SUMMARY
The tuberculosis (TB) bacillus, Mycobacterium tuberculosis (Mtb), and HIV-1 act synergistically; however, the mechanisms by which Mtb exacerbates HIV-1 pathogenesis are not well known. Using in vitro and ex vivo cell culture systems, we show that human M(IL-10) anti-inflammatory macrophages, present in TB-associated microenvironments, produce high levels of HIV-1. In vivo, M(IL-10) macrophages are expanded in lungs of co-infected non-human primates, which correlates with disease severity. Further, HIV-1/Mtb co-infected patients display an accumulation of M(IL-10) macrophage markers (soluble CD163 and MerTK). These M(IL-10) macrophages form direct cell-to-cell bridges, which we identified as tunneling nanotubes (TNTs) involved in viral transfer. TNT formation requires the IL-10/STAT3 signaling pathway, and targeted inhibition of TNTs substantially reduces the enhancement of HIV-1 cell-to-cell transfer and overproduction in M(IL-10) macrophages. Our study reveals that TNTs facilitate viral transfer and amplification, thereby promoting TNT formation as a mechanism to be explored in TB/AIDS potential therapeutics.
Keywords: AIDS, HIV-1, Tuberculosis, Mycobacterium tuberculosis, Co-infection, Macrophage, Monocyte, IL-10, STAT3, Viral spread, Tunneling nanotubes, Biomarker
INTRODUCTION
Worldwide, individuals co-infected with Mycobacterium tuberculosis (Mtb), the agent of tuberculosis (TB), and the AIDS virus, HIV-1, pose particular clinical challenges not only because a significant proportion of co-infected patients remain sputum smear-negative, hampering TB diagnosis, but also because HIV-1 infection makes these individuals more prone to TB reactivation (WHO TB 2016, UNAIDS Report 2016)(Getahun et al., 2007). At the heart of this problem is the synergy between HIV-1 and Mtb which interferes with treatment and promotes the pathogenesis of both pathogens (Diedrich and Flynn, 2011; Diedrich et al., 2016). On the one hand, CD4+ T cell decay and other mechanisms induced by HIV-1 are a leading cause for reactivation of latent TB and progression to active TB disease in AIDS patients (Bell and Noursadeghi, 2018; Tomlinson et al., 2013). On the other hand, clinical and epidemiological data clearly identify TB as a risk factor amplifying HIV-1-associated morbidity and mortality (Toossi, 2003). However, the mechanisms by which Mtb exacerbates HIV-1 infection require further investigation (Charles and Shellito, 2016; Esmail et al., 2018; Toossi, 2003) (Bell and Noursadeghi, 2018). Addressing this issue should help in developing strategies for the attenuation of viral activation in co-infected subjects and for a better control of the AIDS epidemic (Diedrich and Flynn, 2011).
Lung macrophages are the primary host cells for Mtb (O’Garra et al., 2013; Russell et al., 2010). While CD4+ T cells are the major target cells for HIV-1, macrophages, including those in the lungs, are also infected by HIV-1 in humans (Bell and Noursadeghi, 2018; Cribbs et al., 2015) and by simian immunodeficiency virus (SIV) in experimentally infected non-human primates (NHPs) (Avalos et al., 2016). Recent data indicate that macrophages play an important role in HIV-1 pathogenesis (Honeycutt et al., 2017; Honeycutt et al., 2016; Sattentau and Stevenson, 2016) and may also be involved in HIV-1/Mtb co-infection (Khan and Divangahi, 2018; Kuroda et al., 2018). In HIV-1-infected individuals with active TB, for example, macrophages from the lungs and pleural effusions (PE) exhibit high levels of HIV-1 infection (Lawn et al., 2001; Toossi, 2003). Furthermore, Mtb increases the level of HIV-1 infection either in vitro in monocyte-derived macrophages or ex vivo in lung macrophages obtained from patients with HIV-1 (Mancino et al., 1997; Toossi et al., 1997). It is presently unclear how a TB-associated microenvironment renders macrophage more susceptible to HIV-1.
Macrophages display considerable heterogeneity in tissues (Gordon et al., 2014). The broad spectrum of pro- (M1) and anti-inflammatory (M2) activation programs are a manifestion of the different levels of response to HIV-1 and Mtb infections (Cassol et al., 2009; Lugo-Villarino et al., 2011). We have shown that active TB skews human monocyte differentiation towards M2-like macrophages, distinguished by a CD16+CD163+MerTK+ phenotype, as well as by increased immunomodulatory activity and Mtb permissivity (Lastrucci et al., 2015). This phenotype is dependent on the IL-10/STAT3 signaling axis and is closely related to the so-called “M(IL-10)” activation program (Murray et al., 2014). We further reported that the abundance of M(IL-10) cells correlates with TB severity in patients and NHPs (Lastrucci et al., 2015). Herein, we investigated whether the TB-induced M(IL-10) macrophage activation program also plays a role in promoting HIV-1 infection in co-infected individuals.
RESULTS
TB-associated microenvironments increase HIV-1 infection in human macrophages
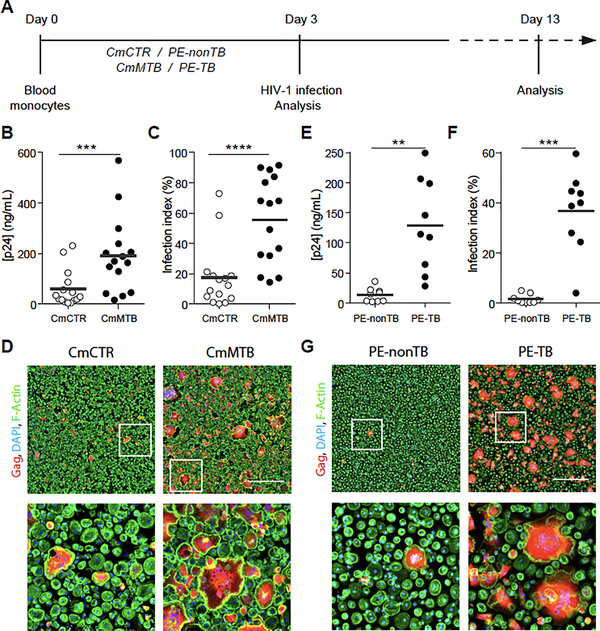
To determine whether HIV-1 replication is modulated in TB-induced M(IL-10) macrophages, we employed our previously described in vitro model (23), which uses conditioned medium from either mock-infected macrophages (CmCTR) or Mtb-infected macrophages (CmMTB). CmMTB triggered primary human monocytes to differentiate into M(IL-10) macrophages, which activated STAT3, as well as acquiring a CD16+CD163+MerTK+PD-L1+ receptor signature, similar to differentiated M(IL-10) macrophages observed in vivo (Figure S1 and (23)). When macrophages treated with either CmCTR or CmMTB were then infected with HIV-1 ADA (Figure 1A) or NLAd8 strain (data not shown) (Raynaud-Messina et al., 2018), we observed a substantial increase in viral replication, as measured by the level of the viral protein p24 in culture supernatants, only in the CmMTB-treated M(IL-10) macrophages (Figures 1B, S2A). In addition, the number of HIV-1 infected cells increased by 3-fold, as measured by the expression of the HIV-1 Gag protein (Figure 1C–D). The M(IL-10) receptor signature was maintained upon HIV1 infection (Figure S1). HIV-1 infection of macrophages enhanced their migration capacity in dense 3-dimensional matrices, together with their fusion potential, forming multinucleated giant cells (MGCs) (Orenstein, 2000; Verollet et al., 2015a; Verollet et al., 2010). We found that these properties, known to contribute to viral dissemination (Verollet et al., 2015a; Verollet et al., 2015b), were further amplified specifically in M(IL-10) macrophages infected with HIV-1 (Figures S2B–E).
Figure 1. (Related to Figure S1, S2, S3). TB-induced microenvironments exacerbate HIV-1 infection of M(IL-10) macrophages.
(A) Representation of the experimental design of two in vitro models. Briefly, monocytes from healthy subjects were treated either with conditioned medium from mock- (CmCTR) or Mtb-infected macrophages (CmMTB), or with pleural effusions (PE) from TB (PE-TB) or non-TB (PE-nonTB) patients for 3 days. Cells were then infected with HIV-1 ADA strain at MOI of 0.1 and kept in culture for at least 10 more days. (B) Vertical scatter plot showing p24 concentration from day 13 supernatants of HIV-1 infected macrophages treated with CmCTR or CmMTB. (C) Vertical scatter plot showing the infection index of day 13 HIV-1 infected macrophages treated with CmCTR or CmMTB. Infection index was calculated as 100 x the ratio of the area covered by Gag+ cells over the total cell area, measured from immunofluorescence (IF) images. (D) Representative IF images of day 13 HIV-1 infected macrophages treated with CmCTR or CmMTB. HIV-1 Gag (red), F-actin (green), DAPI (blue). Scale bar, 500μm. Insets are 4x zooms (lower panels). (E) Vertical scatter plot showing p24 concentration from day 13 supernatants of HIV-1 infected macrophages treated with PE-TB or PE-nonTB. (F) Vertical scatter plot showing the infection index of day 13 HIV-1 infected macrophages treated with PE-TB or PE-nonTB. (G) Representative IF images of day 13 HIV-1 infected macrophages treated with PE-nonTB or PE-TB. HIV-1 Gag (red), F-actin (green), DAPI (blue). Scale bar, 500μm. Insets are 4x zooms (lower panels). Each circle within vertical scatter plots represents a single donor. Mean value is represented as a dark grey line. In this figure, PE-nonTB are parapneumonic PE. Statistical analyses: Two-tailed, Wilcoxon matched-paired signed rank test (B,C), paired t-test (E,F). ** p≤0.005; *** p≤0.0005; **** p≤0.0001.
Pleural effusion (PE) fluid from TB patients (PE-TB) was used as an ex vivo TB-associated microenvironment model (Genoula et al., 2018). PE is observed in up to 30% of TB patients and results from infection-induced local inflammation and recruitment of leukocytes into the pleural space (Vorster et al., 2015). In co-infected patients, the formation of PE is more common than in TB patients and PE contains high viral titers, compared to serum from the same patient (Collins et al., 2002; Toossi, 2003). Unlike control PE obtained from non-TB patients (PE-nonTB), differentiation of macrophages in the presence of PE-TB (Table S1 and Figure 1A) yielded the M(IL-10) phenotype (Figure S3A–B), similar to macrophages isolated from PE-TB (Lastrucci et al., 2015). This M(IL-10) phenotype also correlated with the high level of soluble IL-10 contained in PE-TB compared to PE-nonTB (Figure S3C). We found that cell treatment with PE-TB increased the production of HIV-1 by 6-fold and increased the number of infected macrophages and MGCs (Figures 1E–G, S3D).
Collectively, using CmMTB- and PE-TB-conditioned macrophages as model systems, we show that HIV-1 production by M(IL-10) macrophages is enhanced in a TB-associated microenvironment.
M(IL-10) cells accumulate in co-infected non-human primates and patients
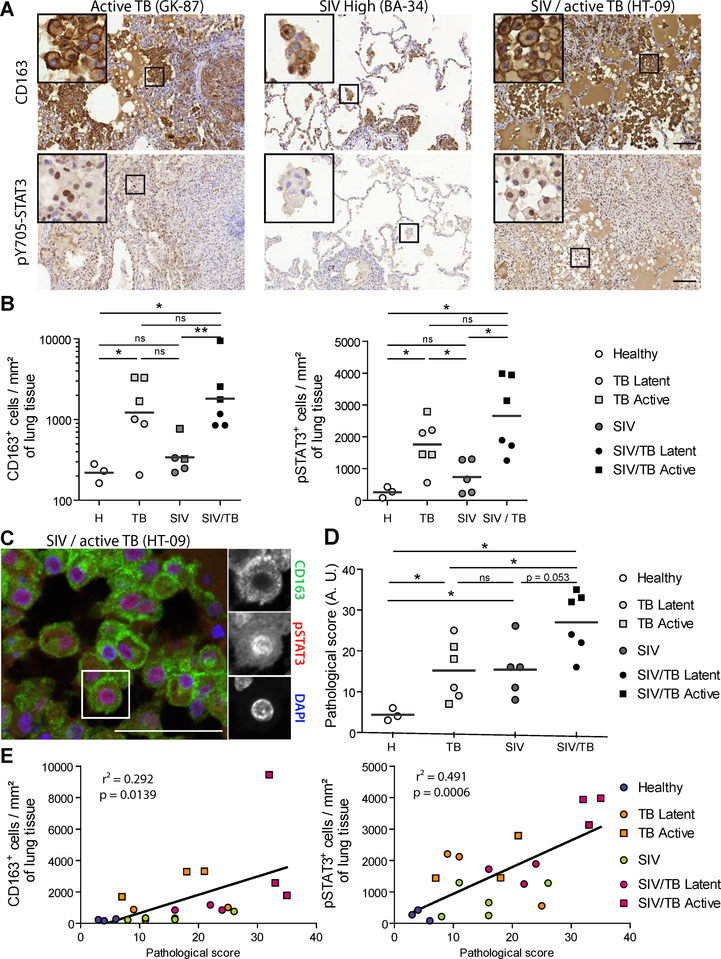
To investigate M(IL-10) cells in HIV-1-Mtb co-infections, we compared, by using CD163 and pSTAT3 staining (Lastrucci et al., 2015), the number of M(IL-10) macrophages in pulmonary samples from NHPs which had been: (i) co-infected with Mtb (active or latent TB) and simian immunodeficiency virus (SIV); (ii) mono-infected with Mtb (active or latent TB); (iii) mono-infected with SIV (Cai et al., 2015; Kuroda et al., 2018); or (iv) uninfected (Tables S2 and S3). Histological staining revealed abundant CD163+ and nuclear pSTAT3+ cells in co-infected NHPs (Figure 2A–B). While the detection of pSTAT3 is not specific to macrophages, double staining analysis demonstrated that most CD163+ alveolar macrophages were also positive for nuclear pSTAT3 (Figure 2C). The increased abundance of CD163+ and pSTAT3+ cells was correlated with the severity of lung histopathology (Table S3) and the gross pathological status of the animals, as analyzed in 30 organs (Figure 2D–E).
Figure 2. (Related to Table S2, S3). Accumulation of M(IL-10) macrophages in the lung of co-infected non-human primates (NHPs) correlates with pathology.
(A)Representative immunohistochemical images illustrate higher number of CD163+ cells and pY705-STAT3+ cells in lung biopsies of SIV-Mtb infected NHPs compared to Mtb or SIV mono-infected NHPs. Scale bar, 100μm. Insets are 4x zooms. (B) Quantification of the number of CD163+ cells (left) and pY705-STAT3+ cells (right) per mm2 of lung tissue of healthy (H), SIV-infected, Mtb-infected and SIV-Mtb-co-infected NHPs. (C) Immunohistochemistry staining of lung biopsy of SIV-Mtb co-infected NHP showing the nuclear localization of pY705-STAT3 (red, center) in CD163 alveolar macrophages (green, top). Nuclei are stained using DAPI (blue, bottom). Scale bar, 50 μm. Insets are 1.3x zooms. (D) Vertical scatter plot showing the pathological scoring of NHPs used in this study (See Table S3). (E) Correlation between CD163+ cells (left) or pY705-STAT3+ cells (right) per mm2 of lung tissue and pathological score in the indicated NHPs. Each symbol within vertical scatter plots represents a single animal. Mean value is represented as a dark grey line. Statistical analyses: Two-tailed Mann-Whitney (B,D). * p≤0.05; ** p≤0.005; ns, not significant.
We have previously shown that CD14+CD16+ circulating monocytes have a predisposition to differentiate into M(IL-10) macrophages in patients with active TB (Lastrucci et al., 2015). Here, we confirmed that CD14+CD16+ monocytes are expanded in the peripheral blood of TB, HIV-1, and co-infected patients, compared to healthy subjects (Balboa et al., 2011; Ellery et al., 2007; Ziegler-Heitbrock, 2007) (Table S1 and Figure S4A). We examined two cell surface markers characteristic of the M(IL-10) phenotype, which are selectively expressed in the monocytic lineage, CD163 and MerTK, and which are both subjected to inflammation-driven shedding (Fabriek et al., 2005; Sather et al., 2007). We found that the soluble forms of these receptors (sCD163 and sMer), but not their membrane-bound forms, were substantially increased in co-infected patients when compared to mono-infected patients and healthy subjects (Figure 3). The amount of these soluble factors correlated with one another (Figure S4B). Receiver Operating Characteristic (ROC) curve analyses for plasma concentration of sCD163 and sMer (Figure S4C–D) suggested both molecules may be useful biomarkers for co-infection.
Figure 3. (Related to Figure S4 and Table S1). Systemic expansion of the M(IL-10) monocyte population in co-infected patients.
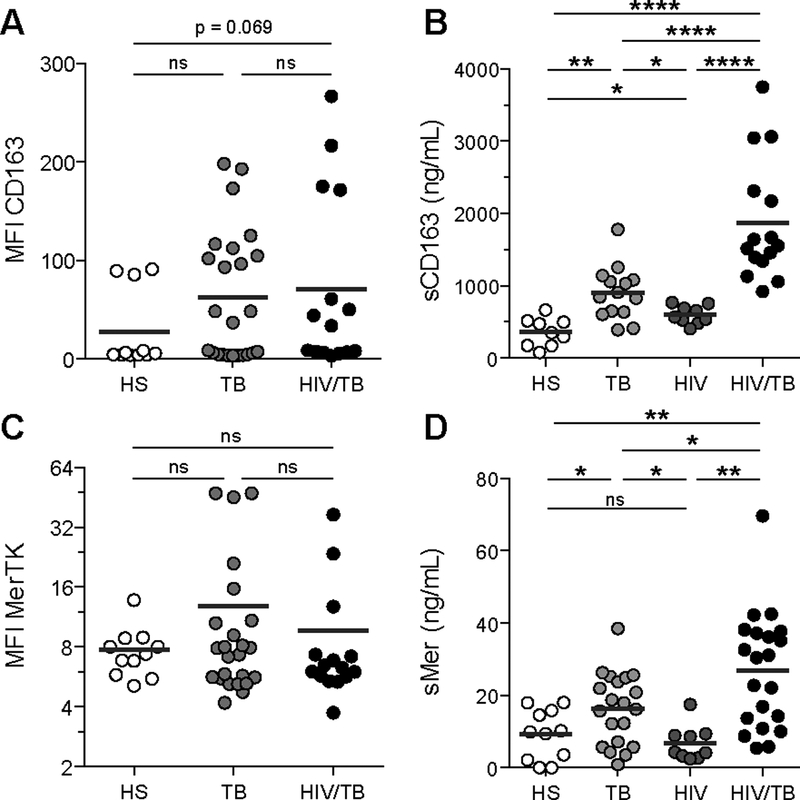
(A) Vertical scatter plots showing the median fluorescence intensity (MFI) of cell-surface marker CD163 on CD14+ monocytes from peripheral blood of healthy subjects, TB patients (TB) and HIV-1/Mtb co-infected patients (HIV/TB). (B) Vertical scatter plots showing the amount of sCD163, the cleaved form of CD163 in the serum of healthy subjects, TB, HIV and HIV/TB patients. (C) Vertical scatter plots showing the MFI of cell-surface marker MerTK on CD14+ monocytes from PB of healthy subjects, TB and HIV/TB patients. (D) Vertical scatter plots showing the amount of sMer, the cleaved form of MerTK in the serum of healthy subjects, TB, HIV and HIV/TB patients. Each circle within vertical scatter plots represents a single donor. Mean value is represented as a dark grey line. Statistical analyses: Two-tailed, Mann-Whitney (A-C), unpaired t-test (D). * p≤0.05; ** p≤0.005; *** p≤0.0005; ns, not significant.
Our findings reveal the extent of the in vivo expansion of TB-induced M(IL-10) cell populations in co-infected individuals. They also identify potential biomarkers for diagnosis and monitoring TB in co-infected patients.
TB-associated microenvironments enhance tunneling nanotube formation
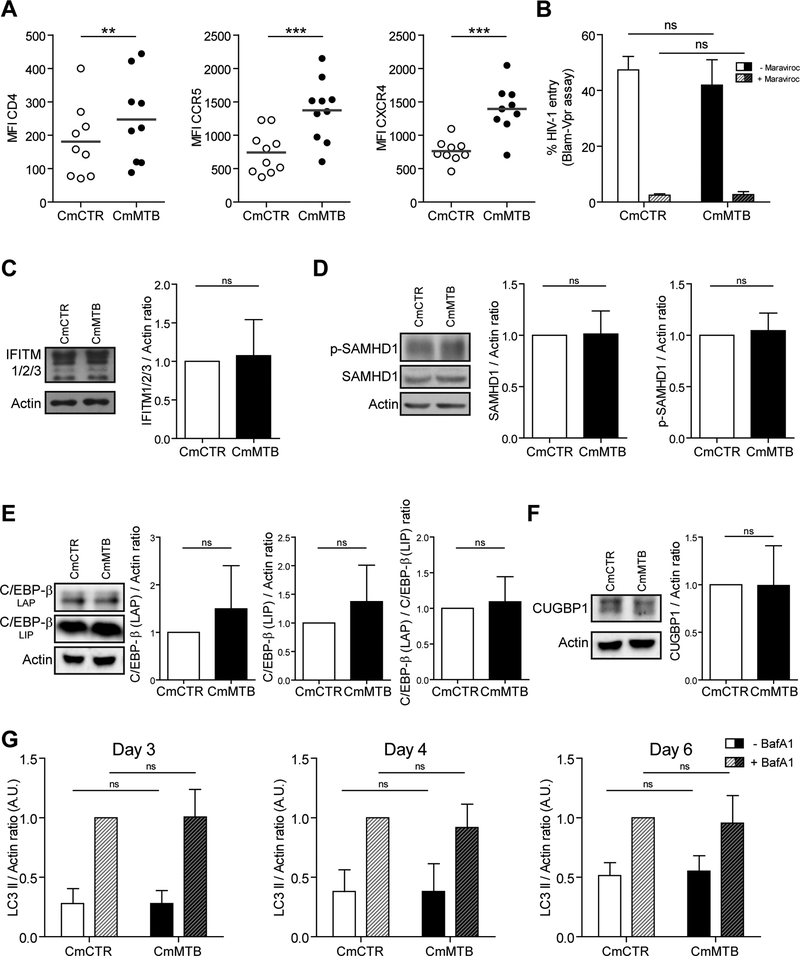
TB-associated microenvironments could increase the level of HIV-1 in M(IL-10) macrophages by modulating viral: (i) entry; (ii) replication; (iii) clearance; (iv) infectivity of the produced virions; and/or (v) cell-to-cell transmission. We tested each of these possibilities in turn. Although cell-surface expression of the HIV-1 entry receptors CD4, CCR5 and CXCR4 was increased in CmMTB-treated cells compared to control cells (Figure 4A), virus entry was unchanged in these cells, as shown using the BlaM-Vpr fusion assay (Cavrois et al., 2002) (Figure 4B). The expression level of several host factors known to be involved in HIV-1 replication (CEBP-β) or restriction (IFITM proteins and SAMHD1) was not modified by CmMTB (Figure 4C–F). As autophagy represents a viral clearance mechanism known to be inhibited by HIV-1 or Mtb infection in macrophages (Espert et al., 2015), we measured the autophagic flux and found it to be similar between CmMTB- and CmCTR-treated cells (Figure 4G). Furthermore, the infectivity of viruses produced in CmMTB conditions was comparable to those produced in CmCTR-treated cells, (7.7 ± 2.1% of p24-positive TZM-bl (Verollet et al., 2015) for CmMTB-treated macrophages vs. 10.3 ± 3.5% for CmCTR-treated cells, n=3; p=0.5066).
Figure 4. (Related to Figure S5). The TB-driven exacerbation of HIV-1 infection in macrophages is not due to modulation in viral entry, HIV-1related activation or restriction factors, or autophagy.
(A) Vertical scatter plots showing the Median Fluorescence Intensity (MFI) of cell-surface receptors involved in HIV-1 entry (CD4, CCR5, CXCR4) on monocytes differentiated for 3 days under the presence of CmCTR and CmMTB. Each circle within vertical scatter plots represents a single donor. Mean value is represented as a dark grey line. (B) Histogram showing the percentage of HIV-1 fusion with CmCTR- (white) or CmMTB (black)-pre-treated cells, as determined using the Blam-Vpr assay in the presence of entry inhibitor Maraviroc (dashed bars). (C) Left: Representative images of Western Blot analysis illustrating the expression of IFITM1/2/3 and Actin as loading control. Right: Quantification of IFITM1/2/3 expressed as a ratio related to actin of monocytes differentiated for 3 days into macrophages under the presence of CmCTR (white) and CmMTB (black). n = 6 donors. (D) Representative images of Western Blot analysis illustrating the expression of SAMHD1 and its phosphorylated version (pSAMHD1), and Actin as loading control (left). Quantification of SAMHD1 (center) pSAMHD1 (right) expressed as a ratio related to actin of monocytes differentiated for 3 days into macrophages under the presence of CmCTR and CmMTB. n = 11 donors. (E) Representative images of Western Blot analysis illustrating the expression of C/EBP-β (LAP), C/EBP-β (LIP), and Actin as loading control (left). Quantification of C/EBP-β (LAP, center left) and C/EBP-β (LIP, center right) expressed as a ratio related to actin, and LAP expressed as a ratio related to LIP (right), of monocytes differentiated for 3 days into macrophages under the presence of CmCTR and CmMTB. n = 9 donors. LAP is an activator of HIV-1-LTR whereas LIP is a repressor of HIV-1-LTR. (F) Representative images of Western Blot analysis illustrating the expression of CUGBP1 and Actin as loading control (left). Quantification of CUGBP1 expressed as a ratio related to actin (right) of monocytes differentiated for 3 days into macrophages under the presence of CmCTR and CmMTB. n = 9 donors. (G) Quantification of LC3-II expression as a ratio to actin of monocytes differentiated for 3 days under the presence of CmCTR and CmMTB at the indicated time points after 2h treatment with Bafilomycin A1 (BafA1) or DMSO as control, as measured by western blot analysis. Uninfected cells at day 3 of the experiment (left, n = 6 donors), and HIV-infected cells at 1 (day 4, center, n = 4 donors) and 3 (day 6, right, n = 6 donors) days post-infection. Each circle within vertical scatter plots represents a single donor. Mean value is represented as a dark grey line. Data in histograms are represented as mean ± SD. * p≤0.05; ** p≤0.005; *** p≤0.0005; **** p≤0.0001.
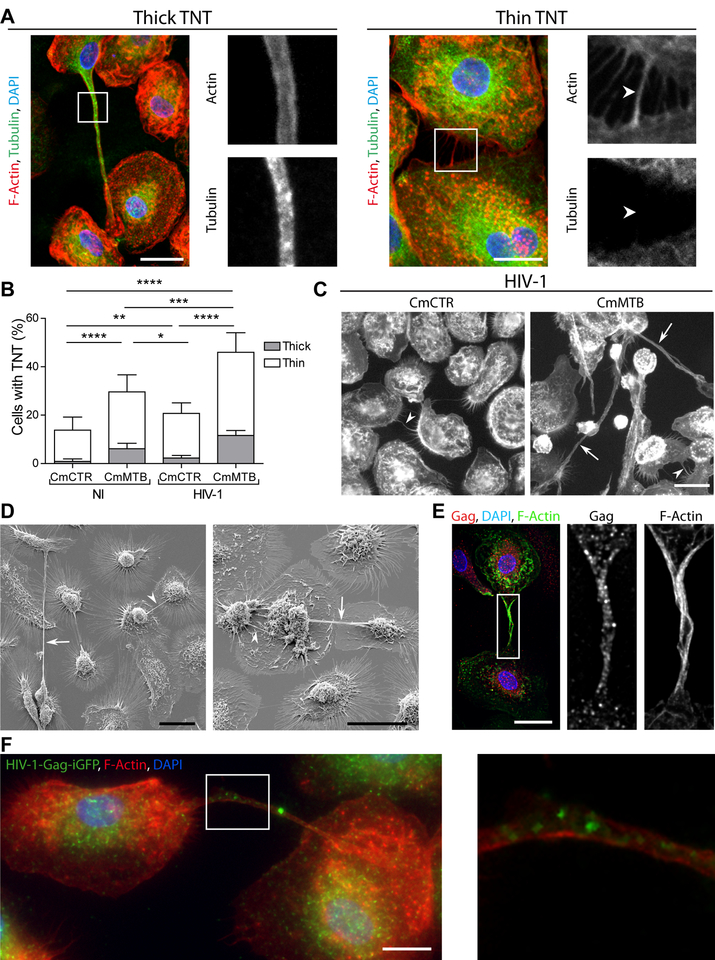
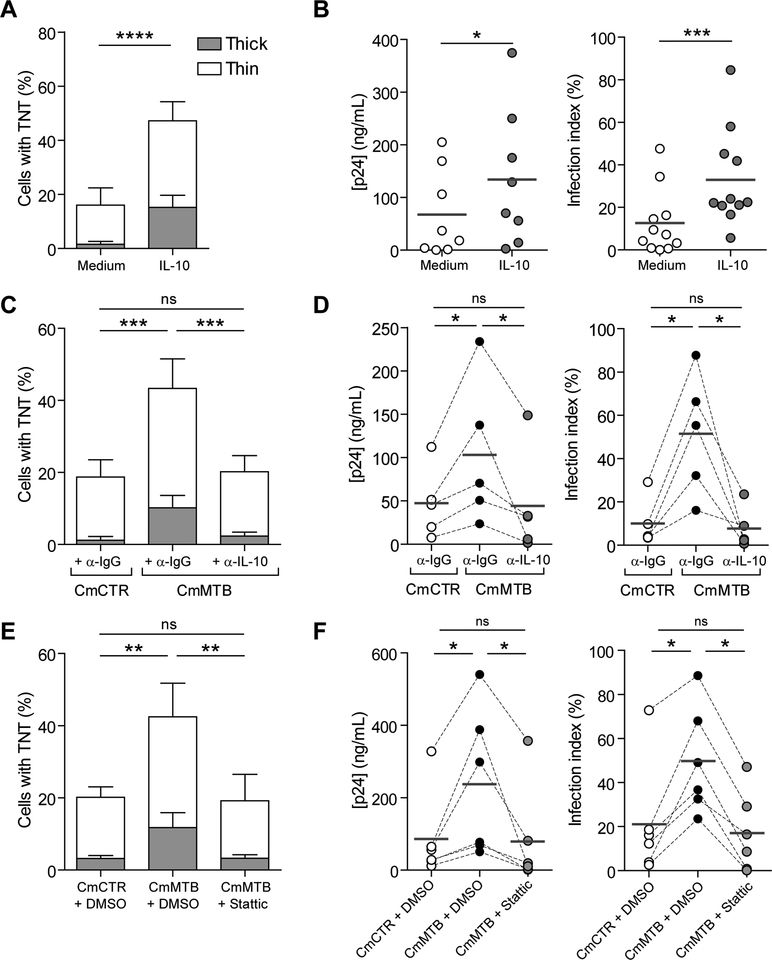
Finally, we investigated whether cell-to-cell virus transfer was influenced by TB-associated microenvironments. Tunneling nanotube (TNT) formation has been proposed as a macrophage-to-macrophage transmission process for host and microbial material (Dupont et al., 2018; Eugenin et al., 2009; Hashimoto et al., 2016; Okafo et al., 2017). Importantly, TNT formation is triggered by HIV-1 infection of macrophages and it has been associated with increase in viral replication (Eugenin et al., 2009; Hashimoto et al., 2016). TNTs are long-range membranous F-actin-containing tubes, not in contact with the extracellular substrate, which are classified into two types based on their thickness and on whether they contain microtubules (Ariazi et al., 2017; Dupont et al., 2018; McCoy-Simandle et al., 2016; Onfelt et al., 2006) (Fig 5A). Treatment with CmMTB increased the percentage of cells forming both types of TNTs by more than 2-fold (Fig 5A–B), compared to CmCTR-treated cells, and HIV-1 infection further amplified this phenomenon (Fig 5B–C).
Figure 5. (Related to Movie 3). TB enhances HIV-1-induced tunneling nanotube (TNT) formation.
(A)Representative immunofluorescences image of macrophages interconnected through thick (left) and thin (right) TNTs. F-Actin (red), Tubulin (green), DAPI (blue). Scale bar, 20 μm. The arrows point at the thin TNT without microtubules. (B) Stacked bars showing the percentage of cells with thick (grey) and thin (white) TNTs of day 6 uninfected or HIV-1-infected macrophages treated with CmCTR or CmMTB. Data in histograms are represented as mean ± SD. (C) Representative wide field Immunofluorescence (IF) images showing F-actin staining in day 6 HIV-1 infected macrophages, treated with CmCTR or CmMTB. Arrows: thick TNTs; arrowheads: thin TNTs. Scale bar, 20μm. (D) Scanning electron microscopy images showing TNTs of day 6 HIV-1 infected macrophages, treated with CmMTB. Arrows : thick TNTs ; arrowheads : thin TNTs. Scale bar, 20μm. (E) Deconvolution microscopy images showing HIV-1 Gag intra-TNT distribution in day 6 HIV-1 infected macrophages, treated with CmMTB. HIV-1 Gag (red), F-actin (green), DAPI (blue). Scale bar, 20μm. Insets are 3x zooms. (F) Immunofluorescence (IF) images showing viral particles in TNT of day 6 HIV-1-Gag-iGFP infected macrophages previously treated with CmMTB. HIV-1-Gag-GFP (green), F-actin (red) and DAPI (blue). Scale bar, 10μm. Inset is 4x zoom. Statistical analyses: Two-tailed paired t-test (B). * p≤0.05; ** p≤0.005; *** p≤0.0005; **** p≤0.0001; ns, not significant.
Using different microscopy approaches, we further characterized macrophage TNTs induced by TB-associated microenvironments. The TNTs are: (i) long (up to 200 μm in length) actin-containing structures connecting two cells, which are above the surface of the substrate (Figures 5D, S5A–B, Movies 1–2 and (Dupont et al., 2018); (ii) positive for M-Sec, a regulator of TNT formation (Hase et al., 2009) (Figure S5C and Movie 4); and (iii) inhibited by cytochalasin D treatment (39 ± 5 % of TNTs in HIV-1-infected CmMTB-treated macrophages vs. 6 ± 4 % upon 2 μM Cytochalasin D treatment, n=3; p=0.0003). We also observed that CmMTB-induced TNTs contained HIV-1 material and particles (Figure 5E–F and Movie 3). Of note, we detected putative TNT-like structures in pulmonary samples from NHPs co-infected with Mtb and SIV, as revealed by hematoxylin and eosin staining, or by immunohistochemistry targeting CD163-positive macrophages (Figure S5D).
Taken together these data indicate that TB-associated microenvironments do not affect the entry, replication, or turnover of HIV-1. Instead, they trigger the formation of TNTs in M(IL-10) macrophages which appear to contain HIV-1 particles.
IL-10/STAT3 promotes TNTs and increased viral production in macrophages
We have previously reported that the expansion of M(IL-10) macrophages relies on the IL-10/STAT3 signaling pathway (Lastrucci et al., 2015). To study how TB-associated microenvironments trigger the formation of TNTs in macrophages, we examined whether IL-10/STAT3 signaling was required for the TNT formation and increased HIV-1 production induced by TB-associated microenvironments. Recombinant IL-10 triggered M(IL-10) macrophage differentiation (Lastrucci et al., 2015) (Figure S6A), increased TNT formation (Figure 6A), and recapitulated the TB-driven expansion of HIV-1 infection, as measured by p24 release, number of infected cells, (Figure 6B) and formation of MGCs (Figure S6B). Depletion of IL10 from CmMTB abolished enhanced TNT formation, the increase in HIV-1 replication in M(IL-10) cells, and the increase in MGCs (Figures 6C–D, S6C). We examined the role of STAT3 activation by pharmacological inhibition with Stattic, which targets the STAT3 SH2 domain to prevent association with upstream kinases and abrogates STAT3 phosphorylation and the associated M(IL-10) phenotype (Lastrucci et al., 2015). Stattic treatment inhibited both the CmMTB-driven TNT formation and the increase in HIV-1 production (Figure 6E,F), along with enhanced cell migration and formation of MGCs (Figures S2E, S6D). Of note, treatment of monocytes with others cytokines prior to HIV-1 infection does not trigger TNT formation significantly, indicating that IL-10 is one of the main factor involved in this process (Figure S6E).
Figure 6. (Related to Figure S2, S6). TB-driven tunneling nanotube (TNT) formation and increased of HIV-1 infection are dependent on the IL10/STAT3 axis.
(A)Stacked bars showing the percentage of cells with thick (grey) and thin (white) TNTs of day 6 HIV-1 infected macrophages untreated or treated with recombinant IL-10 (10ng/mL). (B) Vertical scatter plots showing p24 concentration (left) and infection index (right) of day 13 HIV-1 infected macrophages untreated or treated with recombinant IL-10 (10ng/mL). (C) Stacked bars showing the percentage of cells with thick (grey) and thin (white) TNTs of day 6 HIV-1 infected macrophages treated with IL-10-depleted (α-IL-10) CmMTB and mock depletion controls (α-IgG) of CmCTR and CmMTB. (D) Vertical scatter plots showing p24 concentration (left) and infection index (right) of day 13 HIV-1 infected macrophages treated with IL-10-depleted (α-IL-10) CmMTB and mock depletion controls (α-IgG) of CmCTR and CmMTB. (E) Stacked bars showing the percentage of cells with thick (grey) and thin (white) TNTs of day 6 HIV-1 infected macrophages treated with CmCTR, CmMTB or CmMTB in the presence of the STAT3 activation inhibitor, Stattic (1μM). (F) Vertical scatter plots showing p24 release (left) and infection index (right) of day 13 HIV-1 infected macrophages treated with CmCTR, CmMTB or CmMTB in the presence of the STAT3 activation inhibitor, Stattic (1μM). Statistical analyses: Two-tailed, Wilcoxon matched-paired signed rank test (B right, D right, F), paired t-test (A, B left, C, D left, E). * p≤0.05; ** p≤0.005; *** p≤0.0005; ns, not significant.
Our data demonstrate that TB-associated microenvironments control TNT formation and increase HIV-1 infection in macrophages. In addition, they reveal the IL-10/STAT3 axis as a signaling pathway involved in TNT formation.
TNTs participate in the high viral production in M(IL-10) macrophages
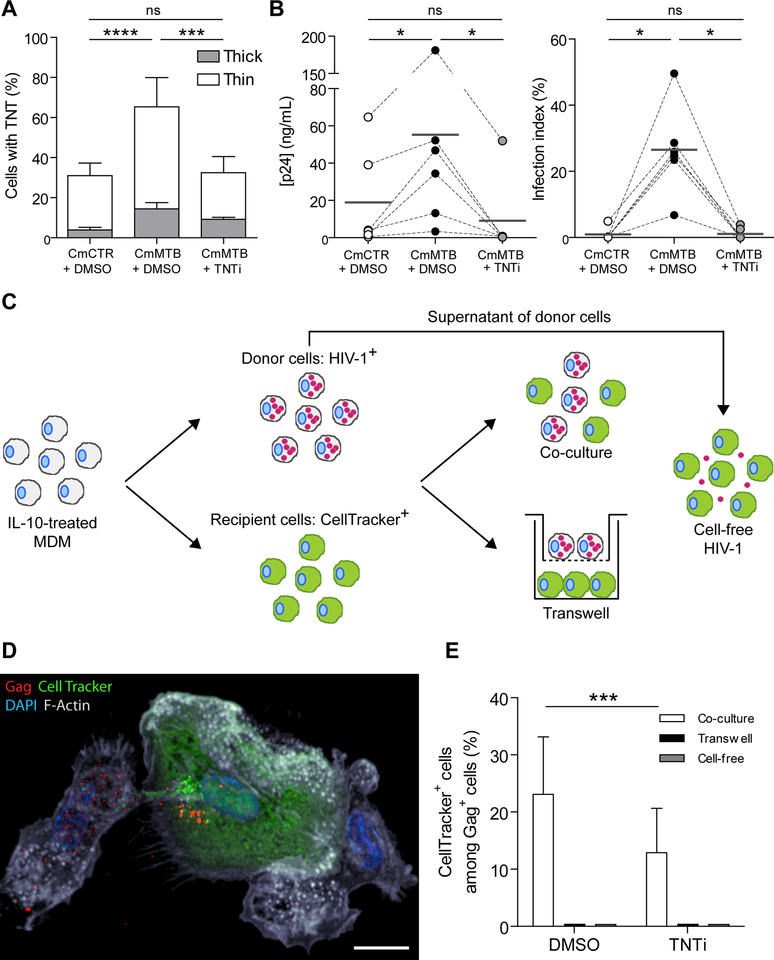
To determine whether TNT formation is involved in the increased HIV-1 production in M(IL-10) macrophages, we used a previously described pharmacological inhibitor of TNT formation, TNTi (Hashimoto et al., 2016). We observed that TNTi inhibited the TNT-mediated transfer of fluorescent material between Di0-stained cells and CellTracker positive cells (Fig S7A), without affecting macrophage viability, or F-actin-dependent processes like podosome formation and phagocytosis (Maridonneau-Parini, 2014) (Figure S7B–E). In TB-induced M(IL-10) macrophages infected with HIV-1, TNTi strongly inhibited TNT formation (Fig. 7A), significantly diminished HIV-1 overproduction (Figure 7B), and reduced MGC formation (Figure S7F).
Figure 7. (Related to Figure S7 and Movies 5 and 6). TB-driven tunneling nanotube (TNT) formation is necessary for increased HIV-1 infection.
(A) Stacked bars showing the percentage of cells with thick (grey) and thin (white) TNTs of day 6 HIV-1 infected macrophages treated with CmCTR, CmMTB or CmMTB in the presence of TNT inhibitor (TNTi, 20μM). (B) Vertical scatter plots showing p24 release (left) and infection index (right) of day 13 HIV-1 infected macrophages treated with CmCTR, CmMTB or CmMTB in the presence of TNTi. (C) Experimental set-up used for the co-culture (also referred to as a transfer assay), transwell and cell-free HIV-1 experiments. IL-10-treated MDM were either infected with HIV-1 NLAD8-VSVG pseudotyped strain at a MOI 5 for 48h (donor cells) or stained with CellTracker (acceptor cells). For the co-culture experiment, donor cells were mixed with acceptor cells at a 1:1 ratio. The transwell assay was designed to separate donor and acceptor cells to investigate the contribution of cell contact to transfer of HIV-1. The experiment was performed as described in the co-culture assay, with the exception that acceptor cells were plated in the well, and then a transwell filter was placed containing the donor cells. Finally, to assess the contribution of cell-free infection in the propagation of HIV-1, the supernatant of donor cells (containing the virions produced during 48h of infection) was incubated with acceptor cells. To assess the contribution of TNTs in HIV-1 transfer, all 3 experiments were performed in the presence or not of TNTi (20μM), and fixed after 24h. The cells were stained for HIV-1 Gag protein and the percentage of CellTracker+ cells among Gag+ cells was quantified. (D) Deconvolution microscopy image showing HIV-1 Gag transfer into CellTracker+ acceptor cell after 24h co-culture. HIV-1 Gag (red), Cell Tracker (green), F-actin (grey), DAPI (blue). Scale bar, 10μm. (E) Stacked bars showing the percentage of CellTracker+ cells among Gag+ cells after 24h co-culture with or without TNTi (20μM). Data in histograms are represented as mean ± SD. Each circle within vertical scatter plots represents a single donor. Mean value is represented as a dark grey line. Statistical analyses: Two-tailed, paired t-test (A, E), Wilcoxon matched-paired signed rank test (B). * p≤0.05; *** p≤0.0005; **** p≤0.0001; ns, not significant.
Unlike in T lymphocytes, the transfer of HIV-1 particles via TNTs has been suggested but not formally demonstrated in macrophages (Dupont et al., 2018; Eugenin et al., 2009; Hashimoto et al., 2016; Okafo et al., 2017). Thus, we set up a co-culture system between M(IL-10) macrophages to assess the transfer of the viral Gag protein from infected donor cells to uninfected recipient cells. Uninfected (recipient, CellTracker+, green) and HIV-1-infected (donor, Gag+, red) macrophages were either co-cultured or separated by a transwell membrane that blocks cell-to-cell connections (Figure 7C). Importantly, TNT quantification showed that productively infected macrophages preferentially formed TNT (38 ± 6 % of TNT formation in Gag+ cells compared to 21 ± 7 % in non-infected CellTracker+ cells, n=5; p=0.0003). After 24 hours in co-culture, we observed that donor macrophages were able to transfer the virus to recipient macrophages, which became HIV-1 positive (Figure 7D and Movies 5–6). By contrast, the transfer of HIV-1 from donor to recipient macrophages was blocked in the transwell cultures (Figure 7E), showing that cell-to-cell contacts are involved in this process. In co-culture experiments, TNTi significantly diminished the capacity of M(IL-10) macrophages to transfer HIV-1 to recipient cells (Figure 7E), indicating that TNT formation is responsible for cell-to-cell viral spread.
These results establish a role for TNTs in spreading the virus between M(IL-10) macrophages, uncovering a key cellular mechanism responsible for HIV-1 overproduction in the context of TB.
DISCUSSION
TB is the most common co-infection among people living with HIV-1 and is a leading cause of AIDS-related deaths. Here, we asked whether TB-associated microenvironments influence the control of HIV-1 infection in human macrophages. We report that M(IL-10) macrophages (CD163+MerTK+CD16+pSTAT3+), which accumulate in TB patients and can be derived in vitro from TB-associated microenvironments, are highly permissive for HIV-1 production. This exacerbation of HIV-1 infection involves the IL-10/STAT3 signaling axis, which controls TNT formation, thus enhancing cell-to-cell transfer of the virus in our experimental systems. In vivo, M(IL-10) cells are more abundant in co-infected individuals compared to mono-infection settings. We have identified potential biomarkers, sCD163 and sMer, in the blood of co-infected patients, which could be used to monitor disease progression, as their expression correlates with the severity of the pathology. All things considered, our study makes three important contributions to the general understanding of how TB exacerbates HIV-1 infection.
First, we show that M(IL-10) macrophages (CD163+pSTAT3+) accumulate in coinfected patients and NHPs. In co-infected NHPs, we observe M(IL-10) macrophages in great abundance in the lung environment, including the alveolar space and the lung interstitial tissue. M(IL-10) macrophage abundance in lungs is correlated with the pathological score of the animals, arguing for the physiological and pathological pertinence of these macrophages. These macrophages are likely driven by the IL-10/STAT-3 signaling pathway and originate from CD14+CD16+ monocytes (Lastrucci et al., 2015), which are increased in the blood of patients with active TB, regardless of their HIV-1 infection status (Ziegler-Heitbrock, 2007). Monocytes are the only circulating leukocytes known to express membrane-bound CD163 and MerTK (Fabriek et al., 2005; Sather et al., 2007). As these membrane receptors are subjected to inflammation-driven shedding (Fabriek et al., 2005; Sather et al., 2007), their soluble form in the blood can be used as a proxy for estimating the abundance of CD14+CD16+ monocytes in the circulation. Here, we show that in HIV-1+ patients with active TB, the plasma levels of both sCD163 and sMer are significantly higher than those found in healthy subjects, HIV-1- or Mtb-infected patients. sCD163 is already described as a clinical indicator of monocyte activation in HIV-1 patients (Burdo et al., 2011), as an indicator of lesions and accumulation of macrophages in the brain of SIV-infected NHPs (Burdo et al., 2010), and as an independent predictor of survival in TB (Knudsen et al., 2005). Although our findings will need to be expanded in larger cohorts, they nevertheless reveal both sCD163 and sMer as potential tools for the diagnosis and disease monitoring in HIV-1/Mtb co-infected patients. Because TB diagnosis in HIV-1-infected individuals remain a major clinical challenge (Getahun et al., 2007), sCD163 and sMer may hold great promise as biomarkers in these populations.
Second, our study reveals that M(IL-10) macrophages are highly susceptible to HIV-1 infection. This is accompanied by enhanced MGC formation and protease-dependent migration capacity, which could play a role in HIV-1 dissemination (Verollet et al., 2015). All these effects are fully dependent on the anti-inflammatory IL-10/STAT3 signaling pathway. Most of our work has been done with the ADA virus which is a useful laboratory strain that can infect efficiently macrophages with low CD4 levels. However, these viruses are rather rare in the blood (Joseph and Swanstrom, 2018). As infected macrophages are found in the lungs (Bell and Noursadeghi, 2018; Cribbs et al., 2015), the characterization of the types of viruses mainly found in this co-infection sites (lung and pleural cavity) (Collins et al., 2002; Nakata et al., 1997; Singh et al., 1999) would be of great interest as we would have the opportunity to assess their effect in our model. Other reports also show that Mtb infection increases HIV-1 replication in vitro (Diedrich and Flynn, 2011; Goletti et al., 1998; Hoshino et al., 2002; Lederman et al., 1994; Zhang et al., 1995). However, unlike our study, they show that Mtb induces a pro-inflammatory environment resulting in the auto-activation of NF-κB, which ultimately binds the HIV-1 long terminal repeat (LTR) and initiates viral transcription in co-infected macrophages (Collins et al., 2002; Goletti et al., 2004; Goletti et al., 1998; Hoshino et al., 2002; Lederman et al., 1994; Mancino et al., 1997; Nakata et al., 1997; Orenstein et al., 1997; Queval et al., 2016; Tanaka et al., 2005; Toossi, 2003; Zhang et al., 1995). Based on both these published observations and ours, we propose that TB exacerbates HIV-1 pathogenesis in a two-step manner: initially, by activating HIV-1 LTR transcription via short-lived pro-inflammatory signals in resident macrophages, and, then, by enhancing HIV-1 infection and spread in newly recruited monocytes predisposed towards the M(IL-10) phenotype, as a result of the anti-inflammatory signals induced by IL-10. This two-step model reconciles previous work done in co-infected cells and the results of our study, which is based on inherent susceptibility to HIV-1 infection observed in CD16+ monocytes (Ellery et al., 2007) and macrophages derived from these cells (Ancuta et al., 2006). In light of recent evidence for macrophages as targets for HIV-1 pathogenesis (Honeycutt et al., 2017; Honeycutt et al., 2016; Sattentau and Stevenson, 2016), together with the renewed appreciation of the importance of IL-10-driven anti-inflammatory program in macrophages (Ip et al., 2017), we believe that our in vitro model to generate M(IL-10) cells will help further characterize how TB influences viral infection of macrophages, and onward viral spread.
Third, based on our findings, we infer that TNT formation are likely a key cellular mechanism by which the IL-10/STAT3 signaling axis increases virus spread in human macrophages in a TB context. TNTs are transient membrane projections (36, 37, 42) that facilitate intercellular communication to allow the transfer of endosomal cargo vesicles, calcium fluxes, and pathogens, such as bacteria, viruses and prions (Davis and Sowinski, 2008; Dupont et al., 2018; Malik and Eugenin, 2016; McCoy-Simandle et al., 2016; Sherer and Mothes, 2008). In T cells, transmission of HIV-1 through tunneling nanotubes is estimated to be approximately 100–1,000 fold more efficient than through classical cell-free viral infection (Sowinski et al., 2008). In macrophages, HIV-1 induces the formation of TNT via the protein Nef (Hashimoto et al., 2016; Xu et al., 2004). We confirmed that Nef is also involved in HIV-1-induced TNT formation in a TB-associated microenvironment (data not shown). Since the transmission of HIV-1 between macrophages via TNTs had not been formally demonstrated, we used here tranwells to separate HIV-1-infected macrophages from uninfected macrophages, and we demonstrated that M(IL-10) macrophages are capable of transferring HIV1 to non-infected macrophages through a cell-to-cell contact-dependent mechanism. While we cannot exclude the contribution of other cell-to-cell contact-dependent mechanisms of virus spread, such as virological synapse, cell fusion, phagocytosis or efferocytosis (Baxter et al., 2014; Bracq et al., 2017; Jolly and Sattentau, 2004; Karaji and Sattentau, 2017), we clearly show that TNTs play a major role in this process. Pharmacological inhibition of TNT formation results in both reduced capacity of M(IL-10) macrophages to transfer viral particles and a substantial decrease of HIV-1 production. TNTs have been observed in several cell types and their formation is stimulated by different factors, including lipopolysaccharide (LPS) and IFN-γ, pathogens and oxidative stress; yet the signaling pathway(s) involved in TNT formation have not been identified (Ariazi et al., 2017; Dupont et al., 2018; Malik and Eugenin, 2016; Zhang, 2011). We reveal the IL-10/STAT3-axis as the main signaling pathway responsible for TNT formation between macrophages, providing an important contribution to this emerging field and paving the way for further elucidating the biology of TNTs. It would be interesting to find out whether TNTs could also form between M(IL-10) macrophages and other cell types, such as T or B cells, to transfer HIV-1 and/or viral material (Xu et al., 2009), which could participate in HIV-1 dissemination and pathogenesis within the co-infection context. Finally, whether TNTs are involved in MGC formation during HIV-1 infection of macrophages is suggested by a correlation between TNT formation and the number of MGC that are formed (r2=0.7 to 0.9). Another aspect that needs further improvement is the in vivo relevance of TNTs. Despite evidence for material transfer via TNT-like structures in vivo (Naphade et al., 2015; Rocca et al., 2017), the current lack of a specific TNT marker prevents the formal demonstration of existence of TNT in vivo. In pulmonary tissue lesions of co-infected NHPs, we observed TNT-like structures between CD163+ macrophages, suggesting that cell-to-cell transfer of HIV-1 via TNTs could also occur in vivo. Indeed, M(IL-10) macrophages are in close proximity to each other, particularly in the lungs of co-infected NHPs, a parameter that is critical for TNT formation and viral transmission. Taken together our results, we propose that enhanced TNT formation induced by TB-associated microenvironments in M(IL-10) macrophages favors HIV-1 cell-to-cell transmission and consequently, could play a key role in the high HIV-1 levels usually observed at anatomical sites of co-infection (Toossi, 2003).
In conclusion, this study highlights the importance of TB-associated microenvironments in shaping the response of macrophages to HIV-1 infection. Our results are highly relevant to the investigation of the role of macrophages in the pathogenesis of Mtb/HIV-1 co-infection, cell signaling pathways involved in TNT formation, and the identification of potential biomarkers to monitor disease progression. Future research will reveal whether HIV-1 production and spread through TB-driven TNT formation occurs among M(IL-10) macrophages only, or also affects macrophage-to-T cell viral transmission (Groot et al., 2008) and whether TNT can be considered as a therapeutic target in co-infected patients.
EXPERIMENTAL PROCEDURES
Experimental models, study approval and subject details for human subjects and NHP samples, bacteria and viruses are described in star METHODS. Methods details are also available in star METHODS.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Geanncarlo Lugo-Vilarino (lugo@ipbs.fr). Sharing of antibodies and other reagents with academic researchers may require UBMTA agreements.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Monocytes from healthy subjects (HS) were provided by Etablissement Français du Sang, Toulouse, France, under contract 21/PLER/TOU/IPBS01/20130042. According to articles L12434 and R124361 of the French Public Health Code, the contract was approved by the French Ministry of Science and Technology (agreement number AC 2009921). Written informed consents were obtained from the donors before sample collection. The sex of HS is unknown.
Tuberculous (TB) patients from the División de Tisioneumonología at the Instituto de Tisioneumonología Vaccarezza-University of Buenos Aires, Argentina and co-infected TB/HIV patients from the Division de SIDA at the Hospital F.J Muñiz, from 2015 to 2016, were diagnosed by the presence of recent clinical respiratory symptoms, abnormal chest radiography and positive sputum smear test for acid-fast bacilli. The research was carried out in accordance with the Declaration of Helsinki (2013) of the World Medical Association, and was approved by the Ethics Committees of the institutes mentioned above (protocol numbers: NIN-1671–12 and proceedings 37/38). Written informed consent was obtained before sample collection. Exclusion criteria included the presence of concurrent infectious diseases or non-infectious conditions (cancer, diabetes, or steroid therapy). Blood samples were collected at 3 to 10 days after start of TB treatment. The diagnosis of tuberculous pleurisy was based on a positive Ziehl–Nielsen stain or Lowestein–Jensen culture from pleural effusion (PE) and/or histopathology of pleural biopsy, and was further confirmed by an Mycobacterium tuberculosis-induced IFN-γ response and an ADA-positive test (Light, 2010). Effusions were classified as exudates according to Light et al. criteria (Light, 2010). PE and peripheral blood (PB) samples were obtained as described previously (Schierloh et al., 2007). PB samples from healthy subject (HS) were provided by the Blood Transfusion Service, Hospital Fernandez, Buenos Aires (agreement number CEIANM-52-5-2012). All HS had received BCG vaccination in childhood and their tuberculin-test status (TTS) was unknown. Clinical features and sex of the experimental groups are summarized in Table S1.
Strategy for randomization and/or stratification. Blood samples from enrolled patients were classified in 3 groups of individuals: 1) TB patients with abnormal chest radiography and positive sputum smear test for acid-fast bacilli, who received less than one week of anti-TB therapy (TB); 2) HIV-1 infected patients with active TB, who received less than one week of anti-TB therapy (HIV/TB); and 3) Bacillus Calmette-Guerin-vaccinated healthy donors (HS). Pleural effusions samples from enrolled patients were classified in 2 groups of individuals: 1) TB pleurisy based on a positive Ziehl–Nielsen stain or Lowestein–Jensen culture from PE and/or histopathology of pleural biopsy, and confirmed by an Mtb-induced IFN-γ response and an adenosine deaminase-positive test; and 2) no-TB pleurisy including the following etiologies: malignant effusions (mesothelioma, lung carcinoma or metastatic patients), parapneumonic effusions and heart failure associated effusions.
Inclusion and Exclusion criteria for blood samples. Inclusion: person >21 years old diagnosed with TB (clinical or laboratory criteria) having HIV+ serology (HIV/TB) or not and who consent to participate. Exclusion: the presence of concurrent infectious diseases, except HIV, or non-infectious conditions (cancer, diabetes, or steroid therapy) and patients who do not provide consent.
Inclusion and Exclusion criteria for pleural effusion samples. Inclusion: person >21 years old having a pleural effusion requiring drainage by therapeutic thoracentesis and who consent to participate. Exclusion: the presence of empyema, pleural effusion that does not require therapeutic drainage and patients who do not provide consent.
The GPower software was used for sample-size estimation and statistics. No replicates and no blinding were done.
Non-Human Primate samples
All animal procedures were approved by the Institutional Animal Care and Use Committee of Tulane University, New Orleans, LA, and were performed in strict accordance with NIH guidelines. The twenty adult rhesus macaques used in this study (Table S2) were bred and housed at Tulane National Primate Research Center (TNPRC). The sex of animals is specified in Table S2. All macaques were infected as previously described (Foreman et al., 2016; Mehra et al., 2011). Briefly, macaques were aerosol-exposed to a low dose (25 CFU implanted) of Mtb CDC1551 and a subset of the macaques were also exposed 9 weeks later to 300 TCID50 of SIVmac239 administrated intravenously in 1mL saline. The control subset received an equal volume of saline intravenously. Criteria for killing included presentation of four or more of the following conditions: (i) body temperatures consistently greater than 2°F above preinfection values for 3 or more weeks in a row; (ii) 15% or more loss in body weight; (iii) serum CRP values higher than 10 mg/mL for 3 or more consecutive weeks, CRP being a marker for systemic inflammation that exhibits a high degree of correlation with active TB in macaques (Kaushal et al., 2012; Mehra et al., 2011); (iv) CXR values higher than 2 on a scale of 0–4; (v) respiratory discomfort leading to vocalization; (vi) significant or complete loss of appetite; and (vii) detectable bacilli in BAL samples.
Bacteria
Mtb H37Rv strain was grown in 7H9 media at 37°C, as described (Lastrucci et al., 2015). Exponentially growing Mtb was centrifuged (460g) and resuspended in PBS (MgCl2, CaCl2 free, Gibco). Clumps were dissociated by 20 passages through a 26-G needle then resuspended in RPMI-1640 containing 10% FBS. Bacterial concentration was determined by measuring OD600.
Viruses
Proviral infectious clones of the macrophage-tropic HIV-1 isolate ADA (Verollet et al., 2015a) was kindly provided by Serge Benichou (Institut Cochin, Paris, France). Virions were produced by transient transfection of 293T cells with proviral plasmids, as previously described (Verollet et al., 2015a). HEK 293 T cells (the sex of original cells is unknown) were maintained in DMEM, 20% FBS, 5% CO2. HIV-1 p24 antigen concentration of viral stocks was assessed by an home-made ELISA. HIV-1 infectious units were quantified, as reported (Verollet et al., 2015a) using TZM-bl cells. TZM-bl cells are Hela cell lines (female origin) that were obtained through NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc). TZM-bl In some experiments, we used Betalactamase (BlaM)-Vpr-containing ADA virus and NLAD8-VSVG pseudotyped viruses and HIV-1 ADA Gag-iGFP strain (see Key Resource Table).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD14 | Biolegend | Clone M5E2 |
| Anti-human CD16 | Biolegend | Clone 3G8 |
| Anti-human CD163 | Biolegend | Clone GHI/61 |
| Anti-human CD184 (CXCR4) | Biolegend | Clone 12G5 |
| Anti-human CD195 (CCR5) | Biolegend | Clone J418F1 |
| Anti-human CD274 (B7-H1, PD-L1) | Biolegend | Clone 29E.2A3 |
| Anti-human CD4 | Biolegend | Clone RPA-T4 |
| Anti-human CD64 | Biolegend | Clone 10.1 |
| Anti-human CD86 | Biolegend | Clone IT2.2 |
| Anti-human HLA-DR | Biolegend | Clone L243 |
| Anti-human MerTK | Biolegend | Clone 590H11G1E3 |
| Anti-human STAT3 | Cell Signaling Technology | Clone D1A5 |
| Anti-human phosphoTyr705-STAT3 | Cell Signaling Technology | Clone D3A7 |
| Anti-human SAMHD1 | Gift from Dr. O Schwartz Institut Pasteur, Paris, France | (Laguette et al., 2011) |
| Anti-human phosphoThr592-SAMHD1 | ProSci | Cat# 8005 |
| Anti-human IFITM1/2/3 | Santa Cruz Biotechnology | Clone FL-125 |
| Anti-human C/EBP-β | Santa Cruz Biotechnology | Clone H-7 |
| Anti-human CUGBP1 | Santa Cruz Biotechnology | Clone 3B1 |
| Anti-human Actin | Sigma-Aldrich | Clone 20–33 |
| Anti-human LC3 | Sigma-Aldrich | Cat# L8918 |
| Anti-human Gag-RD1 | Beckman Coulter | Clone KC57 |
| Anti-human CD163 | Leica/Novocastra | Clone 10D6 |
| LEAF purified anti-human IL-10 | Biolegend | CloneJES3-19F1 |
| LEAF purified rat anti-human IgG2a | Biolegend | Clone RTK2758 |
| Goat anti-rabbit IgG, AlexaFluor 555 | Thermo Fisher Scientific | Cat# A-21430 |
| Goat anti-mouse IgG, AlexaFluor 488 | Thermo Fisher Scientific | Cat# A-10684 |
| Goat anti-Mouse IgG, AlexaFluor 555 | Cell Signaling Technology | Cat# 4409 |
| Goat anti-rabbit IgG, HRP | Thermo Fisher Scientific | Cat# 32460 |
| Goat anti-mouse IgG, HRP | Thermo Fisher Scientific | Cat# 31430 |
| Bacterial and Virus Strains | ||
| M. tuberculosis H37Rv | N/A | N/A |
| HIV-1 ADA strain | Gift from Dr. S Benichou Institut Cochin, Paris, France | (Verollet et al., 2015a) |
| HIV-1 ADA Gag-iGFP strain | Gift from Dr P. Benaroch Institut Pasteur, Paris, France | (Gaudin et al., 2013) |
| HIV-1 NLAD8-VSVG | Gift from Dr. S Benichou Institut Cochin, Paris, France | (Bracq et al., 2017) |
| HIV-1 ADA (BlaM-Vpr) | N/A | N/A |
| Biological Samples | ||
| Buffy Coat | Etablissement Français du Sang, Toulouse, France | N/A |
| Patients-derived pleural effusions | División de Tisioneumonología at the Instituto de Tisioneumonologi’a Vaccarezza-University of Buenos Aires, Argentina | N/A |
| TB patients-derived peripheral blood | División de Tisioneumonología at the Instituto de Tisioneumonologi’a Vaccarezza-University of Buenos Aires, Argentina | N/A |
| TB/HIV patients-derived peripheral blood | Division de SIDA at the Hospital F.J Muñiz, Buenos Aires, Argentina | N/A |
| Healthy subjects-derived peripheral blood | Blood Transfusion Service, Hospital Fernandez, Buenos Aires, Argentina | N/A |
| Histological slides of lung biopsies from rhesus macaques | Tulane National Primate Research Center | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human M-CSF | Peprotech | Cat# 300-25 |
| Stattic | Sigma-Aldrich | Cat# S7947 |
| TNTi | Pharmeks | N/A |
| Cytochalasin D | Sigma-Aldrich | Cat# 22144-77-0 |
| Maraviroc | Sigma-Aldrich | Cat# 376348-65-1 |
| Bafilomycin A1 | Sigma-Aldrich | Cat# 88899-55-2 |
| Critical Commercial Assays | ||
| Mouse anti-human CD14 microbeads | Miltenyi Biotec | Cat# 130-050-201 |
| LS magnetic columns | Miltenyi Biotec | Cat# 130-042-401 |
| Protein G Agarose, Fast Flow | Millipore | Cat# 16-266 |
| Renilla Luciferase Assay | Promega | Cat# E2810 |
| Amersham ECL Prisme Western Blotting Detection Reagent | GE Healthcare | Cat# RPN2232 |
| SuperSignal WestPico Chemiluminescent Substrate | Thermo Scientific | Cat# 34080 |
| Matrigel | BD Bioscience | Cat# 356234 |
| IL-10 ELISA set | BD Bioscience | Cat# 555157 |
| Human CD163 ELISA kit | BD Bioscience | Cat# DY1607-05 |
| Human Mer ELISA kit | BD Bioscience | Cat# DY6488 |
| Human TruStain FcX | Biolegend | Cat#422302 |
| Cell Dissociation Buffer | Thermo Fisher Scientific | Cat# 13151014 |
| Phalloidin AlexaFluor 488 | Thermo Fisher Scientific | Cat# A12379 |
| DAPI | Sigma Aldrich | Cat# D9542 |
| Vybrant® DiO | Thermo Fisher Scientific | Cat# V22886 |
| CellTracker Red CMPTX Dye | Thermo Fisher Scientific | Cat# C34552 |
| CellTracker Green CMFDA Dye | Thermo Fisher Scientific | Cat# C7025 |
| Fluorescence Mounting Medium | Agilent Technologies | Cat# S302380-2 |
| Prolong anti-fade reagent | Thermo Fisher Scientific | Cat# P10144 |
| Experimental Models: Cell Lines | ||
| TZM-bl cell line | NIH AIDS Reagent Program | Cat# 8129 |
| Software and Algorithms | ||
| ImageJ | ImageJ | www.imagej.nih.gov/ij |
| Prism (v5) | GraphPad | www.graphpad.com |
| Photoshop CS3 | Adobe | http://www.adobe.com |
| Illustrator CS3 | Adobe | http://www.adobe.com |
| Huygens Professional Version 16.10 | Scientific Volume Imaging | https://svi.nl/HuygensProfessional |
| FACS DIVA | BD Bioscience | http://www.bdbiosciences.com/ |
| FlowJo 7.6.5 | TreeStar | https://www.flowjo.com/ |
| FCS Express V3 | DeNovo Software | http://www.denovosoftware.com |
| Image Lab | Bio-Rad Laboratories | http://www.bio-rad.com |
Preparation of human monocytes and monocyte-derived macrophages
Monocytes from healthy subjects (HS) were isolated and differentiated into macrophages as previously described (Balboa et al., 2011; Lastrucci et al., 2015; Verollet et al., 2015a). Briefly, purified CD14+ monocytes from HS were differentiated for 5–7 days in RPMI-1640 medium (GIBCO), 10% Fetal Bovine Serum (FBS, Sigma-Aldrich) and human M-CSF (Peprotech) at 20 ng/mL. Differentiated macrophages were used as the cellular source to prepare the CmMTB after infection with Mtb. The cell medium was renewed every 3 or 4 days. For the human samples of peripheral blood and pleural effusion (PE) from nonTB, TB and HIV-1/TB patients and HS, mononuclear cells were isolated by Ficoll Hypaque gradient centrifugation (Pharmacia, Uppsala, Sweden), as described previously (Balboa et al., 2011; Lastrucci et al., 2015). Isolated monocytes from patients were allowed to adhere to 24-well plates, at 5x105 cells/well for 1 h at 37°C in warm RPMI-1640 medium. The medium was then supplemented to a final concentration of 10% FBS and human recombinant M-CSF at 10 ng/mL. Flow cytometry analysis was performed in mononuclear cells, gating within the CD14+ population.
METHODS DETAILS
Chemicals
When stated, monocytes (day 0) are treated with STAT3 activation inhibitor Stattic (1μM, kindly provided by Fabienne Meggetto from CRCT, INSERM Toulouse). In some conditions, monocytes (day 0) were treated with an inhibitor of TNT formation (TNTi, 20μM, Pharmeks), which was renewed after HIV-1 infection. In some experiments, macrophages were treated with Cytochalasin D (Sigma-Aldrich) at 2 μM. DMSO alone was used as vehicle control.
Preparation of conditioned media and cytokine depletion
The preparation of the conditioned medium of Mtb-infected macrophages (CmMTB) has been reported previously (Lastrucci et al., 2015). Briefly, macrophages were infected with Mtb H37Rv at a MOI of 3. The conditioned control medium (CmCTR) was obtained from uninfected macrophages. After incubation at 37°C for 18h, culture media were collected, sterilized by filtration (0.2μm pores) and aliquots were stored at −80°C. For cytokine depletion experiments, CmCTR or CmMTB were incubated with 10 μg/mL of control IgG or neutralizing IL-10 antibody (JES3-19F1, Biolegend) for 1h at 4°C. Afterwards, 50 μg/mL of 50% slurry Protein G agarose beads (EMD Millipore) were added and incubated for 1h at 4°C. Conditioned media were centrifuged (300g) to remove antibody-bead complexes and then filtered (0.2 μm pores) before use. The depletion was controlled by ELISA.
Conditioning of monocytes with the secretome of Mtb-infected macrophages or pleural effusions from TB patients
Freshly isolated CD14+ monocytes from HS were allowed to adhere in the absence of serum (4×105cells/24-well in 50μl or 2×106 cells/6-well in 1.5ml). After 1h of culture, CmMTB or CmCTR supplemented with 20 ng/mL M-CSF and 20% FBS were added to the cells (vol/vol). For experiments with PE, samples were collected in heparin tubes and centrifuged at 300 g for 10 minutes at room temperature without brake. The cell-free supernatant was transferred into new plastic tubes, further centrifuged at 12000 g for 10 minutes and aliquots were stored at −80°C. After having the diagnosis of the PE, pools were prepared by mixing same amounts of individual PE associated to a specific etiology. The pools were de-complemented at 56°C for 30 minutes, and filtered by 0.22 μm in order to remove any remaining debris or residual bacteria. Particularly, we studied 21 patients with PE that were divided according their etiology. First group had 8 patients with tuberculous PE (PE-TB), second group had 8 patients with parapneumonic PE (PE-NPI), and third group had 5 patients with transudates secondary to heart failure (PE-HF). PE-TB or PE-nonTB (PE-NPI and PE-HF) supplemented with 40ng/mL M-CSF and 40% FBS were added to the cells (25% vol/vol). Cells were then cultured for 3 days. Monocytes were also conditioned in presence of 10 ng/mL M-CSF and 10 ng/mL recombinant human Interleukin-10 (IL-10) (PeproTech). Cell-surface expression of macrophage activation markers was measured by flow cytometry using standard procedures detailed in supplemental information.
HIV-1 infection of cells
Cells were infected with HIV-1 ADA strain at day 3 of culture at MOI 0.1. HIV-1 infection and replication were assessed 10 days post-infection by measuring p24-positive cells by immunostaining and the level of p24 released in culture media by ELISA.
Blam-Vpr fusion assay
Experiments were carried out as described previously (Garcia-Perez et al., 2015). Briefly, monocytes adhered on glass coverslips were conditioned for 3 days with CmMTB or CmCTR medium and then collected with cell dissociation buffer (Thermo Fisher Scientific). Then, 1.5 × 105 cells were incubated for 3 h at 37°C with 100 ng p24 of a Betalactamase (BlaM)-vpr-containing ADA virus. Infected cells were next incubated for 2 h with the CCF2/AM dye according to the manufacturer’s instructions (Invitrogen). After washing, cells were fixed in 2% paraformaldehyde. Enzymatic cleavage of CCF2 by BlaM, which causes a blue-shift in the CCF2 fluorescence emission spectrum, was measured by flow cytometry (FACSCanto, BD Biosciences).
Flow cytometry
Staining of conditioned monocytes-to-macrophage population was performed as previously described (Lastrucci et al., 2015). Adherent cells were harvested using Cell Dissociation Buffer (Life Technologies), centrifuged for 5 min at 340xg, incubated in staining buffer (PBS, 2mM EDTA, 0.5% FBS) with a 1:100 dilution of Human TruStain FcX (Biolegend) for 5 minutes at room temperature. Cells were then stained in cold staining buffer for 25 min with fluorophore-conjugated antibodies (See Table S4) and in parallel, with the corresponding isotype control antibody using a general dilution of 1:400. After staining, the cells were washed with cold staining buffer, centrifuged for 5 min at 340g at 4°C (twice), and analyzed by flow cytometry using LSR-II flow cytometer (BD Biosciences) and the associated BD FACSDiva software. Data was then analyzed using the FlowJo 7.6.5 software (TreeStar). The monocyte-to-macrophage population was first gated according to its Forward Scatter (FSC) and Size Scatter (SSC) properties before singlet selection and analysis of the percentage of positive cells and the median fluorescence intensity (MFI) for each staining.
Cells from nonTB, TB and HIV/TB patients and related controls from HS (2 × 105 cells) were labeled as described above and acquired in a FACSAria II cytometer (BD Biosciences) and analyzed using FCS Express V3 software (De Novo Software, Los Angeles, CA, USA).
Immunofluorescence microscopy
Cells were fixed with PFA 3.7%, Sucrose 30mM in PBS. Stainings were performed as described (Verollet et al., 2015a). Briefly, cells were permeabilized with Triton X-100 0,3% for 10 minutes, and saturated with PBS BSA 1% for 30 minutes. Cells were incubated with anti-Gag KC57 antibody (1:100, Beckman Coulter) in PBS BSA 1% for 1 hour, washed and then incubated with Alexa Fluor 555 Goat anti-Mouse IgG secondary antibody (1:1000, Cell Signaling Technology), Alexa Fluor 488 Phalloidin (1:500, Thermo Fisher Scientific) and DAPI (500 ng/mL, Sigma Aldrich) in PBS BSA 1% for 30 minutes. Coverslips were mounted on a glass slide using Fluorescence Mounting Medium (Dako). Slides were visualized with a Leica DM-RB fluorescence microscope or a FV1000 confocal microscope (Olympus). For deconvolution images shown in Figures 5E, S5A and C, macrophages were imaged over nine planes at 100 nm intervals with a 100x/1.49 Nikon objective mounted on a Nikon Eclipse Ti-E and an Hamamatsu Orca flash 4.0 LT sCMOS. All images were deconvolved with Huygens Professional version 16.10 (Scientific Volume Imaging, The Netherlands), using the CMLE algorithm, with SNR:20 and 40 iterations.
For the deconvolution image shown in Figure 7D, imaging was performed using a dragonfly multimodal imaging platform in three-dimensional fast confocal mode using a 63x/1.47 objective and a Zyla 4.2 PLUS sCMOS camera (Andor).
Tunneling nanotubes (TNTs) were detected and counted using Phalloidin and microtubules staining. We quantified both thick and thin nanotubes: thin membrane nanotubes contained only F-actin, whereas thick TNTs contained both F-actin and microtubules.
As HIV-1 infection of macrophages makes them fuse into MGC (Orenstein, 2001; Verollet et al., 2010), the number of infected cells largely underestimates the rate of infection. Thus, to better reflect the rate of infection, we quantified HIV infection index (HIV-stained area divided by total cell area × 100), percentage of nuclei in multinucleated cells and percentage of multinucleated cells. These parameters were assessed by using semi-automatic quantification with home-made Image J macros, allowing the study of more than 5,000 cells per condition in at least five independent donors.
HIV-1 and DiO transfer monitoring
Freshly isolated CD14+ monocytes from HS were allowed to adhere in the absence of serum (2×106 cells/6-well in 1.5ml). After 1h of culture, RPMI medium supplemented with 20 ng/mL M-CSF and 20% FBS and 20ng/mL IL-10 were added to the cells (vol/vol). At day 3, half cells are infected with 120ng p24 of a HIV-1 NLAD8 strain pseudotyped with a VSVG envelope. The other half cells are stained with CellTracker Green CMFDA Dye at day 5 (Thermo Fisher Scientific). Briefly, cells are washed with Mg2+/Ca2+-containing PBS and stained for 30 minutes in Mg2+/Ca2+-containing PBS supplemented with 500 ng/mL CellTracker, then washed with FBS-containing RPMI medium. HIV+ and CellTracker+ were then detached using accutase and co-cultured at a 1:1 ratio. In order to co-culture HIV+ and CellTracker+ cells in conditions that allow their physical separation, HIV+ cells were plated on 6.5-mm transwell filters with 0.4 μm pore polyester membrane insert (Sigma-Aldrich) while CellTracker+ cells were plated on coverslips at the bottom of 24-well plates holding the transwell filters. The cells therefore shared only the culture medium but were unable to physically contact each other. For the cell-free HIV-1 control, CellTracker+ cells were incubated with the supernatant of HIV+ cells containing produced virions. After 24 h, cells were fixed, stained and imaged.
For DiO transfer monitoring, cells were stained either with CellTracker Red CMPTX Dye (Thermo Fisher Scientific) as described above, or with a lypophylic tracer Vybrant® DiO (Thermo Fisher Scientific), according to manufacturer’s instructions. Then, cells were detached using accutase, co-cultured at a 1:1 ratio in 8-well Lab-Tek® chambers for 24h and imaged.
Scanning electron microscopy
Scanning electron microscopy observations were performed as previously described (Lastrucci et al., 2015). Briefly, HIV-1-infected macrophages were fixed in 2.5% glutaraldehyde/3,7% PFA/0.1M sodium cacodylate (pH 7.4), post-fixed in 1% osmium tetraoxide (in 0.2M cacodylate buffer), dehydrated in a series of increasing ethanol. Critical point was dried using carbon dioxide in a Leica EMCPD300. After coating with gold, cells were examined with a FEI Quanta FEG250 scanning electron microscope.
Immunoblot analyses
Total protein lysates were extracted as previously described (Lastrucci et al., 2015; Verollet et al., 2015a). Total proteins were separated through SDS-polyacrylamide gel electrophoresis, transferred and immunoblotted overnight at 4°C with indicated primary antibodies (See Key Resource Table). Secondary antibodies were the following: Anti-rabbit and anti-mouse IgG, HRP-linked Antibody (Cell Signaling Technology). Proteins were visualized with Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare). Chemiluminescence was detected with ChemiDoc Touch Imaging System (Bio-Rad Laboratories) and quantified using Image Lab software (Bio-Rad Laboratories). For the detection of LC3 autophagy protein, monocytes were seeded on 12-well plates at a density of 0.8 × 106 cells by well. After infection and treatment with Bafilomycin A1 (100 nM) or DMSO vehicle control for 2 h, cells were lysed. Proteins harvested were subjected to a SDS-polyacrylamide gel electrophoresis, 4–15% gradient (Biorad). Blots were blocked with 5% dried milk in PBS, incubated with LC3 primary antibody and then with the corresponding horseradish peroxidase-conjugated secondary antibody (ThermoScientific). Staining was detected with SuperSignal WestPico Chemiluminescent Substrate (ThermoScientific), and immunostained proteins were visualized using the ChemidocTouch Imaging System (Biorad). Densitometric LC3-II/actin ratios were measured using the Image J software.
3D migration assays
3D migration assays of cells in Matrigel (10–12 mg/mL, BD Biosciences) were performed as described (Verollet et al., 2015a) and quantified at 48h. Briefly, pictures of cells were taken automatically with a 10X objective at constant intervals using the motorized stage of an inverted microscope (Leica DMIRB, Leica Microsystems). Cells were counted using Image J software as described previously (Verollet et al., 2015a). The number of cells inside the matrix (% of migration measured after 48h of migration) was quantified.
Protein quantifications in cell-free fluids by ELISA
Soluble protein concentration was measured in cell supernatants by ELISA using kits from BD Bioscience (IL-10) and R&D Systems (sCD163 and sMer), according to manufacturer’s instructions.
Cytotoxicity assay
To exclude any impact of cytotoxicity of TNTi used in this study we measured lactate dehydrogenase (LDH) activity released from the cytosol of damaged cells into the supernatant. LDH release was measured using the Cytotoxicity Detection kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. Monocytes were treated with TNTi (20μM) or DMSO as a control for 13 days. Cell-free culture supernatants were collected and incubated with LDH assay solution at 25°C for 30 min. The optical density values were analyzed at 490 nm by subtracting the reference value at 620 nm.
Phagocytosis assay
IgG-opsonized zymosan was prepared by incubating particles in suspension in PBS with human IgG (13 mg/ml, 30 min at 37°C) and washing as described (Le Cabec and Maridonneau-Parini, 1994). The number of particles was counted in a Malassez chamber. FITC-labeled zymosan particles were then added at a MOI of 5 and incubated for 30 minutes at 37°C. Cells were then extensively washed with PBS and fixed in 3,7% PFA. To exclusively quantify phagocytosis and not binding, extracellular zymosan was stained as described (Le Cabec et al., 2000).
Histological analyses
Paraffin embedded tissue samples were sectioned and stained with hematoxylin and eosin for histomorphological analysis. Immunohistochemical staining was performed on paraffin-embedded tissue sections, using anti-CD163 (10D6, mouse mAb, Leica/Novocastra) and anti-pSTAT3 (D3A7 rabbit mAb, Cell Signaling Technology). Immunostaining of paraffin sections was preceded by different antigen unmasking methods. After incubation with primary antibodies, sections were incubated with biotin-conjugated polyclonal anti-mouse or anti-rabbit immunoglobulin antibodies followed by the streptavidin-biotin-peroxidase complex (ABC) method (Vector Laboratories) and then were counter-stained with hematoxylin. Slides were scanned with the Panoramic 250 Flash II (3DHISTECH). Virtual slides were automatically quantified for macrophage distribution as previously described (Lastrucci et al., 2015; Verollet et al., 2015a). For confocal microscopy, samples were stained with primary antibodies as described above and followed by anti-mouse IgG isotype specific or anti-rabbit IgG antibodies labeled with Alexa488 and Alexa555 (Molecular Probes). Samples were mounted with Prolong® Antifade reagent (Molecular Probes) and examined using a 60x/1.40N.A. objective of an Olympus FV1000 confocal microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Information on the statistical tests used, and the exact values of n (number of donors) can be found in the Figure Legends. All statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software Inc.). The statistical tests were chosen according to the following. Two-tailed paired or unpaired t-test was applied on data sets with a normal distribution (determined using Kolmogorov-Smirnov test), whereas two-tailed Mann-Whitney (unpaired test) or Wilcoxon matched-paired signed rank tests were used otherwise. p<0.05 was considered as the level of statistical significance (* p≤0.05; ** p≤0.005; *** p≤0.0005; **** p≤0.0001).
Supplementary Material
Movie 1 (Related to Figure 5A (left) and Figure S5A (left)). Z-stack deconvoluted microscopy images, showing F-actin staining of thick TNT in day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 2 (Related to Figure 5A (right) and Figure S5A (right)). Z-stack deconvoluted microscopy images, showing F-actin staining of thin TNTs in day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 3 (Related to Figure 5E). Z-stack deconvoluted microscopy images, showing HIV-1 Gag (red) and F-actin (green) staining of day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 4 (Related to Figure 5 and Figure S5C). 3D reconstitution of deconvoluted confocal images, showing M-Sec (red) and F-actin (green) staining of day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 5 (Related to Figure 7D). 3D reconstitution of deconvoluted confocal images, showing HIV-1 Gag (red), CellTracker (green), F-actin (gray) and nuclei (blue) of HIV+ macrophages co-cultured with CellTracker+ macrophages for 6h (both populations were previously treated with IL-10, 10ng/mL).
Movie 6 (Related to Figure 7D). Volume rendering of 3D reconstitution of deconvoluted confocal images, showing HIV-1 Gag (red), CellTracker (green), F-actin (gray) and nuclei (blue) of HIV+ macrophages co-cultured with CellTracker+ macrophages for 6h (both populations were previously treated with IL-10, 10ng/mL).
ACKNOWLEDGMENTS
We greatly acknowledge F. Capilla and C. Salon, US006/CREFRE, and M. Ben Nej i for histology analyses; M. Dubois, M. Cazabat, M. Requena and J. Izopet, BiVic facility and P. Constant, F. Levillan, F. Moreau and C. Berrone, IPBS (ASB3 and P3 multi-pathogens), for BSL3 facilities; I. Fourqueaux, CMEAB, for SEM imaging; T. Mangeat, LBCMCP, for deconvolution imaging; and A. Peixoto, IPBS, for imaging and flow cytometry analyses. The authors acknowledge the Genotoul- TRI and ANEXPLO facilities. We thank I. Gaillard and S. Caldirola, Immunology Division of the Hospital de Niños R. Gutiérrez (Buenos Aires), for their technical assistance. We greatly thank I. Staropoli and Z. Zhou, Pasteur Institute, for HIV-1 entry experiments and C. A. Spinner, IPBS, C. Gutierrez, IPBS, A. Moris, Cimi-Paris and N. Iakobachvili, Maastricht University, for critical reading of the manuscript and helpful comments. This work was supported by the Centre National de la Recherche Scientifique, Université Paul Sabatier, the Agence Nationale de la Recherche (ANR 2010–01301, ANR14-CE11-0020-02, ANR16-CE13-0005-01, ANR-11-EQUIPEX-0003), the Agence Nationale de Recherche sur le Sida et les hépatites virales (ANRS2014-CI-2, ANRS2014–049), the ECOS-Sud program (A14S01), the Fondation pour la Recherche Médicale (DEQ2016 0334894; DEQ2016 0334902), the Fondation Bettencourt-Schueller, INSERM Plan Cancer, the Argentinean National Agency of Promotion of Science and Technology (PICT2015–0055) and Alberto J. Roemmers Fundation (2016). We also thank the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. The NHP study was supported by NIH award OD011104 and AI058609 (to the Tulane National Primate Research Center and bridge fund by the Tulane Vice President for Research and awards by the Wetmore TB and Leprosy Foundtion and the Louisiana Board of Regents). SS is supported by Sidaction and RG by ANRS.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have declared that no conflict of interest exists.
REFERENCES
- Ancuta P, Wang J, and Gabuzda D (2006). CD16+ monocytes produce IL-6, CCL2, and matrix metalloproteinase-9 upon interaction with CX3CL1-expressing endothelial cells. Journal of leukocyte biology 80, 1156–1164. [DOI] [PubMed] [Google Scholar]
- Ariazi J, Benowitz A, De Biasi V, Den Boer ML, Cherqui S, Cui H, Douillet N, Eugenin EA, Favre D, Goodman S, et al. (2017). Tunneling Nanotubes and Gap Junctions-Their Role in Long-Range Intercellular Communication during Development, Health, and Disease Conditions. Front Mol Neurosci 10, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos CR, Price SL, Forsyth ER, Pin JN, Shirk EN, Bullock BT, Queen SE, Li M, Gellerup D, O’Connor SL, et al. (2016). Quantitation of Productively Infected Monocytes and Macrophages of Simian Immunodeficiency Virus-Infected Macaques. Journal of virology 90, 5643–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa L, Romero MM, Basile JI, Sabio y Garcia CA, Schierloh P, Yokobori N, Geffner L, Musella RM, Castagnino J, Abbate E, et al. (2011). Paradoxical role of CD16+CCR2+CCR5+ monocytes in tuberculosis: efficient APC in pleural effusion but also mark disease severity in blood. Journal of leukocyte biology 90, 69–75. [DOI] [PubMed] [Google Scholar]
- Baxter AE, Russell RA, Duncan CJ, Moore MD, Willberg CB, Pablos JL, Finzi A, Kaufmann DE, Ochsenbauer C, Kappes JC, et al. (2014). Macrophage Infection via Selective Capture of HIV-1-Infected CD4(+) T Cells. Cell host & microbe 16, 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell LCK, and Noursadeghi M (2018). Pathogenesis of HIV-1 and Mycobacterium tuberculosis co-infection. Nature reviews Microbiology 16, 80–90. [DOI] [PubMed] [Google Scholar]
- Bracq L, Xie M, Lambele M, Vu LT, Matz J, Schmitt A, Delon J, Zhou P, Randriamampita C, Bouchet J, et al. (2017). T cell-macrophage fusion triggers multinucleated giant cell formation for HIV-1 spreading. Journal of virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo TH, Lentz MR, Autissier P, Krishnan A, Halpern E, Letendre S, Rosenberg ES, Ellis RJ, and Williams KC (2011). Soluble CD163 Made by Monocyte/Macrophages Is a Novel Marker of HIV Activity in Early and Chronic Infection Prior to and After Anti-retroviral Therapy. The Journal of infectious diseases 204, 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo TH, Soulas C, Orzechowski K, Button J, Krishnan A, Sugimoto C, Alvarez X, Kuroda MJ, and Williams KC (2010). Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS pathogens 6, e1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Sugimoto C, Liu DX, Midkiff CC, Alvarez X, Lackner AA, Kim WK, Didier ES, and Kuroda MJ (2015). Increased monocyte turnover is associated with interstitial macrophage accumulation and pulmonary tissue damage in SIV-infected rhesus macaques. Journal of leukocyte biology 97, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassol E, Cassetta L, Rizzi C, Alfano M, and Poli G (2009). M1 and M2a polarization of human monocyte-derived macrophages inhibits HIV-1 replication by distinct mechanisms. Journal of immunology 182, 6237–6246. [DOI] [PubMed] [Google Scholar]
- Cavrois M, De Noronha C, and Greene WC (2002). A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol 20, 1151–1154. [DOI] [PubMed] [Google Scholar]
- Charles TP, and Shellito JE (2016). Human Immunodeficiency Virus Infection and Host Defense in the Lungs. Semin Respir Crit Care Med 37, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KR, Quinones-Mateu ME, Wu M, Luzze H, Johnson JL, Hirsch C, Toossi Z, and Arts EJ (2002). Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. Journal of virology 76, 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs SK, Lennox J, Caliendo AM, Brown LA, and Guidot DM (2015). Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS research and human retroviruses 31, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DM, and Sowinski S (2008). Membrane nanotubes: dynamic long-distance connections between animal cells. Nature reviews Molecular cell biology 9, 431–436. [DOI] [PubMed] [Google Scholar]
- Diedrich CR, and Flynn JL (2011). HIV-1/mycobacterium tuberculosis coinfection immunology: how does HIV-1 exacerbate tuberculosis? Infection and immunity 79, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diedrich CR, O’Hern J, and Wilkinson RJ (2016). HIV-1 and the Mycobacterium tuberculosis granuloma: A systematic review and meta-analysis. Tuberculosis (Edinb) 98, 62–76. [DOI] [PubMed] [Google Scholar]
- Dupont M, Souriant S, Lugo-Villarino G, Maridonneau-Parini I, and Verollet C (2018). Tunneling Nanotubes: Intimate Communication between Myeloid Cells. Frontiers in immunology 9, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, Lewin SR, Gorry PR, Jaworowski A, Greene WC, et al. (2007). The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol 178, 6581–6589. [DOI] [PubMed] [Google Scholar]
- Esmail H, Riou C, du Bruyn E, Lai RP, Harley YXR, Meintjes G, Wilkinson KA, and Wilkinson RJ (2018). The Immune Response to Mycobacterium tuberculosis in HIV-1-Coinfected Persons. Annual review of immunology. [DOI] [PubMed] [Google Scholar]
- Espert L, Beaumelle B, and Vergne I (2015). Autophagy in Mycobacterium tuberculosis and HIV infections. Frontiers in cellular and infection microbiology 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin EA, Gaskill PJ, and Berman JW (2009). Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: a potential mechanism for intercellular HIV trafficking. Cellular immunology 254, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabriek BO, Dijkstra CD, and van den Berg TK (2005). The macrophage scavenger receptor CD163. Immunobiology 210, 153–160. [DOI] [PubMed] [Google Scholar]
- Foreman TW, Mehra S, LoBato DN, Malek A, Alvarez X, Golden NA, Bucsan AN, Didier PJ, Doyle-Meyers LA, Russell-Lodrigue KE, et al. (2016). CD4+ Tcell-independent mechanisms suppress reactivation of latent tuberculosis in a macaque model of HIV coinfection. Proceedings of the National Academy of Sciences of the United States of America 113, E5636–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez J, Staropoli I, Azoulay S, Heinrich JT, Cascajero A, Colin P, Lortat-Jacob H, Arenzana-Seisdedos F, Alcami J, Kellenberger E, et al. (2015). A single-residue change in the HIV-1 V3 loop associated with maraviroc resistance impairs CCR5 binding affinity while increasing replicative capacity. Retrovirology 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin R, Berre S, Cunha de Alencar B, Decalf J, Schindler M, Gobert FX, Jouve M, and Benaroch P (2013). Dynamics of HIV-containing compartments in macrophages reveal sequestration of virions and transient surface connections. PloS one 8, e69450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoula M, Marin Franco JL, Dupont M, Kviatcovsky D, Milillo A, Schierloh P, Morana EJ, Poggi S, Palmero D, Mata-Espinosa D, et al. (2018). Formation of Foamy Macrophages by Tuberculous Pleural Effusions Is Triggered by the Interleukin10/Signal Transducer and Activator of Transcription 3 Axis through ACAT Upregulation. Frontiers in immunology 9, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getahun H, Harrington M, O’Brien R, and Nunn P (2007). Diagnosis of smear-negative pulmonary tuberculosis in people with HIV infection or AIDS in resource-constrained settings: informing urgent policy changes. Lancet 369, 2042–2049. [DOI] [PubMed] [Google Scholar]
- Goletti D, Carrara S, Vincenti D, Giacomini E, Fattorini L, Garbuglia AR, Capobianchi MR, Alonzi T, Fimia GM, Federico M, et al. (2004). Inhibition of HIV-1 replication in monocyte-derived macrophages by Mycobacterium tuberculosis. The Journal of infectious diseases 189, 624–633. [DOI] [PubMed] [Google Scholar]
- Goletti D, Weissman D, Jackson RW, Collins F, Kinter A, and Fauci AS (1998). The in vitro induction of human immunodeficiency virus (HIV) replication in purified protein derivative-positive HIV-infected persons by recall antigen response to Mycobacterium tuberculosis is the result of a balance of the effects of endogenous interleukin-2 and proinflammatory and antiinflammatory cytokines. The Journal of infectious diseases 177, 1332–1338. [DOI] [PubMed] [Google Scholar]
- Gordon S, Pluddemann A, and Martinez Estrada F (2014). Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunological reviews 262, 36–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot F, Welsch S, and Sattentau QJ (2008). Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111, 4660–4663. [DOI] [PubMed] [Google Scholar]
- Hase K, Kimura S, Takatsu H, Ohmae M, Kawano S, Kitamura H, Ito M, Watarai H, Hazelett CC, Yeaman C, et al. (2009). M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nature cell biology 11, 1427–1432. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Bhuyan F, Hiyoshi M, Noyori O, Nasser H, Miyazaki M, Saito T, Kondoh Y, Osada H, Kimura S, et al. (2016). Potential Role of the Formation of Tunneling Nanotubes in HIV-1 Spread in Macrophages. J Immunol 196, 1832–1841. [DOI] [PubMed] [Google Scholar]
- Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, Cleary RA, Hudgens MG, Richman DD, and Garcia JV (2017). HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nature medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, et al. (2016). Macrophages sustain HIV replication in vivo independently of T cells. The Journal of clinical investigation 126, 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y, Nakata K, Hoshino S, Honda Y, Tse DB, Shioda T, Rom WN, and Weiden M (2002). Maximal HIV-1 replication in alveolar macrophages during tuberculosis requires both lymphocyte contact and cytokines. The Journal of experimental medicine 195, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip WKE, Hoshi N, Shouval DS, Snapper S, and Medzhitov R (2017). Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SB, and Swanstrom R (2018). The evolution of HIV-1 entry phenotypes as a guide to changing target cells. Journal of leukocyte biology 103, 421–431. [DOI] [PubMed] [Google Scholar]
- Jolly C, and Sattentau QJ (2004). Retroviral spread by induction of virological synapses. Traffic 5, 643–650. [DOI] [PubMed] [Google Scholar]
- Kaushal D, Mehra S, Didier PJ, and Lackner AA (2012). The non-human primate model of tuberculosis. Journal of medical primatology 41, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaji N, and Sattentau QJ (2017). Efferocytosis of Pathogen-Infected Cells. Frontiers in immunology 8, 1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, and Divangahi M (2018). Mycobacterium tuberculosis and HIV Coinfection Brings Fire and Fury to Macrophages. The Journal of infectious diseases. [DOI] [PubMed] [Google Scholar]
- Knudsen TB, Gustafson P, Kronborg G, Kristiansen TB, Moestrup SK, Nielsen JO, Gomes V, Aaby P, Lisse I, Moller HJ, et al. (2005). Predictive value of soluble haemoglobin scavenger receptor CD163 serum levels for survival in verified tuberculosis patients. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 11, 730–735. [DOI] [PubMed] [Google Scholar]
- Kuroda MJ, Sugimoto C, Cai Y, Merino KM, Mehra S, Arainga M, Roy CJ, Midkiff CC, Alvarez X, Didier ES, et al. (2018). High Turnover of Tissue Macrophages Contributes to Tuberculosis Reactivation in Simian Immunodeficiency Virus-Infected Rhesus Macaques. The Journal of infectious diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, and Benkirane M (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastrucci C, Benard A, Balboa L, Pingris K, Souriant S, Poincloux R, Al Saati T, Rasolofo V, Gonzalez-Montaner P, Inwentarz S, et al. (2015). Tuberculosis is associated with expansion of a motile, permissive and immunomodulatory CD16(+) monocyte population via the IL-10/STAT3 axis. Cell research 25, 1333–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn SD, Pisell TL, Hirsch CS, Wu M, Butera ST, and Toossi Z (2001). Anatomically compartmentalized human immunodeficiency virus replication in HLA-DR+ cells and CD14+ macrophages at the site of pleural tuberculosis coinfection. The Journal of infectious diseases 184, 1127–1133. [DOI] [PubMed] [Google Scholar]
- Le Cabec V, Cols C, and Maridonneau-Parini I (2000). Nonopsonic phagocytosis of zymosan and Mycobacterium kansasii by CR3 (CD11b/CD18) involves distinct molecular determinants and is or is not coupled with NADPH oxidase activation. Infection and immunity 68, 4736–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cabec V, and Maridonneau-Parini I (1994). Annexin 3 is associated with cytoplasmic granules in neutrophils and monocytes and translocates to the plasma membrane in activated cells. Biochem J 303 (Pt 2), 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman MM, Georges DL, Kusner DJ, Mudido P, Giam CZ, and Toossi Z (1994). Mycobacterium tuberculosis and its purified protein derivative activate expression of the human immunodeficiency virus. J Acquir Immune Defic Syndr 7, 727–733. [PubMed] [Google Scholar]
- Light RW (2010). Update on tuberculous pleural effusion. Respirology 15, 451–458. [DOI] [PubMed] [Google Scholar]
- Lugo-Villarino G, Verollet C, Maridonneau-Parini I, and Neyrolles O (2011). Macrophage polarization: convergence point targeted by mycobacterium tuberculosis and HIV. Frontiers in immunology 2, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, and Eugenin EA (2016). Mechanisms of HIV Neuropathogenesis: Role of Cellular Communication Systems. Current HIV research 14, 400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancino G, Placido R, Bach S, Mariani F, Montesano C, Ercoli L, Zembala M, and Colizzi V (1997). Infection of human monocytes with Mycobacterium tuberculosis enhances human immunodeficiency virus type 1 replication and transmission to T cells. The Journal of infectious diseases 175, 1531–1535. [DOI] [PubMed] [Google Scholar]
- Maridonneau-Parini I (2014). Control of macrophage 3D migration: a therapeutic challenge to limit tissue infiltration. Immunol Rev 262, 216–231. [DOI] [PubMed] [Google Scholar]
- McCoy-Simandle K, Hanna SJ, and Cox D (2016). Exosomes and nanotubes: Control of immune cell communication. Int J Biochem Cell Biol 71, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra S, Golden NA, Dutta NK, Midkiff CC, Alvarez X, Doyle LA, Asher M, Russell-Lodrigue K, Monjure C, Roy CJ, et al. (2011). Reactivation of latent tuberculosis in rhesus macaques by coinfection with simian immunodeficiency virus. Journal of medical primatology 40, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. (2014). Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Rom WN, Honda Y, Condos R, Kanegasaki S, Cao Y, and Weiden M (1997). Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication in the lung. American journal of respiratory and critical care medicine 155, 996–1003. [DOI] [PubMed] [Google Scholar]
- Naphade S, Sharma J, Gaide Chevronnay HP, Shook MA, Yeagy BA, Rocca CJ, Ur SN, Lau AJ, Courtoy PJ, and Cherqui S (2015). Brief reports: Lysosomal cross-correction by hematopoietic stem cell-derived macrophages via tunneling nanotubes. Stem Cells 33, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, and Berry MP (2013). The immune response in tuberculosis. Annual review of immunology 31, 475527. [DOI] [PubMed] [Google Scholar]
- Okafo G, Prevedel L, and Eugenin E (2017). Tunneling nanotubes (TNT) mediate long-range gap junctional communication: Implications for HIV cell to cell spread. Scientific reports 7, 16660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onfelt B, Nedvetzki S, Benninger RK, Purbhoo MA, Sowinski S, Hume AN, Seabra MC, Neil MA, French PM, and Davis DM (2006). Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol 177, 8476–8483. [DOI] [PubMed] [Google Scholar]
- Orenstein JM (2000). In vivo cytolysis and fusion of human immunodeficiency virus type 1-infected lymphocytes in lymphoid tissue. The Journal of infectious diseases 182, 338–342. [DOI] [PubMed] [Google Scholar]
- Orenstein JM (2001). The macrophage in HIV infection. Immunobiology 204, 598–602. [DOI] [PubMed] [Google Scholar]
- Orenstein JM, Fox C, and Wahl SM (1997). Macrophages as a source of HIV during opportunistic infections. Science 276, 1857–1861. [DOI] [PubMed] [Google Scholar]
- Queval CJ, Song OR, Deboosere N, Delorme V, Debrie AS, Iantomasi R, Veyron-Churlet R, Jouny S, Redhage K, Deloison G, et al. (2016). STAT3 Represses Nitric Oxide Synthesis in Human Macrophages upon Mycobacterium tuberculosis Infection. Scientific reports 6, 29297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca CJ, Goodman SM, Dulin JN, Haquang JH, Gertsman I, Blondelle J, Smith JLM, Heyser CJ, and Cherqui S (2017). Transplantation of wild-type mouse hematopoietic stem and progenitor cells ameliorates deficits in a mouse model of Friedreich’s ataxia. Science translational medicine 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina B, Bracq L, Dupont M, Souriant S, Usmani SM, Proag A, Pingris K, Soldan V, Thibault C, Capilla F, et al. (2018). Bone degradation machinery of osteoclasts: An HIV-1 target that contributes to bone loss. Proceedings of the National Academy of Sciences of the United States of America 115, E2556–E2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DG, Barry CE 3rd, and Flynn JL (2010). Tuberculosis: what we don’t know can, and does, hurt us. Science 328, 852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather S, Kenyon KD, Lefkowitz JB, Liang X, Varnum BC, Henson PM, and Graham DK (2007). A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood 109, 10261033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau QJ, and Stevenson M (2016). Macrophages and HIV-1: An Unhealthy Constellation. Cell host & microbe 19, 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierloh P, Yokobori N, Aleman M, Landoni V, Geffner L, Musella RM, Castagnino J, Baldini M, Abbate E, de la Barrera SS, et al. (2007). Mycobacterium tuberculosis-induced gamma interferon production by natural killer cells requires cross talk with antigen-presenting cells involving Toll-like receptors 2 and 4 and the mannose receptor in tuberculous pleurisy. Infection and immunity 75, 5325–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, and Mothes W (2008). Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends in cell biology 18, 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Kohler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, et al. (2008). Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nature cell biology 10, 211–219. [DOI] [PubMed] [Google Scholar]
- Singh A, Besson G, Mobasher A, and Collman RG (1999). Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood. Journal of virology 73, 6680–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Hoshino Y, Gold J, Hoshino S, Martiniuk F, Kurata T, Pine R, Levy D, Rom WN, and Weiden M (2005). Interleukin-10 induces inhibitory C/EBPbeta through STAT-3 and represses HIV-1 transcription in macrophages. American journal of respiratory cell and molecular biology 33, 406–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson GS, Bell LC, Walker NF, Tsang J, Brown JS, Breen R, Lipman M, Katz DR, Miller RF, Chain BM, et al. (2013). HIV-1 Infection of Macrophages Dysregulates Innate Immune Responses to Mycobacterium tuberculosis by Inhibition of Interleukin-10. The Journal of infectious diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toossi Z (2003). Virological and immunological impact of tuberculosis on human immunodeficiency virus type 1 disease. The Journal of infectious diseases 188, 11461155. [DOI] [PubMed] [Google Scholar]
- Toossi Z, Nicolacakis K, Xia L, Ferrari NA, and Rich EA (1997). Activation of latent HIV-1 by Mycobacterium tuberculosis and its purified protein derivative in alveolar macrophages from HIV-infected individuals in vitro. Journal of acquired immune deficiency syndromes and human retrovirology: official publication of the International Retrovirology Association 15, 325–331. [DOI] [PubMed] [Google Scholar]
- Verollet C, Souriant S, Bonnaud E, Jolicoeur P, Raynaud-Messina B, Kinnaer C, Fourquaux I, Imle A, Benichou S, Fackler OT, et al. (2015a). HIV-1 reprograms the migration of macrophages. Blood 125, 1611–1622. [DOI] [PubMed] [Google Scholar]
- Verollet C, Souriant S, Raynaud-Messina B, and Maridonneau-Parini I (2015b). [HIV-1 drives the migration of macrophages]. Medecine sciences : M/S 31, 730–733. [DOI] [PubMed] [Google Scholar]
- Verollet C, Zhang YM, Le Cabec V, Mazzolini J, Charriere G, Labrousse A, Bouchet J, Medina I, Biessen E, Niedergang F, et al. (2010). HIV-1 Nef triggers macrophage fusion in a p61Hck- and protease-dependent manner. J Immunol 184, 7030–7039. [DOI] [PubMed] [Google Scholar]
- Vorster MJ, Allwood BW, Diacon AH, and Koegelenberg CF (2015). Tuberculous pleural effusions: advances and controversies. Journal of thoracic disease 7, 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Santini PA, Sullivan JS, He B, Shan M, Ball SC, Dyer WB, Ketas TJ, Chadburn A, Cohen-Gould L, et al. (2009). HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nature immunology 10, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Kulkosky J, Acheampong E, Nunnari G, Sullivan J, and Pomerantz RJ (2004). HIV-1-mediated apoptosis of neuronal cells: Proximal molecular mechanisms of HIV-1-induced encephalopathy. Proceedings of the National Academy of Sciences of the United States of America 101, 7070–7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y (2011). Tunneling-nanotube: A new way of cell-cell communication. Communicative & integrative biology 4, 324–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nakata K, Weiden M, and Rom WN (1995). Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication by transcriptional activation at the long terminal repeat. The Journal of clinical investigation 95, 2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L (2007). The CD14+ CD16+ blood monocytes: their role in infection and inflammation. Journal of leukocyte biology 81, 584–592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1 (Related to Figure 5A (left) and Figure S5A (left)). Z-stack deconvoluted microscopy images, showing F-actin staining of thick TNT in day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 2 (Related to Figure 5A (right) and Figure S5A (right)). Z-stack deconvoluted microscopy images, showing F-actin staining of thin TNTs in day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 3 (Related to Figure 5E). Z-stack deconvoluted microscopy images, showing HIV-1 Gag (red) and F-actin (green) staining of day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 4 (Related to Figure 5 and Figure S5C). 3D reconstitution of deconvoluted confocal images, showing M-Sec (red) and F-actin (green) staining of day 6 HIV-1 infected macrophages, treated with CmMTB.
Movie 5 (Related to Figure 7D). 3D reconstitution of deconvoluted confocal images, showing HIV-1 Gag (red), CellTracker (green), F-actin (gray) and nuclei (blue) of HIV+ macrophages co-cultured with CellTracker+ macrophages for 6h (both populations were previously treated with IL-10, 10ng/mL).
Movie 6 (Related to Figure 7D). Volume rendering of 3D reconstitution of deconvoluted confocal images, showing HIV-1 Gag (red), CellTracker (green), F-actin (gray) and nuclei (blue) of HIV+ macrophages co-cultured with CellTracker+ macrophages for 6h (both populations were previously treated with IL-10, 10ng/mL).