Abstract
Epigenetic modifications and regulators represent potential molecular elements which control relevant physiological and pathological features, thereby contributing to the natural history of human disease. These epigenetic modulators can be employed as disease biomarkers, since they show several advantages and provide information about gene function, thus explaining differences among patient endophenotypes. In addition, epigenetic biomarkers can incorporate information regarding the effects of the environment and lifestyle on health and disease, and monitor the effect of applied therapies. Technologies used to analyze these epigenetic biomarkers are constantly improving, becoming much easier to use. Laboratory professionals can easily acquire experience and techniques are becoming more affordable. A high number of epigenetic biomarker candidates are being continuously proposed, making now the moment to adopt epigenetics in the clinical laboratory and convert epigenetic marks into reliable biomarkers. In this review, we describe some current promising epigenetic biomarkers and technologies being applied in clinical practice. Furthermore, we will discuss some laboratory strategies and kits to accelerate the adoption of epigenetic biomarkers into clinical routine. The likelihood is that over time, better markers will be identified and will likely be incorporated into future multi-target assays that might help to optimize its application in a clinical laboratory. This will improve cost-effectiveness, and consequently encourage the development of theragnosis and the application of precision medicine.
Keywords: Epigenetic biomarkers, DNA methylation, histone postranslational modifications, miRNAs
Introduction
Human beings are different because of genetic background, lifestyle, natural history of disease, and environmental factors. Novel medical technologies have become more precise, improving the tools available in clinical laboratory. Epigenetics, genomic, and epigenomic technologies have opened new and promising landscapes for healthcare professionals enabling them to diagnose, and monitor diseases more precisely, effectively, and rapidly. It is obvious that epigenetics is revolutionizing the potential of biomedicine and clinical diagnostics, therefore, contributing to future development of precision medicine.
In routinely clinical practice, during the process of diagnosis and therapy, there are several parameters that must be taken into account to consider a molecule as a reliable biomarker, such as high sensitivity, specificity, and positive predictive value (PPV) [1]. In an ideal clinical landscape, a biomarker should have high sensitivity and specificity, a high area under the curve (AUC) in a receiver-operator characteristic (ROC) analysis, and a high PPV. In this context, epigenetic biomarkers must be robust, routinely affordable, easy to use, and accurate in order to provide better information for patient diagnosis, prognosis, stratification, prediction of future disease development, and treatment response monitoring. Furthermore, to achieve high clinical impact, a biomarker should be accurately measurable across individuals and populations; and of course, be inexpensive [2].
Biomarker candidates are being selected from a huge number of molecules produced by cells and tissues in long-term laboratory tests, observational studies, and clinical trials. Furthermore, the clinical use of new technologies is increasingly being established in clinical laboratories, and currently it is possible to analyze nucleic acids in a wide array of biospecimens (i.e. urine, plasma, serum, milk, fresh, and frozen tissue, formalin-fixed paraffin-embedded (FFPE), etc.). Hence, these epigenetic biomarkers bear obvious potential as clinical tools for diagnosis, prognosis, monitoring disease evolution, and the facilitation of clinical decision-making.
In 2015, some of us defined an epigenetic biomarker as “any epigenetic mark or altered epigenetic mechanism (i) that can be measured in the body fluids or tissues and (ii) that defines a disease (detection); (iii) predicts the outcome of disease (prognostic); (iv) responds to therapy (predictive); (v) monitors responses to therapy or medication (therapy monitoring); and (vi) predicts risk of future disease development (risk) [3].
Considering all these precedents and the dynamic properties of epigenetic biomarkers, we can redefine an epigenetic biomarker as “any epigenetic mark or altered epigenetic mechanism which is stable and reproducible during sample processing and can be measured in the body fluids or primary types of tissue preparations (fresh, frozen, and formalin-fixed paraffin embedded) (i) that predicts risk of future disease development (risk); (ii) defines a disease (detection); (iii) reveals information about natural history of disease (iv) predicts the outcome of disease (prognostic); (v) responds to therapy (predictive); and (vi) monitors responses to therapy or medication (therapy monitoring), and and/or (vii) allows simultaneously conduction of diagnosis and targeted therapy (theragnosis).
Current state of the art of epigenetic biomarkers
Epigenetic modifications across the genome represent orchestrated phenomena which modulate the transcriptional output of the genetic code. In this sense, identifying the aberrant changes in the epigenetic landscape associated with human disease [4] and the factors promoting such alterations provides the potential for new biomarkers that contribute to clinical decisions [5-10].
There are several advantages of an epigenetic biomarker compared to a genetic biomarker. To mention just a few, an epigenetic biomarker can provide relevant information about the gene function in individual cell types, filling clinical gaps by showing to what extent specific genetic programs are controlled. Besides, an epigenetic biomarker can incorporate information from the environment [11] and lifestyle, therefore, explaining, for example, how nutrition and metabolic factors affect health and disease [2]. In this sense, epigenetic biomarkers can provide information about the natural history of the disease, thus being considered as actual bioarchives. Another advantage of epigenetic biomarkers is that, because of their own nature, a wide array of these marks (i.e. microRNAs and post-translational modifications (PTMs) in histones) are extremely stable in fluids (i.e. plasma, serum, urine, saliva, semen, and vaginal secretion, among others) [12-14] and importantly, most of them are also extremely stable in primary types of tissue preparations [i.e. fresh and frozen tissue, dried blood spot (Guthrie cards), and FFPE] [15,16]. In fact, as demonstrated in one of our studies, using accidentally thawed samples and FFPE tissues [15], we found that miRNAs were highly stable molecules in samples of compromised quality. In other studies, Bulla et al. demonstrated that DNA integrity in plasma EDTA samples at 4, −20, and −80 °C was not altered and it was possible to perform DNA methylation analysis using a 22-gene panel, reporting no significant impact on any measure regardless of storage conditions [17]. Furthermore, Joo and collaborators demonstrated that DNA from archived dried blood spots was suitable for genome-wide DNA methylation profiling using Infinum Human Methylation 450 Beadchip (Illumina, San Diego, CA) [18]. Nonetheless, novel methods of sample preservation that guarantee stability and quality of epigenetic marks are continuously being developed, as will be discussed in the next section.
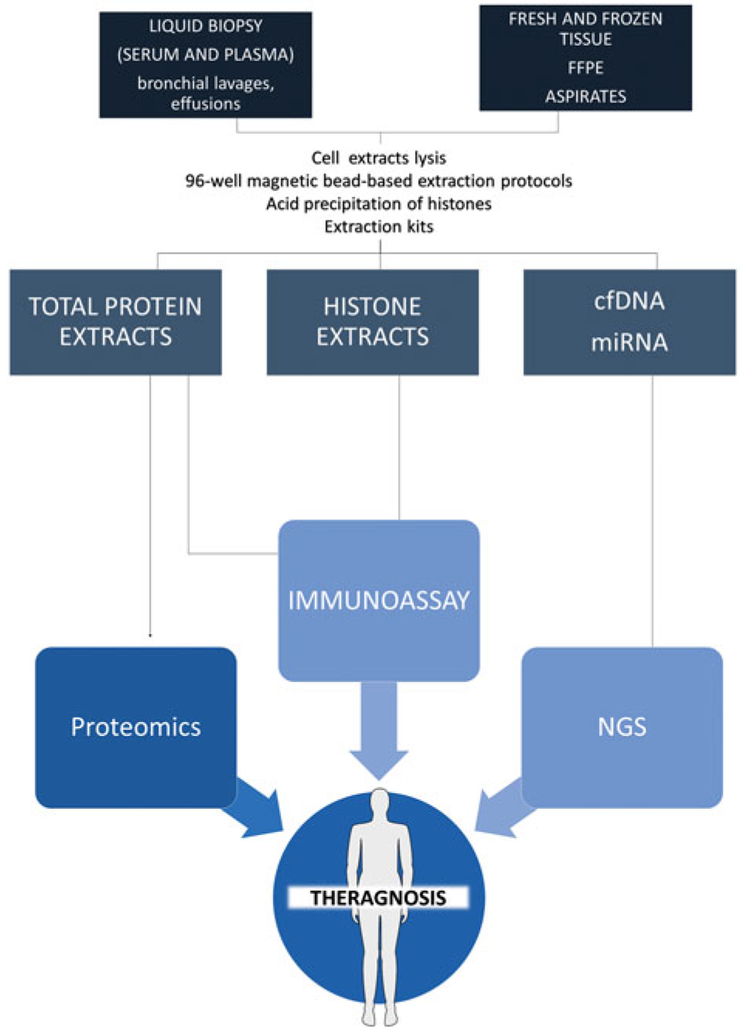
This research is showing the potential of epigenetic biomarkers to provide valuable information about disease diagnosis, prognosis, and treatment monitoring [19]. Furthermore, an epigenetic biomarker may allow simultaneous conduction of diagnosis and targeted therapy, therefore, contributing to theragnosis (Figure 1). In clinical practice, besides biomarker stability, there are some other common issues in biomarker assessment. One of the most critical is the clinical evidence required to assess test performance and cost effectiveness, to ensure the incorporation of new biomarkers in the clinical routine [20]. A marker needs to be measurable in the clinical laboratory in an acceptable window of time without the need for special handling conditions [20]. In addition, although new biomarkers are appearing, in the particular case of epigenetic markers there is a necessity to adapt the methods of analysis to the nature of the biomarker (i.e. bisulfite treatment for DNA methylation analysis, or exosome isolation/separation for circulating miRNA analysis) or to the new technologies able to perform high-throughput experiments (i.e. microarrays and next generation sequencing (NGS) to measure DNA methylation or microRNAs, HPLC coupled to mass spectrometry to detect PTMs in histones, etc.). Finally, it is important to stress that the potential purpose of the use of a new biomarker is its use to address an unmet clinical need and, therefore, the laboratory assay should be developed to measure this biomarker using a validated test in a clinical trial in the easiest and most cost-effective manner.
Figure 1.
Epigenetic biomarkers (cf(methylated)DNA, histones, and miRNAs) analyzed from multiple biospecimens (i.e. liquid biopsy, fresh tissue, and FFPE tissue) may allow simultaneous conduction of diagnosis and targeted therapy, therefore, contributing to theragnosis and precision medicine.
Many clinical laboratories are developing new and emerging technologies for clinical lab applications. In some cases, such efforts are focused on specific or modified protocols and modified self-laboratory-developed procedures to be performed within the lab’s own facilities. In other cases, clinical laboratories are also collaborating with in vitro diagnostics (IVD) manufacturers to develop or improve future generation of instruments and related tests and kits, giving them advanced insight into such products long before they are ready for commercialization. Furthermore, IVD companies may work on a series of user-friendly kits based on the detection of epigenetic biomarkers in order to further improve the diagnostics of several diseases and facilitate the access of the clinical laboratories to these tests. It is also noteworthy that clinical laboratories are establishing important collaborative efforts with academic and research institutions in order to co-develop applications of basic biological knowledge in terms of epigenetic biomarkers transference to the clinical ambit.
Improvement of extraction and purification methods
There exists different sources to obtain the material to analyze epigenetic biomarkers, and we have focused our attention on two biospecimens which are mostly used for IVD: FFPE tissue and liquid biopsy (specifically blood-derived samples, i.e. serum and plasma).
Formalin-fixed paraffin-embedded biospecimens
A problem in the clinical pathology services around the world is managing biopsies that are frequently preserved as FFPE as a standard procedure for retrospective studies, which are important for evaluating treatment history and disease outcomes. The main problem is the low quantity and quality of nucleic acids and proteins due to degradation [21]. These limitations are a challenge when we are interested in purifying nucleic acids for NGS. However, the optimization of current procedures, improvement of protocols, and the release of new commercial purification kits are allowing effective nucleic acid (DNA/RNA/miRNA) extraction from FFPE tissues for their use in different high-throughput platforms [22]. Fortunately, miRNAs are so small that challenges inherent to longer RNA analyses, such as crosslinking and fragmentation stemming from formalin fixation, do not arise [23]. In this regard, miRNA expression profiling, as with epigenetic biomarkers, is more accurate for pathologic diagnosis in FFPE tissues than mRNA expression and transcriptomic markers, due to its robustness against the consequence of formalin fixation [24].
Although methods are continuously being improved, NGS procedures for FFPE samples are currently a challenge because it is necessary to avoid nucleic acid fragmentation and the generation of DNA and RNA artifacts during preservation, which result from chemical modifications generated during the extraction procedure [25]. The process to repair nucleic acid damage is tedious and costly in time because it requires enzyme-catalyzed nucleic acid treatment to repair the DNA obtained from FFPE [26-28]. In this regard, Haile et al. optimized and automated 96-well magnetic bead-based extraction protocol which allows the construction of libraries from 100 ng of total nucleic acid, and the subsequent sequencing of DNA, RNA, and microRNAs [29].
There exist also a number of commercial magnetic bead-based protocols including: AxyMag FFPE (DNA/RNA/miRNA) kit (Corning Inc., Corning, NY), ALINE FFPE Magnapure kit (ALINE Biosciences, Woburn, MA), Agencourt FormaPure kit (Beckman Coulter, Brea, CA) and VERSANT Tissue Preparation Reagents Kit (Siemens, Munich, Germany). However, VERSANT kit used in an automated system for nucleic acid extraction was only evaluated by qRT-PCR (Bio-Rad, Hercules, CA) for gene expression and mass spectrometry for single nucleotide polymorphism analysis [30,31], and we have not found evidence of its use in high throughput technologies. Agencourt FormaPure kit was previously evaluated manually, assessing RNA from long-term stored FFPE samples (>10 years old) using qRT-PCR [32] and gene expression microarrays [33,34]. For this kit, the manufacturers confirmed that small RNAs are not lost during the protocol of extraction, thus increasing its possibilities. This assertion was validated by Haile et al., who met the input-requirements for NGS studies performed in small RNA obtained from FFPE clinical samples. They confirmed that when the protocols are optimized, the kit allows efficient removal of paraffin, reversal of formaldehyde crosslinks, and extraction of RNA (and importantly conserving miRNAs) and genomic DNA, providing nucleic acids with the sufficient quality and quantities for library construction and NGS. In addition, Haile et al. developed a series of protocols to maintain data quality obtained from NGS [29].
Liquid biopsy and biological fluids
Liquid biopsy refers to the analysis of components that can be isolated or purified and analyzed from a blood sample. For example, in cancer these components are circulating tumor cells (CTCs), cell-free circulating DNA (cfDNA), and exosomes (a part of the secreted microvesicles which can transport microRNAs, long non-coding RNAs, and double-stranded DNA) [35-38]. Furthermore, it is possible to isolate other epigenetic markers from blood samples, such as free microRNAs (miRNAs) and circulating histones and nucleosomes.
Circulating cell-free DNA.
Circulating cell-free DNA (cfDNA) has been found in various pathological conditions like cancer, autoimmune diseases, infectious diseases, stroke, sepsis, trauma, and pregnancy. The quantity of cfDNA is generally very low in healthy subjects (less than 5 ng/ml of plasma) and increases up to 8–10 times in subjects affected by a neoplastic disease. The isolation and quantification of cfDNA from body fluids represents a challenge due to their small quantity and fragmented nature. Furthermore, the extraction and purification stage is critical for the development of reproducible, standardized methods for cfDNA isolation, including quality controls for measurement of extraction efficiency, fragment size bias, and yield [39].
Devonshire et al. investigated the efficiency of methods for extraction of cfDNA from plasma and demonstrated how a spike-in containing fragment sizes relevant to cfDNA can be used to assess recovery of differently-sized DNA. They demonstrated that the extraction efficiency of the kits was better for QIAamp® circulating nucleic acid kit (Qiagen, Hilden, Germany) and QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) while FitAmp™ plasma/serum DNA isolation kit (Epigentek, Hopkinton, MA) showed the lowest efficiency. Furthermore, the QIAamp® circulating nucleic acid and NucleoSpin® Plasma XS kit (Macherey-Nagel, Bethlehem, PA) gave a better representation of smaller DNA fragments in the extract than the QIAamp DNA blood mini kit [39].
cfDNA is fragmented by nature and is found in normal healthy individuals. Tumor-derived cfDNA can differ in fragment size profile, which depends on the cellular process causing its release into circulation [40,41]. Interestingly, it has been shown that fetal-derived cfDNA (mostly shorter than 500 bp) is often more fragmented than maternal cfDNA (500–1000 bp) therefore, requiring further separation approaches in order to improve recovery of the fetal-derived fraction [42]. To improve the yield of the nucleic acid purification it would be recommended to design and use kits that allow the parallel purification and efficient recovery of DNA, RNA, and smallRNAs (i.e. miRNAs).
There are some commercial kits designed for circulating nucleic acids purification (Table 1). However, these kits are recommended for research use only, and not intended for diagnostic or therapeutic application. Qiagen also offers in its portfolio Diagnostic sample preparation kits (DSP), products that can be used manually or in an automated way using QIAcube® (Qiagen, Hilden, Germany) which complies with ISO13485 (for quality management systems requirement for regulatory purposes) and Food and Drug Administration (FDA) Quality System Regulations 21 CFR820, and are in compliance with good manufacturing processes (GMPs) and international IVD standards. For example, the QIAsymphony DSP Circulating DNA Kit (Qiagen, Hilden, Germany) is intended for in vitro diagnostic use, and it allows fully automated and simultaneous purification of human cfDNA from 2 to 4 ml of human plasma and urine, but this kit requires the QIAsymphony SP instrument (Qiagen, Hilden, Germany).
Table 1.
Commercial kits and main characteristics of six kits designed for cfDNA isolation and purification from biofluids.
| Kit name | Manufacturer | Additional and optional steps |
Advantages | Disadvantages | Protocol time |
|---|---|---|---|---|---|
| QIAamp® circulating nucleic acid kit | Qiagen | Proteinase K digestion treatment Recommended DNase I treatment for RNA downstream studies |
Allows purification and concentration of free-circulating DNA, RNA, miRNA, and viral nucleic acids from human plasma, serum, urine, or other cell-free body fluids (4–8 ml) exist in format DSP for IVD use | Requires 4ml of plasma | 3 h |
| NucleoSpin® Plasma XS kit | Macherey-Nagel | Proteinase K digestion treatment | Allows nucleic acid purification and concentration from serum, plasma, and bronchial lavage (up to 240 μl), also it allows to purify DNA from buccal swabs and dried blood spots specially designed purification column allowing elution volumes as low as 5 μl | - | 30min-1h |
| FitAmp™ plasma/serum DNA isolation kit | Epigentek | Proteinase K digestion treatment | Allows DNA isolation from plasma, serum and other body fluids (500 μl) | Low efficiencies compared with other kits [39] | 15-30 min |
| QIAamp DNA blood mini kit | Qiagen | Protease or proteinase K digestion treatment | Allows nucleic acid purification from serum, plasma and urine (from 200 μl), also it allows to purify DNA from buccal swabs and dried blood spots exist in format DSP for IVD use | - | 2-3 h |
| Maxwell 16 blood DNA kit | Promega | Allows nucleic acid purification from 500 μl. It can be used with the Maxwell® 16 Blood DNA purification system (IVD) which compiles with EU directive for in vitro diagnostic medical devices. |
The original protocol is designed for whole blood DNA isolation but not for plasma or serum. | 30 min-1h | |
| Automated QIAsymphony DSP circulating DNA kit | Qiagen | Allows purification of free-circulating DNA, from human plasma, serum, and urine (2-4 ml) 96 simultaneous samples in 6 h | Requires the QIAsymphony SP instrument | 6h |
QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany), the NucleoSpin® Plasma XS kit (Macherey Nagel, Düren, Germany), the FitAmp™ plasma/serum DNA isolation kit (Epigentek, Farmingdale, NY); QIAamp DNA blood mini kit (Qiagen, Hilden, Germany), and Maxwell 16 blood DNA kit (Promega, Madison, WI).
All of these kits have been used for cfDNA methylation studies previously. For example, cfDNA was isolated from 400 μL of plasma using the QIAamp MinElute Virus Spin Kit (Qiagen, Hilden, Germany) [43] with good quality starting material to perform bisulfite treatment and MiSeq Sequencing. In another study, cfDNA was extracted from 800 μl aliquots of serum using the Maxwell 16 blood DNA kit (Promega, Madison, WI) [44], and the yield and quality obtained was good enough to perform Illumina Infinium HumanMethylation 27 BeadChip (Illumina, San Diego, CA). Importantly, the Maxwell 16 blood DNA kit can be used with Maxwell® 16 Blood DNA Purification System (IVD), which complies with EU Directive 98/79/EC on IVD medical devices and is used in conjunction with the Maxwell® 16 IVD Instrument. The QIAamp DNA Micro Kit (Qiagen, Hilden, Germany) can be used to analyze DNA methylation by whole genome bisulfite sequencing, but it requires an additional step of adding 1 μg of carrier RNA, during the protocol for isolation of “genomic DNA from small volumes of blood” [45]. However, QIAamp Circulating NA kit (Qiagen, Hilden, Germany) requires between 4–8ml of plasma to obtain enough nucleic acid (above 100 ng) for downstream applications, such as gene library preparation for NGS experiments and qRT-PCR experiments. We can also use QIAamp DSP Circulating NA kit (Qiagen, Hilden, Germany) which is intended for IVD use and nucleic acid purification (cfDNA, RNA, and miRNAs) as described above. For NucleoSpin® Plasma XS kit (Macherey-Nagel), up to 100 ng cfDNA per mL of plasma with efficient isolation of fragmented DNA in a range of 50–1000 bp can been obtained [46], which is enough for downstream NGS applications.
Circulating miRNAs.
miRNAs can also be detected in body fluids or liquid biopsy, and because some of them show altered levels in pathologic situations, new expectancy about their potential use as biomarkers has been generated in recent years [19]. One of the challenges associated with circulating miRNA diagnostics is lower efficiency and yield of miRNA isolation from blood plasma and serum as compared to miRNA isolation from cells and tissues [47]. miRNAs and pre-miRNAs are found as free-circulating molecules, packaged into microparticles (exosomes, microvesicles, and apoptotic bodies), [48-50] associated with lipoprotein complexes (high-density lipoprotein, HDL) [51] or associated with miRNA-mediated RNA silencing proteins (such as Argonaute2 and Ago2 protein) [52] to avoid miRNA degradation.
Depending on the protocol used it is possible to isolate free circulating miRNAs, protein-bound miRNAs, only microvesicles-associated miRNAS, or total miRNA present in a blood sample [53].
There are no standardized methods to extract exosomes and different procedures are found in the literature. Therefore, exosomes and their application in clinical settings are still an active area of research. The most used protocol relies on ultracentrifugation in sucrose gradient. However, purification of exosomes using ultracentrifugation is still complicated to be implemented in the clinical setting. Because of the necessity of new optimized procedures for exosome isolation, some companies are commercializing user-friendly kits to improve the time, quality, yield, and reproducibility of exosome extraction. Some examples are listed in Table 2.
Table 2.
Commercial kits and main characteristics of six kits designed for miRNA isolation and purification from biofluids.
| Kit name | Manufacturer | Phenol/chloroform extraction step |
Advantages | Protocol time |
|---|---|---|---|---|
| miRNeasy serum/plasma kit | Qiagen | Yes | 2–4h | |
| miRCURY™ RNA isolation kit biofluids | Exiqon | No | Carrier MS2 RNA or yeast tRNA that increases the miRNA concentration | 2–3h |
| mirVana™ PARIS™ kit | Ambion | No | Allows isolating both small and large RNAs in one or two fractions | 2–4h |
| NucleoSpin® miRNA plasma | Macherey-Nagel | No | Allows isolating both small and large RNAs in one or two fractions | 1.5–2.5 h |
| Plasma/serum circulating RNA purification kit | Norgen Biotek | Yes | N/A | 2–4h |
| Direct-zol RNA MiniPrep | Zymo Research | Yes | N/A | 2–2.5 h |
| PAXgene Blood RNA Kit | Qiagen | N/A | FDA-cleared, CE-marked | 2–2.5 h (It depends of if the method is fully automated with QIAcube) |
miRNeasy serum/plasma kit (Qiagen, Hilden, Germany); miRCURY™ RNA isolation kit biofluids (Exiqon, Vedbaek, Denmark); mirVana™ PARIS™ kit (Ambion, Austin, TX); NucleoSpin® miRNA plasma (Macherey Nagel, Düren, Germany) and plasma/serum circulating RNA purification kit (slurry format) (Norgen Biotek, Thorold, Canada); Direct-zol RNA MiniPrep (Zymo Research, Irvine, CA); PAXgene Blood RNA Kit (Qiagen, Hilden, Germany). N/A: information not available.
Interest in circulating miRNA has largely intensified as growing evidence accumulates demonstrating that these molecules are potential descriptors and indicators of a wide variety of biological conditions. Several obstacles must be surmounted during ongoing, successful development of miRNA signatures of disease and other conditions, and among others, miRNA isolation and purification is a key process. Not only because of the efficient extraction needed to ensure maximum recovery of RNA from low volumes of input fluid, but for the correct identification and analysis of an epigenetic biomarkers based on miRNAs, and to avoid biased results. Furthermore, other key issues to take into account when analyzing circulating miRNAs consist of time of blood sample processing and plasma or serum storage. In fact, it is highly recommended that the blood extracted in EDTA tubes should be processed within 6 h [54]; and, if possible, within 2–4 h from collection, because blood cells, including erythrocytes, can release miRNAs. This contributes to the over-representation of some miRNAs when these are purified from plasma or serum and in turn alters miRNA biomarker signatures [55]. Regarding miRNA stability during long-term storage, Mraz et al. demonstrate, using a panel of 29 miRNAs, the high stability of these over a period of 10 months at −80 °C [56]. More recent studies have also demonstrated that miRNAs are stable in archived serum samples frozen at −25 °C for at least 40 years [57] and stable at −20 °C for at least 2–4 years [58], demonstrating the high stability of these molecules.
Tan et al. compared the performance of five commercial miRNA extraction kits against each other, using the median and coefficient of variation (CV) of Cq values of the cel-miR-39 and cel-miR-54 Spike-in controls added into plasma samples after the addition of lysis buffer [59]. Low Cq value is an indicator for high recovery while small CV suggests less extraction bias across samples analyzed. The results obtained demonstrated that four of these kits (but not the kit of Norgen Biotek, Thorold, Canada) were comparably good in recovering the Spike-in controls, and the miRCURY™ biofluids RNA isolation kit (Exiqon, Hørsholm, Denmark) and miRNeasy® serum/lasma kits (Qiagen, Hilden, Germany) were comparably efficient in recovering miRNA spike-in controls from plasma samples. Furthermore, extraction bias among samples was the lowest in the NucleoSpin® miRNA Plasma (Macherey Nagel), as it had the smallest CV of Cq values for the spike-in controls evaluated. There are critical issues to take into account when analyzing circulating miRNAs, such as sample handling and processing, blood cell contamination during sample preparation, and lack of consensus for data normalization [53,60]. Another study performed by McAlexander et al. suggested a slightly lower recovery by miRNeasy® serum/plasma kit when compared with miRCURY™ RNA biofluids kit [61]. More recently, El-Khoury et al. assessed the yield of several miRNA purification kits to purify circulating miRNAs. They observed that although miRCURY™ biofluids RNA isolation Kit provided highly pure miRNAS, miRNeasy® kit permitted a better miRNA detection in plasma samples, despite a less pure extracted RNA [62]. The miRNAeasy® serum/plasma kit showed the best performance and yield to obtain purified miRNAs which allows library construction and smallRNA-seq by NGS [63].
In addition to these commercial kits for miRNA isolation from biofluids, PAXgene Blood RNA Kit (Qiagen) is one of a few kits which is suitable for IVD, when used in PAXgene Blood RNA tubes. This system consists of PAXgene Blood RNA Tubes for blood collection, stabilization, and transport, and the PAXgene Blood RNA Kit for silica-membrane-based total RNA isolation and purification using a spin-column format. It has shown its utility to preserve the quality of RNA after long-term freezing PAXgene RNA stabilized whole blood samples [64]. miRNA purification can be carried out manually, using a microcentrifuge, or in an automated-manner using the QIAcube system (Qiagen, Hilden, Germany). The CE-IVD-marked tubes are described for RNA purification kit, but not for PAXgene blood miRNA kit. The CE-IVD-marked PAXgene kits provide exact performance specifications, assuring highly reliable RNA purification for in vitro purposes. Furthermore, the kit is intended for total RNA purification and stabilization from total blood samples, but not for plasma or serum samples; therefore, it is not suitable for circulating miRNAs purification.
Histones and nucleosomes.
The use of histones as biomarkers for disease relies upon two main possibilities: the analysis of histone PTMs and their variations in the context of disease, or the mere presence of histones in extracellular locations (mainly in the bloodstream). In the latter, analysis of histone PTMs is also a valuable tool for diagnosis and/or prognosis of certain diseases, as was previously discussed by García Giménez [19] and Reddy et al. [65]. In the case of tissue and cell extracts, histone proteins can be isolated by procedures that ensure nuclear and cytosolic separation, varying lysis buffer conditions and homogenization/sonication steps. These methods can be manually prepared in the laboratory by a combination of salt and acid precipitation steps [66], but several histone-specific extraction and purification kits (for FFPE, frozen tissue and biological fluids) can also be found in the market (see Table 3). Most of these kits are designed to produce a quick isolation of histone core proteins with easy manipulation, obtaining acceptable yields although not avoiding co-purification of other nuclear proteins, being functional analysis performed by western blotting the main downstream application. Kits specialized in obtaining whole chromatin or combinations of DNA and proteins (i.e. nucleosomes) are designed for ChIP analysis, but their use for epigenetic regulation studies is far from an application as epigenetic biomarkers of clinical relevance. In fact, there are difficulties underlying the methods to isolate histone proteins with contamination by other nuclear proteins and components. Most purification kits and methods require a high density of cells that can be obtained by homogenization of tissues or isolation of blood cells; however, detection of free or circulating histones or analysis of histone PTMs in blood requires exclusive isolation of the plasma or serum fraction. There exists scarce methods to purify histones from biological fluids, although some alternatives have been previously used to concentrate this fraction. One alternative is the use of Amicon-Ultra-15 10k centrifugal filters to concentrate histones, as previously used in urine samples [67]. More recently, Reddy et al. have developed a method called dual acid extraction protocol (DAE) for efficient isolation of histones from serum [65]. The method is basically the same as those described by Shechter et al. [66], with the exception of the use of the hypotonic lysis buffer which is not required for serum samples.
Table 3.
Commercial kits and main characteristics of seven kits designed for whole chromatin or core histones isolation and purification from different biological sources.
| Kit name | Manufacturer | Biological source | Downstream uses |
|---|---|---|---|
| Chromatrap® FFPE ChIP | Chromatrap | FFPE tissue | qPCR, sequencing |
| ChIP-IT® FFPE | Active Motif | FFPE tissue | qPCR, sequencing |
| EPIXTRACT® kits | Enzo | Mammalian cells and tissues, plasma - serum, nuclear proteins | Functional studies (WB, PTMs, enzymatic analysis) |
| Histone extraction kit | Abcam | Mammalian cells and tissues | Functional studies (WB, PTMs analysis) |
| EpiQuik total histone extraction kit | Epigentek | Mammalian cells and tissues | Functional studies (WB, PTMs analysis) |
| ChromaFlash chromatin extraction kit | Epigentek | Cultured cells, fresh, and frozen tissue | ChIP, in vitro protein-DNA binding assays and nuclear enzyme assays |
| EZExtract™ core histone isolation kit | BioVision | Tissues and cultured cells | Protein profiling, post-translational modification and epigenetic analyses |
Chromatrap® (Porvair Sciences, Norfold, UK); ChIP-IT® (Active Motif. Inc. Carlsbad, CA); EPIXTRACT® (Enzo Life Sciences Inc. Farmingdale, NY); Histone extraction kit (Abcam, Cambridge, UK); EpiQuik and ChromaFlash (Epigentek Group INC. Farmingdale, NY); EZExtract™ (BioVision INc., Milpitas, CA).
Diagnosis or monitoring of diseases, such as solid tumors, utilizing circulating epigenetic biomarkers in body fluids of patients will prove to be a very powerful tool for cancer diagnosis or therapy monitoring in addition to avoiding surgical biopsy. Novel methods on the analysis of circulating histones as epigenetic biomarkers in disease will be described in the following section.
Improvement of methods for preparation of samples for epigenetic biomarkers analysis
Sample preparation is one of the most critical steps in epigenetic research, and of course becomes highly relevant in clinical diagnostics. Efficient and high quality isolation of nucleic acids, chromatin, and histones for downstream analyses, such as DNA methylation studies and miRNA analysis, will ensure that the highest quality results can be obtained from clinical samples. Therefore, refined and improved methods are constantly being investigated.
DNA methylation
Bisulfite conversion of DNA is a required step in some of the procedures used for DNA methylation analysis. This is a problem because bisulfite-converted DNA possesses the following limitations: 1) the amount of bisulfite-converted DNA obtained from some biospecimens is often too small to permit extensive DNA methylation analyses. Therefore, sometimes the use of some additional sample material is required. This is not always available and may lead to irreproducible experimental conditions. This can be bypassed by introducing an amplification step to produce more DNA for further analysis. This amplification must be carried out after the bisulfite conversion, because genomic DNA amplified before this treatment can lose its epigenetic modifications during the amplification process; 2) bisulfite-converted DNA can undergo significant degradation upon storage, lowering the sensitivity of DNA methylation assays and making this DNA unsuitable for further analysis if it is not used immediately after bisulfite treatment [68]. Fortunately, during recent years several kits have become commercially available allowing further improvement of the quality of the conversion as well as the stability of bisulfite-converted DNA.
For DNA methylation, Holmes et al. studied the performance of different commercial kits for DNA bisulfite conversion, evaluating different biospecimens, such as FFPE, aspirates, bronchial lavages, effusions, plasma, serum, and urine. In this study, Holmes and her team compared the performance of the following kits: EpiTect Fast FFPE Bisulfite Kit, EpiTect Bisulfite Kit, EpiTect Fast DNA Bisulfite Kit (Qiagen), EZ DNA Methylation-Gold Kit, EZ DNA Methylation-Direct Kit, EZ DNA Methylation-Lightning Kit (Zymo Research, Irvine, CA), innuCONVERT Bisulfite All-In-One Kit, innuCONVERT Bisulfite Basic Kit, and innuCONVERT Bisulfite Body Fluids Kit (Analytik Jena AG, Jena, Germany) [69]. The results obtained demonstrated that all kits yielded highly pure DNA suitable for PCR analyses and good conversion of unmethylated cytosines. The innuCONVERT Bisulfite All-In-One Kit exhibited the highest versatility regarding different input biospecimens (i.e. pure extracted DNA, fresh or frozen tissue, FFPE tissue, cell lines, urine sediment, and cellular fractions of bronchial aspirates, pleural effusions, and ascites), and the highest volumes of bisulfite-converted DNA recovered at concentrations of 500 pg to 10 μg [69]. Furthermore, the innuCONVERT Bisulfite Body Fluids Kit (Analytik Jena AG) was the only commercially available kit enabling the preparation of bisulfite DNA from high volume (up to 3 ml) body fluid (blood plasma and serum) [69]. Bisulfite-treated DNA samples were stable for at least 4 weeks when stored at −80–4 °C when prepared using the above-mentioned kits. This result supports previous findings by Dietrich et al., where bisulfite-converted DNA was stable more than 2 years at −20 °C [70].
More recently, Jung et al. have detailed several methods and protocols for sample preparation, subsequent bisulfite conversion, and sample clean-up for several types from a wide array of biospecimens used in clinical settings. Furthermore, they describe that the cytosine-free fragment (CFF) allows for simultaneous quantification of bisulfite-converted and total DNA, and, therefore, the determination of bisulfite conversion efficiency. The Mer9 real-time PCR assay amplifies the bisulfite-converted sequence of the repetitive element Mer9 and enables the accurate quantification of minute DNA amounts, as occurs in FFPE and in body fluids [71].
miRNAs
RT-qPCR has become one of the most relevant tools in existent molecular diagnostics in clinical laboratories. Therefore, this is one of the most available tools to analyze miRNAs in different kinds of biospecimens during clinical diagnostics. However, samples used as a source of miRNAs must pass quality controls to avoid high degradation of nucleic acids, or contamination of other miRNAs released by other cells or tissues distinct to those required for diagnosis. For example, serum/plasma hemolysis has been reported as a source of significant variation in miRNA profiles, potentially affecting levels of a significant number of proposed biomarker microRNAs. Hemolysis can occur in different steps of sampling until the rest of circulatory components are removed. Serum/plasma samples can be checked for hemolysis by measuring the absorbance peak at 414nm [72], the hemolysis ratio [73], the H-score [74], or using the Harboe method, which measures the concentration of hemoglobin [75]. The most rapid method to check the hemolysis is using the H-score. Absorbance measured at 414nm using a NanoDrop™ 1000 spectrophotometer (Thermo Scientific, Scoresby, Victoria, Australia) that is higher than 0.2 suggests high levels of hemolysis, therefore, the samples should not be used for miRNA analysis if the target miRNAs may be released by red blood cells. In addition, if the original serum or plasma sample is no longer accessible and only purified RNA is available, an alternative method to identify putative hemolyzed samples is to use miRNAs known to be rich in erythrocytes (like miR-451) and those not influenced by hemolysis (like miR-23a-3p) [76]. Shah et al. compared several methods to evaluate hemolysis in plasma/serum samples to be used in miRNA analysis. They found that the ratio of miR-451a-miR-23a-3p [delta Cq (miR-23a-3p-miR-451a)] proposed by Blondal et al. [76] was found to be the most sensitive method to detect low levels of hemolysis, and should be included in the assay during miRNA analysis as quality control [77].
More recently, Pizzamiglio et al. propose a calibration curve for the classification of plasma samples in terms of percentage of red blood cells that allow application of the appropriate statistical methods that correct miRNA expression [78].
On the other hand, RT-qPCR equipment is available in most clinical laboratories worldwide because the technique provides high sensitivity, is cost-efficient, and allows multiplexing and a wide array of applications are available. However, typical RT-qPCR assays require reference genes to normalize Cq values obtained for the target gene, in that case miRNAs. Several authors have tried to find the most powerful miRNAs that can serve as endogenous controls in miRNA analysis from biofluids. In Table 4, some examples of miRNA used as endogenous reference miRNAs for data normalization are shown.
Table 4.
Proposed miRNAs used as endogenous control in RT-qPCR assays for detecting circulating miRNAs in liquid biopsy.
| Reference gene | Biofluid | Disease in which reference miRNA was used as endogenous control | Reference |
|---|---|---|---|
| miR-146b-5p, miR-142-3p, and miR-24 | Serum | Vaccinated healthy donors | [150] |
| miR-17, miR-126, miR-484 | Serum | Hepatitis C infection | [150] |
| miR-26a, miR-221, and miR-22 | Serum | Hepatitis B virus (HBV)-infected | [151] |
| miR-16 and miR-93 | Serum | Gastric cancer | [152] |
| miR-16 | Plasma | Friedreich’s ataxia | [63] |
| Serum | Fibromyalgia syndrome | [153] | |
| miR-25-3p | Serum | Osteoporosis | [154] |
| miR-106b-5p, miR-93-5p, and miR-25-3p | Serum | Colorectal cancer | [155] |
| miR-320a and miR-486-5p | Plasma | Human sepsis and systemic inflammatory response syndrome | [156] |
miRNA detection using RT-qPCR allows for detection of a few molecules per ml of biological fluid as detection limit thereby increasing the possibility of use of this technique to identify circulating miRNAs as biomarkers. Another important aspect is the quantitation of exosomal miRNAs, due to the scarce validation of endogenous controls for exosomal, miRNA-relative quantitation. In order to bypass this problem, the development of new techniques that allow absolute quantitation, such as droplet digital PCR (ddPCR), will revolutionize the application of miRNAS as disease biomarkers.
Histones
Analysis of histone variants and histone PTMs in research has been traditionally performed by a combination of molecular biology techniques that mostly include western blotting, chromatin immunoprecipitation (ChIP), mass spectrometry, and ELISA assays. Generally, it is a combination of different techniques that provides the most relevant information about the specific code of histone variants and modifications in a particular cellular context. Nonetheless, these techniques are barely used in clinical routine or have limited diagnostic/prognostic application. It should be taken into account that all purification methods are headed to the isolation of histone or nucleosomal samples with analytical purposes, and are mainly utilized in research. These techniques are still far from producing samples adequate for high-throughput analysis and quick application into clinical routine. However, several improvements are being made in other directions, mainly making use of the powerful capabilities of modern mass spectrometry technology, as we will discuss further on.
Epigenetic biomarkers (currently used in clinical protocols and approved by the FDA)
A search in the PubMed database (July 30 2017) retrieves 5793 hits for the term “epigenetic biomarker”, 7930 for “DNA methylation biomarker”, 5325 for “histone biomarker” and 11404 for “miRNA (microRNA) biomarker”. The past 30 years have produced a high number of publications claiming the next big thing for epigenetic biomarkers. In fact, recent epigenetic investigations are providing new potential biomarkers for a wide array of diseases and disorders, going beyond cancer, to include obesity and metabolic diseases [79], male infertility [80], neuropsychiatric disorders [81], etc. However, the number of novel biomarkers which have reached widespread clinical acceptance and implementation is relatively small, and an even smaller number of IVD has been cleared by the FDA or European Medicines Agency (EMA).
FDA approved IVD tests based on epigenetic biomarkers
Diagnostic kits based on DNA methylation
Epi proColon. Epi proColon® 2.0 CE (Epigenomics AG, Berlin, Germany) is a blood-based test that using a real-time PCR system and MethyLight assay detects methylated Septin9 in DNA obtained from peripheral blood samples. It is designed to aid in the early detection of colorectal cancer (CRC). However, a positive result obtained in the test should always be verified by other invasive diagnostic procedures, such as colonoscopy or sigmoidoscopy [82]. Across studies, Epi proColon® 2.0 CE discriminated between patients with CRC and healthy controls with a sensitivity of 75–81% with a specificity for CRC versus healthy individuals of 96–99%. Two of the case-controlled studies also compared the performance of Epi proColon® 2.0 CE with that of the fecal immunochemical test (FIT) [83] or the blood-based marker carcinoembryonic antigen (CEA) test (which is not recommended for CRC screening) and the guaiac-based fecal occult blood test (gFOBT) [84]. A clinical trial performed in China also assessed the clinical performance of the test for detection of adenomas [83]. Furthermore, Jin et al. did not find significant differences between left versus the right side CRC [83]. In both the Hungarian and Chinese studies, the sensitivity shown by the test at different stages of the cancer progression was different. In fact, the sensitivity was lower for stage I than for stages II, III, or IV in CRCs. Recently, Lamband Dhillon [82] compared the clinical performance of Epi proColon® 2.0 CE showing the high sensitivity and specificity of the test in three case-control studies and one prospective study.
Furthermore, Chinese guidelines [85,86] recommend the plasma Septin9 DNA methylation assay and the fecal occult blood test for use in early CRC screening. However, guidelines for CRC screening in Europe [87] and Asia Pacific [88] do not include Epi proColon 2.0 CE yet, and recommend the use of gFOBT or quantitative FIT for noninvasive screening. Currently, colonoscopy is considered the gold standard approach to detect CRC, and new screening technologies are not recommended yet.
Cologuard® is a stool-DNA-based, noninvasive colorectal cancer screening test. One advantage of the Cologuard® (Cologuard®, Exact Sciences, Madison, WI) kit is that it is simple, and able to increase the likelihood of colon neoplasia being present when positive. Because cellular exfoliation of DNA into stool occurs continuously, it is possible to detect molecular markers derived from the neoplastic cells in the colon, which made the detection of CRCs more accurate [89,90]. Cologuard® utilizes a multi-target approach to detect specific DNA methylation and hemoglobin in feces associated with CRC, as well as pre-malignant colorectal neoplasia. Cologuard® is different from single tests based on the detection of blood in feces, which may not be reliable for detecting polyps or lesions that bleed intermittently. This test measures the specific methylated gene targets including N-Myc Downstream-Regulated Gene 4 (NDRG4) and the Bone Morphogenetic Protein 3 (BMP3). NDRG4 and BMP3 have been shown to be hypermethylated in CRC. The Cologuard test also requires bisulfite conversion of non-methylated cytosine residues to uracil in the DNA sequence, to enable sensitive detection of hypermethylated NDRG4 and BMP3. Recently, the FDA approved NDRG4 and BMP3 methylation assay as a new test (Cologuard® stool-DNA-based test that screens for CRC with a sensitivity of 92.3% and specificity 86.6% [91].
Cologuard® is based in a multi-targeted stool DNA test, therefore, requires more time in a clinical lab to provide a result. Cologuard® utilizes a multi-target approach to detect DNA and hemoglobin (fecal immunochemical test, FIT) markers associated with CRC, as well as pre-malignant colorectal neoplasia [92]. As described above, it detects promoter CpG islands (NDRG4 and BMP3) and 7 KRAS mutations in Exon 2 (accounting for 98% of KRAS mutations) with an ELISA immunoassay to detect human hemoglobin (FIT) as previously reported [93,94]. Finally, an algorithm derived from these assays will determine the test result.
NDRG4 and BMP3 methylation are detected using methylation-specific PCR. In a separate analysis, DNA copy number changes that are associated with adenoma to carcinoma progression are also assessed for the identification of high-risk adenomas [95]. The presence of two or more of the seven chromosomal changes (8p21-pter, 15q11-q21, 17p12–13, and 18q12–21, and gains in 8q23-qter, 13q14–31, and 20q13) defines a high-risk adenoma. Finally, the FIT assay OC-sensor® is performed to detect hemoglobin presence in the stool (Eiken Chemical Co., Tokyo, Japan) and FOB Gold® (Sentinel, Milan, Italy).
A comparison to determine the performance characteristics of Cologuard® with the well-established noninvasive FIT in the detection of CRC and advanced adenomas as an alternative for colonoscopy surveillance, demonstrated the use of Cologuard® or FIT is cost-effectiveness and has sufficient sensitivity and specificity to be used as an alternative for colonoscopy [92]. However, as described above, this test requires epigenetic (NDRG4 and BMP3 methylation, genetic (chromosomal changes), and biochemical (hemoglobin detection) determinations to provide a diagnosis.
Epigenetic diagnostic tests performed at clinical laboratories
Although only two of the assays for methylation-based diagnostics have achieved approval by authorities, some commercial epigenetic-based tests are available for clinical laboratories. In particular, a test for neurodevelopmental disorders of genomic imprinting is commercially offered for Prader-Willi syndrome (15q11−13; PWS, OMIM#176270; ORPHA739), Angelman syndrome (15q11−13; AS, OMIM#105839; ORPHA72) and Beckwith-Wiedemann syndrome (11p15; BWS, OMIM#130650; ORPHA116), among others. Other application of epigenetic biomarkers which is gaining much interest in clinical diagnostic laboratories is noninvasive prenatal testing (NIPT), also known as prenatal cell-free DNA (pcfDNA), to screen for certain chromosomal abnormalities in a fetus. During prenatal cell-free DNA screening, DNA from the mother and fetus is extracted from a maternal blood sample and screened for the increased chance of specific chromosome problems, such as Down syndrome (Trisomy 21, DS, OMIM# 190685; ORPHA870), Patau syndrome (Trisomy 13; PS; ORPHA 3378), and Edwards syndrome (Trisomy 18; ES; ORPHA 3380). This screening can also provide information about fetal sex (chromosomes X and Y) and rhesus (Rh) blood type. Recently, differently-methylated regions are being used to differentiate maternal from fetal cfDNA [96-98], e.g. the levels of methylated HLCS and methylated RASSF1A were simultaneously measured by multiplex qPCR [96,97]. Similarly, Lee et al., 2013 tested the methylation of maspin gene (Serpin peptidase inhibitor, clade B (ovalbumin), member 5; SERPINB5; 18q21.33) in maternal plasma from 66 pregnant women, 11 carrying fetuses with trisomy 18, and 55 carrying normal fetuses [98]. They found that the median unmethylated-maspin concentrations were significantly elevated in women with trisomy 18 fetuses compared with controls. This suggests that unmethylated-maspin and methylated-maspin concentrations may be useful as potential biomarkers for NIPT of fetal trisomy 18 in the first trimester of pregnancy, irrespective of the sex and genetic variations of the fetus. In general, most conventional techniques used in clinical laboratories for diagnosing these disorders are based on methylation sensitive restriction enzymes, and chemical treatment of DNA. These techniques will be described in the next section.
Conversely, in our search, we did not find any kit based on the detection of circulating miRNAs approved by the FDA. However, there exists few assays under the oversight provided by the Clinical Laboratory Improvement Amendments (CLIA) which ensures accurate, reliable, and reproducible testing in clinical laboratories [99]. Some examples are ThyGenX® Oncogene Panel and ThyraMIR™ developed by Interpace® Diagnostic Inc. (Parsippany, NJ) and the wide array of miRView tests developed by Rosetta® Genomics (Rehovot, Israel). CLIA certification recognizes some laboratories as qualified to perform high-complexity testing used for clinical purposes. However, final diagnosis and optimal patient management are the responsibility of the referring physician or health care provider.
Briefly, the ThyraMIR™ microRNA Classifier and the ThyGenX® Oncogene Panel each consist of markers strongly associated with thyroid cancer, and whose detection in preoperative thyroid nodule aspirations has been shown to be highly predictive of thyroid cancer [100,101]. miRNA expression in residual total nucleic acid samples is measured by RT-qPCR using a custom-designed microRNA PCR Panel based on the expression levels of 10 miRNA genes: miR-29 b-1–5p, miR-31–5p, miR-138–1–3p, miR-139–5p, miR-146 b-5p, miR-155, miR-204–5p, miR-222–3p, miR-375, and miR-551 b-3p [102] and the evaluation of their levels using a proprietary algorithm. In a prospective study of thyroid nodules with indeterminate cytology (N = 109), combined testing with ThyGenX® and ThyraMIR™ produced a good sensitivity of 89% (95% confidence interval [CI], 73–97%) and a specificity of 85% (95% CI, 75–92%) [102].
miRView assays from RosettaGX performed in Rosetta Genomics laboratories which are CAP-accredited (College of American Pathologists (CAP’s) Laboratory Accreditation Program) and CLIA-certified, with state licenses in California, Florida, Maryland, New York, Pennsylvania, and Rhode Island, are provided in their headquarters. There exists different miRView assays for cancer diagnostic. Rosetta GX Cancer Origin consists on a miRNA-based array designed to provide a diagnosis for cancers of unknown origin. It utilizes proprietary technology developed by Rosetta Genomics for extraction and quantification of miRNAs from FFPE samples. A total of 64 miRNAs in a custom-designed array from Agilent Technologies is used to analyze specific miRNA profiles. The assay relies on two classifiers to determine the tissue of origin, a binary decision tree, and a k-nearest-neighbor (KNN) [103,104]. Both classifiers assign one out of 42 tissues of cancer origin, based on the normalized expression of 64 miRNAs [103], although in the commercial web page the company claims differentiation of up to 49 cancer types. In an independent validation set study (N = 509) the overall accuracy of the assay was high (85%), with 82% of the samples producing a single predicted origin with 90% accuracy. In addition, inter-laboratory reproducibility was assessed by processing RNA from the training and validation samples independently and blindly in two Rosetta Genomics laboratories (Philadelphia and Israel) [103].
Rosetta GX MI-lung™ is designed to classify and diagnose lung cancer patients. In this test, samples from FFPE blocks from resection or biopsies and cell blocks from cytological procedures, including FNA biopsy, bronchial brushing, and bronchial washing are used to determine the expression level of eight miRNA biomarkers by using a RT-qPCR analysis. These biomarkers are used to differentiate small cell lung cancer (SCLC), carcinoid, squamous non-small cell lung cancer (NSCLC), and non-squamous NSCLC [105]. Validation determined the test’s sensitivity to be 93.7% (95% CI, 90.8–95.8%) and specificity >90%, for the identification of the four main histological subtypes of lung tumors [105].
Finally, Rosetta GX MI-Kidney™ is intended to provide a differential diagnosis with fast, standardized classification using renal tumor FFPE or biopsy samples that are used to determine expression levels of 24 miRNAs using custom-designed arrays from Agilent Technologies. This molecular diagnostic test allows clinicians to differentiate four main histological types of primary kidney tumors: benign oncocytoma and the three most common subtypes of renal cell carcinomas (clear cell, papillary, and chromophobe) with an overall accuracy of 95%, therefore, avoiding unnecessary surgeries. In addition, inter-laboratory concordance of 100% demonstrates the robustness of the assay [106].
Regarding histones, there are no FDA-approved diagnostic methods based on the detection of neither circulating histones, nor specific histone PTMs detected in tissues. However, there exists a histone ELISA kit (http://www.rapidtest.com) designed for the diagnosis of autoimmune diseases. In particular, the kit allows semi-quantification of autoantibodies against histones in human serum. Since the presence of histones can be used in the differential diagnosis between systemic lupus erythematosus or other rare autoimmune disorders which also show nuclear proteins in serum (i.e. scleroderma or other connective tissue diseases), the potential of the kit relies upon combination with other diagnostic tools. However, it is still an indirect measure of the presence of histones in serum, and provides no information about specific histone variants or PTMs.
Technologies for epigenetic biomarker analysis in clinical laboratories
Medical tests applied in clinical laboratories are usually part of a more complex clinical process including anamnesis. Study of the signs and symptoms of the patient and analysis of test results to propose a diagnosis, and help guide treatment decisions, which include a variety of medical actions and therapies. Current and future technologies may help facilitate the determination of newly implemented biomarkers by clinical laboratory professionals. There is, to date, no completely defined set of criteria which should be used to define a realistic biomarker. Among other criteria, Collinson defined a series of interesting questions for evaluating the evidence base and cost-effectiveness for the clinical use of new appearing biomarkers [20].
In this section, we describe the most common techniques used in clinical laboratories with potential use in the determination of epigenetic biomarkers and by possible contribution to the effectiveness of the new biomarkers.
DNA methylation
DNA methylation assays have been used in clinical laboratory routine for diagnosis of rare syndromes, as we have described previously. Some of these techniques, which are being used in the clinical diagnosis, are based on methylation-sensitive restriction enzymes. However, one of the major problems of techniques based on methylation-sensitive restriction enzymes are that restriction fragments generated are analyzed by semi-quantitative methods, such as Southern-blot or PCR+electrophoresis procedures which are inherently linked to eye-reading of the gel and lack automation. This problem has led to the development of alternative techniques, such as multiplex ligation-dependent probe amplification (MLPA) assays, or other tests based on the chemical conversion of DNA using bisulfite treatment and qRT-PCR-based reactions, such as single and multiplex MethyLight, methylation-sensitive-high resolution melting (MS-HRM), and pyrosequencing. Some studies highlight MS-HRM and pyrosequencing as the techniques with best performance, reproducibility, and sensitivity [107,108]. Detailed description of each of these techniques is presented in the book Epigenetic Biomarkers and Diagnostics, edited by José Luis García-Giménez [109].
Methylation Specific-MLPA
Methylation-specific MLPA (MS-MLPA) is a semi-quantitative method for methylation profiling. MS-MLPA is a variant of the MLPA technique in which copy number detection is combined with the use of a methylation-sensitive restriction enzyme. MS–MLPA PCR products are usually analyzed on an automated sequencer (capillary electrophoresis) [110]. To calculate the gene dosage, the analysis of results is based on the comparison between the signals of undigested samples and undigested controls. Calculation of the methylation level of probes containing the HhaI restriction site is performed comparing the signals of restriction digested samples with undigested samples. The analysis of raw data may be carried out using a free software provided by the manufacturers (Coffalyser®, MLPA-Holland, Amsterdam, Netherlands) or using Excel-based in-house program (such Meth-HULP version 1.1 and others) (Romanelli et al., 2011). Currently, there are commercial kits to detect methylation status of loci for PWS/AS, BWS/Silver Russell syndrome, GNAS albright hereditary osteodystrophy, pseudohypoparathyroidism, uniparental disomy of the region 6q24, 7p12, 7q32, and 14q32, transient neonatal diabetes mellitus, gliomas, and retinoblastoma, which are commercialized by different companies, such as MRC-Holland (Amsterdam, Netherlands), Prevention Genetics Inc. (Marshfield, WI), Premier Biosoft (Palo Alto, CA) and Eurofins Biomnis (Dublin, Ireland), among others.
MethyLight
MethyLight or quantitative real-time MS-PCR was first developed by Eads et al. in 1999 [111]. This technique is based on standard methyl-specific PCR technology using TaqMan™ fluorescent probes in the amplification of bisulfite-converted DNA. Primers and probes for MethyLight assays can be acquired from different companies (i.e. Thermo Fisher, Integrated DNA Technologies Inc., Roche, etc.). Since the distinguishing method is through qPCR amplification, high sensitivity (up to a single-nucleotide resolution) may be achieved while maintaining high specificity [112]. Furthermore, MethyLight is capable of detecting very low frequencies of hypermethylated alleles, detecting 1 methylated allele among 10,000 unmethylated alleles, or a detection frequency of 0.01%. MethyLight has been used to simultaneously analyze the methylation of DNA obtained from FFPE [113,114] and also from cfDNA [115], as well as from urine sediments [116].
In 2010, He et al. developed a new multiplex MethyLight technique which is able to simultaneously analyze methylation of multiple genes in the same DNA sample depending only in the amount of bisulfite-converted DNA available and the number of fluorescent detectors of the qPCR machine [117]. Bapat et al. introduced the use of the ALU element as reference and internal control in the procedure. Multiplex MethyLight is highly accurate and reproducible when compared with singleplex MethyLight. It further reduces the requirement of initial input DNA for the analysis of multiple genes, and hence, it is cost-effective. Furthermore, multiplex MethyLight has successfully been applied to measure DNA methylation in FFPE tissue, fresh frozen tissue, tissue biopsy, and urine samples [118]. MethyLight is also the technology used in the EpiPro Colon test commercialized by Epigenomics Inc.
MS-HRM
High-resolution melting analysis is a technique based on the DNA dissociation behavior, from double-stranded DNA (dsDNA) to single-stranded DNA (ssDNA) with increasing temperature. Methylated and unmethylated DNA acquires different sequences after bisulfite treatment, resulting in PCR products with markedly different melting profiles. Reliable and sensitive HRM results require the capture of many data points per run and a high well-to-well precision. The central basis for measuring DNA methylation with HRM (methylation sensitive high-resolution melting analysis, MS-HRM) is that changes are introduced into DNA during the bisulfite conversion step, depending on the degree of methylation, and these changes can be measured by examining the melting behavior of the DNA. When used with bisulfite-converted DNA standards, HRM can also be used to estimate the proportion of methylation within a sample [119]. Wojdacz and Dobrovic have developed well-described protocols for DNA methylation analysis using MS-HRM, and reduction of PCR bias to favor amplification of the methylated template [120-122]. Because HRM analysis requires short PCR products, which can easily be generated from a large number of different samples, it can be robustly applied in the methylation studies of archival material, such as FFPE samples. Besides being a fast and cost-effective tool, MS-HRM is an interesting technique suitable for clinical laboratories [123]. Although HRM is not locus-specific, it has practical applications and it is widely used for diagnosis of genetic imprinting diseases, such as PWS, AS [124], and BWS [125,126].
Furthermore, a comparison of laboratory techniques to detect methylation patterns in the two main imprinting loci in BWS patients demonstrated that a combination of two tests (HRM + MS – MLPA or pyrosequencing+MS – MLPA) is the most useful approach for clinical diagnosis in BWS [126].
Pyrosequencing
Pyrosequencing is based on sequential addition and incorporation of nucleotides, using the so-called “sequencing-by-synthesis” principle, which can be quantitated through conversion of naturally released inorganic pyrophosphate into a light signal in real time [127,128]. During the process of pyrosequencing, the released pyrophosphate is used in a sulfurylase reaction releasing ATP that is subsequently used by luciferase to oxidize luciferin, generating light, which is detected. Since in this method the order of addition of nucleotides is known, the template sequence can be determined. Pyrosequencing has a wide range of applications including detection of SNPs, insertion/deletions, gene copy number, and DNA methylation [112]. Several pyrosequencing platforms are available today, such as PyroMark Q24, PyroMark Q24 Advanced, or PyroMark Q96 ID from Qiagen, or the 454 Sequencing from Roche, which is a high-throughput NGS platform (e.g. GS FLX+System9). A comparison of these platforms is presented in the chapter by Florea A. M. in Epigenetic Biomarkers and Diagnostics [129].
Pyrosequencing is a reliable, fast, and high-throughput technique that can simultaneously analyze up to 96 bisulfite-converted DNA samples in approximately 4 h. This technique has been previously used in the analysis of DNA methylation in FFPE, blood and cfDNA [126,129-131].
miRNAs
The use of microarrays or NGS panels designed for miRNA analysis are time-consuming and usually expensive. In this regard, these tools are very useful for biomedical research when exploring or investigating mechanisms involved in disease. But usefulness is compromised for the analysis of single or few miRNAs as biomarkers.
The sensitivity and specificity of RT-qPCR based tests that have been cleared by the FDA for IVD are very high, compared to other FDA-cleared assays which use different methods. However, even with RT-qPCR, false negative results can occur due to improper or poor clinical specimen collection, or from poor handling of a specimen after collection and before testing. Moreover, false positive results can occur by lab contamination, bad preparation of samples, or even bad selection of the selected reference miRNA (see previous section). Therefore, RT-qPCR requires of several controls in order to ensure the quality of the miRNA diagnostic assays.
qRT-PCR and droplet digital PCR seem the most feasible tools to be used for the analysis of circulating miRNAs, although these techniques require a knowledge of primer design and some technical aspects are difficult to convey to medical personnel. However, we envision that in the near future the appearance of new IVD kits based on the detection of miRNAs using PCR-based methods will further improve the incorporation of these techniques to clinical laboratories. Furthermore, the cost of the last ddPCR suggests that these platforms will require more time to be implanted in clinical settings.
In the coming years, the introduction of innovative platforms for NGS – like the currently widely used Illumina platforms – and particularly their use for small-RNAseq and the parallel development of sophisticated computational methods for miRNA analysis will further improve the potential of clinical laboratories in the use of this type of epigenetic biomarkers for clinical diagnostics.
Histones and histones PTMs
Direct measuring of histones by the use of immunoassays in plasma samples is the standard way to detect circulating histones, although novel mass spectrometry based methods are acquiring relevance, as we will discuss later on. Regarding the analysis of certain histone PTMs as biomarkers for disease, examples of these are the use of citrullinated histone H3 as a biomarker for immune diseases, such as systemic lupus erythematous or rheumatoid arthritis [132] or the use of acetylated histones H3 and H4 to monitor cancer treatment with histone deacetylase inhibitors (HDACi) [133]. Since histone modifiers, such as methyltransferases and demethylases are being incorporated into clinical trials for cancer therapy [134], methods to detect those modifications are required to evaluate treatment efficiency. In this sense, technical limitations of ELISA kits, or low reproducibility, are the main bottle-necks to provide confident value as biomarkers to immune assays. Thus, although clinical routine includes several ELISA kits in the differential diagnosis of immunological disorders that produce release of nuclear proteins to the bloodstream, novel methods are being developed that provide higher specificity. For instance, mass spectrometry proteomic tools are becoming widely used in clinical facilities, with a predicted expansion in the upcoming years [135] mainly due to the fact that they can reduce costs by replacing expensive antibody-based techniques [136]. These analytical methods provide a powerful diagnostic potential when coupled to other manipulations of biological samples. Recent work by Noberini et al. shows that the analysis of histone PTMs can be performed in FFPE tissue, with similar efficiency as that obtained from the analysis of frozen tissue samples [137]. In this context, histone PTMs could be used as biomarkers for breast cancer with the benefits of using long-term stored biological samples in hospital archives [138]. The potential of mass spectrometry can also be applied to liquid biopsies as a source of histone proteins, as mentioned above. Novel methods have been developed to improve the efficiency of histone isolation from serum that constitute a good source for histone PTM analysis in combination with MS techniques [65]. A good example of the monitoring of treatments performed by MS-analysis of histone PTMs is the evaluation of changes in the histone PTM profile in leukemia cell lines sensitive to decitabine [139]. Noteworthy, we have been able to use targeted mass spectrometry to detect circulating histones in untreated plasma samples, to serve as biomarkers for prediction of the fatal outcome in septic shock patients [140], proving to be a very promising tool for clinicians to better assess decisions in sepsis management, which still harbors plenty of difficulties in ICUs worldwide [141]. Nevertheless, mass spectrometry based methods, although very well established in a research context, should still be accurately adjusted and evaluated for many of these clinical uses in order to develop their true and maximum potential [135].
Challenges in the analysis of epigenetic biomarkers in clinical samples
Although this review shows the striking advances and the improvements made during the latest years in terms of development and implantation of epigenetic biomarkers into clinical practice and routine, there is still a long way ahead. Several challenges remain unmet, mainly related to the novelty of these biomarkers, and the intrinsic difficulty to substitute already standardized methodologies. The biggest challenge, however, is related to the particularities of epigenetic regulatory mechanisms. These novel biological parameters require a very deep knowledge of the underlying biological epigenetic regulatory mechanisms in health and disease. Furthermore, it is noteworthy that both clinical laboratory technicians and clinical practitioners will be required to increase their knowledge on new epigenetic technologies and platforms, and receive specific training to manage the analysis and interpret the results. However, some efforts led by the research community, pharmaceutical industry, IVD and medical device companies, and clinicians exist to implement epigenetic biomarkers into clinical routine. Of course, this effort also requires of the generation, not only of new biomarkers, but also of the commercialization of new, easy-to-use assays, appropriate procedures to preserve epigenetic biomarkers in biospecimens, and standardized operation procedures during sample processing and sample analysis. There is also a notable technical bottleneck for many of the current applications, since not all hospitals or clinical laboratories possess the technical facilities required for a standard application of detection methods that involve, for instance, mass spectrometry or NGS. Important to note is the fact that all of these issues will likely be addressed in the near future, given the direction of modern medicine toward a more personalized and precise practice. Precision medicine aims to improve, via theragnosis and predicting and monitoring treatments, the safety and efficacy of medical treatments in order to save on healthcare costs. Accordingly, a number of precision medicine initiatives have been announced in USA and in European countries to revolutionize healthcare systems. In this regard, epigenomic technologies will empower clinical laboratory results to refine clinical decisions. Amidst all this, the regulatory environment for in vitro diagnostic technologies is changing rapidly and, therefore, revisions of directives and regulations on medical in vitro diagnostic devices should be prepared accordingly [142].
Conclusions and perspective
The applications for epigenetic biomarkers are spreading and becoming regularly present in clinical laboratories, which contributes to the screening, diagnosis, and monitoring of all types of health conditions. However, new products, kits, and tests sometimes advance lab capabilities more rapidly than others in a particular area. In the coming years, epigenetics will generate new diagnostics strategies. In this regard, adaptation of new technologies and methods to a clinical laboratory environment may ensure the adoption of epigenetic biomarkers in the diagnostic process. Easy-to-use, labor-efficient, uncomplicated, and inexpensive methods are needed to evaluate the diagnostic power of new biomarkers in the experiments that normally involve a large number of samples, as occurs in a clinical laboratory. In this regard, RT-qPCR systems seem to be the candidates for being the more versatile systems for gene-specific DNA-methylation analyses, miRNA measurement, and even PTM in histone studies [some new applications for RT-qPCR based on the application of proximity ligation assay (PLA) technology are appearing to detect protein levels, see (Swartzman et al. [143]). Furthermore, the introduction of digital PCR and new applications, such as digital MethyLight will further improve the potential of clinical laboratories, for example, increasing the sensitivity for DNA methylation detection up to three times, and requiring low amounts of input of genetic material. In the near future, the standardization of isolation and purification methods and the simplification of the protocols and procedures required to use new technologies, together with the generation of point-of-care (POC) diagnostic tools, may produce the greatest impact on clinical laboratories. POC testing remains a major interest among investigators, and an important market for IVD manufacturers. A future ideal development for the easy implementation of miRNA analysis for clinical settings would be the generation of POC diagnostics. Some research is focused in the development of a platform for miRNA detection using total internal reflection fluorescence (TIRF)-based microarray system. This will consist of an initial PCR reaction coupled to a microfluidic microarray-based system using a photodiode to detect the fluorescent signals [144]. Other POC developments are based on slot waveguide based Mach-Zehnder interferometer-miRNA detection system capable of detecting multiple miRNAs rapidly and simultaneously in a label-free and real-time manner [145].
Regarding the legal issues and standardization of these novel protocols, kits and techniques, a regulatory legal network is required to provide high quality epigenetics-based IVD tests, and FDA approval and CE marking processes may ensure that patients will be diagnosed with safer, higher quality and more effective devices. In this regard, revised policies and procedures of IVD regulatory notified bodies in the USA and Europe have discussed the need of pre-market presentation of high quality evidence and the post-market surveillance of the clinical safety and performance of IVD medical devices [146-148]. Additionally, expanded oversight of clinical laboratories through a revised CLIA process has also been proposed [149].
Finally, technologic advances in techniques, such as NGS and mass spectrometry will allow clinical laboratory professionals to offer new testing and IVD kits that will provide more information than ever before and often within a time frame that will allows rapid patient care and better outcomes.
Acknowledgments
J. L. G-G thanks INCLIVA and GVA for the starting grants (GV/2014/132) and AES2016 (ISCIII) for grant number PI16/01036, co-financed by the European Regional Development Fund (ERDF). J. L. G-G. and FVP thank Grand Challenges Canada and the Spanish Ministry of Economy and Competitiveness, ISCIII through CIBERER (Consorcio Centro de Investigación en Red del Instituto de Salud Carlos III, CIBER-ISCIII and INGENIO2010). L. P-C thanks GVA for the starting grants (GV/2016/029) MEC, ISCIII, FEDER for grant PT13/0010/0004. T.O.T acknowledges grant support from the NIH (R01 CA178441 and R01 CA204346) as well as the American Institute for Cancer Research (316184). P.L. has a grant from AES2016 (ISCIII) PI15/01481.
Footnotes
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Lech G, Slotwinski R, Slodkowski M, et al. Colorectal cancer tumour markers and biomarkers: recent therapeutic advances. World J Gastroenterol. 2016;22: 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Andersen AM, Dogan MV, Beach SR, et al. Current and future prospects for epigenetic biomarkers of substance use disorders. Genes. 2015;6:991–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].García-Giménez JL, Ushijima T, Tollefsbol TO. Epigenetic biomarkers: new findings, perspectives, and future directions in diagnostics In: García-Giménez JL, editor. Epigenetic biomarkers and diagnostics. Amsterdam: Elsevier Inc.;2016. [Google Scholar]
- [4].Toraño EG, Petrus S, Fernandez AF, et al. Global DNA hypomethylation in cancer: review of validated methods and clinical significance. Clin Chem Lab Med. 2012;50:1733–1742. [DOI] [PubMed] [Google Scholar]
- [5].Hu Y, Li P, Hao S, et al. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med. 2009;47: 923–929. [DOI] [PubMed] [Google Scholar]
- [6].Balgkouranidou I, Chimonidou M, Milaki G, et al. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin Chem Lab Med. 2016;54:1385–1393. [DOI] [PubMed] [Google Scholar]
- [7].Gao H, Zhang N, Lu F, et al. Circulating histones for predicting prognosis after cardiac surgery: a prospective study. Interact Cardiovasc Thorac Surg. 2016;23:681–687. [DOI] [PubMed] [Google Scholar]
- [8].Vrtačnik P, Marc J, Ostanek B. Epigenetic mechanisms in bone. Clin Chem Lab Med. 2014;52:589–608. [DOI] [PubMed] [Google Scholar]
- [9].Markopoulou S, Nikolaidis G, Liloglou T. DNA methylation biomarkers in biological fluids for early detection of respiratory tract cancer. Clin Chem Lab Med. 2012;50:1723–1731. [DOI] [PubMed] [Google Scholar]
- [10].Heichman KA, Warren JD. DNA methylation biomarkers and their utility for solid cancer diagnostics. Clin Chem Lab Med. 2012;50:1707–1721. [DOI] [PubMed] [Google Scholar]
- [11].Relton CL, Hartwig FP, Davey Smith G. From stem cells to the law courts: DNA methylation, the forensic epigenome and the possibility of a biosocial archive. Int J Epidemiol. 2015;44:1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Glinge C, Clauss S, Boddum K, et al. Stability of circulating blood-based microRNAs - pre-analytic methodological considerations. PLoS One. 2017; 12:e0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Park NJ, Zhou H, Elashoff D, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15: 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zubakov D, Boersma AW, Choi Y, et al. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Legal Med. 2010;124: 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Peiró-Chova L, Peña-Chilet M, López-Guerrero JA, et al. High stability of microRNAs in tissue samples of compromised quality. Virchows Arch. 2013;463: 765–774. [DOI] [PubMed] [Google Scholar]
- [16].Patnaik SK, Mallick R, Yendamuri S. Detection of microRNAs in dried serum blots. Anal Biochem. 2010;407:147–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bulla A, De Witt B, Ammerlaan W, et al. Blood DNA yield but not integrity or methylation is impacted after long-term storage. Biopreserv Biobank. 2016;14: 29–38. [DOI] [PubMed] [Google Scholar]
- [18].Joo JE, Wong EM, Baglietto L, et al. The use of DNA from archival dried blood spots with the Infinium HumanMethylation450 array. BMC Biotechnol. 2013; 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].García-Gimenez JL. Epigenetic biomarkers and diagnostics. Vol. 1 Cambridge, MA: Academic Press; 2015. [Google Scholar]
- [20].Evidence Collinson P. and cost effectiveness requirements for recommending new biomarkers. EJIFCC. 2015;26:183–189. [PMC free article] [PubMed] [Google Scholar]
- [21].Mojica WD, Hou T, Sykes D, et al. Front-end genomics: using an alternative approach for the recovery of high-quality DNA from core needle biopsies. J Clin Pathol. 2017;70:488–493. [DOI] [PubMed] [Google Scholar]
- [22].Kotorashvili A, Ramnauth A, Liu C, et al. Effective DNA/RNA co-extraction for analysis of microRNAs, mRNAs, and genomic DNA from formalin-fixed paraffin-embedded specimens. PLoS One. 2012;7:e34683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Medeiros F, Rigl CT, Anderson GG, et al. Tissue handling for genome-wide expression analysis: a review of the issues, evidence, and opportunities. Arch Pathol Lab Med. 2007;131:1805–1816. [DOI] [PubMed] [Google Scholar]
- [24].Klopfleisch R, Weiss AT, Gruber AD. Excavation of a buried treasure-DNA, mRNA, miRNA and protein analysis in formalin fixed, paraffin embedded tissues. Histol Histopathol. 2011;26:797–810. [DOI] [PubMed] [Google Scholar]
- [25].Roberts L, Bowers J, Sensinger K, et al. Identification of methods for use of formalin-fixed, paraffin-embedded tissue samples in RNA expression profiling. Genomics. 2009;94:341–348. [DOI] [PubMed] [Google Scholar]
- [26].Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61:64–71. [DOI] [PubMed] [Google Scholar]
- [27].Hosein A, Cocciardi S, Jayanthan J, et al. The use of the Illumina FFPE Restoration Protocol to obtain suitable quality DNA for SNP-based CGH–a pilot study. Hered Cancer Clin Pract. 2012;10:A85. [Google Scholar]
- [28].Mouttham N, Klunk J, Kuch M, et al. Surveying the repair of ancient DNA from bones via high-throughput sequencing. Biotechniques. 2015;59:19–25. [DOI] [PubMed] [Google Scholar]
- [29].Haile S, Pandoh P, McDonald H, et al. Automated high throughput nucleic acid purification from formalin-fixed paraffin-embedded tissue samples for next generation sequence analysis. PLoS One. 2017;12:e0178706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bohmann K, Hennig G, Rogel U, et al. RNA extraction from archival formalin-fixed paraffin-embedded tissue: a comparison of manual, semiautomated, and fully automated purification methods. Clin Chem. 2009;55:1719–1727. [DOI] [PubMed] [Google Scholar]
- [31].Hennig G, Gehrmann M, Stropp U, et al. Automated extraction of DNA and RNA from a single formalin-fixed paraffin-embedded tissue section for analysis of both single-nucleotide polymorphisms and mRNA expression. Clin Chem. 2010;56:1845–1853. [DOI] [PubMed] [Google Scholar]
- [32].Ribeiro-Silva A, Zhang H, Jeffrey SS. RNA extraction from ten year old formalin-fixed paraffin-embedded breast cancer samples: a comparison of column purification and magnetic bead-based technologies. BMC Mol Biol. 2007;8:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fedorowicz G, Guerrero S, Wu TD, et al. Microarray analysis of RNA extracted from formalin-fixed, paraffin-embedded and matched fresh-frozen ovarian adenocarcinomas. BMC Med Genomics. 2009;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kasaian K, Wiseman SM, Thiessen N, et al. Complete genomic landscape of a recurring sporadic parathyroid carcinoma. J Pathol. 2013;230:249–260. [DOI] [PubMed] [Google Scholar]
- [35].Yap TA, Lorente D, Omlin A, et al. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20:2553–2568. [DOI] [PubMed] [Google Scholar]
- [36].Mabert K, Cojoc M, Peitzsch C, et al. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol. 2014;90:659–677. [DOI] [PubMed] [Google Scholar]
- [37].Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101: 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406:6499–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang BG, Huang HY, Chen YC, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–3968. [PubMed] [Google Scholar]
- [41].Giacona MB, Ruben GC, Iczkowski KA, et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. [DOI] [PubMed] [Google Scholar]
- [42].Jorgez CJ, Bischoff FZ. Improving enrichment of circulating fetal DNA for genetic testing: size fractionation followed by whole gene amplification. Fetal Diagn Ther. 2009;25:314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li Z, Guo X, Tang L, et al. Methylation analysis of plasma cell-free DNA for breast cancer early detection using bisulfite next-generation sequencing. Tumor Biol. 2016;37:13111–13119. [DOI] [PubMed] [Google Scholar]
- [44].Zhai R, Zhao Y, Su L, et al. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia. 2012;14:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Legendre C, Gooden GC, Johnson K, et al. Whole-genome bisulfite sequencing of cell-free DNA identifies signature associated with metastatic breast cancer. Clin Epigenet. 2015;7:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schmidt B, Weickmann S, Witt C, et al. Improved method for isolating cell-free DNA. Clin Chem. 2005;51:1561–1563. [DOI] [PubMed] [Google Scholar]
- [47].Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50:298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009; 2:ra81. [DOI] [PubMed] [Google Scholar]
- [49].Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- [50].Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Moret I, Sanchez-Izquierdo D, Iborra M, et al. Assessing an improved protocol for plasma microRNA extraction. PLoS One. 2013;8:e82753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- [55].Duttagupta R, Jiang R, Gollub J, et al. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6:e20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mraz M, Malinova K, Mayer J, et al. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390:1–4. [DOI] [PubMed] [Google Scholar]
- [57].Rounge TB, Lauritzen M, Langseth H, et al. MicroRNA biomarker discovery and high-throughput DNA sequencing are possible using long-term archived serum samples. Cancer Epidemiol Biomarkers Prev. 2015;24:1381–1387. [DOI] [PubMed] [Google Scholar]
- [58].Grasedieck S, Scholer N, Bommer M, et al. Impact of serum storage conditions on microRNA stability. Leukemia. 2012;26:2414–2416. [DOI] [PubMed] [Google Scholar]
- [59].Tan GW, Khoo AS, Tan LP. Evaluation of extraction kits and RT-qPCR systems adapted to high-throughput platform for circulating miRNAs. Sci Rep. 2015;5:9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cheng HH, Yi HS, Kim Y, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8:e64795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].McAlexander MA, Phillips MJ, Witwer KW. Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front Genet. 2013;4:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].El-Khoury V, Pierson S, Kaoma T, et al. Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci Rep. 2016;6:19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Seco-Cervera M, Gonzalez-Rodriguez D, Ibanez-Cabellos JS, et al. Circulating miR-323-3p is a biomarker for cardiomyopathy and an indicator of phenotypic variability in Friedreich’s ataxia patients. Sci Rep. 2017;7:5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Debey-Pascher S, Hofmann A, Kreusch F, et al. RNA-stabilized whole blood samples but not peripheral blood mononuclear cells can be stored for prolonged time periods prior to transcriptome analysis. JMD. 2011;13:452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Reddy D, Khade B, Pandya R, et al. A novel method for isolation of histones from serum and its implications in therapeutics and prognosis of solid tumours. Clin Epigenet. 2017;9:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shechter D, Dormann HL, Allis CD, et al. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. [DOI] [PubMed] [Google Scholar]
- [67].Bohmann K, Evans A, Gilbert MT, et al. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol. 2014;29:358–367. [DOI] [PubMed] [Google Scholar]
- [68].Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM):a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Holmes EE, Jung M, Meller S, et al. Performance evaluation of kits for bisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urine. PLoS One. 2014;9:e93933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dietrich D, Hasinger O, Banez LL, et al. Development and clinical validation of a real-time PCR assay for PITX2 DNA methylation to predict prostate-specific antigen recurrence in prostate cancer patients following radical prostatectomy. J Mol Diagn. 2013;15: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jung M, Uhl B, Kristiansen G, et al. Bisulfite conversion of DNA from tissues, cell lines, buffy coat, FFPE tissues, microdissected cells, swabs, sputum, aspirates, lavages, effusions, plasma, serum, and urine. Methods Mol Biol. 2017;1589:139–159. [DOI] [PubMed] [Google Scholar]
- [72].Kirschner MB, Kao SC, Edelman JJ, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zanutto S, Pizzamiglio S, Ghilotti M, et al. Circulating miR-378 in plasma:a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer. 2014;110:1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Appierto V, Callari M, Cavadini E, et al. A lipemia-independent NanoDrop(®)-based score to identify hemolysis in plasma and serum samples. Bioanalysis. 2014;6:1215–1226. [DOI] [PubMed] [Google Scholar]
- [75].MacLellan SA, MacAulay C, Lam S, et al. Pre-profiling factors influencing serum microRNA levels. BMC Clin Pathol. 2014;14:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Blondal T, Jensby Nielsen S, Baker A, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59: S1–S6. [DOI] [PubMed] [Google Scholar]
- [77].Shah JS, Soon PS, Marsh DJ. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS One. 2016;11:e0153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pizzamiglio S, Zanutto S, Ciniselli CM, et al. A methodological procedure for evaluating the impact of hemolysis on circulating microRNAs. Oncol Lett. 2017;13:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].van Dijk SJ, Tellam RL, Morrison JL, et al. Recent developments on the role of epigenetics in obesity and metabolic disease. Clin Epigenet. 2015;7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ferlin A, Foresta C. New genetic markers for male infertility. Curr Opin Obstet Gynecol. 2014;26: 193–198. [DOI] [PubMed] [Google Scholar]
- [81].Bakulski K, Halladay A, Hu VW. Epigenetic research in neuropsychiatric disorders: the “tissue issue”. Curr Behav Neurosci Rep. 2016;3:264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lamb YN, Dhillon S. Epi proColon(®) 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther. 2017;21:225–232. [DOI] [PubMed] [Google Scholar]
- [83].Jin P, Kang Q, Wang X, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol. 2015;30:830–833. [DOI] [PubMed] [Google Scholar]
- [84].Toth K, Sipos F, Kalmar A, et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One. 2012;7:e46000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chinese Society of Digestive Endoscopy of the Chinese Medical Association OECotCA-CA. Chinese early colorectal cancer screening and endoscopic diagnosis and treatment guidelines. Chin J Dig Endosc. 2015;32:341–360. [Google Scholar]
- [86].Epigenomics AG. Epigenomics and BioChain announce the inclusion of their proprietary blood-based septin9 test in the Chinese screening guideline for colorectal cancer. Berlin: Epigenomics AG; 2015. [Google Scholar]
- [87].European Colorectal Cancer Screening Guidelines Working G, von Karsa L, Patnick J, et al. European guidelines for quality assurance in colorectal cancer screening and diagnosis: overview and introduction to the full supplement publication. Endoscopy. 2013;45:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sung JJ, Ng SC, Chan FK, et al. An updated Asia Pacific Consensus Recommendations on colorectal cancer screening. Gut. 2015;64:121–132. [DOI] [PubMed] [Google Scholar]
- [89].Quintero E, Castells A, Bujanda L, et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med. 2012;366:697–706. [DOI] [PubMed] [Google Scholar]
- [90].Morikawa T, Kato J, Yamaji Y, et al. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129:422–428. [DOI] [PubMed] [Google Scholar]
- [91].MDXHealth. MDxHealth (R): MDxHealth Licensee Exact Sciences Receives FDA Approval for its Cologuard Colon Cancer Screening Assay. 2014. Available from: http://mdxhealth.com/press-release/mdxhealth-r-mdxhealth-licensee-exact-sciences-receives-fda-approval-its-cologuard
- [92].van Lanschot MC, Carvalho B, Coupe VM, et al. Molecular stool testing as an alternative for surveillance colonoscopy: a cross-sectional cohort study. BMC Cancer. 2017;17:07–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;371:187–188. [DOI] [PubMed] [Google Scholar]
- [94].Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. [DOI] [PubMed] [Google Scholar]
- [95].Hermsen M, Postma C, Baak J, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–1119. [DOI] [PubMed] [Google Scholar]
- [96].Lim JH, Lee DE, Kim KS, et al. Non-invasive detection of fetal trisomy 21 using fetal epigenetic biomarkers with a high CpG density. Clin Chem Lab Med. 2014;52:641–647. [DOI] [PubMed] [Google Scholar]
- [97].Lim JH, Lee DE, Park SY, et al. Disease specific characteristics of fetal epigenetic markers for non-invasive prenatal testing of trisomy 21. BMC Med Genomics. 2014;7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lee DE, Kim SY, Lim JH, et al. Non-invasive prenatal testing of trisomy 18 by an epigenetic marker in first trimester maternal plasma. PLoS One. 2013;8:e78136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kaul KL, Sabatini LM, Tsongalis GJ, et al. The case for laboratory developed procedures: quality and positive impact on patient care. Acad Pathol. 2017;4:2374289517708309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ferraz C, Eszlinger M, Paschke R. Current state and future perspective of molecular diagnosis of fine-needle aspiration biopsy of thyroid nodules. J Clin Endocrinol Metab. 2011;96:2016–2026. [DOI] [PubMed] [Google Scholar]
- [101].Giordano TJ, Beaudenon-Huibregtse S, Shinde R, et al. Molecular testing for oncogenic gene mutations in thyroid lesions: a case-control validation study in 413 postsurgical specimens. Hum Pathol. 2014;45:1339–1347. [DOI] [PubMed] [Google Scholar]
- [102].Kitano M, Rahbari R, Patterson EE, et al. Evaluation of candidate diagnostic microRNAs in thyroid fine-needle aspiration biopsy samples. Thyroid. 2012;22: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Meiri E, Mueller WC, Rosenwald S, et al. A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist. 2012;17:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin.Nat Biotechnol. 2008;26:462–469. [DOI] [PubMed] [Google Scholar]
- [105].Gilad S, Lithwick-Yanai G, Barshack I, et al. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn. 2012;14:510–517. [DOI] [PubMed] [Google Scholar]
- [106].Spector Y, Fridman E, Rosenwald S, et al. Development and validation of a microRNA-based diagnostic assay for classification of renal cell carcinomas. Mol Oncol. 2013;7:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118: 4201–4211. [DOI] [PubMed] [Google Scholar]
- [108].Migheli F, Stoccoro A, Coppede F, et al. Comparison study of MS-HRM and pyrosequencing techniques for quantification of APC and CDKN2A gene methylation. PLoS One. 2013;8:e52501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].García-Giménez JL, Ushijima T, Tollefsbol TO. Epigenetic biomarkers and diagnostics Boston: Academic Press; 2016. Chapter 1-epigenetic biomarkers: new findings, perspectives, and future directions in diagnostics. p. 1–18. [Google Scholar]
- [110].Nygren AO, Ameziane N, Duarte HM, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28: E32–10734209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhao FBB The role of methylation-specific PCR and associated techniques in clinical diagnostics In: García-Giménez JL, editor. Epigenetic biomarkers and diagnostics. Translational epigenetic series; Vol. 1 New York (NY): Academic Press;2015. p. 155–173. [Google Scholar]
- [113].Liu L, Kron KJ, Pethe VV, et al. Association of tissue promoter methylation levels of APC, TGFbeta2, HOXD3 and RASSF1A with prostate cancer progression. Int J Cancer. 2011;129:2454–2462. [DOI] [PubMed] [Google Scholar]
- [114].Olkhov-Mitsel E, Van der Kwast T, Kron KJ, et al. Quantitative DNA methylation analysis of genes coding for kallikrein-related peptidases 6 and 10 as biomarkers for prostate cancer. Epigenetics. 2012;7: 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Muller HM, Widschwendter A, Fiegl H, et al. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- [116].Reinert T, Borre M, Christiansen A, et al. Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS One. 2012;7:e46297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].He Q, Chen HY, Bai EQ, et al. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet. 2010;202:1–10. [DOI] [PubMed] [Google Scholar]
- [118].Olkhov-Mitsel E, Zdravic D, Kron K, et al. Novel multiplex MethyLight protocol for detection of DNA methylation in patient tissues and bodily fluids. Sci Rep. 2014;4:4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Wojdacz TK. Methylation-sensitive high-resolution melting in the context of legislative requirements for validation of analytical procedures for diagnostic applications. Expert Rev Mol Diagn. 2012; 12:39–47. [DOI] [PubMed] [Google Scholar]
- [120].Wojdacz TK, Borgbo T, Hansen LL. Primer design versus PCR bias in methylation independent PCR amplifications. Epigenetics. 2009;4:231–234. [DOI] [PubMed] [Google Scholar]
- [121].Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008;3:1903–1908. [DOI] [PubMed] [Google Scholar]
- [122].Wojdacz TK, Dobrovic A. Melting curve assays for DNA methylation analysis. Methods Mol Biol. 2009; 507:229–240. [DOI] [PubMed] [Google Scholar]
- [123].Worm J, Aggerholm A, Guldberg P. In-tube DNA methylation profiling by fluorescence melting curve analysis. Clin Chem. 2001;47:1183–1189. [PubMed] [Google Scholar]
- [124].White HE, Hall VJ, Cross NC. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin Chem. 2007;53: 1960–1962. [DOI] [PubMed] [Google Scholar]
- [125].Tenorio J, Romanelli V, Martin-Trujillo A, et al. Clinical and molecular analyses of Beckwith-Wiedemann syndrome: comparison between spontaneous conception and assisted reproduction techniques. Am J Med Genet A. 2016;170:2740–2749. [DOI] [PubMed] [Google Scholar]
- [126].Romanelli V, Meneses HN, Fernandez L, et al. Beckwith-Wiedemann syndrome and uniparental disomy 11p: fine mapping of the recombination breakpoints and evaluation of several techniques. Eur J Hum Genet. 2011;19:416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. [DOI] [PubMed] [Google Scholar]
- [128].Tost J, Gut IG. Analysis of gene-specific DNA methylation patterns by pyrosequencing technology. Methods Mol Biol. 2007;373:89–102. [DOI] [PubMed] [Google Scholar]
- [129].Florea A-M. Pyrosequencing and its application in epigenetic clinical diagnostics In: García-Giménez JL, editor. Epigenetic biomarkers and diagnostics. Translational Epigenetic Series; Vol. 1 New York (NY): Academic Press; 2015. p. 175–194. [Google Scholar]
- [130].Tang Q, Cheng J, Cao X, et al. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin Epigenet. 2016;8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zmetakova I, Danihel L, Smolkova B, et al. Evaluation of protein expression and DNA methylation profiles detected by pyrosequencing in invasive breast cancer. Neoplasma. 2013;60:635–646. [DOI] [PubMed] [Google Scholar]
- [132].Frede A, Neuhaus B, Klopfleisch R, et al. Colonic gene silencing using siRNA-loaded calcium phosphate/PLGA nanoparticles ameliorates intestinal inflammation in vivo. J Control Release. 2016;222:86–96. [DOI] [PubMed] [Google Scholar]
- [133].Rigby L, Muscat A, Ashley D, et al. Methods for the analysis of histone H3 and H4 acetylation in blood. Epigenetics. 2012;7:875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Morera L, Lubbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenet. 2016;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Vogeser M, Seger C. Quality management in clinical application of mass spectrometry measurement systems. Clin Biochem. 2016;49:947–954. [DOI] [PubMed] [Google Scholar]
- [136].Hetu PO, Robitaille R, Vinet B. Successful and cost-efficient replacement of immunoassays by tandem mass spectrometry for the quantification of immuno-suppressants in the clinical laboratory. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;883:95–101. [DOI] [PubMed] [Google Scholar]
- [137].Noberini R, Uggetti A, Pruneri G, et al. Pathology tissue-quantitative mass spectrometry analysis to profile histone post-translational modification patterns in patient samples. Mol Cell Proteomics. 2016;15:866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Noberini R, Pruneri G, Minucci S, et al. Mass-spectrometry analysis of histone post-translational modifications in pathology tissue using the PAT-H-MS approach. Data Brief. 2016;7:188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Zhang C, Suo J, Katayama H, et al. Quantitative proteomic analysis of histone modifications in decitabine sensitive and resistant leukemia cell lines. Clin Proteom. 2016;13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].García-Giménez JL, Romá-Mateo C, Carbonell N, et al. A new mass spectrometry-based method for the quantification of histones in plasma from septic shock patients. Sci Rep. 2017;7:10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Reinhart K, Daniels R, Kissoon N, et al. Recognizing sepsis as a global health priority - a WHO resolution. N Engl J Med. 2017;377:414–417. [DOI] [PubMed] [Google Scholar]
- [142].Council Directive 93/42/EEC of 14 June 1993 concerning medical devices (OJ L 169, 12.7.1993, p. 1–43). http://ec.europa.eu/growth/single-market/european-standards/harmonised-standards/medical-devices/. Available from: http://ec.europa.eu/consumers/sectors/medical-devices/files/revision_docs/2007-47-en_en.pdf
- [143].Swartzman E, Shannon M, Lieu P, et al. Expanding applications of protein analysis using proximity ligation and qPCR. Methods. 2010;50:S23–S26. [DOI] [PubMed] [Google Scholar]
- [144].Vaca L. Point-of-care diagnostic tools to detect circulating microRNAS as biomarkers of disease. Sensors. 2014;14:9117–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Liu Q, Shin Y, Kee JS, et al. Mach-Zehnder interferometer (MZI) point-of-care system for rapid multiplexed detection of microRNAs in human urine specimens. Biosens Bioelectron. 2015;71:365–372. [DOI] [PubMed] [Google Scholar]
- [146].Institute of Medicine (US). Committee on the Public Health Effectiveness of the FDA 510(k) Clearance Process Medical devices and the public’s health: the FDA 510(k) clearance process at 35 years. Washington (DC): National Academies Press; 2011. [PubMed] [Google Scholar]
- [147].Samson D, Schoelles KM. Chapter 2: medical tests guidance (2) developing the topic and structuring systematic reviews of medical tests:utility of PICOTS, analytic frameworks, decision trees, and other frameworks. J Gen Intern Med. 2012;27(1): S11–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Standards for reporting of diagnostic accuracy. Clin Chem. 2003;49:1–6. [DOI] [PubMed] [Google Scholar]
- [149].Association for Molecular Pathology. Proposal for modernization of CLIA regulations for laboratory developed testing procedures (LDPs) 2015. Available from: http://www.amp.org/advocacy/documents/AMPCLIAmodernizationproposalFINAL8.14.15.pdf
- [150].Marabita F, de Candia P, Torri A, et al. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 2016;17:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Zhu HT, Dong QZ, Wang G, et al. Identification of suitable reference genes for qRT-PCR analysis of circulating microRNAs in hepatitis B virus-infected patients. Mol Biotechnol. 2012;50:49–56. [DOI] [PubMed] [Google Scholar]
- [152].Song J, Bai Z, Han W, et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. [DOI] [PubMed] [Google Scholar]
- [153].Masotti A, Baldassarre A, Guzzo MP, et al. Circulating microRNA profiles as liquid biopsies for the characterization and diagnosis of fibromyalgia syndrome. Mol Neurobiol. 2017;54:7129–7136. [DOI] [PubMed] [Google Scholar]
- [154].Chen J, Li K, Pang Q, et al. Identification of suitable reference gene and biomarkers of serum miRNAs for osteoporosis. Sci Rep. 2016;86:36347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Niu Y, Wu Y, Huang J, et al. Identification of reference genes for circulating microRNA analysis in colorectal cancer. Sci Rep. 2016;6:1935611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Caserta S, Kern F, Cohen J, et al. Circulating plasma microRNAs can differentiate Human Sepsis and Systemic Inflammatory Response Syndrome (SIRS). Sci Rep. 2016;6:28006. [DOI] [PMC free article] [PubMed] [Google Scholar]