Abstract
Mixotrophic species use both organic and mineral carbon sources. Some mixotrophic plants combine photosynthesis and a nutrition called mycoheterotrophy, where carbon is obtained from fungi forming mycorrhizal symbiosis with their roots. These species can lose photosynthetic abilities and evolve full mycoheterotrophy. Besides morphological changes, the latter transition is associated with a deep alteration of the plastid genome. Photosynthesis-related genes are lost first, followed by housekeeping genes, eventually resulting in a highly reduced genome. Whether relaxation of selective constraints already occurs for the plastid genome of mixotrophic species, which remain photosynthetic, is unclear. This is partly due to the difficulty of comparing plastid genomes of autotrophic, mixotrophic, and mycoheterotrophic species in a narrow phylogenetic framework. We address this question in the orchid tribe Neottieae, where this large assortment of nutrition types occurs. We sequenced 13 new plastid genomes, including 9 mixotrophic species and covering all 6 Neottieae genera. We investigated selective pressure on plastid genes in each nutrition type and conducted a phylogenetic inference of the group. Surprisingly, photosynthesis-related genes did not experience selection relaxation in mixotrophic species compared with autotrophic relatives. Conversely, we observed evidence for selection intensification for some plastid genes. Photosynthesis is thus still under purifying selection, maybe because of its role in fruit formation and thus reproductive success. Phylogenetic analysis resolved most relationships, but short branches at the base of the tree suggest an evolutionary radiation at the beginning of Neottieae history, which, we hypothesize, may be linked to mixotrophy emergence.
Keywords: Mycorrhiza, mycoheterotrophy, mixotrophy, phylogeny, Neottieae, plastome
Introduction
Mixotrophic organisms grow by using both inorganic and organic carbon resources, either simultaneously or successively, and therefore represent intermediates between autotrophy and heterotrophy (Selosse et al. 2017; Tĕšitel et al. 2018). Land plants widely associate with fungi through mycorrhizal symbiosis and usually exchange their photosynthates for mineral nutrients (Smith and Read 2008). Repeatedly, plants evolved the ability to gain organic compounds also from their mycorrhizal fungi, a mixotrophic nutrition also called partial mycoheterotrophy (Selosse and Roy 2009; Hynson et al. 2013; Tĕšitel et al. 2018). We hereafter use the word mixotrophy for these plants that combine photosynthetic and mycorrhizal carbon sources. In many mixotrophic lineages from different families, species eventually lost photosynthesis and became fully mycoheterotrophic (Leake 1994; Merckx, Freudenstein, et al. 2013). Traits of mycoheterotrophy such as chlorophyll loss, reduction in leaf surface area, development of short clumpy roots, or production of reserve-less “dust” seeds have been first studied (Merckx, Mennes, et al. 2013). An increasing amount of molecular data now clarifies the genetic traits linked with the evolution to mycoheterotrophy.
The plastid genomes (or plastomes) of 24 fully mycoheterotrophic species have been sequenced to date (Graham et al. 2017; Petersen et al. 2018). Their comparative analysis has resulted in a model of sequential plastid genome degradation associated with mycoheterotrophy, which parallels what has been observed for heterotrophic parasitic plants (Funk et al. 2007; Barrett and Davis 2012; Barrett et al. 2014; Wicke et al. 2016; Graham et al. 2017). Genes encoding subunits of the nicotinamide adenine dinucleotide H dehydrogenase (NDH)-like complex, which regulates excessive electron flow in the plastidial electron transfer chain, are the first to be lost, followed by most photosynthesis-related genes. Rubisco large subunit (rbcL), adenosine triphosphate (ATP)-synthase, and plastid-encoded RNA polymerase (PEP) genes are often retained in the early stage of photosynthesis loss. Eventually, degradation encompasses housekeeping genes involved in plastid translation and other specific functions. This model summarizes well how the plastid genome evolves in mycoheterotrophic and parasitic plant as they become more and more dependent on organic carbon uptake (Wicke et al. 2016; reviewed in Graham et al. [2017] and Wicke and Naumann [2018]).
Due to the few data available, still little is known about what occurs in the evolutionary steps prior to mycoheterotrophy, that is, in mixotrophic species that retain photosynthetic abilities. Whether these species already experience relaxed selective pressure on photosynthesis remains unclear in the general case. This is a challenging question because of the scarcity of lineages that have retained together autotrophic, mixotrophic, and mycoheterotrophic species. Known examples include the genus Burmannia L. (Burmanniaceae; Bolin et al. 2017), the tribe Pyroleae (Ericaceae; Lallemand et al. 2016), and the tribe Neottieae (Orchidaceae; Selosse and Roy 2009; Gonneau et al. 2014). In the leafless genus Corallorhiza Gagnebin (Orchidaceae), where no full autotroph occurs, comparison of plastomes from mixotrophic (green) and mycoheterotrophic (nongreen) species showed very limited gene loss in the former and relaxed selective constraints on photosynthesis and ATP-synthase genes in the latter (Barrett et al. 2014). Consistently, the selective regime does not differ between mixotrophic Corallorhiza and autotrophic outgroups (Barrett et al. 2014). The absence of autotrophic Corallorhiza as reference yet prevents firm conclusions regarding selective regime in mixotrophs. In the orchid genus Cymbidium Sw., C. macrorhizon is leafless but still holds chlorophyll in the stem and fruits and the importance of its photosynthesis for fruiting has recently been demonstrated (Suetsugu et al. 2018). The plastome of this mixotrophic species shows no gene loss except for a few NDH genes and is mostly under purifying selection (Kim et al. 2018). The two genera Corallorhiza and Cymbidium where photosynthesis already undergoes relaxed selection in mixotrophic species (Barrett et al. 2014) call for extensive analyses in other lineages.
We sequenced plastomes of mixotrophs in Neottieae, an orchid tribe mostly found in northern hemisphere forests. Neottieae include 6 genera and about 200 species (WCSP 2018) with various nutrition types: Palmorchis Barb. Rodr. (all putative autotrophs), Neottia Guett. (many mycoheterotrophs and some probable mixotrophs; Tĕšitelová et al. 2012; Yagame et al. 2016; Schiebold et al. 2018), Epipactis Zinn (numerous mixotrophs and some autotrophs; Lallemand et al. 2018), Cephalanthera Rich. (some mycoheterotrophs and many mixotrophs), Limodorum Boehm. (all mixotrophs; Girlanda et al. 2006), and Aphyllorchis Blume (all mycoheterotrophs; Roy et al. 2009). Early studies on mycoheterotrophic and mixotrophic Neottieae contributed to our understanding of the ecophysiology and evolution of plants feeding on fungal carbon (Selosse et al. 2002; Gebauer and Meyer 2003; Julou et al. 2005). The transition to mixotrophy in this group is consistently linked to a shift in type of mycorrhizal fungi (supplementary table S1, Supplementary Material online). Autotrophic orchids associate with a polyphyletic group of saprotrophic and endophytic fungi, the rhizoctonias (Dearnaley et al. 2012), whereas mixotrophic Neottieae use ectomycorrhizal fungi from different genera, which simultaneously form mycorrhizas with surrounding trees (Selosse and Martos 2014). For the purpose of this study, we had to assign a given nutrition type to each species investigated. Even if there is a continuum between autotrophy and mixotrophy (Jacquemyn et al. 2017), we decided to consider a species as mixotrophic when it dominantly associated with ectomycorrhizal fungi and showed an enrichment in 13C compared with surrounding autotrophs, a usual marker of fungal carbon gain (Hynson et al. 2013; see supplementary table S1, Supplementary Material online, for references regarding species nutrition).
Plastome sequencing of some Neottieae species by Feng et al. (2016) has yielded important results: 1) new hypotheses regarding the controversial phylogenetic relationships between Neottieae genera (Bateman et al. 2005; Roy et al. 2009; Górniak et al. 2010; Xiang et al. 2012), 2) patterns of gene loss in mycoheterotrophs matching the model described above, and 3) no relationship between the chlorophyll level (presence or absence based on visual estimation) and the plastome degradation in the leafless Neottia clade. Their sampling, however, included only four out of six Neottieae genera and one mixotrophic species (Cephalanthera longifolia (L.) Fritsch), limiting the phylogenetic resolution and the analysis of plastome evolution during the emergence of mixotrophy and mycoheterotrophy. Here, we sequenced 13 additional Neottieae plastomes, including representatives of the 2 remaining genera, as well as 9 mixotrophic species (table 1). We investigated how gene content and selective pressure are related to nutrition type and carried out a tentative plastome-based phylogenetic reconstruction of Neottieae.
Table 1.
Length, GC Content, and Genes Lost of the 13 Neottieae Plastomes Sequenced
| Species a | Accession | Length |
GC Content |
Gene Loss
b
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | LSC | SSC | IR | Total | LSC | SSC | IR | Pseudogene/Undetected | ||
| P. pabstii | MH590357 | 163,909 | 90,710 | 18,823 | 27,188 | 37.3 | 35 | 31 | 43.3 | |
| Ce. damasonium | MH590345 | 161,699 | 88,720 | 19,085 | 26,947 | 37.3 | 35.1 | 30.7 | 43.2 | |
| Ce. longibracteata | MH590346 | 161,986 | 88,888 | 19,138 | 26,980 | 37.2 | 35 | 30.6 | 43.1 | |
| Ce. rubra | MH590347 | 162,277 | 88,814 | 19,199 | 27,132 | 37.2 | 35 | 30.6 | 43.1 | |
| E. albensis | MH590348 | 159,763 | 87,237 | 18,782 | 26,872 | 37.3 | 35.1 | 30.7 | 43.2 | |
| E. atrorubens | MH590349 | 159,790 | 87,237 | 18,803 | 26,875 | 37.3 | 35.1 | 30.7 | 43.3 | |
| E. gigantea | MH590350 | 158,977 | 87,101 | 18,664 | 26,606 | 37.3 | 35.2 | 30.8 | 43.2 | |
| E. helleborine | MH590351 | 159,822 | 87,313 | 18,785 | 26,862 | 37.3 | 35.1 | 30.7 | 43.2 | |
| E. microphylla | MH590352 | 159,236 | 86,706 | 18,784 | 26,873 | 37.4 | 35.2 | 30.7 | 43.3 | ndhD, ndhE, ndhF, ndhH |
| E. palustris | MH590353 | 159,134 | 87,114 | 18,702 | 26,659 | 37.4 | 35.2 | 30.8 | 43.2 | |
| E. purpurata | MH590354 | 159,864 | 87,246 | 18,786 | 26,916 | 37.3 | 35.1 | 30.7 | 43.2 | |
| L. abortivum | MH590355 | 128,822 | 85,544 | 15,099 | 27,102 | 36.3 | 35.1 | 30.4 | 43.1 | ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK, cemA |
| N. cordata | MH590356 | 147,034 | 82,416 | 12812 | 25903 | 37.5 | 35.1 | 29.3 | 43.4 | ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
Complete genera names are Palmorchis, Cephalanthera, Epipactis, Limodorum, and Neottia.
A gene was presumed pseudogenized if a frameshift indel resulted in a reduction in protein length higher than 25% of its original length.
Plastome Structure, Gene Content, and Selective Regime
None of the plastomes of the 13 photosynthetic species sequenced displayed loss of large chromosome regions (>5 kb) or changes in synteny compared with related autotrophic Neottieae (Feng et al. 2016). Their sizes and GC contents were similar, with the exception of Neottiacordata and Limodorumabortivum which have smaller genomes as a consequence of NDH gene loss (table 1). A given gene was presumed pseudogenized if a frameshift indel resulted in a reduction in protein length higher than 25% of its original length. It was considered physically lost when it failed to pass the 77% similarity threshold (compared with closest available reference species) used for gene annotation and was thus undetected. Gene loss (pseudogenization or undetected) was restricted to the NDH complex in a few species (table 1): N. cordata (10 genes lost, ndhI with 148 amino acids vs. 182 in autotrophic Neottia species may also be nonfunctional), L. abortivum (all genes) and Epipactismicrophylla (4 genes). An exception was cemA (chloroplast envelope membrane protein), which is lost in L. abortivum similarly to some mixotrophic Corallorhiza (Barrett et al. 2014), which may indicate a nonessential photosynthetic function. A 9-amino-acid deletion and a 28-amino-acid insertion occurred respectively in rpoC1 for Cephalantherarubra and accD for N. cordata. However, we did not find any stop codons and/or frameshift indels in their sequence, and accD is known to be highly variable in length (Kim and Lee 2004; Gurdon and Maliga 2014; Wicke and Naumann 2018), suggesting that both proteins may still be functional in these species.
The selective regime was analyzed for five main gene subsets (as in Graham et al. [2017]): photosynthesis-related, NDH, ATP, PEP, and housekeeping (table 2). We used two different methods, both based on ω, the ratio of nonsynonymous substitutions per nonsynonymous site (dN) to synonymous substitutions per synonymous site (dS). ω provides insights into selection intensity but is also sensitive to population size and other parameters such as time of divergence (Ohta and Ina 1995). The first analysis was done with the PAML program, which allows to compute different ω values for different groups according to their nutrition type. Although this method provides useful information, it is not sufficient to confidently conclude about intensification or relaxation of selective pressure. A higher ω value can especially be a consequence of either increased positive selection or relaxation (Wertheim et al. 2015). We used the program RELAX as a second method to refine our analysis. RELAX allows more subtle estimation of ω variations by considering different categories of sequence sites that can evolve under different selective constraints (Wertheim et al. 2015).
Table 2.
Analysis of Selective Pressure for Different Sets of Genes Conserved among Autotrophic and Mixotrophic Neottieae
| Genes and Models | PAML Analysis |
RELAX Analysis |
||||
|---|---|---|---|---|---|---|
| np a | ln L b | Λ c | P Value | k d | P Value | |
| Photosynthesis e | ||||||
| M0: ω0 = 0.11 | 43 | −33,266.07 | ||||
| M1: ω0 = 0.12, ωN = 0.11 | 44 | −33,265.49 | 1.17 | 0.28 | 1.03 | 0.67 |
| M2: ω0 = 0.12, ωp = 0.15, ωN* = 0.10 | 45 | −33,263.31 | 4.35 | 0.04 | 1.13 | 0.44 |
| M3: ω0 = 0.12, ωp = 0.15, ωNa = 0.11, ωNx = 0.09 | 46 | −33,263 | 0.62 | 0.43 | 0.91 | 1 |
| rbcL | ||||||
| M0: ω0 = 0.12 | 43 | −2,687.57 | ||||
| M1: ω0 = 0.21, ωN = 0.09 | 44 | −2,685.94 | 3.26 | 0.07 | 0.57 | 0.03 |
| M2: ω0 = 0.20, ωp = 0.52, ωN* = 0.04 | 45 | −2,676.02 | 19.85 | 10−5 | 5.24 | 10−4 |
| M3: ω0 = 0.20, ωp = 0.52, ωNa = 0.05, ωNx = 0.01 | 46 | −2,675.11 | 1.82 | 0.18 | 10.97 | 0.04 |
| NDH f | ||||||
| M0: ω0 = 0.21 | 29 | −18,772.43 | ||||
| M1: ω0 = 0.18, ωN = 0.22 | 30 | −18,771.82 | 1.22 | 0.27 | 1.63 | 0.22 |
| M2: ω0 = 0.18, ωp = 0.16, ωN* = 0.24 | 31 | −18,770.10 | 3.44 | 0.06 | 1.61 | 0.10 |
| M3: ω0 = 0.18, ωp = 0.16, ωNa = 0.24, ωNx = 0.23 | 32 | −18,770.09 | 0.02 | 0.90 | 0.57 | 0.77 |
| ATP g | ||||||
| M0: ω0 = 0.13 | 43 | −10,149.71 | ||||
| M1: ω0 = 0.15, ωN = 0.13 | 44 | −10,149.43 | 0.56 | 0.45 | 1.08 | 0.49 |
| M2: ω0 = 0.15, ωp = 0.12, ωN* = 0.13 | 45 | −10,149.4 | 0.05 | 0.82 | 0.72 | 0.16 |
| M3: ω0 = 0.15, ωp = 0.12, ωNa = 0.13, ωNx = 0.12 | 46 | −10,149.36 | 0.09 | 0.76 | 1.01 | 0.93 |
| PEP h | ||||||
| M0: ω0 = 0.26 | 43 | −22,109.79 | ||||
| M1: ω0 = 0.23, ωN = 0.26 | 44 | −22,109.33 | 0.92 | 0.34 | 0.93 | 0.83 |
| M2: ω0 = 0.23, ωp = 0.19, ωN* = 0.28 | 45 | −22,107.93 | 2.8 | 0.09 | 1.98 | 0.04 |
| M3: ω0 = 0.23, ωp = 0.19, ωNa = 0.30, ωNx = 0.25 | 46 | −22,107.14 | 1.59 | 0.21 | 0.69 | 0.39 |
| Housekeeping i | ||||||
| M0: ω0 = 0.26 | 43 | −25,635.43 | ||||
| M1: ω0 = 0.23, ωN = 0.27 | 44 | −25,634.46 | 1.94 | 0.16 | 1.16 | 0.56 |
| M2: ω0 = 0.22, ωp = 0.30, ωN* = 0.26 | 45 | −25,634.21 | 0.49 | 0.48 | 1.1 | 0.75 |
| M3: ω0 = 0.22, ωp = 0.30, ωNa = 0.27, ωNx = 0.25 | 46 | −25,633.98 | 0.46 | 0.5 | 1.18 | 0.02 |
| matK | ||||||
| M0: ω0 = 0.35 | 43 | −4,121.08 | ||||
| M1: ω0 = 0.39, ωN = 0.34 | 44 | −4,120.92 | 0.33 | 0.56 | 1.11 | 0.7 |
| M2: ω0 = 0.39, ωp = 0.37, ωN* = 0.34 | 45 | −4,120.9 | 0.05 | 0.83 | 1.34 | 1 |
| M3: ω0 = 0.39, ωp = 0.37, ωNa = 0.45, ωNx = 0.23 | 46 | −4,118.08 | 5.63 | 0.02 | 0.48 | 0.52 |
| ycf1 + ycf2 | ||||||
| M0: ω0 = 0.68 | 43 | −27,092.79 | ||||
| M1: ω0 = 0.78, ωN = 0.65 | 44 | −27,091.67 | 2.23 | 0.14 | 1.12 | 0.46 |
| M2: ω0 = 0.78, ωp = 0.57, ωN* = 0.66 | 45 | −27,091.38 | 0.59 | 0.44 | 1.1 | 1 |
| M3: ω0 = 0.78, ωp = 0.57, ωNa = 0.75, ωNx = 0.55 | 46 | −27,088.8 | 5.16 | 0.02 | 1.13 | 0.13 |
Number of parameters for the model.
Log-likelihood of the data for the model.
Log-likelihood ratio test statistic used to compute P value.
Selection intensity parameter.
ccsA, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, rbcL, ycf3, and ycf4.
ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, and ndhK; species analyzed were Cephalanthera damasonium, Ce. longibracteata, Ce. longifolia, Ce. rubra, Epipactis albensis, E. atrorubens, E. gigantea, E. helleborine, E. palustris, E. purpurata, Neottia fugongensis, N. ovata, Palmorchis pabstii, Sobralia callosa, and Calanthe triplicata.
atpA, atpB, atpE, atpF, atpH, and atpI.
rpoA, rpoB, rpoC1, and rpoC2.
accD, clpP, infA, matK, rpl2, rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36, rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps15, rps16, rps18, and rps19.
For PAML analysis, four nested models allowing the calculation of different ω values for some subsets of branches were defined: M0, the null model with one ω for the whole tree; M1, which allowed ω to differ between outgroups (ω0) and Neottieae (ωN); M2, same as M1 but allowing a different ω for Palmorchis (ωp) and the remaining Neottieae (ωN*; below noted as Neottieae* = Neottieae without Palmorchis); M3, same as M2 but allowing ω to differ between autotrophic (ωNa) and mixotrophic Neottieae* (ωNx, see fig. 1B for species considered as mixotrophic). Models were compared by likelihood ratio tests following this scheme: M1 versus M0, M2 versus M1, and M3 versus M2. For all subsets, successive comparisons of the four models of ω variation among branches did not show a better fit for one model against another one, except for the photosynthesis genes for M2 versus M1 (table 2). M2 allowed different ω for outgroups (ω0 = 0.12), Palmorchispabstii (ωp = 0.15), and the remaining Neottieae (ωN* = 0.10; table 2). These results were not supported by the RELAX analysis, which indicated instead intensification of selective constraints for PEP genes comparing Neottieae* and Palmorchis (k = 1.98; P = 0.04) and for housekeeping genes comparing mixotrophic and autotrophic Neottieae* (k = 1.18; P = 0.02; table 2). But one should consider that these results are not adjusted for multiple comparisons.
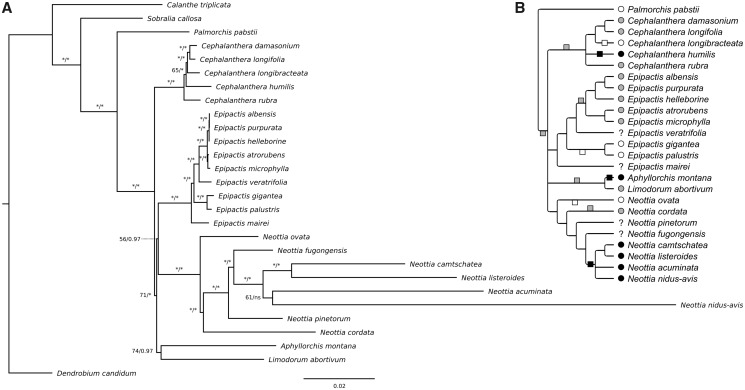
Fig. 1.
—(A) Phylogeny of Neottieae based on whole plastome analysis. Numbers above or to the left of branches represent bootstrap values (1,000 replicates) from ML analysis (left, stars indicate numbers above 95) and posterior probabilities from Bayesian inference (right, stars indicate probabilities above 0.98), ns means that the branch was not recovered in the Bayesian analysis. Scale bar: number of substitutions per site. (B) Two scenarios of mixotrophy and mycoheterotrophy evolution among Neottieae, that is, autotrophy (changes above the line) versus mixotrophy (changes below the line) of the Neottieae (excl. Palmorchis) ancestor. The tree is a consensus of the three analyses carried out on different sets of species and plastid DNA data (see supplementary fig. S1, Supplementary Material online, for other analyses). Nutrition type evolution is mapped considering the most parsimonious scenario in each case. Circles in front of species names indicate autotrophy (white), mixotrophy (gray), or mycoheterotrophy (black) and squares on the branches indicate shifts to these nutrition types in each scenario. Nutrition type evolution is mapped considering the most parsimonious scenario in each case. Boxes on the branches show changes occurring in both scenarios.
The same analysis carried out for individual photosynthesis-related genes showed a variation in selective regime only for rbcL (table 2 for rbcL, other genes shown in supplementary table S2, Supplementary Material online). rbcL data fitted models M2 much better than M1. In M2, ω was lower for Neottieae* (ωN* = 0.04) compared with outgroups (ω0 = 0.20) and P. pabstii (ωp = 0.52; table 2). This was supported by the RELAX analysis, which showed an intensification of selective constraints in Neottieae* compared with Palmorchis on the one hand (k = 5.24; P = 10−4), and in mixotrophic compared with autotrophic Neottieae* on the other hand (k = 10.97; P = 0.04). Thus, rbcL experiences stronger purifying selection in Neottieae*, and more particularly in mixotrophic species. For individual nonphotosynthesis genes, PAML analysis showed that both matK and ycf1 + ycf2 fitted M3 better than M2, with higher ω for autotrophic (ωNa = 0.45, resp. 0.75) compared with mixotrophic Neottieae* (ωNx = 0.23, resp. 0.55; table 2). This was, however, not supported by the RELAX analysis (table 2).
To summarize, we did not find evidence for relaxed selection in plastomes of mixotrophic species. By contrast, both PAML and RELAX analyses showed stronger selective pressure in mixotrophic Neottieae* compared with Palmorchis for rbcL.
Plastome Evolution in Mixotrophic Neottieae
The observed loss of NDH genes in three plastomes confirms that this gene complex is sensitive to evolutionary decay in Neottieae (Feng et al. 2016) and more generally in orchids (Barrett et al. 2014; Luo et al. 2014; Kim et al. 2015; Lin et al. 2017; Kim et al. 2018) and some other photosynthetic lineages (Wicke and Naumann 2018). Interestingly, it does not correlate with species nutrition. For example, the 11 genes were lost in L. abortivum and N. cordata (if ndhI is indeed nonfunctional). The first is mixotrophic and highly dependent on fungal carbon because its photosynthesis does not even compensate for its respiration (Girlanda et al. 2006); the second displays a more versatile use of fungal resources (Schiebold et al. 2018), which may even be optional (Těšitelová et al. 2015). Similarly, E. microphylla is the only species of the mixotrophic Epipactis section (E. albensis, E. atrorubens, E. helleborine, and E. purpurata) (Jin et al. 2014) for which NDH gene loss occurred, whereas some losses have been reported in E. veratrifolia and E. mairei (Feng et al. 2016) that are supposed autotrophic (pending more investigations). This suggests a species-specific decay of NDH genes in orchids, independently of their autotrophic or mixotrophic habit. In addition, we did not observe any evidence for intensification or relaxation of selective pressure on these genes (table 2). We suppose that relaxation on NDH genes occurs in the whole orchid clade, making any difference among orchid species hardly detectable. Changes in nutrition type do not seem to alter this pattern.
Photosynthesis-related genes neither are lost nor experience relaxed selective constraints in mixotrophic species but, in Neottieae*, they showed a slightly lower ω; rbcL was even under stronger purifying selection compared with P. pabstii (table 2). Some parts of the plastomes unrelated to photosynthesis also experienced higher selective constraints in mixotrophs, as shown by 1) lower ω for matK and ycf1 + ycf2 in mixotrophic than in autotrophic Neottieae* and 2) the RELAX analyses of PEP and housekeeping genes (displaying k > 1). These results are concordant with what has been reported for mixotrophic Corallorhiza, which 1) retain all photosynthesis-related genes except cemA and psbM, 2) display apparently unimpaired photosynthesis, and 3) show no difference in plastome selective regime compared with autotrophic references (Barrett et al. 2014). This also agrees with the retention of purifying selection on plastidial genes observed in the mixotrophic C.macrorhizon (Kim et al. 2018). However, our results contrast with other kinds of mixotrophy where relaxed purifying selection has been observed for photosynthesis, PEP and ATP-synthase genes, namely 1) in carnivorous plants (psa, atp, and rpo genes in Wicke et al. [2014]) and 2) in obligate hemiparasites (psa, psb, pet, atp, and rbcL in Petersen et al. [2015]; photosynthesis and atp genes in Wicke et al. [2016]). For this latter trophic strategy, retention of selective constraints on photosynthesis genes has nevertheless been reported for photosynthesis genes in Cuscuta (McNeal et al. 2007) and also for some housekeeping genes in Viscum (accD, cemA, clpP, and ycf2, Petersen et al. 2015). However, these comparisons between studies have to be taken with caution given the differences in scales among them or in a given study (individual genes vs. gene subsets).
In Neottieae, depending on the plastid genes considered, mixotrophic species thus feature either no change or a lowly supported increase in selective pressure compared with other mixotrophic groups such as Corallorhiza, carnivorous or parasitic plants (Barrett et al. 2014; Wicke et al. 2014, 2016; Petersen et al. 2015). Photosynthesis therefore seems to remain advantageous in mixotrophic Neottieae, and this is congruent with physiological evidence for the importance of photosynthetic carbon in fruit development and reproduction in these species (Roy et al. 2013; Bellino et al. 2014; Gonneau et al. 2014; Lallemand et al. 2019). We hypothesize that the retention of genes and purifying selection in the plastome of C.macrorhizon (Kim et al. 2018) indicates a similar crucial role of photosynthesis in reproduction, as illustrated by Suetsugu et al. (2018).
Neottieae Phylogeny and Mycoheterotrophy Evolution
Phylogenetic analysis of the 28 available plastomes confirmed Neottieae monophyly with Palmorchis in sister position to the remaining species (fig. 1A). Neottia, Epipactis, and Cephalanthera genera were each monophyletic and most intrageneric relationships were well defined. Aphyllorchis and Limodorum were sister genera but with mild support from maximum likelihood (ML) analysis (bootstrap = 74, fig. 1A). However, very short branches with low support prevented strong conclusions about intergeneric relationships (fig. 1A). Because mycoheterotrophic Neottia species display accelerated evolutionary rates (see long branches in fig. 1A), we removed them to increase alignment quality: We then obtained a congruent tree with better support for most but not all clades (supplementary fig. S1A, Supplementary Material online). In particular, Neottia, Epipactis, Aphyllorchis, and Limodorum clustered together in a monophyletic group sister to Cephalanthera with better support from ML analysis (bootstrap = 87 vs. 71). When restricting the analysis to the plastidial coding sequences (CDS) of nonmycoheterotrophic species, that is, excluding noncoding regions to allow better alignment, Limodorum was sister to Epipactis but Neottia no longer clustered with these genera in a monophyletic group (supplementary fig. S1B, Supplementary Material online). Combining these results suggests that at least Cephalanthera is sister clade to the rest of Neottieae. Analysis based on ITS (internal transcribed spacer of nuclear ribosomal DNA, including ITS1-5.8S rRNA-ITS2) confirmed Aphyllorchis and Limodorum as sister genera but failed to clarify further intergeneric relationships (supplementary fig. S1C, Supplementary Material online).
Phylogenetic relationships between the Neottieae genera have long been controversial and studies using different species and marker combinations yielded contradictory results, without any clear intergeneric relationships (ITS and the plastid trnL intron in Bateman et al. [2005]; ITS, trnS-G spacer, and rbcL in Roy et al. [2009]; ITS, Xdh, rbcL, matK, psaB, and trnL-F spacer in Xiang et al. [2012]; ITS, rbcL, and matK in Zhou and Jin [2018]). More confusingly, the phylogeny of Feng et al. (2016), based on a subset of the plastomes we analyzed, disagrees with our results, displaying Cephalanthera and Aphyllorchis as sister genera, and Epipactis and Neottia as successive sisters. This inconsistency may result from the difference in the number and diversity of species included in the analysis. We added 13 species and 2 new genera (Palmorchis and Limodorum) but failed to reproduce their results when considering only their subset of species, suggesting that this is rather due to different phylogenetic methods and/or to evolutionary signals conveyed by the plastome regions used. Feng et al. (2016) selected conserved plastome regions, whereas we made a dual analysis of whole plastome alignment and CDS-restricted alignment. Moreover, we chose not to filter or manually edit alignments following Tan et al. (2015) for accurate phylogenetic inferences (see also Hallas et al. 2017).
Neottieae intergeneric relationships thus remain difficult to estimate confidently. Our plastome-based analysis showed that the branches separating these genera are very short (<10−3 substitutions per site). Thus, the common ancestor of Neottieae* (i.e., Neottieae with exclusion of Palmorchis) likely experienced rapid speciation, with little time for informative mutations to accumulate (Glor 2010). Such a fast evolution of ancestral lineage makes it prone to hybridization and incomplete lineage sorting (Glor 2010; Wang et al. 2014), so that independent loci (e.g., nuclear and plastidial markers) may encapsulate different phylogenetic stories and make any combined analysis unreliable. Another possibility is heterotachy (Fitch and Markowitz 1970), a variation (here a decrease) in evolutionary rate in the ancestral lineage of Neottieae*. Arbitrating between fast speciation and heterotachy would be particularly challenging given the very short branch lengths. Confident reconstruction of early speciation in Neottieae will be complicated, but different gene trees could provide interesting insights into early Neottieae evolution and possible hybridization events.
We also wanted to compare two possible evolution of nutrition in Neottieae*. Based on our plastid phylogeny, we display two hypotheses, namely 1) an autotrophic ancestor and 2) a mixotrophic ancestor for Neottieae* (see Materials and Methods for reconstruction of intermediate characters). Assuming an autotrophic common ancestor, mixotrophy evolved at least four times (fig. 1B, gray boxes above the branches). Conversely, assuming a mixotrophic common ancestor would give three reversions to autotrophy (fig. 1B; white boxes below the branches). Each scenario implies two emergences of mycoheterotrophy, but unfortunately, the lack of information about the trophic status of some taxa (such as E. mairei or Neottiafugongensis) and the absence of several mycoheterotrophic clades limit our analysis. For example, green Neottia other than N. cordata or Epipactis species outside the Epipactis section may actually show mixotrophic nutrition. Assuming a mixotrophic common ancestor implies one change less than an autotrophic one but includes reversion to autotrophy. Although unlikely at first glance, reversion is allowed because mixotrophy is flexible and displays a continuum from autotrophy to mycoheterotrophy (Jacquemyn et al. 2017). Most importantly, the selective pressure maintaining photosynthesis genes reported here allows a reversion from mixotrophy to autotrophy, because potential for autotrophy remains intact in mixotrophic Neottieae. The question whether mixotrophy is a plesiomorphy of Neottieae thus remains open, pending for analyses of more species.
A final speculation comes at this point: If mixotrophy turns out to be indeed the ancestral state of Neottieae, could it have been a key factor in the fast early diversification speculated above? Mixotrophy allows plants to grow in light-limited forest understory and so to occupy a broad new ecological niche (Tĕšitel et al. 2018). Orchids show important dispersion abilities and most mixotrophic Neottieae do not seem very selective regarding the fungi they associate with (Tĕšitelová et al. 2012). One can therefore speculate that an early mixotrophic Neottieae lineage could have rapidly colonized distant and heterogeneous forest patches, with ability to keep or loose photosynthetic abilities, thus favoring radiation.
Materials and Methods
DNA Extraction and Sequencing
Total DNA was extracted from the leaves or fruits of the following Neottieae species (table 1 and supplementary table S1, Supplementary Material online): Palmorchis pabstii Veyret, N.cordata (L.) Rich., E.albensis Nováková & Rydlo, E. atrorubens (Hoffm.) Besser, E. gigantea Douglas ex Hook., E. helleborine (L.) Crantz, E. microphylla (Ehrh.) Sw., E. palustris (L.) Crantz, E. purpurata Sm., Cephalanthera damasonium (Mill.) Druce, Ce. longibracteata Blume, Ce.rubra (L.) Rich., and L.abortivum (L.) Sw. The following extraction kits and protocols were used (supplementary table S3, Supplementary Material online): DNeasy Plant Mini Kit (Qiagen) with final elution in distilled water, NucleoSpin Plant II kit (Macherey-Nagel), CTAB extraction as in Porebski et al. (1997).
Given low yield due to DNA degradation in some old samples, the Accel-NGS 1S Plus DNA Library Kit for Illumina with the 1S Plus Indexing kit (Swift Biosciences Inc., Ann Arbor, MI) was used for preparation of libraries, following the manufacturer’s instructions. We did so for the set of samples handled in EPGV laboratory (supplementary table S3, Supplementary Material online). For other samples, handled in Moscow laboratory, the TruSeq DNA sample preparation kit (Illumina, USA) and the NEBNext Ultra DNA kit (New England Biolabs, USA) were used for preparation of libraries (supplementary table S3, Supplementary Material online). Paired-end sequencing on multiplexed libraries was performed either on Illumina HiSeq 2000, Illumina HiSeq 2500 using the rapid run mode, and Illumina HiSeq 4000 or Illumina MiSeq, depending on the samples (different material available in the different teams involved in the study; supplementary table S3, Supplementary Material online). Raw sequences for Ce. longibracteata, Ce. rubra, E. albensis, E. atrorubens, E. gigantea, E. microphylla, E. palustris, E. purpurata, and P. pabstii were submitted to the NCBI database: BioProject PRJNA484137, SRA accession number ongoing.
Read Cleaning, Plastome Assembly, and Annotation
Redundant reads were removed with a homemade C++ script. The redundancies were sought by pairs read1–read2, the redundancy with the best Phred score was kept. Trimming of the low-quality 3′ end was done using a homemade C++ script, one base after the other until reaching a base with a Phred score > 30 or until the mean Phred score of the read was above 30. Pairs with at least one read shorter than 30 bp after this step or with N were removed. Read number before and after cleaning are indicated in supplementary table S4, Supplementary Material online.
For samples that were prepared with the Accel-NGS 1S Plus DNA Library Kit, adapter trimming was conducted using BBDuk Trimmer plugin version 37.64 in Geneious version 11.1.3 (http://www.geneious.com; last accessed September 2018; Kearse et al. 2012) with the following parameters: adapters = Illumina Truseq DNA adapters, ktrim = r, k = 27, hdist = 1, edist = 0, mink = 6, minlength = 10, trimbyoverlap = t, and minoverlap = 6. Addition of a low complexity tail to the 3′ end of fragments was part of the library preparation kit and had to be removed from the beginning of reads 2. Following the manufacturer’s recommendations, trimming an additional 10 bp at the 5′ end was done for all reads.
De novo assembly was done differently depending on the species (supplementary table S3, Supplementary Material online). 1) A subset of 25% of the reads was assembled using the Geneious algorithm with medium-low sensitivity parameters. Resulting contigs were mapped on the plastome of Neottia ovata (L.) Bluff & Fingerh. (KU551271), Cephalantera longifolia (L.) Fritsch (KU551263), or E.veratrifolia Boiss. & Hohen (KU551267), depending on the species. Contigs corresponding to the plastome were then dissolved and reassembled with medium sensitivity parameters to increase assembly quality. The resulting contigs were again mapped to the reference genome and large single copy (LSC), inverted repeat B (IRB), and small single copy (SSC) regions were isolated. IRB was duplicated and reverse complemented to obtain inverted repeat A (IRA). Concatenation of LSC, IRB, SSC, and IRA led to the final plastome sequence. 2) The CLC Genomics Workbench de novo assembly procedure was used. In this latter case, reads were first trimmed with the following settings: quality limit = 0.01 (corresponds to Q ≥ 20), remove adapters = yes (Illumina Truseq adapters), discard short reads = yes (limit 50 nt for MiSeq data, 25 nt for HiSeq data). Then de novo assembly was performed with the following settings: automatic bubble size = yes, minimum contig length = 1,000 bp, automatic word size = yes, perform scaffolding = yes, and auto-detect paired distances = yes. Contigs of plastid origin were selected based on the results of BLAST search with Phalaenopsis aphrodite plastome used as query. Contigs corresponding to LSC, IR, and SSC were concatenated accordingly and resulting sequences were verified and corrected by performing the gap closure method using back mapping of the reads. IRs were duplicated and reverse complemented to obtain the final parts (IRA) of the plastomes.
Coverage values of the contigs used for assembly are indicated in supplementary table S4, Supplementary Material online. Depending on the species, the phylogenetically closest reference plastome was used for annotations transfer in Geneious with a 77% similarity threshold that turned out, after preliminary attempts, to be low enough to detect similar gene sequences and sufficiently high to avoid false positives. Manual correction and addition of missing information was then carried out for each plastome. Annotated plastome sequences were submitted to GenBank under the accession numbers MH590345–MH590357 (table 1).
Phylogenetic Inferences
Including the 12 already published sequences, we ended up with a set of 25 Neottieae plastomes and added 3 outgroup species from the same subfamily Epidendroideae (supplementary table S1, Supplementary Material online). The four mycoheterotrophic Neottia were either kept or removed to test whether their rapidly evolving plastomes (much higher than the two other mycoheterotrophs; fig. 1A) and the increased length and quality of the alignment obtained without them alter phylogeny reconstruction. The second copy of the inverted repeat region (IRA) was removed from all plastomes and the resulting sequences were aligned a first time using the Mauve progressive algorithm, as implemented in Geneious (parameters set to automatically calculate seed weight and minimum locally collinear block (LCB) score, compute LCBs, full alignment), to detect potential rearrangements (Darling et al. 2004). The LCBs obtained were individually aligned a second time using the MAFFT online service (Katoh et al. 2017) with automatic selection of alignment strategy, gap opening penalty set to 3.0, offset value set to 0.1 and other parameters left as default. The resulting alignments were then concatenated. Model selection was done with the ModelFinder algorithm (Kalyaanamoorthy et al. 2017) implemented in the IQ-TREE program (Nguyen et al. 2015). The model with lowest Bayesian information criterion score was used for ML analysis with IQ-TREE (Nguyen et al. 2015). The first model with the lowest Bayesian information criterion compatible with MrBayes v3.2 parameters (Ronquist et al. 2012) was chosen for Bayesian inference with this program. A complementary analysis using the nuclear marker ITS, including ITS1-5.8S rRNA-ITS2, was done using available Neottieae sequences (supplementary table S1, Supplementary Material online).
On the plastid phylogeny (fig. 1B), we displayed two possible evolution of nutrition based on the hypotheses of 1) an autotrophic ancestor or 2) a mixotrophic ancestor for Neottieae*. Intermediate characters for other nodes were proposed in order to minimize the number of transitions between trophic types (autotrophic, mixotrophic, or mycoheterotrophic), that is, the most parsimonious scenario under each hypothesis, considering the nutritional status of extant species (supplementary table S1, Supplementary Material online). A given species was considered as mixotrophic when it dominantly associated with ectomycorrhizal fungi and showed an enrichment in 13C compared with surrounding autotrophs, a usual marker of fungal carbon gain (Hynson et al. 2013; see supplementary table S1, Supplementary Material online, for references regarding species nutrition).
Selective Regime Analyses
All CDS were extracted from the same set of plastomes used for phylogeny reconstruction, except the six mycoheterotrophic species. Except for NDH complex, genes that were lost or pseudogenized in at least one of the remaining species were discarded from subsequent analyses (see table 2 for the remaining genes). For NDH, because some species had lost all the genes, we rather only kept for subsequent analysis only the 15 species, which retained the 11 NDH genes (table 2). CDS were aligned based on their amino acid sequences with subsequent backtranslation using the AlignTranslation function (Wright 2015) of the DECIPHER package (Wright 2016) in the R environment for statistical computing (R Development Core Team 2007). Functional gene groups were built by concatenating CDS for photosynthesis, NDH, ATP-synthase, PEP, and housekeeping genes (table 2). ycf1 and ycf2 were combined and analyzed independently because of their specific evolutionary rates (Barrett et al. 2014). In addition, some genes were analyzed individually when long enough for confident parameter estimation: ccsA, psaA, psaB, psbA, psbB, psbC, psbD, rbcL, accD, and matK (see supplementary table S2, Supplementary Material online). ω, the ratio of nonsynonymous substitutions per nonsynonymous site (dN) to synonymous substitutions per synonymous site (dS), was used to estimate selective pressure for different sets of genes. The codeml program implemented in PAML version 4.9 (Yang 2007) was used to compute ω values, with the branch model and the tree shown in figure 1B. The codon frequency parameter was set to F3X4. Four nested models allowing the calculation of different ω values for some subsets of branches were defined: M0, the null model with a unique ω for the whole tree; M1, which allowed ω to differ between outgroups (ω0) and Neottieae (ωN); M2, same as M1 but allowing a different ω for Palmorchis (ωp) and the remaining Neottieae (ωN*); M3, same as M2 but allowing ω to differ between autotrophic (ωNa) and mixotrophic Neottieae* (ωNx, see fig. 1B for species considered as mixotrophic). Models were compared by likelihood ratio tests following this scheme: M1 versus M0, M2 versus M1, and M3 versus M2. The statistic of the test (Λ) was calculated as two times the difference in model likelihood and compared with a chi-squared distribution with degrees of freedom equaling the difference in parameters number between the two models. P values were obtained using the function pchisq of the R stats package.
We completed the PAML analysis with a specific test dedicated to the detection of changes in selective pressure (relaxation or intensification). This was carried out with the program RELAX, available at the Datamonkey webserver http://datamonkey.org/relax; last accessed March 6, 2019. RELAX allows more subtle estimation of ω variations by considering different categories of sequence sites that can evolve under different selective constraints (Wertheim et al. 2015). ω distributions are estimated for a test (T) and a reference (R) subset of branches and a selection intensity parameter k such that ωT = is introduced. The program then compares the goodness of fit of the data to a branch-site evolutionary model with k = 1 (null model) or k being a free parameter (alternative model). When the alternative model shows a better fit following a likelihood ratio test, k < 1(resp. k > 1) indicates a relaxation (resp. an intensification) of selective constraints in the test group. Three different tests were done with three different designs of test versus reference groups, copying what has been done with the PAML nested analysis (see supplementary fig. S2, Supplementary Material online, for group designs).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We acknowledge Aurélie Chauveau for preparation of the libraries, Élodie Marquand and Aurélie Canaguier for data processing, two anonymous referees and Dennis Lavrov for their corrections, and David Marsh for English corrections. This work was funded by the Fondation de France (funds from the Fondation Ars Cuttoli & Paul Appell, project name : EVOLUTION ET PHYSIOLOGIE DES PLANTES QUI SE NOURRISSENT DE CHAMPIGNONS (MYCOHETEROTROPHIE)) and the National Science Center (Poland; grant no. 2015/18/A/NZ8/00149). We thank the Commissariat à l’Énergie Atomique et aux Énergies Alternatives – Institut de Génomique/Centre National de Génotypage for conducting the DNA quality control and for giving the INRA-EPGV group access to their Illumina Sequencing Platform. Plastomes of Ce. damasonium and L. abortivum were sequenced with support of budgetary subsidy to IITP RAS (Laboratory of Plant Genomics). We thank the MSU Herbarium (MW) for providing a sample of Ce. longibracteata Blume MW0047886 and “Service de Systématique Moléculaire” (UMS2700 MNHN/CNRS) for granting access to its technical platform.
Author Contributions
Conceptualization: F.L. and M.-A.S.; formal analysis: F.L.; investigation: F.L., M.L., M.M., I.L.C, A.B., and M.-C.L.P.; resources: M.L., I.L.C, A.B., E.Z., M.M., and M.-C.L.P.; data curation: F.L., M.L., M.M., M.J., and A.B.; writing—original draft: F.L.; writing—review and editing; F.L., M.L., E.Z., E.D., M.J., M.-C.L.P., and M.-A.S; supervision: M.-C.L.P. and M.-A.S.; funding acquisition: M.-A.S.
Data deposition: This project has been deposited at GenBank under the accession MH590345-MH590357; at NCBI BioProject under the accession PRJNA484137; at NCBI SRA under the accession SRP160956; at TreeBASE under the accession 24919; at the GitHub repository https://github.com/flallema/Neottieae_plastomes.
Literature Cited
- Barrett CF, Davis JI.. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99(9):1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Bateman RM, Hollingsworth PM, Squirrel J, Hollingsworth M.. 2005. Tribe Neottieae In: Pridgeon A, Cribb PJ, Chase MW, Rasmussen FN, editors. Genera Orchidacearum Volume 4: Epidendroideae, Part 1. Oxford, New York: Oxford University Press. [Google Scholar]
- Bellino A, et al. 2014. Nutritional regulation in mixotrophic plants: new insights from Limodorum abortivum. Oecologia 175(3):875–885. [DOI] [PubMed] [Google Scholar]
- Bolin JF, Tennakoon KU, Majid MBA, Cameron DD.. 2017. Isotopic evidence of partial mycoheterotrophy in Burmannia coelestis (Burmanniaceae). Plant Species Biol. 32(1):74–80. [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearnaley JDW, Martos F, Selosse M-A.. 2012. Orchid mycorrhizas: molecular ecology, physiology, evolution and conservation aspects In: Hock B, editor. Fungal Associations. The Mycota (a comprehensive treatise on fungi as experimental systems for basic and applied research). Vol. 9 Berlin/Heidelberg (Germany: ): Springer; p. 207–230. [Google Scholar]
- Feng Y-L, et al. 2016. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol Evol. 8(7):2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM, Markowitz E.. 1970. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 4(5):579–593. [DOI] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K.. 2007. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7:45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer G, Meyer M.. 2003. 15N and 13C natural abundance of autotrophic and myco-heterotrophic orchids provides insight into nitrogen and carbon gain from fungal association. New Phytol. 160(1):209–223. [DOI] [PubMed] [Google Scholar]
- Girlanda M, et al. 2006. Inefficient photosynthesis in the Mediterranean orchid Limodorum abortivum is mirrored by specific association to ectomycorrhizal Russulaceae. Mol Ecol. 15(2):491–504. [DOI] [PubMed] [Google Scholar]
- Glor RE. 2010. Phylogenetic insights on adaptive radiation. Annu Rev Ecol Evol Syst. 41(1):251–270. [Google Scholar]
- Gonneau C, et al. 2014. Photosynthesis in perennial mixotrophic Epipactis spp. (Orchidaceae) contributes more to shoot and fruit biomass than to hypogeous survival. J Ecol. 102(5):1183–1194. [Google Scholar]
- Górniak M, Paun O, Chase MW.. 2010. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol Phylogenet Evol. 56(2):784–795. [DOI] [PubMed] [Google Scholar]
- Graham SW, Lam VKY, Merckx V.. 2017. Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 214(1):48–55. [DOI] [PubMed] [Google Scholar]
- Gurdon C, Maliga P.. 2014. Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula. DNA Res 21: 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallas JM, Chichvarkhin A, Gosliner TM.. 2017. Aligning evidence: concerns regarding multiple sequence alignments in estimating the phylogeny of the Nudibranchia suborder Doridina. R Soc Open Sci. 4(10):171095.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynson NA, et al. 2013. The physiological ecology of mycoheterotrophy In: Merckx VSFT, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 297–342. [Google Scholar]
- Jacquemyn H, et al. 2017. Mycorrhizal associations and trophic modes in coexisting orchids: an ecological continuum between auto- and mixotrophy. Front Plant Sci. 8:1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X-H, et al. 2014. The evolution of floral deception in Epipactis veratrifolia (Orchidaceae): from indirect defense to pollination. BMC Plant Biol. 14:63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julou T, et al. 2005. Mixotrophy in orchids: insights from a comparative study of green individuals and nonphotosynthetic individuals of Cephalanthera damasonium. New Phytol. 166(2):639–653. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler AV, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform bbx 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Shin C-H, Sun H, Kim J-H.. 2018. Sequencing of the plastome in the leafless green mycoheterotroph Cymbidium macrorhizon helps us to understand an early stage of fully mycoheterotrophic plastome structure. Plant Syst Evol. 304(2):245–258. [Google Scholar]
- Kim HT, et al. 2015. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS One 10(11):e0142215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-J, Lee H-L.. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11(4):247–261. [DOI] [PubMed] [Google Scholar]
- Lallemand F, Robionek A, Courty P-E, Selosse M-A.. 2018. The 13C content of the orchid Epipactis palustris (L.) Crantz responds to light as in autotrophic plants. Bot Lett. 165:265–273. [Google Scholar]
- Lallemand F, et al. 2016. The elusive predisposition to mycoheterotrophy in Ericaceae. New Phytol. 212(2):314–319. [DOI] [PubMed] [Google Scholar]
- Lallemand F, et al. 2019. Mixotrophic orchids do not use photosynthates for perennial underground organs. New Phytol. 221(1):12–17. [DOI] [PubMed] [Google Scholar]
- Leake JR. 1994. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 127(2):171–216. [DOI] [PubMed] [Google Scholar]
- Lin C-S, et al. 2017. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J 90: 994–1006. [DOI] [PubMed] [Google Scholar]
- Luo J, et al. 2014. Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS One 9(6):e99016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW.. 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx VSFT, Freudenstein JV, et al. 2013. Taxonomy and classification In: Merckx VSFT, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 19–101. [Google Scholar]
- Merckx VSFT, Mennes CB, Peay KG, Geml J.. 2013. Evolution and diversification In: Merckx VSFT, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 215–244. [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Ina Y.. 1995. Variation in synonymous substitution rates among mammalian genes and the correlation between synonymous and nonsynonymous divergences. J Mol Evol. 41(6):717–720. [DOI] [PubMed] [Google Scholar]
- Petersen G, Cuenca A, Seberg O.. 2015. Plastome evolution in hemiparasitic mistletoes. Genome Biol Evol. 7(9):2520–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, Zervas A, Pedersen HÆ, Seberg O.. 2018. Genome reports: contracted genes and dwarfed plastome in mycoheterotrophic Sciaphila thaidanica (Triuridaceae, Pandanales). Genome Biol Evol. 10(3):976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR.. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15. [Google Scholar]
- R Development Core Team. 2007. R: a language and environment for statistical computing.
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, et al. 2009. Two mycoheterotrophic orchids from Thailand tropical dipterocarpacean forests associate with a broad diversity of ectomycorrhizal fungi. BMC Biol. 7:51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, et al. 2013. Why do mixotrophic plants stay green? A comparison between green and achlorophyllous orchid individuals in situ. Ecol Monogr 83:95–117. [Google Scholar]
- Schiebold J-I, Bidartondo MI, Lenhard F, Makiola A, Gebauer G.. 2018. Exploiting mycorrhizas in broad daylight: partial mycoheterotrophy is a common nutritional strategy in meadow orchids. J Ecol. 106(1):168–178. [Google Scholar]
- Selosse M-A, Charpin M, Not F.. 2017. Mixotrophy everywhere on land and in water: the grand écart hypothesis. Ecol Lett. 20(2):246–263. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Martos F.. 2014. Do chlorophyllous orchids heterotrophically use mycorrhizal fungal carbon? Trends Plant Sci. 19(11):683–685. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Roy M.. 2009. Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci. 14(2):64–70. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Weiß M, Jany J-L, Tillier A.. 2002. Communities and populations of sebacinoid basidiomycetes associated with the achlorophyllous orchid Neottia nidus-avis (L.) L.C.M. Rich. and neighbouring tree ectomycorrhizae. Mol Ecol. 11(9):1831–1844. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ.. 2008. Mycorrhizal symbiosis. London: Academic Press. [Google Scholar]
- Suetsugu K, Ohta T, Tayasu I.. 2018. Partial mycoheterotrophy in the leafless orchid Cymbidium macrorhizon. Am J Bot. 105(9):1595–1596. [DOI] [PubMed] [Google Scholar]
- Tan G, et al. 2015. Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst Biol 64: 778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tĕšitel J, Těšitelová T, Minasiewicz J, Selosse M-A.. 2018. Mixotrophy in Land Plants: why to Stay Green? Trends Plant Sci. 23(8):656–659. [DOI] [PubMed] [Google Scholar]
- Tĕšitelová T, Tĕšitel J, Jersáková J, RÍhová G, Selosse M-A.. 2012. Symbiotic germination capability of four Epipactis species (Orchidaceae) is broader than expected from adult ecology. Am J Bot. 99(6):1020–1032. [DOI] [PubMed] [Google Scholar]
- Těšitelová T, et al. 2015. Two widespread green Neottia species (Orchidaceae) show mycorrhizal preference for Sebacinales in various habitats and ontogenetic stages. Mol Ecol. 24(5):1122–1134. [DOI] [PubMed] [Google Scholar]
- Wang W, Li H, Chen Z.. 2014. Analysis of plastid and nuclear DNA data in plant phylogenetics-evaluation and improvement. Sci China Life Sci. 57(3):280–286. [DOI] [PubMed] [Google Scholar]
- WCSP. 2018. World checklist of selected plant families. Facilitated by the Royal Botanic Gardens, Kew. Available from: http://wcsp.science.kew.org/ (accessed February 17, 2018).
- Wertheim JO, et al. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Naumann J.. 2018. Molecular evolution of plastid genomes in parasitic flowering plants In: Chaw S-M, Jansen RK, editors. Advances in botanical research. Vol. 85 Cambridge: Academic Press; p. 315–347. [Google Scholar]
- Wicke S, Schäferhoff B, dePamphilis CW, Müller KF.. 2014. Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol Biol Evol. 31(3):529–545. [DOI] [PubMed] [Google Scholar]
- Wicke S, et al. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 113(32):9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ES. 2015. DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinformatics 16:322.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ES. 2016. Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 8(1):352–359. [Google Scholar]
- Xiang X-G, et al. 2012. Phylogenetic placement of the enigmatic orchid genera Thaia and Tangtsinia: evidence from molecular and morphological characters. Taxon 61(1):45–54. [Google Scholar]
- Yagame T, et al. 2016. Fungal partner shifts during the evolution of mycoheterotrophy in Neottia. Am J Bot. 103(9):1630–1641. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhou T, Jin X.. 2018. Molecular systematics and the evolution of mycoheterotrophy of tribe Neottieae (Orchidaceae, Epidendroideae). PhytoKeys 94:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.