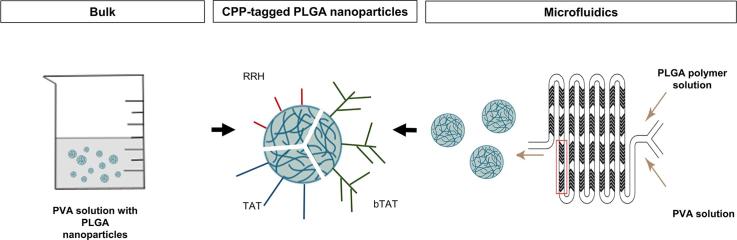
Graphical abstract
Keywords: Nanoprecipitation, FTIR, Bio-nano interactions, Cell-penetrating peptides, PLGA nanoparticles
Abstract
The efficient and reproducible production of nanoparticles using bulk nanoprecipitation methods is still challenging because of low batch to batch reproducibility. Here, we optimize a bulk nanoprecipitation method using design of experiments and translate to a microfluidic device to formulate surface-modified poly-lactic-co-glycolic (PLGA) nanoparticles. Cell-penetrating peptides (CPPs) with a short, long linear or branched architecture were used for the surface modification of PLGA nanoparticles. The microfluidics method was more time efficient than the bulk nanoprecipitation method and allowed the formulation of uniform PLGA nanoparticles with a size of 150 nm, a polydispersity index below 0.150 and with better reproducibility in comparison to the bulk nanoprecipitation method. After surface modification the size of CPP-tagged PLGA nanoparticles increased to 160–180 nm and the surface charge of the CPP-tagged PLGA nanoparticles varied between −24 mV and +3 mV, depending on the architecture and concentration of the conjugated CPP. Covalent attachment of CPPs to the PLGA polymer was confirmed with FTIR by identifying the formation of an amide bond. The conjugation efficiency of CPPs to the polymeric PLGA nanoparticles was between 32 and 80%. The development and design of reproducible nanoformulations with tuneable surface properties is crucial to understand interactions at the nano-bio interface.
1. Introduction
Efficient production of nanoparticles for drug delivery applications is increasingly important to ensure that the benefits of these delivery systems are translated from ‘bench to bedside’. Nanoprecipitation is one of the most commonly used techniques for the preparation of polymeric nanoparticles and is suitable to encapsulate proteins, hydrophobic and poorly water soluble drugs (Banik et al., 2016, Danhier et al., 2012, Jung et al., 2000). Polymeric nanoparticles are usually prepared using a bulk beaker method with volumes up to 50 mL (Fessi et al., 1989). The nanoprecipitation method involves mixing of an organic, water-miscible solvent like acetone, ethanol or acetonitrile with an aqueous solution (Masood, 2016, Yadav and Sawant, 2010). Surfactants like the non-ionic polyvinyl alcohol (PVA) are added to the aqueous phase to act as a stabilizer and to reduce the interfacial tension between the organic and aqueous phase (Tefas et al., 2015). Mixing the organic and aqueous phases leads to a reduction in polymer solubility and nanoparticles assemble due to nucleation and subsequent growth of the polymer (Karnik et al., 2008). Nanoprecipitation is characterized by the production of nanoparticles with a narrow size distribution (polydispersity index below 0.2) and the potential to scale up the technique to an industrial scale (Danaei et al., 2018, Galindo-Rodríguez et al., 2005, Masood, 2016). The main drawback, however of bulk preparation methods is the variation in nanoparticle size from batch to batch due to the limited control over fluid dynamics during nanoparticle formation and aggregation of the polymeric nanoparticles (Donno et al., 2017, Jara et al., 2018). This variability in nanoparticle size can lead to variability in uptake following administration and therefore, undesirable inconsistencies in therapeutic effect.
A recent advance in the production of nanoparticles that offers superior nanoparticle size uniformity is microfluidics (Valencia et al., 2012). Liquid reagents are introduced into a microfluidics chip at pre-specified total flow rates and flow rate ratios, which are expressed as the combined flow rate of the liquid reagents and the ratio between the aqueous and organic phase, respectively. Using microfluidics, nanoliter amounts of liquid reagents are rapidly mixed in a microchannel in a highly controlled way (Garg et al., 2016). In order to increase the mixing efficiency, passive mixers are incorporated into the microchannel (Capretto et al., 2013). The staggered herringbone mixer (SHM) is a common passive mixer and is described as an in-floor groove in the microchannel with the shape of an asymmetric herringbone (Belliveau et al., 2012). Using the SHM configuration, mixing is induced by chaotic advection and contributes to an increased mixing efficiency and reduced mixing times (Belliveau et al., 2012, Stroock et al., 2002).
The biodegradable and biocompatible polymer poly (lactic-co-glycolic acid) (PLGA) is the most commonly used polymer for the formulation of nanomedicines (Danhier et al., 2012). The size and polydispersity of PLGA nanoparticles can be optimized using microfluidic devices by using a high total flow rate of the two miscible solvents and a high flow rate ratio between the aqueous and organic phase, while using polymers with a low molecular weight to produce monodisperse nanoparticles below 180 nm in size (Chiesa et al., 2018, Donno et al., 2017, Morikawa et al., 2018).
Cell-penetrating peptides (CPPs) show great potential to facilitate enhanced cell uptake of therapeutic compounds (Nel et al., 2009, Vasconcelos et al., 2015). The translocation ability applies to the CPP on its own and also for conjugates of the CPPs with different cargos like nanoparticles and macromolecules such as peptides, proteins and DNA oligomers (Feiner-Gracia et al., 2018, Kristensen et al., 2015, Trehin et al., 2004). The internalisation of CPPs is proposed to occur via direct translocation or endocytic pathways (Birch et al., 2018) and is influenced by the structure and concentration of the CPP, the cargo and the cell type (Cardoso et al., 2012, Hoyer et al., 2012). Several strategies for the application of CPPs have been described in the literature and include a co-administration of CPPs with the drug, formation of a covalent bond between CPP and drug, as well as a surface modification of nanoparticles with CPPs (Liu et al., 2013). Surface modification of polymeric nanoparticles to enhance delivery of macromolecules is a prudent approach that does not alter the bioactivity of the encapsulated macromolecule (Jain and Jain, 2015, Liu et al., 2013).
We have exploited our discovery that histidine co-polymerizes with polymeric nanoparticles to covalently attach CPPs to the nanoparticle wall (Chiu et al., 2015, Kafka et al., 2009). Our previous work has shown that a short peptide, RRH (arginine-arginine-histidine), increased the cellular uptake of RRH-modified poly(ethyl-cyanoacrylate) (PECA) nanoparticles in Caco-2 cells by 30% in comparison to unmodified PECA nanoparticles (Chiu et al., 2015). Other studies have shown that dendrimers based on the CPP Tat (47–57) with a multi-branched topology containing arginine-rich branches showed higher uptake with a 10–15 times higher florescence intensity in HeLa cells in comparison to the linear Tat (Eggimann et al., 2014). To the best of our knowledge branched CPPs have not yet been conjugated to polymeric nanoparticles to enhance their cellular uptake.
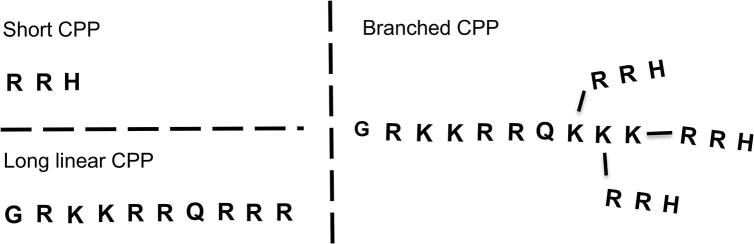
The aim of this study was to optimize a bulk nanoprecipitation method for the formulation of PLGA nanoparticles using a design of experiments approach. The bulk nanoprecipitation method was then compared with a microfluidics method to evaluate the uniformity of the formulated PLGA nanoparticles. Further, PLGA nanoparticles prepared with the microfluidics method were surface-modified with CPPs of different architectures (Fig. 1) to alter their physicochemical properties. The modified nanoparticles were characterized and can potentially be used as a drug delivery system to enhance uptake after oral administration.
Fig. 1.
Illustration of the architecture of the short (RRH), the long linear (TAT) and the branched CPP (bTAT: modified TAT backbone and three RRH branches). Amino acid single letter code is used (R = arginine, H = histidine, G = glycine, K = lysine, Q = glutamine).
2. Materials and methods
2.1. Materials
Poly(dl-lactide-co-glycolide) (PLGA) 50:50, ester terminated, MW 15,980 Da, was supplied by Durect Lactel (Cupertino, USA). Acetonitrile (ACN) (HPLC grade) was supplied by Merck (Darmstadt, Germany). Mowiol® 4-88 (polyvinyl alcohol, PVA, MW 31,000), 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) (purity > 99.5%), N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC) (purity >98%), trifluoroacetic acid (TFA, HPLC grade, purity >99%) and GRKKRRQK(RRH)K(RRH)K(RRH) (bTAT, custom synthesized, MW 2661.2 Da, purity >95%) were purchased from Sigma–Aldrich (St. Louis, USA). Snake Skin™ dialysis tubing (10 kDa MWCO) and sulfo-N-hydroxysulfosuccinimide (sulfo-NHS) were purchased from Thermo Fisher Scientific Australia Pty (Scoresby, Australia). The peptides RRH (MW 467.3 Da, purity >98%) and GRKKRRQRRR (TAT (48–57), MW 1396.7 Da, purity >98%) were custom synthesized by GLS (Shanghai, China) and used as supplied. Distilled, ultra-pure water was produced using a Milli-Q water Millipore Purification System™ (Billerica, MA, USA).
In this work, CPPs with three different architectures were designed for conjugation to PLGA nanoparticles to be able to investigate how the architecture of the CPPs influences cell uptake (Fig. 1). The short RRH was chosen from our previous work (Chiu et al., 2015), the long linear CPP was the widely investigated TAT peptide and for the third, we designed a branched TAT (bTAT), which has a modified TAT backbone with three RRH side branches (Fig. 1).
2.2. Methods
2.2.1. Design of experiments to optimize preparation of PLGA nanoparticles using a bulk nanoprecipitation method
A design of experiments study was used to identify the optimal concentration of PLGA, PVA and solvent evaporation time for the applied bulk nanoprecipitation method. MODDE GO 12 software (Umetrics, Umeå, Sweden) was used for the design and analysis of the experiments. After definition of the input parameters, a full factorial (2-levels) design with a center point for objective screening was used to define parameters for the individual experiments (Table 1). The selected response parameters for the PLGA nanoparticles were size and polydispersity index (PDI). For the validation of how well the model fits the data (R2) and to validate the ability to predict new data (Q2), analysis of variance (ANOVA) was performed. Further statistical testing included, testing the regression model for significance to obtain a probability value (p) and a lack of fit test to estimate the error of the model.
Table 1.
Input parameters for the design of experiments study to optimize the formulation of PLGA nanoparticles using a bulk nanoprecipitation method.
| Parameters | Level |
||
|---|---|---|---|
| −1 | 0 | 1 | |
| PLGA concentration (mg/mL) | 2 | 13.5 | 25 |
| PVA concentration (% w/v) | 0.5 | 2.75 | 5 |
| Stirring time (h) | 0.5 | 2.25 | 4 |
2.2.2. Bulk nanoprecipitation method for preparation of PLGA nanoparticles
PLGA nanoparticles were prepared using a bulk nanoprecipitation method based on the description by Fessi et al. (1989). Briefly, an organic PLGA solution (10 mg/mL in ACN) was added dropwise to a beaker containing 10 mL PVA (2% w/v in ultra-pure water) and the solvent was evaporated during magnetic stirring for 2 h and 15 min. The resulting PLGA nanoparticles were washed twice with ultra-pure water by centrifugation at 17,900g for 10 min (Eppendorf 5417 C centrifuge) at room temperature and stored as suspension in ultra-pure water at 4 °C until required with a maximum storage time of 7 days.
2.2.3. Microfluidics method for preparation of PLGA nanoparticles
PLGA nanoparticles were prepared using the NanoAssemblr® Benchtop Device (Precision NanoSystems, Vancouver, Canada). The aqueous PVA (2% w/v) solution was mixed with the organic PLGA solution (10 mg/mL in ACN) at a flow rate ratio (aqueous:organic) of 6:1 and a total flow rate of 10 mL/min (Streck et al., 2019). PLGA nanoparticles were dialyzed overnight against ultra-pure water at room temperature using Snake Skin™ dialysis tubing (10 kDa MWCO) and stored as suspension in ultra-pure water at 4 °C until required with a maximum storage time of 7 days.
2.2.4. Preparation of CPP-tagged PLGA nanoparticles
Prior to surface-modification, both types of PLGA nanoparticle suspensions produced with the bulk nanoprecipitation and the microfluidics method as described in 2.2.2, 2.2.3 were dialyzed overnight against ultra-pure water at room temperature using Snake Skin™ dialysis tubing (10 kDa MWCO).
A zero-length crosslinking reaction for the conjugation of CPPs to the surface of PLGA nanoparticles was adapted from Egusquiaguirre et al. (2015) with some modifications. Briefly, 500 µL of the PLGA nanoparticle suspension was diluted with HEPES buffer (0.025 M, pH 6.4) to 1.5 mL and the nanoparticle suspension was gently stirred while adding 250 µL EDC (1.5 mM in ultra-pure water) and 250 µL sulfo-NHS (2 mM in ultra-pure water). The stirring continued for 30 min at room temperature, before the solution was separated by ultra-centrifugation at 42,800g for 15 min at 4 °C. The pellet containing the activated nanoparticles was re-suspended and diluted with HEPES buffer (0.025 M, pH 6.4) to a volume of 1.1 mL. To this suspension, 100 µL of the CPP solutions with varying concentrations of RRH (10, 50 and 75 mM), TAT (2.9, 5.7, and 8.6 mM) and bTAT (4.5 mM and 6 mM) were added. The mixture was left to incubate overnight at 4 °C and was then separated by ultra-centrifugation at 42,800g for 15 min at 4 °C. The supernatant was aspirated and kept at 4 °C until HPLC analysis and the nanoparticle pellet was stored at 4 °C until required with a maximum storage time of 7 days.
2.2.5. Characterization of PLGA and CPP-tagged PLGA nanoparticles
Dynamic light scattering (DLS) was used to measure the Z-average diameter and PDI of the nanoparticles (Malvern® Nano ZS, Model Zen 3600, Malvern Instruments, Malvern, UK) equipped with a 633 nm laser and 173° detections optics. The laser Doppler electrophoresis technique was used to measure zeta potential of the nanoparticles with the same instrument. All samples were diluted to an appropriate concentration and measurements of three individual batches were taken in triplicate.
The morphologies of the PLGA and CPP-tagged PLGA nanoparticles prepared using the bulk nanoprecipitation method and the microfluidics method were investigated using transmission electron microscopy (TEM). First, the carbon film on a 300 mesh copper grid was made hydrophilic by glow discharge in air (Aebi and Pollard, 1987). Afterwards, 10 µL of the PLGA nanoparticle suspension was placed on the grid for 1 min and excess suspension was blotted before the grid was washed twice with ultra-pure water and stained to contrast the specimen with phosphotungstic acid (1% w/v, pH 6.8). The samples were viewed on a Phillips CM100 BioTWIN TEM (Philips Electron Optics, Eindhoven, The Netherlands) and the micrographs were recorded using a MegaView3 camera (Soft Imaging System, Münster, Germany).
2.2.6. Investigation of binding between CPPs and PLGA
To investigate any interactions between the CPPs and PLGA, Fourier-transform infrared spectroscopy (FTIR) spectra of PLGA polymer, CPPs and PLGA-CPP conjugates were recorded with a Varian 3100 FTIR (Varian, Palo Alto, USA) using transmission mode and compared. The spectra were obtained at wavenumbers between 400 and 4000 cm−1 with 64 scans per sample and a resolution of 4 cm−1. The PLGA-CPP conjugates were prepared using the zero-length crosslinking reaction. The PLGA polymer was dissolved in ACN (10 mg/mL) and to a 1 mL solution of the polymer 250 μL EDC (1.5 mM in ultra-pure water) and 250 μL of sulfo-NHS (2 mM in ultra-pure water) were added. The mixture was gently stirred on a magnetic stirring plate for 30 min and then precipitated by adding 3 mL of ultra-pure water and separated by centrifugation at 3220g for 20 min at 8 °C (Karve et al., 2011). The supernatant was removed and the pellet re-suspended in ACN and 200 μL of CPP solutions (50 mM RRH, 8.6 mM TAT and 4.5 mM bTAT) were added and incubated while gently stirred at room temperature for 30 min. The PLGA-CPP conjugates were precipitated again as described above and the pellet was dissolved in ACN at a 1:10 ratio (v/v).
The PLGA polymer was dissolved in ACN at a concentration of 10 mg/mL and the CPPs were dissolved in 20 μL ultra-pure water and then diluted with ACN to the intended concentration (50 mM RRH, 8.6 mM TAT and 4.5 mM bTAT) before analysis.
2.2.7. Conjugation efficiency of CPPs tagged to PLGA nanoparticles
The conjugation efficiency of the CPPs was obtained indirectly from the amount of CPPs remaining in the supernatant. The supernatant was collected after overnight incubation of the activated nanoparticles with the different concentrations of the three CPPs as described in Section 2.2.4.
CPP concentrations in the supernatant were quantified with validated RP-HPLC methods using the Agilent™ series 1200 HPLC system (Agilent Technologies, Santa Clara, USA) with a reverse phase column (HiChrom ultrasphere 5 ODS, 250 × 4.6 mm, 5 µm particle size, 300 Å pore size (HiChrom, Theale, UK)) and a UV detector (G1314B) at wavelengths between 214 and 220 nm. The aqueous mobile phase contained 0.1% (v/v) TFA in ultra-pure water and the organic mobile phase contained 0.1% (v/v) TFA in ACN. The CPPs in the supernatant were eluted by gradient (0–7% over 25 min for RRH, 0–13% over 37 min for TAT and 12–22% over 25 min for bTAT) at a flow rate of 0.6 mL/min. The CPP concentration was quantified using peak integration and an individual standard curve was produced for each CPP. The limits of quantification and the limits of detection are summarized in Table 2.
Table 2.
The limit of quantification and the limit of detection for the three different HPLC assays to quantify the concentration of each of the CPPs.
| Limit of quantification (LoQ) µg/mL | Limit of detection (LoD) µg/mL | |
|---|---|---|
| RRH | 5.4 | 1.8 |
| TAT | 3.1 | 1.0 |
| bTAT | 2.7 | 0.9 |
Eq. (1) was used to calculate the conjugation efficiency of the different CPPs used in the study. The conjugation efficiency was obtained by calculating the difference between the total concentration of CPPs added to the nanoparticle suspension and the measured concentration of CPPs in the supernatant after conjugation.
| (1) |
2.2.8. Statistical analysis
All experiments were performed in triplicate and results are presented as mean ± standard deviation (SD). Statistical analysis for design of experiments was performed using one-way analysis of variance (ANOVA) with MODDE GO 12 software. Comparisons between groups were made by performing a Student’s t test. The reported p-values were considered statistically significant at p < 0.05.
3. Results and discussion
3.1. Optimal PLGA and stabilizer concentration for the formulation of PLGA nanoparticles determined by design of experiments
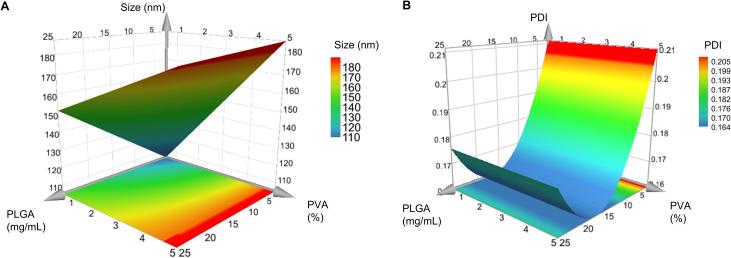
The size of nanoparticles plays a crucial role in cellular uptake and formulation of PLGA nanoparticles with a size below 200 nm facilitating cellular uptake via endocytosis (Sharma et al., 2015). In a design of experiments study the influence of PLGA concentration, stabilizer concentration and stirring time on the size and PDI of PLGA nanoparticles was investigated. A response surface plot was used for model interpretation and represents the combined effect of alterations of the input factors on the output response (Bairagi et al., 2018, Yadav and Sawant, 2010). The response surface plot obtained for size of PLGA nanoparticles prepared using a bulk nanoprecipitation method showed that a low concentration of PVA (≥ 2%) and a low concentration of PLGA (<15 mg/mL) was advantageous for the production of PLGA nanoparticles with a size of approximately 150 nm (Fig. 2A). The significant factors influencing the size of PLGA nanoparticles were the concentration of PLGA polymer, the concentration of PVA and the interaction term PLGA * PVA. Stirring time was found to have no significant influence on the size and PDI of PLGA nanoparticles and a stirring time of 2 h and 15 min, equal to the centre point of the design of experiments, was selected for the preparation of PLGA nanoparticles. With increasing concentrations of PLGA and PVA, size increased to approximately 200 nm. This observation was similar to other studies that reported an increase in nanoparticle size of PLGA nanoparticles when increased concentrations of PLGA (Sonam et al., 2014, Tefas et al., 2015) and PVA (Patel et al., 2016, Sonam et al., 2014) were used.
Fig. 2.
Response surface plots for the size (nm) (A) and polydispersity index (PDI) (B) of nanoparticles as a function of PLGA and PVA concentration for preparation of PLGA nanoparticles using the bulk nanoprecipitation method.
The PDI of the formulated PLGA nanoparticles was highly dependent on the PLGA concentration and was not influenced by the PVA concentration (Fig. 2B). The response surface plot showed that the PDI of PLGA nanoparticles formulated using the bulk nanoprecipitation method was the lowest between 10 and 22 mg/mL of the PLGA polymer and increased towards higher PDI values outside of this PLGA concentration range. This is in contrast to literature reporting on increased PDI with increasing polymer concentration (Patel et al., 2016). The PDI indeed increased with PLGA polymer concentrations above 20 mg/mL, but also at concentrations below 5 mg/mL of the PLGA polymer, which might be due to the instability of the formed PLGA nanoparticles at the low polymer concentration and a subsequent nanoparticle aggregation (Fig. 2B).
The partial least square model of size and PDI for the formulation of PLGA nanoparticles was analyzed with ANOVA to describe the quality of the model (Table 3). The values for R2 and Q2 (Table 3) were above the recommended values from the literature (R2 > 0.75, Q2 > 0.60) (Mandenius and Brundin, 2008) and provide support that the models of size and PDI produced with a bulk nanoprecipitation method were able to fit the data and can be used to predict new data. The p-values for the regression of the models were <0.05 and lack of fit p-values were either above or approximating 0.25, indicating that both models were significant and statically good at a 95% confidence interval.
Table 3.
Summary of results for the statistical analysis of the size and polydispersity index models for the bulk nanoprecipitation method to produce PLGA nanoparticles.
| R2 | Q2 | Regression p | Lack of fit | Significant | |
|---|---|---|---|---|---|
| Size | 0.963 | 0.806 | >0.001 | 0.442 | Yes |
| Polydispersity index | 0.869 | 0.763 | 0.006 | 0.234 | Yes |
The response surface plots for the size and PDI of the experimental data were used to select the parameters to produce optimized nanoformulations. The selected optimized parameters from the response surface plot for further experiments were a PLGA concentration of 10 mg/mL and a PVA concentration of 2% (w/v).
3.2. Characterization of PLGA nanoparticles
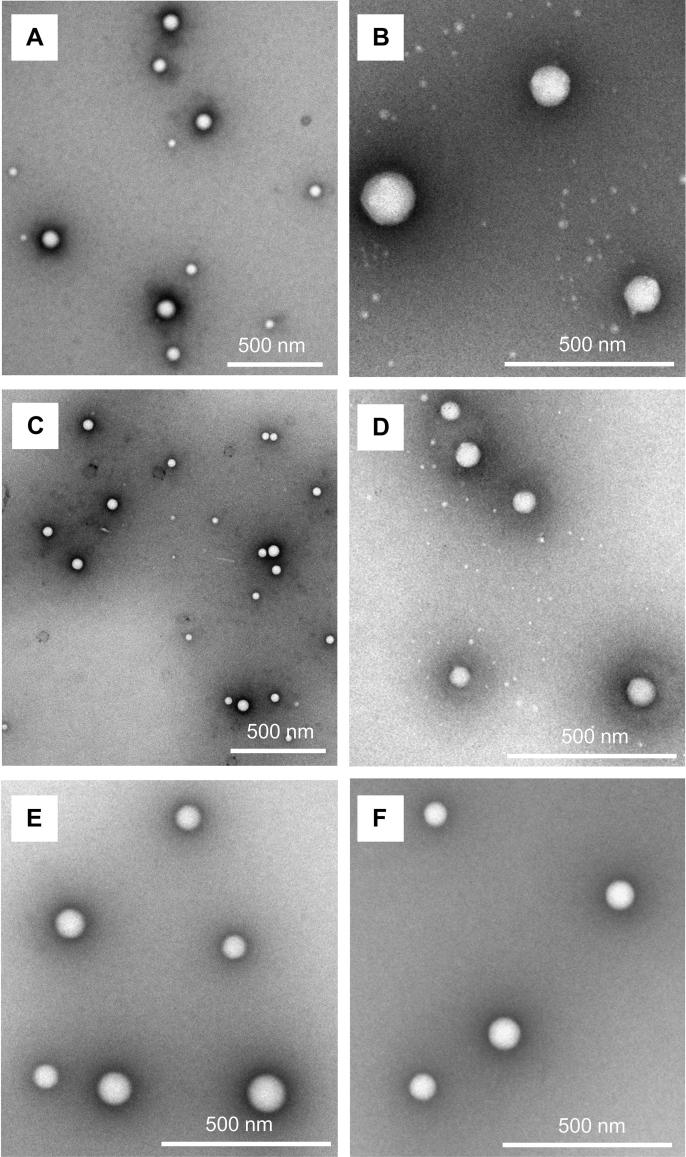
PLGA nanoparticles prepared using the bulk nanoprecipitation method had a significantly larger size compared to PLGA nanoparticles prepared using the microfluidics method (Table 4). PLGA nanoparticles formulated with both methods were monodisperse, but the PDI of the PLGA nanoparticles was higher when prepared with the microfluidics method. Patel et al. (2016) reported that a decrease in PDI was obtained with an increasing ratio between the aqueous and organic solutions from 3:1 to 6:1 for the formulation of cromolyn-loaded PLGA using a bulk nanoprecipitation method. A similar observation was made in the present study where the bulk nanoprecipitation method with a higher ratio between the aqueous and organic phase (20:1) yielded nanoparticles with a lower PDI compared to the microfluidics method, where the ratio between the aqueous and organic phase was 6:1. Using a higher ratio between the aqueous and organic phase during the preparation of polymeric nanoparticles results in a greater dilution of the organic polymer solution and the formed nanoparticles are less likely to aggregate. The higher ratio of aqueous to organic phase in the bulk nanoprecipitation method did not reduce the size of PLGA nanoparticles and the formation of nanoparticles >180 nm might be due to a polymer concentration above the critical polymer concentration (Lepeltier et al., 2014). The zeta potential of PLGA nanoparticles prepared with the two methods were similar (Table 4) and a negative surface charge below −20 mV suggested the presence of negatively charged carboxyl groups on the surface of the PLGA nanoparticles (Vasconcelos et al., 2015). The TEM micrographs revealed that PLGA nanoparticles with a spherical shape were obtained with both preparation methods (Fig. 3A and C).
Table 4.
Characterization of PLGA and TAT-tagged PLGA nanoparticles prepared with the optimized parameters for the bulk nanoprecipitation method and with the microfluidics method. Data are means ± SD (n = 3 independent batches). *p-value <0.05 in comparison to the respective bulk method.
| Size ± SD (nm) | PDI ± SD | Zeta potential ± SD (mV) | Conjugation efficiency (%) | |
|---|---|---|---|---|
| Unmodified PLGA nanoparticles | ||||
| Bulk | 184.0 ± 3.9 | 0.110 ± 0.007 | −23.0 ± 0.8 | N/A |
| Microfluidics | 151.2 ± 1.2* | 0.149 ± 0.014 | −22.2 ± 0.5 | N/A |
| TAT-tagged PLGA nanoparticles | ||||
| Bulk | 201.9 ± 12.5 | 0.136 ± 0.057 | −6.0 ± 3.4 | 55.3 ± 0.7 |
| Microfluidics | 172.4 ± 2.6* | 0.164 ± 0.014 | −4.3 ± 0.8 | 56.3 ± 1.0 |
Fig. 3.
TEM micrographs of unmodified PLGA (A) and TAT-tagged PLGA nanoparticles (B) prepared using a bulk nanoprecipitation method and unmodified PLGA (C), RRH- (D), TAT- (E) and bTAT-tagged PLGA nanoparticles (F) prepared using a microfluidics method.
With both preparation methods, the size of the PLGA nanoparticles increased with the conjugation of TAT (Table 4), suggesting successful conjugation of the CPP to the surface of the PLGA nanoparticles (Vasconcelos et al., 2015). The TAT-tagged PLGA nanoparticles produced using the microfluidics method were significantly smaller than TAT-tagged PLGA nanoparticles produced using the bulk nanoprecipitation method (Table 4). Further, the TAT-tagged PLGA nanoparticles prepared with the bulk nanoprecipitation method showed a size of 200 nm with a high standard deviation indicating a low batch to batch reproducibility (Table 4). The zeta potential of TAT-tagged PLGA nanoparticles prepared in this study was between −4 and −6 mV (Table 4). Although there was a shift in zeta potential to become less negative after conjugation of TAT, the overall negative zeta potential indicated that negatively charged carboxyl groups of the PLGA polymer were still present on the surface and that the surface density of TAT was not sufficient to achieve a positive surface charge (Gullotti and Yeo, 2012). The obtained surface charge of the TAT-tagged PLGA nanoparticles in the present study was within the range of −7 to +3 mV published in the literature for TAT conjugated to PLGA-PEG or PLGA nanoparticles with a size larger than 200 nm (Gartziandia et al., 2016, Xu et al., 2013). The conjugation efficiency of TAT conjugated to the PLGA nanoparticles was not different after production of the nanoparticles using the bulk nanoprecipitation or microfluidics method, with 55.3 ± 0.7% and 56.3 ± 1.0% conjugation efficiency, respectively (Table 4). However, since the PLGA nanoparticles prepared with the microfluidics method had a smaller size, more surface area of the nanoparticles might be covered with TAT to mask the negative charges of the carboxyl group of PLGA resulting in a slightly less negative zeta potential (Table 4). The morphology of the CPP-tagged PLGA nanoparticles did not change with the conjugation of TAT to PLGA nanoparticles prepared using the bulk nanoprecipitation or microfluidics method (Fig. 3B and E). Similarly, the conjugation of RRH and bTAT to PLGA nanoparticles prepared using the microfluidics method did not influence the morphology of the nanoparticles (Fig. 3D and F). The microfluidics method was selected for further investigations of the utility of this method for producing CPP-tagged PLGA nanoparticles due to the smaller nanoparticle size with better reproducibility.
3.3. Investigation of binding between CPPs and PLGA polymer
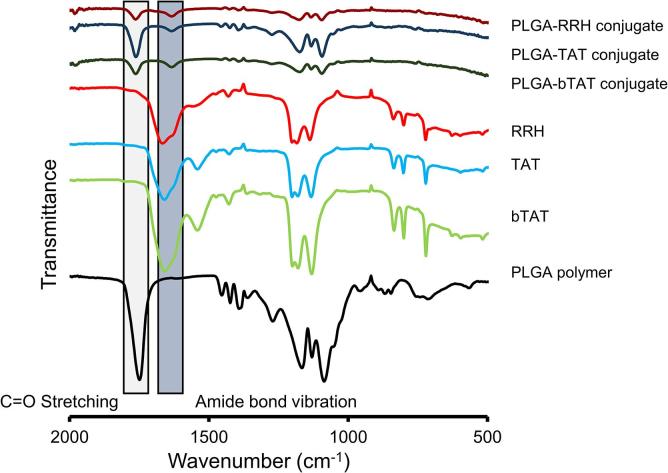
FTIR was used for the physicochemical characterization of the PLGA polymer, the three different CPPs and PLGA-CPP conjugates. The FTIR spectrum of the PLGA polymer alone showed the characteristic C O stretch of the carboxyl group at 1749 cm−1 and C–O ester stretches between 1167 and 1086 cm−1 (Fig. 4). The individual CPPs showed bands related to the amide I and II bond vibration at 1667–1659 and 1541–1560 cm−1, respectively (Fig. 4). The amide bond I and II vibrations were associated with the C O stretching and N–H bending of the amide bonds between the individual amino acids of the CPPs (Haris and Severcan, 1999). In addition, C–O ester stretch vibrations from the C-terminal amino acid of the CPPs were observed between 1202 and 1130 cm−1 (Fig. 4).
Fig. 4.
FTIR spectra of PLGA polymer, the individual cell-penetrating peptides (RRH, TAT, bTAT) and PLGA-CPP conjugates.
PLGA nanoparticles tagged with CPPs were analyzed using FTIR, however the FTIR spectrum showed broad and weak FTIR bands, which made assignment of wavenumbers difficult. Therefore, PLGA-CPP conjugates prepared by covalently attaching CPPs to the PLGA polymer were used to identify any interactions between the PLGA polymer and the CPPs. The carboxyl group of the PLGA polymer and the primary amine of the CPPs were utilized to form a covalent bond using a zero-length crosslinking reaction (Hermanson, 2013). Conjugation between the CPPs and PLGA polymer was confirmed with the observation of an amide I bond vibration in the CPP-PLGA conjugates at approximately 1635 cm−1. The shift of the amide I bond vibration to a lower wavelength in comparison to the amide I bond vibration in RRH, TAT and bTAT (1667–1659 cm−1) provided evidence of the successful formation of a covalent bond between these two components (Fig. 4). The FTIR spectra of the PLGA-CPP conjugates also showed a C O stretch at 1763 cm−1 for the carboxyl group of the PLGA polymer indicating that there were unconjugated carboxyl groups present.
3.4. Characteristics of CPP-tagged PLGA nanoparticles depends on the CPP architecture
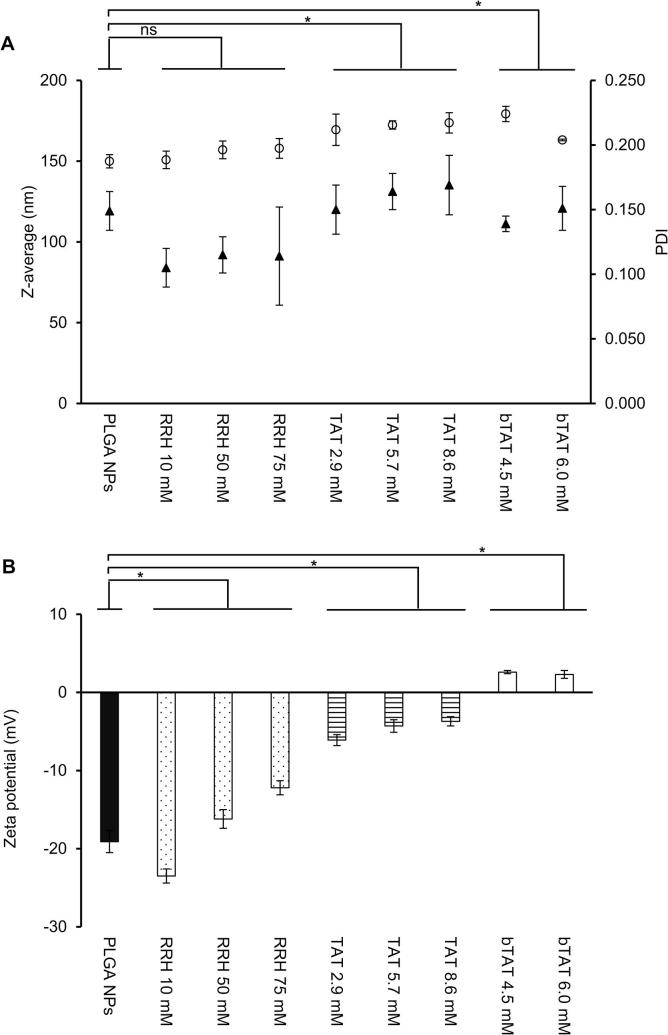
The influence of the CPP architecture on the characteristics of CPP-tagged PLGA nanoparticles was investigated by using a zero-length crosslinking reaction to conjugate CPPs to the surface of PLGA nanoparticles produced using microfluidics. PLGA nanoparticles had an average size of 150 nm and a zeta potential of −19 mV (Fig. 5A and B). With conjugation of the three different CPPs the size of the nanoparticles increased, but the increase in size was independent of the concentration of CPP (Fig. 5A). The slight increase in size of the CPP-tagged PLGA nanoparticles could be attributed to the presence of CPPs. Further, nanoparticles may increase in size due to swelling caused by the more hydrophilic surface of the CPP-tagged PLGA nanoparticles compared to PLGA nanoparticles (Feiner-Gracia et al., 2018).
Fig. 5.
Size, PDI (A) and zeta potential (B) of PLGA nanoparticles and CPP-tagged PLGA nanoparticles with different concentrations of CPPs prepared using the post-microfluidics conjugation approach. Size is displayed as circles and triangles represent PDI. Data are means ± SD (n = 3 independent batches). *p-value < 0.05 for the comparison of CPP-tagged PLGA nanoparticles with PLGA nanoparticles, ns = not significant.
The influence of different CPP concentrations on the physicochemical characteristics of CPP-tagged PLGA nanoparticles was evaluated to design nanoformulations with a diameter below 200 nm and a neutral zeta potential to be suitable for future investigations as oral drug delivery system. Studies have shown PLGA nanoparticles with a size below 200 nm were taken up by Caco-2 cells to a higher extent compared to larger nanoparticles and that the surface charge of nanoformulations influences the electrostatic interactions with the negatively charged cell membrane (Danhier et al., 2012, Gaumet et al., 2009). The zeta potential of RRH-tagged PLGA nanoparticles shifted from −24 to −12 mV with increasing concentrations of RRH (Fig. 5B). The conjugation of TAT resulted in the formation of slightly negative TAT-tagged PLGA nanoparticles with a surface charge of −4 mV (Fig. 5B). After conjugation of bTAT to the PLGA nanoparticles the zeta potential was slightly positive (+3 mV, Fig. 5B). The change in zeta potential can be explained by the architecture of the CPPs. The CPPs are characterized by a short, long linear and branched architecture (Fig. 1). The number of positive charges in the peptide sequence is increased with a longer and more complex architecture. RRH contains 2 positive charges in the peptide sequence, whereas TAT and bTAT contain 8 and 12 positive charges, respectively. The increase in positive charges from RRH > TAT > bTAT resulted in positively charged bTAT-tagged PLGA nanoparticles after the post-microfluidics conjugation approach. According to the PDI values all CPP-tagged PLGA nanoparticles showed a monodisperse size distribution (Fig. 5A) and no aggregation was observed. This is in contrast to the literature, where highly negatively or positively charged nanoparticles with a zeta potential below −30 and above +30 mV are believed to be more stable because of stronger repulsion forces than between neutral charged nanoparticles with a zeta potential between −10 and +10 mV (Sonam et al., 2014, Tefas et al., 2015, Vasconcelos et al., 2015).
3.5. Conjugation efficiency of CPPs with different architectures on PLGA nanoparticles
The amount of CPPs binding to the surface of PLGA nanoparticles depends on the number of available carboxyl groups and the orientation of the CPP. The number of binding sites on a single PLGA nanoparticle with a size of 155 nm was estimated to be 16,000. For this calculation the ratio between the lactic and glycolic acid groups (50:50) and their Connolly surface area was considered (Connolly, 1983). The three different CPPs have multiple primary amines that are likely to act as a binding site depending on their position in the CPP sequence (Fig. 1). An estimate of the amount of CPPs covalently attached to the surface available carboxyl groups of the PLGA nanoparticles was quantified by calculating the conjugation efficiency. The conjugation efficiency for RRH, TAT and bTAT depended on the concentration of the CPPs added (Table 5). The amount of RRH conjugated to the PLGA nanoparticles increased by 12-fold when the concentration of RRH was increased from 10 to 50 mM. A further increase in the RRH concentration to 75 mM resulted in a slight decrease in conjugation efficiency, suggesting that at 50 mM there was a saturation of the surface available carboxyl groups on the PLGA nanoparticles that can react with the primary amines in RRH. The concentration of TAT conjugated to PLGA nanoparticles increased in a linear manner with increasing concentrations of the CPP. The bTAT-tagged PLGA nanoparticles showed a similar concentration of bTAT conjugated to the surface of the PLGA nanoparticles as TAT (Table 5). These results can be related to the structure of the CPPs. The short CPP RRH showed the highest concentration of CPP on the surface, which is related to the amount of RRH that was added and the shorter amino acid sequence in comparison to the other two CPPs. The concentrations of TAT and bTAT conjugated to the PLGA nanoparticles were similar and the amount of CPP that can be conjugated might be limited because of the more complex and branched architecture of the CPPs, which induced steric hindrance between individual CPP molecules. With the conjugation of TAT and bTAT to the surface of PLGA nanoparticles, it is possible that not all available binding sites would be used due to the orientation of the CPPs. TAT has primary amines along the amino acid sequence and can, therefore, bind in an orientation perpendicular to the nanoparticle surface or with a more tangential orientation in relation to the nanoparticle surface. A tangential orientation of the CPP may cover other binding sites on the surface of the PLGA nanoparticles since the tangential orientation covers more surface area. The same applies to the bTAT, which has primary amino acids in the long linear backbone and the short branches.
Table 5.
Conjugation efficiencies and the equivalent concentration of the CPPs tagged to PLGA nanoparticles (NPs) using different concentrations of CPPs prepared with a post-microfluidics conjugation approach. Data are means ± SD (n = 3 independent batches). *p-value <0.05 in comparison to the conjugation efficiency of 8.6 mM TAT.
| CPP concentration (mM) | Conjugation efficiency (%) ± SD | CPP concentration (µM) ± SD |
|---|---|---|
| RRH-tagged PLGA NPs | ||
| 10 | 32.4 ± 0.8 | 0.25 ± 0.03 |
| 50 | 74.4 ± 4.5 | 3.10 ± 0.19 |
| 75 | 58.5 ± 6.0 | 3.66 ± 0.37 |
| TAT-tagged PLGA NPs | ||
| 2.9 | 57.6 ± 0.5 | 0.150 ± 0.001 |
| 5.7 | 56.3 ± 1.0 | 0.29 ± 0.01 |
| 8.6 | 55.5 ± 0.8 | 0.43 ± 0.01 |
| bTAT-tagged PLGA NPs | ||
| 4.5 | 79.8 ± 0.3* | 0.327 ± 0.001 |
| 6.0 | 45.4 ± 0.7 | 0.248 ± 0.004 |
4. Conclusion
The design of experiments study of the bulk nanoprecipitation method showed that the PLGA polymer concentration and the surfactant concentration influenced the size and PDI of PLGA nanoparticles. The optimized concentrations of the PLGA polymer and surfactant were used in the microfluidics method and the size of PLGA nanoparticles was reduced compared to the size of PLGA nanoparticles prepared with the bulk nanoprecipitation method. The successful functionalization of CPPs to the PLGA polymer was shown by the formation of amide bonds. The CPP architecture determined the physicochemical characteristics and especially the zeta potential of the CPP-tagged PLGA nanoparticles. The surface charge was tuned from negatively charged RRH- and TAT-tagged PLGA nanoparticles to slightly positively charged bTAT-tagged PLGA nanoparticles with the introduction of a branched CPP architecture. The CPP-tagged PLGA nanoparticles with tuned surface properties have the potential to be an effective drug delivery system intended for oral administration of drugs.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge Dr Woravimol Krittaphol for assistance and advice with development of the HPLC methods and Sanjay Patel for help with the HPLC method for branched CPP bTAT. The TEM micrographs were obtained at the Otago Micro and Nanoscale Imaging facility with assistance of Richard Easingwood. This work was supported by the New Zealand Pharmacy Education Research Foundation [Reference No. 301].
References
- Aebi U., Pollard T.D. A glow discharge unit to render electron microscope grids and other surfaces hydrophilic. J. Electron Microsc. Tech. 1987;7:29–33. doi: 10.1002/jemt.1060070104. [DOI] [PubMed] [Google Scholar]
- Bairagi U., Mittal P., Singh J., Mishra B. Preparation, characterization, and in vivo evaluation of nano formulations of ferulic acid in diabetic wound healing. Drug Dev. Ind. Pharm. 2018;44:1783–1796. doi: 10.1080/03639045.2018.1496448. [DOI] [PubMed] [Google Scholar]
- Banik B.L., Fattahi P., Brown J.L. Polymeric nanoparticles: the future of nanomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2016;8:271–299. doi: 10.1002/wnan.1364. [DOI] [PubMed] [Google Scholar]
- Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucl. Acids. 2012;1 doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch D., Christensen M.V., Staerk D., Franzyk H., Nielsen H.M. Stereochemistry as a determining factor for the effect of a cell-penetrating peptide on cellular viability and epithelial integrity. Biochem. J. 2018;475:1773–1788. doi: 10.1042/bcj20180155. [DOI] [PubMed] [Google Scholar]
- Capretto L., Carugo D., Mazzitelli S., Nastruzzi C., Zhang X. Microfluidic and lab-on-a-chip preparation routes for organic nanoparticles and vesicular systems for nanomedicine applications. Adv. Drug Deliv. Rev. 2013;65:1496–1532. doi: 10.1016/j.addr.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Cardoso A.M.S., Trabulo S., Cardoso A.L., Lorents A., Morais C.M., Gomes P., Nunes C., Lúcio M., Reis S., Padari K., Pooga M., Pedroso de Lima M.C., Jurado A.S. S4(13)-PV cell-penetrating peptide induces physical and morphological changes in membrane-mimetic lipid systems and cell membranes: implications for cell internalization. Biochim. Biophys. Acta. 2012;1818:877–888. doi: 10.1016/j.bbamem.2011.12.022. [DOI] [PubMed] [Google Scholar]
- Chiesa E., Dorati R., Modena T., Conti B., Genta I. Multivariate analysis for the optimization of microfluidics-assisted nanoprecipitation method intended for the loading of small hydrophilic drugs into PLGA nanoparticles. Int. J. Pharm. 2018;536:165–177. doi: 10.1016/j.ijpharm.2017.11.044. [DOI] [PubMed] [Google Scholar]
- Chiu J.Z., Tucker I.G., McLeod B.J., McDowell A. Arginine-tagging of polymeric nanoparticles via histidine to improve cellular uptake. Eur. J. Pharm. Biopharm. 2015;89:48–55. doi: 10.1016/j.ejpb.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Connolly M. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Danaei M., Dehghankhold M., Ataei S., Hasanzadeh Davarani F., Javanmard R., Dokhani A., Khorasani S., Mozafari M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics. 2018;10:57. doi: 10.3390/pharmaceutics10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhier F., Ansorena E., Silva J.M., Coco R., Le Breton A., Preat V. PLGA-based nanoparticles: an overview of biomedical applications. J. Control. Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- Donno R., Gennari A., Lallana E., De La Rosa J.M.R., d’Arcy R., Treacher K., Hill K., Ashford M., Tirelli N. Nanomanufacturing through microfluidic-assisted nanoprecipitation: advanced analytics and structure-activity relationships. Int. J. Pharm. 2017;534:97–107. doi: 10.1016/j.ijpharm.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Eggimann G.A., Blattes E., Buschor S., Biswas R., Kammer S.M., Darbre T., Reymond J.-L. Designed cell penetrating peptide dendrimers efficiently internalize cargo into cells. Chem. Commun. 2014;50:7254–7257. doi: 10.1039/C4CC02780A. [DOI] [PubMed] [Google Scholar]
- Egusquiaguirre S.P., Manguan-Garcia C., Pintado-Berninches L., Iarriccio L., Carbajo D., Albericio F., Royo M., Pedraz J.L., Hernandez R.M., Perona R., Igartua M. Development of surface modified biodegradable polymeric nanoparticles to deliver GSE24.2 peptide to cells: a promising approach for the treatment of defective telomerase disorders. Eur. J. Pharm. Biopharm. 2015;91:91–102. doi: 10.1016/j.ejpb.2015.01.028. [DOI] [PubMed] [Google Scholar]
- Feiner-Gracia N., Dols-Perez A., Royo M., Solans C., Garcia-Celma M.J., Fornaguera C. Cell penetrating peptide grafting of PLGA nanoparticles to enhance cell uptake. Eur. Polym. J. 2018;108:429–438. doi: 10.1016/j.eurpolymj.2018.09.026. [DOI] [Google Scholar]
- Fessi H., Puisieux F., Devissaguet J.P., Ammoury N., Benita S. Nanocapsule formation by interfacial polymer deposition following solvent displacement. Int. J. Pharm. 1989;55:R1–R4. doi: 10.1016/0378-5173(89)90281-0. [DOI] [Google Scholar]
- Galindo-Rodríguez S.A., Puel F., Briançon S., Allémann E., Doelker E., Fessi H. Comparative scale-up of three methods for producing ibuprofen-loaded nanoparticles. Eur. J. Pharm. Sci. 2005;25:357–367. doi: 10.1016/j.ejps.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Garg S., Heuck G., Ip S., Ramsay E. Microfluidics: a transformational tool for nanomedicine development and production. J. Drug Target. 2016;24:821–835. doi: 10.1080/1061186X.2016.1198354. [DOI] [PubMed] [Google Scholar]
- Gartziandia O., Egusquiaguirre S.P., Bianco J., Pedraz J.L., Igartua M., Hernandez R.M., Préat V., Beloqui A. Nanoparticle transport across in vitro olfactory cell monolayers. Int. J. Pharm. 2016;499:81–89. doi: 10.1016/j.ijpharm.2015.12.046. [DOI] [PubMed] [Google Scholar]
- Gaumet M., Gurny R., Delie F. Localization and quantification of biodegradable particles in an intestinal cell model: the influence of particle size. Eur. J. Pharm. Sci. 2009;36:465–473. doi: 10.1016/j.ejps.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Gullotti E., Yeo Y. Beyond the imaging: limitations of cellular uptake study in the evaluation of nanoparticles. J. Control. Release. 2012;164:170–176. doi: 10.1016/j.jconrel.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haris P.I., Severcan F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. J. Mol. Catal. B: Enzyme. 1999;7:207–221. doi: 10.1016/S1381-1177(99)00030-2. [DOI] [Google Scholar]
- Hermanson G.T. In: Bioconjugate Techniques. third ed. Audet J., Preap M., editors. Academic Press; 2013. Zero-length crosslinkers; pp. 259–273. [DOI] [Google Scholar]
- Hoyer J., Schatzschneider U., Schulz-Siegmund M., Neundorf I. Dimerization of a cell-penetrating peptide leads to enhanced cellular uptake and drug delivery. Beilstein J. Org. Chem. 2012;8:1788–1797. doi: 10.3762/bjoc.8.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Jain S.K. L-Valine appended PLGA nanoparticles for oral insulin delivery. Acta Diabetol. 2015;52:663–676. doi: 10.1007/s00592-015-0714-3. [DOI] [PubMed] [Google Scholar]
- Jara M.O., Catalan-Figueroa J., Landin M., Morales J.O. Finding key nanoprecipitation variables for achieving uniform polymeric nanoparticles using neurofuzzy logic technology. Drug Deliv. Transl. Res. 2018;8:1797–1806. doi: 10.1007/s13346-017-0446-8. [DOI] [PubMed] [Google Scholar]
- Jung T., Kamm W., Breitenbach A., Kaiserling E., Xiao J.X., Kissel T. Biodegradable nanoparticles for oral delivery of peptides: is there a role for polymers to affect mucosal uptake? Eur. J. Pharm. Biopharm. 2000;50:147–160. doi: 10.1016/S0939-6411(00)00084-9. [DOI] [PubMed] [Google Scholar]
- Kafka A.P., Kleffmann T., Rades T., McDowell A. Histidine residues in the peptide d-Lys6-GnRH: potential for copolymerization in polymeric nanoparticles. Mol. Pharm. 2009;6:1483–1491. doi: 10.1021/mp900043e. [DOI] [PubMed] [Google Scholar]
- Karnik R., Gu F., Basto P., Cannizzaro C., Dean L., Kyei-Manu W., Langer R., Farokhzad O.C. Microfluidic platform for controlled synthesis of polymeric nanoparticles. Nano Lett. 2008;8:2906–2912. doi: 10.1021/nl801736q. [DOI] [PubMed] [Google Scholar]
- Karve S., Werner M.E., Cummings N.D., Sukumar R., Wang E.C., Zhang Y.-A., Wang A.Z. Formulation of diblock polymeric nanoparticles through nanoprecipitation technique. JoVE. 2011:e3398. doi: 10.3791/3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen M., de Groot A.M., Berthelsen J., Franzyk H., Sijts A., Nielsen H.M. Conjugation of cell-penetrating peptides to parathyroid hormone affects its structure, potency, and transepithelial permeation. Bioconjug. Chem. 2015;26:477–478. doi: 10.1021/bc5005763. [DOI] [PubMed] [Google Scholar]
- Lepeltier E., Bourgaux C., Couvreur P. Nanoprecipitation and the “Ouzo effect”: application to drug delivery devices. Adv. Drug Deliv. Rev. 2014;71:86–97. doi: 10.1016/j.addr.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Liu X., Liu C., Zhang W., Xie C., Wei G., Lu W. Oligoarginine-modified biodegradable nanoparticles improve the intestinal absorption of insulin. Int. J. Pharm. 2013;448:159–167. doi: 10.1016/j.ijpharm.2013.03.033. [DOI] [PubMed] [Google Scholar]
- Mandenius C.F., Brundin A. Bioprocess optimization using design-of-experiments methodology. Biotechnol. Prog. 2008;24:1191–1203. doi: 10.1002/btpr.67. [DOI] [PubMed] [Google Scholar]
- Masood F. Polymeric nanoparticles for targeted drug delivery system for cancer therapy. Mater. Sci. Eng. C: Mater. Biol. Appl. 2016;60:569–578. doi: 10.1016/j.msec.2015.11.067. [DOI] [PubMed] [Google Scholar]
- Morikawa Y., Tagami T., Hoshikawa A., Ozeki T. The use of an efficient microfluidic mixing system for generating stabilized polymeric nanoparticles for controlled drug release. Biol. Pharm. Bull. 2018;41:899–907. doi: 10.1248/bpb.b17-01036. [DOI] [PubMed] [Google Scholar]
- Nel A.E., Mädler L., Velegol D., Xia T., Hoek E.M.V., Somasundaran P., Klaessig F., Castranova V., Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009;8:543. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- Patel R.R., Chaurasia S., Khan G., Chaubey P., Kumar N., Mishra B. Cromolyn sodium encapsulated PLGA nanoparticles: an attempt to improve intestinal permeation. Int. J. Biol. Macromol. 2016;83:249–258. doi: 10.1016/j.ijbiomac.2015.11.084. [DOI] [PubMed] [Google Scholar]
- Sharma G., Sharma A.R., Nam J.-S., Doss G.P.C., Lee S.-S., Chakraborty C. Nanoparticle based insulin delivery system: the next generation efficient therapy for Type 1 diabetes. J. Nanobiotechnol. 2015;13:74. doi: 10.1186/s12951-015-0136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonam, Chaudhary H., Kumar V. Taguchi design for optimization and development of antibacterial drug-loaded PLGA nanoparticles. Int. J. Biol. Macromol. 2014;64:99–105. doi: 10.1016/j.ijbiomac.2013.11.032. [DOI] [PubMed] [Google Scholar]
- Streck S., Clulow A.J., Mørck Nielsen H., Rades T., Boyd B.J., McDowell A. The distribution of cell-penetrating peptides on polymeric nanoparticles prepared using microfluidics and elucidated with small angle X-ray scattering. J. Colloid Interface Sci. 2019;555:438–448. doi: 10.1016/j.jcis.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Stroock A.D., Dertinger S.K., Ajdari A., Mezic I., Stone H.A., Whitesides G.M. Chaotic mixer for microchannels. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- Tefas L.R., Tomuta I., Achim M., Vlase L. Development and optimization of quercetin-loaded PLGA nanoparticles by experimental design. Clujul Med. 2015;88:214–223. doi: 10.15386/cjmed-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trehin R., Krauss U., Beck-Sickinger A.G., Merkle H.P., Nielsen H.M. Cellular uptake but low permeation of human calcitonin-derived cell penetrating peptides and Tat(47–57) through well-differentiated epithelial models. Pharm. Res. 2004;21:1248–1256. doi: 10.1023/B:PHAM.0000033013.45204.c3. [DOI] [PubMed] [Google Scholar]
- Valencia P.M., Farokhzad O.C., Karnik R., Langer R. Microfluidic technologies for accelerating the clinical translation of nanoparticles. Nat. Nanotechnol. 2012;7:623. doi: 10.1038/nnano.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos A., Vega E., Perez Y., Gomara M.J., Garcia M.L., Haro I. Conjugation of cell-penetrating peptides with poly(lactic-co-glycolic acid)-polyethylene glycol nanoparticles improves ocular drug delivery. Int. J. Nanomed. 2015;10:609–631. doi: 10.2147/ijn.S71198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Kona S., Su L.-C., Tsai Y.-T., Dong J.-F., Brilakis E.S., Tang L., Banerjee S., Nguyen K.T. Multi-ligand poly(l-lactic-co-glycolic acid) nanoparticles inhibit activation of endothelial cells. J. Cardiovasc. Transl. Res. 2013;6:570–578. doi: 10.1007/s12265-013-9460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav K.S., Sawant K.K. Modified nanoprecipitation method for preparation of cytarabine-loaded PLGA nanoparticles. AAPS Pharm. Sci. Tech. 2010;11:1456–1465. doi: 10.1208/s12249-010-9519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]