Abstract
Introduction
Chronic obstructive pulmonary disease (COPD) is a lung disease closely related to exposure to exogenous substances. CYP2B6 can activate many exogenous substances, which in turn affect lung cells. The aim of this study was to assess the association of single-nucleotide polymorphisms (SNPs) in CYP2B6 with COPD risk in a Chinese Han population.
Materials and methods
Genotypes of the five candidate SNPs in CYP2B6 were identified among 318 cases and 508 healthy controls with an Agena MassARRAY method. The association between CYP2B6 polymorphisms and COPD risk was evaluated using genetic models and haplotype analyses.
Results
In allele model, we observed that rs4803420 G and rs1038376 A were related to COPD risk. And rs4803420 G/T and G/T-T/T were related to a decreased COPD risk compared to GG genotype in the co-dominant and dominant models, respectively. When comparing with the AA genotype, rs1038376 A/T and A/T-T/T were associated with an increased COPD risk in the co-dominant and dominant models, respectively. Further gender stratification co-dominant and dominant models analysis showed that genotype G/T and G/T-T/T of rs4803420, and genotype A/T and A/T-T/T of rs1038376 were significantly associated with COPD risk compared to the wide type in males and females, while allele C of rs12979270 was only associated with COPD risk in females. Smoking status stratification analysis showed that rs12979270 C was significantly associated with an increased COPD risk under the allele model compared with allele A in the smoking subgroup. Haplotype analysis showed that haplotype GTA and TAA were related to COPD risk.
Conclusion
Our data is the first to demonstrate that CYP2B6 polymorphisms may exert effects on COPD susceptibility in the Chinese Han population.
Keywords: CYP2B6, chronic obstructive pulmonary disease, COPD, gene polymorphisms, case–control study
Introduction
Chronic obstructive pulmonary disease (COPD) is a leading cause of morbidity and mortality worldwide, characterized by persistent airflow limitation that is usually progressive and associated with an enhanced chronic inflammatory response in the airways and the lung to noxious particles or gases.1,2 It was reported that 210 million people were affected worldwide and caused more than 4 million people deaths per year, 90% of which occurred in low- and middle-income countries.3 Among them, 9–10% of the adult patients and 8.2% of the patients in 40 years old age group in China suffer from COPD.4–6 Based on data from the World Health Organization, COPD will become the fourth most prevalent disease and the fourth most common cause of death by 2035.7 At the same time, COPD also led to economic and social burden that is substantial and increasing.8
The pathophysiology of COPD involved in complex interactions among several factors. These factors included tobacco, marijuana, indoor pollution (eg, biomass cooking and heating in poorly ventilated dwellings), and occupational exposure to organic and inorganic dust, chemicals and smoke, as well as genetic vulnerability.9–11 It is worth noting that COPD is a lung disease intimately linked to exposure to exogenous substances. Although many of the exogenous compounds are not inherently dangerous, they have deleterious effect on lung cells after enzymatic activation. And these activation reactions are mainly catalyzed by enzymes in cytochrome P450 (CYP) superfamily.12
Cytochrome P450, family 2, subfamily B, polypeptide 6 (CYP2B6) is a member of the cytochrome P450 superfamily. It has been reported that the CYP2B6 enzyme also played an important role in the biological transformation of many xenobiotics, such as cyclophosphamide, ifosfamide, ketamine, propofol, bupropion, nevirapine, efavirenz, and some carcinogens such as aflatoxin B1.13–15 Moreover, if xenobiotics are converted into their bioactive forms as precancer pathogens, they may have irreversible reactions with DNA, leading to mutations and chromosomal aberrations.16 At present, CYP2B6 mRNA17 and its protein expression18 level were found in human lung. Immunohistochemistry assay detected that CYP2B6 was also expressed in human Clara cells secreted by a large number of non-ciliated secretory cells in the bronchiolar epithelium of the mammalian lung.19 Another reporter found its mRNA was also expressed in bronchial and peripheral lung.20 The catalytic activity associated with CYP2B6 was also detected in human lung.21 To sum up, these studies suggest that CYP2B6 may be closely related to the occurrence of lung disease by activating some exogenous compounds inhaled into the lungs.
Hainan is a multiethnic mix province, while Han and Li are the two main ethnicities, and the incidence of COPD in the Hainan population is higher than that of other regions of the People s Republic of China.2,22 COPD is closely related to exposure to exogenous substances. And the association between CYP2B6 variant and COPD susceptibility has not been investigated in the Hainan population so far. Therefore, in the present study, we firstly investigated the connection of five CYP2B6 gene polymorphisms (rs2099361, rs4803418, rs4803420, rs1038376, and rs12979270) and the risk of COPD in the Hainan population. Our study is supposed to provide more evidence for CYP2B6 in COPD pathogenesis and contribute to early COPD risk estimation among the individuals of Chinese ancestry.
Materials and methods
Ethics statement
The study was approved by the ethics committee of Hainan Province People’s Hospital of China, in accordance with the principles of the Helsinki Declaration and following national and international guidelines. Each participant was informed both in writing and verbally of the procedures and purpose of our research and signed a written informed consent before donating 5 mL venous blood for further analyses.
Research participants
This case–control study consisted of 318 patients and 508 controls, and they were consecutively recruited between July 2016 and July 2018 at the Hainan Province People’s Hospital of China. All subjects in our study were unrelated individuals recruited from Hainan province, China. All patients were diagnosed with COPD based on patient history, physical examination, and pulmonary function, according to the criteria of the National Heart, Lung and Blood Institute (NHLBI)/WHO Global initiative for Chronic Obstructive Lung Disease (GOLD).23 The inclusion criteria for COPD patients were the post-bronchodilator forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) of less than 70% and the FEV1 of less than 80% predicted. COPD severity was classified according to the GOLD guidelines for predicting FEV1 percent: mild (>80%), moderate (50–80%), severe (30–50%), or very severe (<30%). The exclusion criteria for COPD patients were as follows: 1) patients unable to perform lung function tests; 2) had other significant respiratory diseases such as asthma, congestive heart failure, cystic fibrosis, and tuberculosis.
The control group was randomly selected healthy individuals. The control group excluded individuals suffering from any organic disease by clinical examination, chest X-ray examination, and laboratory investigations. Written informed consent was obtained from each participant prior to enrollment in the study.
Data collection
Five milliliters of peripheral blood samples from each participant were collected by a specialized technician and stored into tubes containing ethylenediaminetetraacetic acid (EDTA) for anticoagulation. Genomic DNA was isolated from whole blood samples using the GoldMag DNA Purification Kit (GoldMag Co. Ltd, Xi’an City, China) according to the manufacturer’s instructions, and the purity and concentration of DNA were measured by the NanoDrop2000 (Thermo Scientific, Waltham, MA, USA). The isolated DNA was stored at 80°C until analysis. All subjects signed a written informed consent in this study.
SNP selection and genotyping
This study focused on the relationship between CYP2B6 SNPs and COPD risk in Chinese Han population. Based on the 1000 Genomes database (http://www.internationalgenome.org/), we finally identified five candidate polymorphisms (rs2099361, rs4803418, rs4803420, rs1038376, and rs12979270) for the present case–control study. Each SNP had the minor allele frequency (MAF)>0.05 in global population from the 1000 Genome Project. Agena MassARRAY Assay Design 4.0 software was used to design the primers for amplification and extension reactions. Genotyping was carried out by two laboratory personnel in a double-blinded fashion using the Agena MassARRAY system (Agena, San Diego, CA, USA.).24 The primers used for the five SNPs are shown in Table 1.
Table 1.
Primers used for this study
| SNP_ID | 2nd PCR | 1st PCR | UEP DIR | UEP SEQ |
|---|---|---|---|---|
| rs12979270 | ACGTTGGATGCTGTTGTAGACCACAGTCAC | ACGTTGGATGTGATAGACGGAAGCAGTAGG | F | gCTGCTGTAGTCTTCCCC |
| rs4803420 | ACGTTGGATGTGAAAACATTTGGAGCTAGG | ACGTTGGATGAGCTGGTATCACAGGCGTC | R | ggggaTGACATCAGGAGTTCAAGA |
| rs2099361 | ACGTTGGATGCACAATGGACAATGTCAACG | ACGTTGGATGTTTGTACATCCAGTGGGCTC | F | gtcacCGTAGTTGAGTGATTCTTTAC |
| rs1038376 | ACGTTGGATGCAACATGAAAGCAACCAGAC | ACGTTGGATGTTTAGTAGGGACATGTTGGC | R | caccTTATGCCTGTAATATCAGCACTT |
| rs4803418 | ACGTTGGATGGCAAAGAGAAATTGAGGCAG | ACGTTGGATGTCTCTCTGGTTCTCACTTGC | F | gggcGGCAGAGAAAATTAGAGAGA |
Abbreviations: PCR, polymerase chain reaction; UEP, unextended mini-sequencing primer.
Statistical analysis
Statistical analysis was performed with Microsoft Excel and SPSS 18.0 (SPSS, Chicago, IL, USA). Welch’s t-tests (for continuous variables) and the Chi square test (χ2 test, for categorical variables) were used to assess the differences in the distribution of demographic characteristics of case and control groups. SNP frequencies in the control subjects were evaluated for departure from Hardy–Weinberg Equilibrium (HWE) by the Fisher’s exact test. Allele and genotype frequencies of each SNP in patient and control groups were compared by a χ2 test. The associations between these SNPs and COPD risk were assessed by calculating odds ratios (ORs) and 95% confidence intervals (CIs) using logistic regression analysis with adjustments for age and gender. Multiple inheritance models (codominant, dominant, recessive, and log-additive) were generated by PLINK software (http://www.cog-genomics.org/plink2/) to estimate the relationship between each SNP and COPD risk.25,26 Finally, we used the Haploview software package (version 4.2) for pairwise linkage disequilibrium (LD) and haplotype construction, and used SHEsis software for haplotype association analyses. All P-values were two-sided, and P<0.05 was considered statistically significant.
Results
Demographic characteristics
The basic characteristics of cases and controls are summarized in Table 2. The study included 318 patients with COPD (238 males and 75 females; age at diagnosis: 64.56±8.49 years) and 508 healthy controls (337 males and 171 females; age at diagnosis: 64.52±10.40 years). There was a statistically significant difference in the distributions of gender and age between the case group and the control group (P<0.05). The average of patients’ BMI was slightly higher than that of the control group, and there was no statistically significant difference (P>0.05). Pulmonary function (FEV1/FVC) of COPD cases and controls was 0.56 (±0.05) and 0.79 (±0.04), respectively. Pulmonary function (FEV1/FVC) in the COPD cases was worse than that in the control subjects. The number of non-smokers was larger than that of smokers (p=0.082), but there was no significant difference in the distribution of smoking status.
Table 2.
Characteristics of patients with COPD and the control individuals
| Variable (s) | Case, N (%) | Control, N (%) | P-value |
|---|---|---|---|
| (N=313) | (N=508) | ||
| Gender, N (%) | 0.004a | ||
| Male | 238 (76.0%) | 337 (66.3%) | |
| Female | 75 (24.0%) | 171 (33.7%) | |
| Age, year (mean ± SD) | 71.80±10.09 | 60.05±6.48 | <0.001b |
| Smoking status, N (%) | 0.082b | ||
| Smoking | 147 (47.0%) | 216 (42.5%) | |
| Non-smoking | 164 (52.4%) | 292 (57.5%) | |
| BMI, kg/m2 | 24.67 (±4.62) | 24.35 (±4.58) | 0.587b |
| FEV1/FVC | 0.56 (±0.05) | 0.79 (±0.04) |
Notes: aP-values were calculated by two-side chi-squared tests; bP-values were calculated from sample’s t-tests; P<0.05 indicates statistical significance.
Abbreviations: BMI, body mass index; FEV1, forced expiratory volume in 1 second; FVC, forced volume capacity.
HWE and SNPs alleles
The detailed information of candidate SNPs in CYP2B6 are demonstrated in Table 3. All SNPs were in accordance with HWE in the controls (P>0.05) except for rs4803418 excluded from subsequent analyses. The call rate for all SNPs was >98.40% among the COPD cases and the controls, which was considered high enough to perform association analyses. The minor allele of each SNP was assumed as a risk factor for COPD compared to the wild-type allele. Using χ2 tests and ORs (95% CI), we identified two significant SNP variants associated with the risk of COPD. Among them, the frequency of “G” of rs4803420 (0.158 vs 0.234) and frequency of “A” of rs1038376 (0.220 vs 0.124) were significant between the healthy control group and the COPD patients. Compared to allele “T”, the minor allele “G” of rs4803420 was found related to a decrease the risk of COPD (OR=0.61, 95% CI=0.47–0.80, P<0.000). Similarly, allele “A” of rs1038376 was significantly correlated with an increased risk of COPD compared to allele “T” (OR=2.00, 95% CI=1.53–2.61, P<0.000).
Table 3.
Basic information about candidate SNPs and association with the risk of COPD in allele model
| SNP | Chr | Position | Gene (s) | Role | Alleles | Frequency (MAF) | Call rate (%) | p-HWE | OR (95% CI) | P-value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||||
| rs2099361 | 19 | 40992443 | CYP2B6 | Intronic | G/T | 0.283 | 0.296 | 99.40% | 0.136 | 0.94 (0.75–1.17) | 0.567 |
| rs4803418 | 19 | 41005898 | CYP2B6 | Intronic | C/G | 0.425 | 0.477 | 100.00% | 0.008 | 0.81 (0.66–0.99) | 0.039 |
| rs4803420 | 19 | 41017655 | CYP2B6 | UTR3 | G/T | 0.158 | 0.234 | 100.00% | 0.174 | 0.61 (0.47–0.80) | 2.13E-04* |
| rs1038376 | 19 | 41018104 | CYP2B6 | UTR3 | A/T | 0.220 | 0.124 | 100.00% | 0.152 | 2.00 (1.53–2.61) | 2.38E-07* |
| rs12979270 | 19 | 41018226 | CYP2B6 | UTR3 | A/C | 0.313 | 0.280 | 98.40% | 0.095 | 1.17 (0.94–1.46) | 0.157 |
Notes: P-values were calculated with Pearson’s χ2 tests; *P<0.05 and P values shown in bold are statistically significant.
Abbreviations: SNP, single-nucleotide polymorphism; Alleles A/B, minor/major alleles; MAF, minor allele frequency; OR, odds ratio; 95% CI, 95% confidence interval; HWE, Hardy–Weinberg equilibrium.
Associations between genotype frequencies and COPD risk
We also assessed the association between these SNPs and COPD risk using four genetic models (codominant, dominant, recessive, and log-additive) by logistic regression analysis. The results of the various genetic models are displayed in Table 4. Compared with homozygous “G/G” genotype, genotypes “G/T” and “G/T-T/T” of rs4803420 were predisposition to the development of COPD under the codominant (G/T vs G/G, OR=0.43, 95% CI: 0.29–0.64, P<0.000) and dominant (G/T-T/T vs G/G, OR=0.47, 95% CI: 0.32–0.69, P<0.000) models. Genotypes “A/T” and “A/T-T/T” of rs1038376 were associated with COPD risk according to the analysis of codominant model (A/T vs A/A, OR=2.40, 95% CI: 1.62–3.57, P<0.000) and dominant model (A/T-T/T vs A/A, OR=2.48, 95% CI: 1.68–3.65, P<0.000), respectively. Moreover, rs4803420 was correlated with a decreased risk of COPD based the log-additive model (OR=0.59, 95% CI: 0.43–0.82, P=0.002) and rs1038376 was associated with an increased risk of COPD under the log-additive model (OR=2.34, 95% CI: 1.63–3.36, P<0.000). No significant associations between COPD risk and rs2099361 and rs12979270 were found.
Table 4.
Logistic regression analysis of the association between prominent SNPs and COPD risk
| SNP | Model | Genotype | Control | Case | Without adjustment | With adjustment | ||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |||||
| rs2099361 | Codominant | A/A | 242 (48.1%) | 164 (52.7%) | 1.00 | 1.00 | ||
| A/C | 224 (44.5%) | 118 (37.9%) | 0.78 (0.58–1.05) | 0.098 | 0.96 (0.66–1.39) | 0.810 | ||
| C/C | 37 (7.4%) | 29 (9.3%) | 1.16 (0.68–1.96) | 0.587 | 1.12 (0.58–2.13) | 0.743 | ||
| Dominant | A/A | 242 (48.1%) | 164 (52.7%) | 1.00 | 1.00 | |||
| A/C-C/C | 261 (51.9%) | 147 (47.2%) | 0.83 (0.63–1.10) | 0.200 | 0.98 (0.69–1.40) | 0.917 | ||
| Recessive | A/A-A/C | 466 (92.6%) | 282 (90.6%) | 1.00 | 1.00 | |||
| C/C | 37 (7.4%) | 29 (9.3%) | 1.30 (0.78–2.15) | 0.318 | 1.14 (0.61–2.13) | 0.686 | ||
| Log-additive | – | – | – | 0.94 (0.75–1.17) | 0.564 | 1.01 (0.77–1.33) | 0.924 | |
| rs4803420 | Co-dominant | G/G | 291 (57.5%) | 226 (72.2%) | 1.00 | 1.00 | ||
| G/T | 193 (38.1%) | 75 (24.0%) | 0.50 (0.36–0.69) | 2.01E-05* | 0.43 (0.29–0.64) | 3.11E-05* | ||
| T/T | 22 (4.4%) | 12 (3.8%) | 0.70 (0.34–1.45) | 0.339 | 0.89 (0.38–2.12) | 0.799 | ||
| Dominant | G/G | 291 (57.5%) | 226 (72.2%) | 1.00 | 1.00 | |||
| G/T-T/T | 215 (42.5%) | 87 (27.8%) | 0.52 (0.38–0.71) | 2.58E-05* | 0.47 (0.32–0.69) | 9.25E-05* | ||
| Recessive | G/G -G/T | 484 (95.6%) | 301 (96.2%) | 1.00 | 1.00 | |||
| T/T | 22 (4.4%) | 12 (3.8%) | 0.88 (0.43–1.80) | 0.720 | 1.18 (0.50–2.76) | 0.706 | ||
| Log-additive | – | – | – | 0.61 (0.47–0.79) | 2.36E-04* | 0.59 (0.43–0.82) | 0.002* | |
| rs1038376 | Co-dominant | A/A | 386 (76.0%) | 186 (59.4%) | 1.00 | 1.00 | ||
| A/T | 118 (23.2%) | 116 (37.1%) | 2.04 (1.50–2.78) | 6.68E-06* | 2.40 (1.62–3.57) | 1.37E-05* | ||
| T/T | 4 (0.8%) | 11 (3.5%) | 5.71 (1.79–18.16) | 0.003* | 4.35 (0.97–19.63) | 0.056 | ||
| Dominant | A/A | 386 (76.0%) | 186 (59.4%) | 1.00 | 1.00 | |||
| A/T-T/T | 122 (24.0%) | 127 (40.6%) | 2.16 (1.59–2.93) | 6.76E-07* | 2.48 (1.68–3.65) | 4.35E-06* | ||
| Recessive | A/A-A/T | 504 (99.2%) | 302 (96.5%) | 1.00 | 1.00 | |||
| T/T | 4 (0.8%) | 11 (3.5%) | 4.59 (1.45–14.54) | 0.010* | 3.38 (0.76–15.05) | 0.111 | ||
| Log-additive | – | – | – | 2.11 (1.59–2.79) | 1.78E-07* | 2.34 (1.63–3.36) | 4.41E-06* | |
| rs12979270 | Co-dominant | A/A | 249 (50.2%) | 142 (46.1%) | 1.00 | 1.00 | ||
| A/C | 216 (43.6%) | 139 (45.1%) | 1.13 (0.84–1.52) | 0.425 | 1.09 (0.75–1.58) | 0.641 | ||
| C/C | 31 (6.3%) | 27 (8.8%) | 1.53 (0.88–2.66) | 0.135 | 1.48 (0.69–3.14) | 0.313 | ||
| Dominant | A/A | 249 (50.2%) | 142 (46.1%) | 1.00 | 1.00 | |||
| A/C-C/C | 247 (49.8%) | 166 (53.9%) | 1.18 (0.89–1.57) | 0.259 | 1.13 (0.79–1.62) | 0.496 | ||
| Recessive | A/A-A/C | 465 (93.7%) | 281 (91.2%) | 1.00 | 1.00 | |||
| C/C | 31 (6.3%) | 27 (8.8%) | 1.44 (0.84–2.47) | 0.182 | 1.41 (0.68–2.94) | 0.355 | ||
| Log-additive | – | – | – | 1.19 (0.94–1.49) | 0.143 | 1.15 (0.86–1.54) | 0.353 | |
Notes: P-values were calculated by unconditional logistic regression adjusted for age and gender; *P<0.05 and P values shown in bold are statistically significant.
Abbreviations: SNP, single-nucleotide polymorphism; Alleles A/B, minor/major alleles; MAF, minor allele frequency; OR, odds ratio; 95% CI, 95% confidence interval; HWE, Hardy–Weinberg equilibrium.
Stratification analysis by gender
In accordance with the stratified analysis by gender, we found that CYP2B6 variants rs4803420 and rs1038376 were significantly correlated with COPD risk in both males and females, and rs12979270 was only significantly correlated with COPD risk in females, as shown in Table 5. Rs4803420 allele “T” was associated with a reduced COPD risk compared with allele “G” in allele model (males: OR=0.66, 95% CI=0.49–0.90, P=0.007; females: OR=0.48, 95% CI=0.28–0.82, P=0.008). Compared with homozygous “T/T” genotype, genotypes “G/T” and “G/T-T/T” of rs4803420 were observed to be associated with a reduced the susceptibility of COPD among males and females based on codominant model (males: OR=0.45, 95% CI=0.27–0.73, P=0.001; females: OR=0.36, 95% CI=0.18–0.74, P=0.006) and dominant model (males: OR=0.49, 95% CI=0.30–0.78, P=0.003; females: OR=0.40, 95% CI=0.21–0.79, P=0.008). Rs4803420 was also related to a reduced the risk of COPD under the log-additive model (males: OR=0.61, 95% CI=0.41–0.91, P=0.017; females: OR=0.52, 95% CI=0.29–0.93, P=0.026). Similarly, rs1038376 allele “A” was associated with an increased COPD risk compared with allele “T” in allele model (males: OR=1.70, 95% CI=1.24–2.34, P=0.001; females: OR=3.00, 95% CI=1.82–4.93, P<0.000). Compared with homozygous “A/A” genotype, genotypes “A/T” and “A/T-T/T” rs1038376 were found to be associated with an increased the susceptibility of COPD among males and females based on codominant model (males: OR=1.79, 95% CI=1.09–2.94, P=0.022; females: OR=4.32, 95% CI=2.25–8.32, P<0.000) and dominant model (males: OR=1.91, 95% CI=1.18–3.10, P=0.009; females: OR=4.24, 95% CI=2.22–8.11, P<0.000). Rs1038376 was also related to an enhanced the risk of COPD based on the log-additive model (males: OR=1.89, 95% CI=1.21–2.94, p=0.005; females: OR=3.82, 95% CI=2.04–7.15, P<0.000). Genetic model analysis also revealed that the allele “C” of rs12979270 was linked to an increased risk of COPD in females compared with allele “A” under the allele model (OR=1.55, 95% CI: 1.03–2.36, P=0.039).
Table 5.
Relationship of prominent SNPs with the COPD risk stratified by gender
| SNP | Model | Genotype | Males | Females | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Case | OR (95% CI) | P-value | Control | Case | OR (95% CI) | P-value | |||
| rs4803420 | Co-dominant | G/G | 194 (57.7%) | 168 (70.6%) | 1.00 | 97 (57.1%) | 58 (77.3%) | 1.00 | ||
| G/T | 129 (38.4%) | 61 (25.6%) | 0.45 (0.27–0.73) | 0.001* | 64 (37.7%) | 14 (18.7%) | 0.36 (0.18–0.74) | 0.006* | ||
| T/T | 13 (3.9%) | 9 (3.8%) | 0.95 (0.32–2.85) | 0.928 | 9 (5.3%) | 3 (4.0%) | 0.74 (0.18–3.06) | 0.681 | ||
| Dominant | G/G | 194 (57.7%) | 168 (70.6%) | 1.00 | 97 (57.1%) | 58 (77.3%) | 1.00 | |||
| G/T-T/T | 142 (42.3%) | 70 (29.4%) | 0.49 (0.3–0.78) | 0.003* | 73 (42.9%) | 17 (22.7%) | 0.40 (0.21–0.79) | 0.008* | ||
| Recessive | G/G -G/T | 323 (96.1%) | 229 (96.2%) | 1.00 | 161 (94.7%) | 72 (96.0%) | 1.00 | |||
| T/T | 13 (3.9%) | 9 (3.8%) | 1.24 (0.42–3.68) | 0.693 | 9 (5.3%) | 3 (4.0%) | 1.01 (0.25–4.10) | 0.994 | ||
| Log-additive | – | – | – | 0.61 (0.41–0.91) | 0.017* | – | – | 0.52 (0.29–0.93) | 0.026* | |
| Allele | T | 517 (76.9%) | 397 (83.4%) | 1.00 | 258 (75.9%) | 130 (96.7%) | 1.00 | |||
| G | 155 (23.1%) | 79 (16.6%) | 0.66 (0.49–0.90) | 0.007* | 82 (24.1%) | 20 (13.3%) | 0.48 (0.28–0.82) | 0.008* | ||
| rs1038376 | Co-dominant | A/A | 251 (74.5%) | 150 (63.0%) | 1.00 | 135 (79.0%) | 36 (48.0%) | 1.00 | ||
| A/T | 83 (24.6%) | 78 (32.8%) | 1.79 (1.09–2.94) | 0.022* | 35 (20.5%) | 38 (50.7%) | 4.32 (2.25–8.32) | 1.19E-05* | ||
| T/T | 3 (0.9%) | 10 (4.2%) | 5.30 (0.82–34.32) | 0.080 | 1 (0.6%) | 1 (1.3%) | 2.30 (0.11–47.29) | 0.588 | ||
| Dominant | A/A | 251 (74.5%) | 150 (63.0%) | 1.00 | 135 (79.0%) | 36 (48.0%) | 1.00 | |||
| A/T-T/T | 86 (25.5%) | 88 (37.0%) | 1.91 (1.18–3.10) | 0.009* | 36 (21.0%) | 39 (52.0%) | 4.24 (2.22–8.11) | 1.25E-05* | ||
| Recessive | A/A-A/T | 334 (99.1%) | 228 (95.8%) | 1.00 | 170 (99.4%) | 74 (98.7%) | 1.00 | |||
| T/T | 3 (0.9%) | 10 (4.2%) | 4.50 (0.70–28.81) | 0.113 | 1 (0.6%) | 1 (1.3%) | 1.41 (0.07–28.72) | 0.823 | ||
| Log-additive | – | – | – | 1.89 (1.21–2.94) | 0.005* | – | – | 3.82 (2.04–7.15) | 2.89E-05* | |
| Allele | A | 585 (86.8%) | 378 (79.4%) | 1.00 | 305 (89.2%) | 110 (73.3%) | 1.00 | |||
| T | 89 (13.2%) | 98 (20.6%) | 1.70 (1.24–2.34) | 0.001* | 37 (10.8%) | 40 (26.7%) | 3.00 (1.82–4.93) | 8.43E-06* | ||
| rs12979270 | Co-dominant | A/A | 163 (49.1%) | 113 (48.1%) | 1.00 | 86 (52.4%) | 29 (39.7%) | 1.00 | ||
| A/C | 148 (44.6%) | 104 (44.2%) | 0.99 (0.62–1.57) | 0.952 | 68 (41.5%) | 35 (480%) | 1.41 (0.75–2.67) | 0.284 | ||
| C/C | 21 (6.3%) | 18 (7.7%) | 0.95 (0.34–2.61) | 0.918 | 10 (6.1%) | 9 (12.3%) | 2.77 (0.91–8.46) | 0.074 | ||
| Dominant | A/A | 163 (49.1%) | 113 (48.1%) | 1.00 | 86 (52.4%) | 29 (39.7%) | 1.00 | |||
| A/C-C/C | 169 (50.9%) | 122 (51.9%) | 0.98 (0.63–1.54) | 0.936 | 78 (47.6%) | 44 (60.3%) | 1.57 (0.85–2.88) | 0.147 | ||
| Recessive | A/A-A/C | 311 (93.7%) | 217 (92.3%) | 1.00 | 154 (93.9%) | 64 (87.7%) | 1.00 | |||
| C/C | 21 (6.3%) | 18 (7.7%) | 0.95 (0.35–2.57) | 0.927 | 10 (6.1%) | 9 (12.3%) | 2.33 (0.8–6.79) | 0.121 | ||
| Log-additive | – | – | – | 0.98 (0.67–1.43) | 0.918 | – | – | 1.56 (0.97–2.51) | 0.069 | |
| Allele | A | 474 (71.4%) | 330 (70.2%) | 1.00 | 240 (73.2%) | 93 (63.7%) | 1.00 | |||
| C | 190 (28.6%) | 140 (29.8%) | 1.06 (0.82–1.37) | 0.691 | 88 (26.8%) | 53 (36.3%) | 1.55 (1.03–2.36) | 0.039* | ||
Notes: P-values were calculated by unconditional logistic regression analysis with adjustments for age; *P<0.05 and P values shown in bold are statistically significant.
Abbreviations: SNP, single-nucleotide polymorphism; Alleles A/B, minor/major alleles; MAF, minor allele frequency; OR, odds ratio; 95% CI, 95% confidence interval.
Stratification analysis by smoking status
When stratified analysis by smoking status, the correlation between candidate SNP and COPD risk was listed in Table 6. We only found that allele “C” of rs12979270 was significantly associated with an increased risk of COPD under the allele model compared with allele “A” in the smoking subgroup (OR=1.49, 95% CI: 1.08–2.07, P=0.016).
Table 6.
Relationship between CYP2B6 gene polymorphisms and the risk of COPD stratified by smoking status
| SNP | Model | Genotype | Smoking | Non-smoking | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Case | OR (95% CI) | P-value | Control | Case | OR (95% CI) | P-value | |||
| rs12979270 | Co-dominant | A/A | 114 (53.8%) | 61 (42.1%) | 1.00 | 135 (47.5%) | 81 (50.3%) | 1.00 | ||
| A/C | 88 (41.5%) | 70 (48.3%) | 1.51 (0.79–2.88) | 0.213 | 128 (45.1%) | 67 (41.6%) | 0.81 (0.5–1.31) | 0.397 | ||
| C/C | 10 (4.7%) | 14 (9.7%) | 3.36 (0.84–13.49) | 0.088 | 21 (7.4%) | 13 (8.1%) | 0.82 (0.31–2.16) | 0.687 | ||
| Dominant | A/A | 114 (53.8%) | 61 (42.1%) | 1.00 | 135 (47.5%) | 81 (50.3%) | 1.00 | |||
| A/C-C/C | 98 (46.2%) | 84 (57.9%) | 1.64 (0.88–3.08) | 0.121 | 149 (52.5%) | 80 (49.7%) | 0.81 (0.51–1.29) | 0.383 | ||
| Recessive | A/A-A/C | 202 (95.3%) | 131 (90.3%) | 1.00 | 263 (92.6%) | 148 (91.9%) | 1.00 | |||
| C/C | 10 (4.7%) | 14 (9.7%) | 2.73 (0.71–10.45) | 0.144 | 21 (7.4%) | 13 (8.1%) | 0.91 (0.35–2.32) | 0.837 | ||
| Log-additive | – | – | – | 1.65 (0.97–2.79) | 0.063 | – | – | 0.86 (0.59–1.25) | 0.425 | |
| Allele | A | 316 (74.5%) | 192 (66.2%) | 1.00 | 398 (70.1%) | 229 (71.1%) | 1.00 | |||
| C | 108 (25.5%) | 98 (33.8%) | 1.49 (1.08–2.07) | 0.016* | 170 (29.9%) | 93 (28.9%) | 0.95 (0.7–1.28) | 0.742 | ||
Notes: P-values were calculated by unconditional logistic regression analysis with adjustments for gender and age; *P<0.05 and P values shown in bold are statistically significant.
Abbreviations: SNP, single-nucleotide polymorphism; Alleles A/B, minor/major alleles; MAF, minor allele frequency; OR, odds ratio; 95% CI, 95% confidence interval.
Haplotype analyses
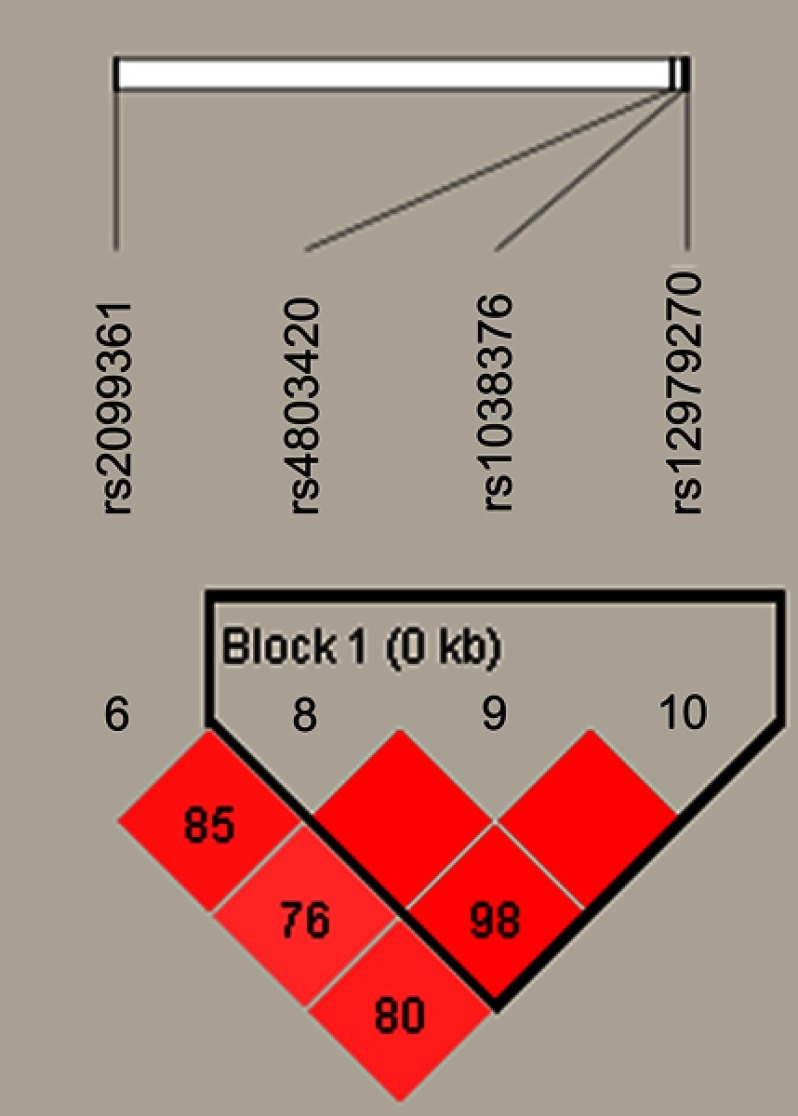
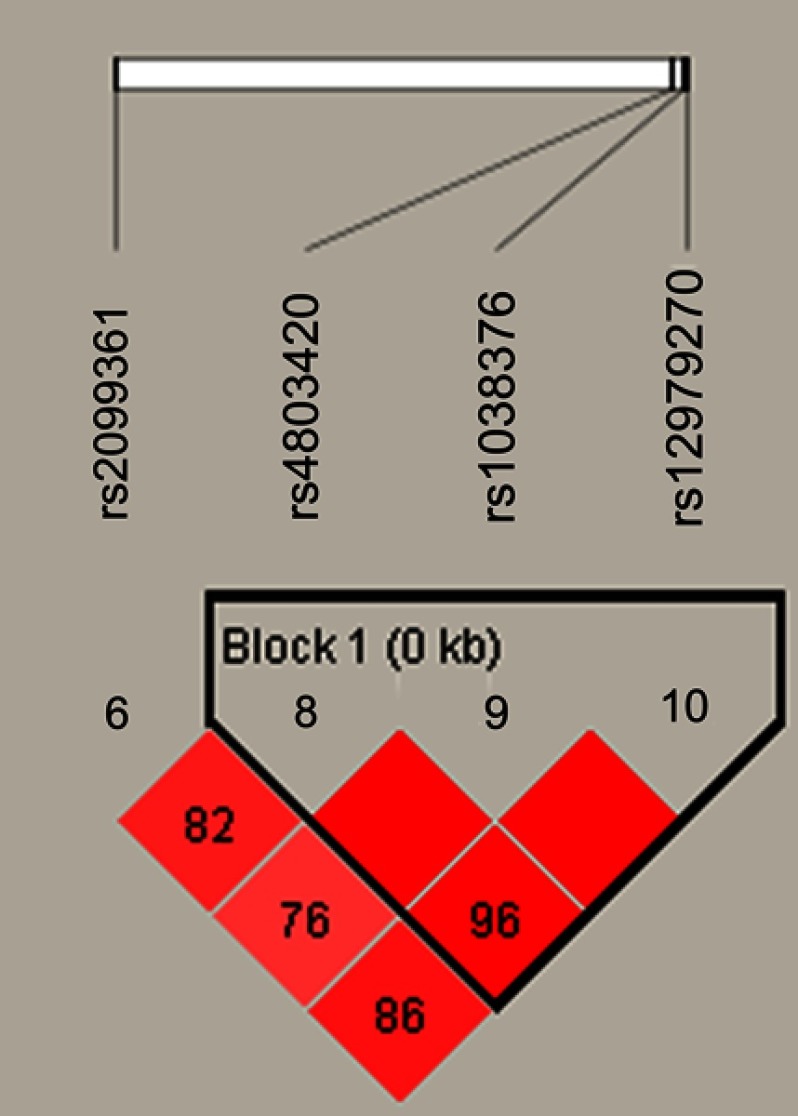
Figure 1 lists the linkage disequilibrium (LD) block of selected SNPs in the CYP2B6 gene. Figures 2 and 3 exhibit the linkage disequilibrium (LD) blocks of selected SNPs in CYP2B6 gene among males and females population, respectively. The haplotype frequency constituted by SNPs in CYP2B6 gene and its association with COPD risk in overall and gender stratification are shown in Table 7. The haplotype “GTA” constructed by rs4803420|rs1038376|rs12979270 can reduce the risk of COPD (OR=0.45; 95% CI=0.31–0.65; P<0.000), while haplotype “TAA” was significantly associated with an increased the risk of COPD (OR=1.69; 95% CI=1.22–2.34; P=0.002). The haplotype “GTA” has a protective effect on the risk of COPD among males (OR=0.56; 95% CI=0.35–0.87; P=0.011) and females (OR=0.27; 95% CI=0.15–0.52; P<0.000). Conversely, haplotype “TAA” was related to an increased the risk of COPD in males (OR=1.60; 95% CI=1.06–2.39; P=0.024) and females (OR=1.99; 95% CI=1.12–3.54; P=0.020). The haplotype “GAA” was only associated with an increased risk of COPD in females (OR=2.15; 95% CI=1.28–3.62; P=0.004).
Figure 1.
The haplotype block map constructed by candidate SNPs in CYP2B6.
Notes: Block 1 includes rs4803420, rs1038376, and rs12979270; the linkage disequilibrium between two SNPs is indicated by standardized r2 (red boxes).
Figure 2.
The haplotype block diagram constructed by candidate SNPs in CYP2B6 in males.
Notes: Block 1 includes rs4803420, rs1038376, and rs12979270; the linkage disequilibrium between two SNPs is indicated by standardized r2 (red boxes).
Figure 3.
The haplotype block diagram constructed by candidate SNPs in CYP2B6 in females.
Notes: Block 1 includes rs4803420, rs1038376, and rs12979270; the linkage disequilibrium between two SNPs is indicated by standardized r2 (red boxes).
Table 7.
Haplotype frequencies of CYP2B6 gene polymorphisms and the association with COPD risk
| SNP | Haplotype | Freq (Case) | Freq (Control) | Without adjusted | With adjusted | |||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |||||
| Total | rs4803420|rs1038376|rs12979270 | GAC | 0.313 | 0.278 | 1.20 (0.96–1.51) | 0.114 | 1.17 (0.87–1.58) | 0.290 |
| GTA | 0.779 | 0.873 | 0.49 (0.37–0.64) | 4.93E-07* | 0.45 (0.31–0.65) | 1.69E-05* | ||
| TAA | 0.839 | 0.764 | 1.63 (1.25–2.12) | 2.97E-04* | 1.69 (1.22–2.34) | 0.002* | ||
| GAA | 0.695 | 0.642 | 1.28 (1.03–1.60) | 0.026* | 1.24 (0.94–1.63) | 0.133 | ||
| Males | rs4803420|rs1038376|rs12979270 | GAC | 0.298 | 0.284 | 1.08 (0.82–1.41) | 0.597 | 1.00 (0.69–1.47) | 0.992 |
| GTA | 0.794 | 0.866 | 0.59 (0.42–0.81) | 0.001* | 0.56 (0.35–0.87) | 0.011* | ||
| TAA | 0.832 | 0.769 | 1.50 (1.11–2.04) | 0.009* | 1.60 (1.06–2.39) | 0.024* | ||
| GAA | 0.672 | 0.651 | 1.10 (0.86–1.42) | 0.454 | 1.00 (0.72–1.39) | 0.997 | ||
| Females | rs4803420|rs1038376|rs12979270 | GAC | 0.363 | 0.265 | 1.62 (1.05–2.49) | 0.029* | 1.58 (0.98–2.54) | 0.060 |
| GTA | 0.733 | 0.889 | 0.28 (0.16–0.49) | 1.01E-05* | 0.27 (0.15–0.52) | 5.69E-05* | ||
| TAA | 0.863 | 0.753 | 2.05 (1.20–3.51) | 0.009* | 1.99 (1.12–3.54) | 0.020* | ||
| GAA | 0.767 | 0.624 | 2.18 (1.35–3.52) | 0.001* | 2.15 (1.28–3.62) | 0.004* | ||
Notes: Block comprised the three closely linked SNPs rs4803420, rs1038376, and rs12979270; P-values were calculated with Pearson’s χ2 tests; *P<0.05 and P values shown in bold are statistically significant.
Abbreviations: OR, odds ratio; 95% CI, 95% confidence interval.
Discussion
In this hospital-based case–control study, we genotyped five polymorphisms of CYP2B6 and evaluated their correlations with the risk of COPD in the Hainan population of China. A remarkable result was showed that rs4803420 was associated with a reduced risk of COPD, while rs1038376 and rs12979270 had adverse effect on COPD risk. Further gender stratification analysis revealed that rs4803420 and rs1038376 were significantly correlated with COPD risk in males and females, while rs12979270 was only related to COPD risk in females. Our findings indicate that polymorphic sites in the CYP2B6 may play a crucial role in the development of COPD.
The CYP2B6 gene is located on chromosome 19q13.2 and comprises 9 exons.27 The CYP2B6 enzyme catalyzes many reactions involved in drug metabolism and synthesis of cholesterol, steroids, and other lipids. In the previous studies, CYP2B6 was mainly expressed in the liver and lung.17,18 We also used HaploReg v4.1 database to predict the function of selected SNPs in CYP2B6 gene, revealing that rs4803420 and rs1038376 may affect the expression of CYP2B6 (Table S1). Recently, diseases associated with CYP2B6 included poor metabolism, acute myeloid leukemia, acute lymphoblastic leukemia, and acute frontal sinusitis.15,28
Table S1.
SNP functional annotation in HaploReg v4.1 database
| SNP | Chr | Role | Gene (s) | HaploReg |
|---|---|---|---|---|
| rs2099361 | 19 | Intronic | CYP2B6 | Promoter histone marks, Enhancer histone marks, Motifs changed |
| rs4803420 | 19 | UTR3 | CYP2B6 | Motifs changed |
| rs1038376 | 19 | UTR3 | CYP2B6 | DNAse, Motifs changed, GRASP QTL hits |
| rs12979270 | 19 | UTR3 | CYP2B6 | Enhancer histone marks, DNAse, Motifs changed |
Abbreviations: SNP, single-nucleotide polymorphism; Ref, reference; Alt, alternation; eDTL, expression quantitative trait loci.
CYP2B6 gene is highly polymorphic, and a total of 63 alleles have been identified so far. Previous studies have shown that some CYP2B6 gene variants had certain effect on its expression and activity, and they were also related to the biotransformation of different drugs. For example, rs3745274 in CYP2B6 gene was related to the decrease of the gene expression and the CYP2B6 activity in the liver.29,30 CYP2B6 516G > T SNP changed the amino acid residues of glutamine to histidine,31 which may lead to a decreased CYP2B6 enzyme activity in the liver.30 CYP2B6*6 was the most common variant associated with functional changes in CYP2B6, which led to adverse effect on the function of the mRNA and protein after causing aberrant splicing.30 The GT genotype frequency of CYP2B6 G15631T polymorphism loci was higher in acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL)
In our study, we firstly found that rs4803420 had a protective effect on the development of COPD. And rs1038376 and rs12979270 may be a risk factor for the incidence of COPD. And rs12979270 was only correlated with an increased COPD risk in females after gender stratification analysis. Smoking status stratification analysis showed that rs12979270 was associated with an increased COPD risk in smokers, while this SNP was not related to COPD risk in nonsmokers. Given that CYP2B6 gene variants had certain effect on its expression and activity, we speculated that CYP2B6 SNPs might affect the occurrence of COPD by changing the expression or activity of CYP2B6 gene. Larger samples are needed to confirm the overall results by functional experiment, including bioinformatics tools, to further elaborate the function of CYP2B6 in the development COPD, contributing to the early diagnosis and targeted treatment of COPD.
Conclusion
In summary, our study is the firstly provided substantial basic evidence that gene polymorphisms in CYP2B6 were related to the susceptibility of COPD in the Hainan population of China. These results may provide new view for the assessment, prevention, and prognosis of COPD risk.
Supplementary material
Acknowledgment
This work was supported by the National Natural Science Foundation of China (No. 81660013 and No. 81860015) and Key Research and Development Plan of Hainan province (No. ZDYF2018116). We are grateful to the patients and control individuals for providing blood samples. We also appreciate all editors and reviewers for their patience and suggestions for this work.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Singh D, Agusti A, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: The GOLD science committee report 2019. Eur Respir J. 2019;53:1900164. doi: 10.1183/13993003.00164-2019 [DOI] [PubMed] [Google Scholar]
- 2.Ding Y, Niu H, Yang H, et al. EGLN2 and RNF150 genetic variants are associated with chronic obstructive pulmonary disease risk in the Chinese population. Int J Chron Obstruct Pulmon Dis. 2015;10:145–151. doi: 10.2147/COPD.S73031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monick MM, Beach SR, Plume J, et al. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet. 2012;159b(2):141–151. doi: 10.1002/ajmg.b.32021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halbert RJ, Natoli JL, Gano A, Badamgarav E, Buist AS, Mannino DM. Global burden of COPD: systematic review and meta-analysis. Eur Respir J. 2006;28(3):523–532. doi: 10.1183/09031936.06.00124605 [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Li L, Xiao J, et al. Association of ADAM33 gene polymorphisms with COPD in a northeastern Chinese population. BMC Med Genet. 2009;10:132. doi: 10.1186/1471-2350-10-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong M, Wang J, Guo M, Zhou Q, Lu W. TRPM8 genetic variations associated with COPD risk in the Chinese Han population. Int J Chron Obstruct Pulmon Dis. 2016;11:2563–2571. doi: 10.2147/COPD.S109026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. doi: 10.1371/journal.pmed.0030442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global initiative for chronic obstructive lung disease (GOLD) workshop summary. Am J Respir Crit Care Med. 2001;163(5):1256–1276. doi: 10.1164/ajrccm.163.5.2101039 [DOI] [PubMed] [Google Scholar]
- 9.Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5(3):e1000421. doi: 10.1371/journal.pgen.1000421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bengt ZL, Xinjun L, Jan S, Kristina S. Familial transmission of chronic obstructive pulmonary disease in adoptees: a Swedish nationwide family study. BMJ Open. 2015;5(4):e007310. doi: 10.1136/bmjopen-2014-007310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman EK, Vestbo J, Agusti A, et al. Opportunities and challenges in the genetics of COPD 2010: an International COPD genetics conference report. Copd. 2011;8(2):121–135. doi: 10.3109/15412555.2011.558864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson DR, Koymans L, Kamataki T, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6(1):1–42. [DOI] [PubMed] [Google Scholar]
- 13.Lamba V, Lamba J, Yasuda K, et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther. 2003;307(3):906–922. doi: 10.1124/jpet.103.054866 [DOI] [PubMed] [Google Scholar]
- 14.Court MH, Duan SX, Hesse LM, Venkatakrishnan K, Greenblatt DJ. Cytochrome P-450 2B6 is responsible for interindividual variability of propofol hydroxylation by human liver microsomes. Anesthesiology. 2001;94(1):110–119. doi: 10.1097/00000542-200101000-00021 [DOI] [PubMed] [Google Scholar]
- 15.Hodgson E, Rose RL. The importance of cytochrome P450 2B6 in the human metabolism of environmental chemicals. Pharmacol Ther. 2007;113(2):420–428. doi: 10.1016/j.pharmthera.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 16.Code EL, Crespi CL, Penman BW, Gonzalez FJ, Chang TK, Waxman DJ. Human cytochrome P4502B6: interindividual hepatic expression, substrate specificity, and role in procarcinogen activation. Drug Metab Dispos. 1997;25(8):985–993. [PubMed] [Google Scholar]
- 17.Thum T, Borlak J. Gene expression in distinct regions of the heart. Lancet. 2000;355(9208):979–983. doi: 10.1016/S0140-6736(00)99016-0 [DOI] [PubMed] [Google Scholar]
- 18.Gervot L, Rochat B, Gautier JC, et al. Human CYP2B6: expression, inducibility and catalytic activities. Pharmacogenetics. 1999;9(3):295–306. [PubMed] [Google Scholar]
- 19.Demura SA, Kogan EA, Goryachkina VL. [Chronic diseases, precancer, and cancer of the lung, which are associated with pathology of the club cells of respiratory and terminal bronchioles]. Arkh Patol. 2018;80(5):63–68. doi: 10.17116/patol20188005163 [DOI] [PubMed] [Google Scholar]
- 20.Mace K, Bowman ED, Vautravers P, Shields PG, Harris CC, Pfeifer AM. Characterisation of xenobiotic-metabolising enzyme expression in human bronchial mucosa and peripheral lung tissues. Eur J Cancer. 1998;34(6):914–920. doi: 10.1016/s0959-8049(98)00034-3 [DOI] [PubMed] [Google Scholar]
- 21.Hukkanen J, Pelkonen O, Hakkola J, Raunio H. Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit Rev Toxicol. 2002;32(5):391–411. doi: 10.1080/20024091064273 [DOI] [PubMed] [Google Scholar]
- 22.Ding Y, Yang D, Xun X, et al. Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Hainan population: a case-control study. Int J Chron Obstruct Pulmon Dis. 2015;10:7–13. doi: 10.2147/COPD.S73042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogelmeier CF, Criner GJ, Martinez FJ, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. 2017;195(5):557–582. doi: 10.1164/rccm.201701-0218PP [DOI] [PubMed] [Google Scholar]
- 24.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009; 60: 2.12.1–2.12.18. doi: 10.1002/0471142905.hg0212s60 [DOI] [PubMed] [Google Scholar]
- 25.Ding Y, Yang D, Zhou L, et al. Variants in multiple genes polymorphism association analysis of COPD in the Chinese Li population. Int J Chron Obstruct Pulmon Dis. 2015;10:1455–1463. doi: 10.2147/COPD.S86721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6(2):121–133. doi: 10.1038/nprot.2010.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics. 2007;8(7):743–759. doi: 10.2217/14622416.8.7.743 [DOI] [PubMed] [Google Scholar]
- 28.Berkoz M, Yalin S. Association of CYP2B6 G15631T polymorphism with acute leukemia susceptibility. Leuk Res. 2009;33(7):919–923. doi: 10.1016/j.leukres.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 29.Hoffman SM, Nelson DR, Keeney DS. Organization, structure and evolution of the CYP2 gene cluster on human chromosome 19. Pharmacogenetics. 2001;11(8):687–698. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann MH, Blievernicht JK, Klein K, et al. Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325(1):284–292. doi: 10.1124/jpet.107.133306 [DOI] [PubMed] [Google Scholar]
- 31.Tong K, He ML, Lin CK, et al. The implications of a high allelic frequency of CYP2B6 G516T in ethnic Chinese persons. Clin Infect Dis. 2006; 43(4):541–542; author reply 542–544. doi: 10.1086/505984 [DOI] [PubMed] [Google Scholar]