar]; This paper described the first of a series of human MERTK mutations that result in retinal dystrophies.
- 152.Audo I et al. MERTK mutation update in inherited retinal diseases. Hum Mutat 39, 887–913 (2018). [DOI] [PubMed] [Google Scholar]
- 153.Ghazi NG et al. Treatment of retinitis pigmentosa due to MERTK mutations by ocular subretinal injection of adeno-associated virus gene vector: results of a phase I trial. Hum Genet 135, 327–343 (2016). [DOI] [PubMed] [Google Scholar]
- 154.Burstyn-Cohen T et al. Genetic dissection of TAM receptor-ligand interaction in retinal pigment epithelial cell phagocytosis. Neuron 76, 1123–1132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nandrot EF & Finnemann SC Lack of alphavbeta5 integrin receptor or its ligand MFG-E8: distinct effects on retinal function. Ophthalmic Res 40, 120–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ruggiero L, Connor MP, Chen J, Langen R & Finnemann SC Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5−/− or Mfge8−/− mouse retina. Proc Natl Acad Sci U S A 109, 8145–8148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper presented evidence that localized PtdSer externalization specifies the localized phagocytic excision of photoreceptor outer segments.
- 157.Hong S, Dissing-Olesen L & Stevens B New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol 36, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thion MS & Garel S Microglia Under the Spotlight: Activity and Complement-Dependent Engulfment of Synapses. Trends Neurosci 41, 332–334 (2018). [DOI] [PubMed] [Google Scholar]
- 159.Schafer DP et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper described a role for complement decoration of pre-synaptic neuronal elements that are phagocytosed by microglia.
- 160.Presumey J, Bialas AR & Carroll MC Complement System in Neural Synapse Elimination in Development and Disease. Adv Immunol 135, 53–79 (2017). [DOI] [PubMed] [Google Scholar]
- 161.Sekar A et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gyorffy BA et al. Local apoptotic-like mechanisms underlie complement-mediated synaptic pruning. Proc Natl Acad Sci U S A 115, 6303–6308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bhattacharyya S et al. Enveloped Viruses Disable Innate Immune Responses in Dendritic Cells by Direct Activation of TAM Receptors. Cell Host Microbe 14, 136–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moller-Tank S & Maury W Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology 468–470, 565–580 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schrijvers DM, De Meyer GR, Kockx MM, Herman AG & Martinet W Phagocytosis of apoptotic cells by macrophages is impaired in atherosclerosis. Arterioscler Thromb Vasc Biol 25, 1256–1261 (2005). [DOI] [PubMed] [Google Scholar]
- 166.Yurdagul A Jr., Doran AC, Cai B, Fredman G & Tabas IA Mechanisms and Consequences of Defective Efferocytosis in Atherosclerosis. Front Cardiovasc Med 4, 86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Grabiec AM & Hussell T The role of airway macrophages in apoptotic cell clearance following acute and chronic lung inflammation. Semin Immunopathol 38, 409–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Baumann I et al. Impaired uptake of apoptotic cells into tingible body macrophages in germinal centers of patients with systemic lupus erythematosus. Arthritis Rheum 46, 191–201 (2002). [DOI] [PubMed] [Google Scholar]
- 169.Schoumacher M & Burbridge M Key Roles of AXL and MER Receptor Tyrosine Kinases in Resistance to Multiple Anticancer Therapies. Curr Oncol Rep 19, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]