Graphical abstract
Abbreviations: AC, ascending colon; DC, descending colon; FOS, fructooligosaccharides; GALT, gut associated lymphoid tissue; GOS, galactooligosaccharides; M-SHIME, mucosal Simulator of the Human Intestinal Microbial Ecosystem; OTU, operational taxonomic unit; qPCR, quantitative polymerase chain reaction; SCFA, short-chain fatty acid; TC, transverse colon; XOS, xylooligosaccharides
Keywords: Faecalibacterium prausnitzii, Endotoxemia, Fructooligosaccharides, Galactooligosaccharides, Xylooligosaccharides, Obesity
Abstract
A standardized in vitro simulation of the human gastrointestinal tract (M-SHIME®) was used to assess the effect of repeated daily administration of a synbiotic formulation, containing five spore-forming Bacillus strains and a prebiotic fiber blend, on the microbial activity and composition of three simulated human subjects. Firstly, while confirming recent findings, deeper phylogenetic insight was obtained in the resident M-SHIME® microbiota, demonstrating that the model maintains a diverse and representative, colon region-specific luminal and mucosal microbial community. Supplementation of the synbiotic concept increased microbial diversity in the distal colon areas, whereas specific enhancement of Bacillaceae levels was observed in the ascending colon suggesting a successful engraftment of the Bacillus spores, which probably resulted in a stimulatory effect on, among others, Bifidobacteriaceae, Lactobacillaceae, Prevotellaceae, Tannerellaceae and Faecalibacterium prausnitzii contributing directly or indirectly to stimulation of acetate, propionate and butyrate production. When compared with a previous study investigating the Bacillus strains, the generated data suggest a synergistic effect on the intestinal microbiota for the synbiotic formulation. Given the fact that the probiotic strains have been shown to impact post-prandial metabolic endotoxemia in human individuals, it might be interesting to further investigate the efficacy of the synbiotic concept in protecting against obesity-related disorders.
1. Introduction
A strategy to improve intestinal health is the use of prebiotics, probiotics and synbiotics. While prebiotics have been defined as ‘non-digestible substrates that are selectively used by the gut microbial community, thereby conferring a health benefit for the host’ (Gibson et al., 2017), probiotics consider the administration of live microorganisms that ‘when administered in adequate amounts, confer a health benefit on the host’ (Hill et al., 2014). A synbiotic formulation combines both concepts often aiming to provide synergistic effects in the gastrointestinal tract as compared to the activity of the single substrates and strains (Schrezenmeir and de Vrese, 2001). The most commonly used prebiotics in synbiotic formulations comprise fibers such as inulin, fructooligosaccharides (FOS), galactooligosaccharides (GOS) and xylooligosaccharides (XOS), while the mostly used probiotic strains include Lactobacillus spp., Bifidobacterium spp., Saccharomyces boulardii and Bacillus coagulans (Pandey et al., 2015). Synbiotic supplementation has been linked with several human health benefits, such as improvement of atopic dermatitis (Farid et al., 2011), reduction of serum lipid profile in patients with type-2 diabetes (Shakeri et al., 2014), alleviation of digestive complaints in patients with gastrointestinal disorders (Fujimori et al., 2009, Šmid et al., 2016) and changes in anthropometric measurements in obese individuals (Ipar et al., 2015, Safavi et al., 2013).
Bacillus spp. have been widely used as probiotic ingredient in both animals and humans as many of these species are adapted to survive in the host gastrointestinal tract (Hong et al., 2005). Indeed, Bacillus species are able to form endospores, which protect the bacterial cell to gastric acidity, followed by germination and further proliferation of the micro-organism in the small intestine (Cartman et al., 2008, Casula and Cutting, 2002, Tam et al., 2006). Such germination is a prerequisite for certain health benefits as shown by Rhee et al. which reported stimulation of gut-associated lymphoid tissue (GALT) development in infant rabbits upon germination of orally administered Bacillus subtilis spores (Rhee et al., 2004). Next to immunomodulation, germinating spores of Bacillus subtilis var. Natto have been shown to secrete nattokinase, a peptidase involved in fibrinolysis (Sumi et al., 1995). Recently, it was reported that supplementation of a blend of five spore-forming Bacillus strains reduced post-prandial metabolic endotoxemia in human individuals, likely by modulation of the gut microbial community (McFarlin et al., 2017).
Recently, our research group evaluated the gut-modulatory effect of the same probiotic formulation as used by McFarlin et al. in a validated in vitro simulation of the human gastrointestinal tract for three different adult donors (Duysburgh et al., 2019, McFarlin et al., 2017). Bifidobacterium levels increased for all three donors tested, while a donor-dependent modulation of metabolic activity was revealed. Indeed, significant stimulation of health-related metabolites acetate, propionate and butyrate production was observed upon probiotic supplementation. Further, donor-dependent effects were observed at microbial community level, with two of the three donors stimulating Faecalibacterium prausnitzii, whereas the final donor enriched Akkermansia muciniphila upon probiotic administration. Interestingly, this related to observed in vivo findings by Everard et al. who showed that stimulation of Akkermansia muciniphila reduced high-fat diet induced metabolic endotoxemia (Everard et al., 2013), while González-Sarrías et al. showed a significant association between decreased endotoxemia and increased abundance of Faecalibacterium and Odoribacter in obese individuals (Gonzalez-Sarrias et al., 2018).
In the present study, we aimed at evaluating the synbiotic effect of the aforementioned probiotic mix of five strains in combination with a functional fiber blend, in order to elucidate potential synergistic effects on microbial metabolic activity and community composition of the same three human individuals that were tested in the preceding in vitro study (Duysburgh et al., 2019). For this purpose, the validated Mucosal Simulator of the Human Intestinal Microbial Ecosystem (M-SHIME®) was used, which allowed to evaluate the synbiotic effect of the test product in three consecutive colon regions, both consisting of a luminal and mucosal environment (Possemiers et al., 2004, Van den Abbeele et al., 2012).
2. Materials and methods
2.1. Chemicals and test product
Unless otherwise stated, all chemicals were obtained from Sigma-Aldrich (Overijse, Belgium). Microbiome Labs (Glenview, USA) provided the synbiotic formulation, consisting of MegaSporebiotic, a proprietary probiotic mixture of Bacillus indicus (HU36), Bacillus subtilis (HU58), Bacillus coagulans SC-208, Bacillus licheniformis and Bacillus clausii SC-109 spores, and MegaPrebiotic, a proprietary prebiotic blend of fructooligosaccharides from green and gold kiwifruit (Livaux™ and ACTAZIN™), xylooligosaccharides from corn cob (PreticX™) and galactooligosaccharides from cow milk (Bimuno®). The daily synbiotic dose contained 8 * 109 Bacillus spores, which corresponds to two capsules of MegaSporeBiotic, and 3775 mg of prebiotic blend.
2.2. Simulator of the human intestinal microbial ecosystem (SHIME®)
The reactor setup represented the different regions of the human gastrointestinal tract and was adapted from the SHIME® (ProDigest and Ghent University, Belgium) as previously described by Molly et al. (1993). The SHIME® setup consists of a series of five reactors simulating the stomach, small intestine and finally three colon compartments that upon inoculation with the fecal microbiota from a healthy human individual represent the ascending (AC), transverse (TC) and descending (DC) colon. During the current study, three SHIME® experiments were conducted in parallel, differing in fecal microbiota used. Fecal inoculum was prepared from three different human adults (male, 35y; female, 29y and male, 34y) as reported by Possemiers et al. (2004). Further, nutritional medium, retention times, pH and temperature settings were adopted from Possemiers et al. (2004). In order to simulate both the luminal and mucus-associated microbial community, a mucosal compartment was included in the colon reactors by addition of mucin beads as described by Van den Abbeele et al. (2012). The experimental design of the SHIME® run included a two-week initiation period in order to obtain a stable microbial community (Van de Wiele et al., 2015), followed by a two-week control period for baseline measurements and a four-week treatment period during which the synbiotic test product was administered once daily with the nutritional medium.
2.3. Microbial metabolic activity
Samples for microbial metabolic activity were collected three times per week during the control and treatment period from each colon compartment. Short-chain fatty acid (SCFA) measurements, including acetate, propionate, butyrate and branched SCFA (isobutyrate, isovalerate and isocaproate), were performed as reported by De Weirdt et al. (2010). Lactate concentrations were determined using a commercially available enzymatic assay kit (R-Biopharm, Darmstadt, Germany) according to manufacturer’s instructions. Ammonium analysis was performed using a KjelMaster K-375 device (Büchi, Hendrik-Ido-Ambacht, The Netherlands). Briefly, addition of 32% NaOH liberated ammonium from the sample in the form of volatile ammonia, which was further distilled into a 2% boric acid solution. The ammonium in the distillate was quantified titrimetrically with a 0.02 M HCl solution.
2.4. Microbial community analysis
During the control and treatment period, samples for microbial community analysis were collected once per week from each colon reactor. Total DNA was isolated as described by Boon et al. (2003), with some minor modifications. Luminal DNA originated from pelleted bacterial cells obtained from 1 mL sample, while mucosal DNA was extracted from 0.25 g mucin agar collected from the mucus beads. A Fastprep-24 device (MP BioMedicals, Illkirch, France) was used for homogenization, which was conducted twice for 40 s at 4 m/s with a resting period of 5 min between shakings.
Microbial community profiling of each colon compartment was established by 16S-targeted Illumina sequencing. Aliquots of the original genomic DNA extract were send out to LGC Genomics GmbH (Berlin, Germany) for library preparation and sequencing on an Illumina Miseq platform with v3 chemistry using the primers as reported by Klindworth et al. (2013), with modification of the reverse primer (785Rmod; 5′-GAC TAC HVG GGT ATC TAA KCC-3′) to increase coverage, which amplified the 16S rRNA gene V3-V4 hypervariable regions.
Subsequently, quantitative polymerase chain reaction (qPCR) assays for Lactobacillus spp., Bifidobacterium spp., Akkermansia muciniphila and Faecalibacterium prausnitzii were completed using a QuantStudio 5 Real-Time PCR system (Applied Biosystems, Foster City, CA USA). Each sample was analysed in technical triplicate and outliers (more than 1 CT difference) were removed. The qPCR assay for Lactobacillus spp. was previously described by Furet et al. (2009), while the qPCR for Bifidobacterium spp. was performed as reported in Rinttilä et al. (2004). The qPCR assays for Akkermansia muciniphila and Faecalibacterium prausnitzii were conducted as described by Collado et al., 2007, Sokol et al., 2009, respectively.
2.5. Statistics
Student’s T-tests for pairwise comparisons were performed for assessment of normally distributed data of the different control and treatment weeks for metabolic markers and microbial community parameters. Differences were considered significant if p < 0.05.
For the 16S-targeted Illumina sequencing, read assembly and cleanup was derived from the MiSeq procedure (Kozich et al., 2013, Schloss and Westcott, 2011). Briefly, mothur (v. 1.39.5) was utilized to assemble reads into contigs, perform alignment-based quality filtering (alignment to the mothur-reconstructed SILVA SEED alignment, v. 123), remove chimeras, assign taxonomy using a naïve Bayesian classifier (Wang et al., 2007) and SILVA NR v128 and cluster contigs into operational taxonomic units (OTUs) at 97% sequence similarity. All sequences classified as Archaea, Chloroplasts, Eukaryota and Mitochondria were omitted, as well as sequences that could not be classified, even not at (super) Kingdom level. For each OTU, representative sequences were picked as the most abundant sequence within that OTU. Finally, the reciprocal Simpson diversity index was calculated as reviewed by Bent and Forney (2008).
3. Results
3.1. Colon-region specific microbial metabolism and composition in the SHIME®
Corresponding colon compartments of the three parallel SHIME® setups inoculated with fecal microbiota of three different donors showed highly comparable SCFA levels during the control period (Fig. S1). Therefore, for optimal visualization of donor-independent colon-region specific colonization, averages over the three units were calculated. This revealed that during the control period, the levels of microbial metabolic markers (SCFA, lactate and ammonium) were colon region-specific, with the levels of most parameters significantly increasing between subsequent colonic environments (Table 1). Lactate concentrations were very low.
Table 1.
Colon-region specific colonization of metabolic activity. Average concentration of acetate (mM), propionate (mM), butyrate (mM), lactate (mM), branched SCFA (mM) and ammonium (mg/L) in the ascending (AC), transverse (TC) and descending colon (DC) of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) during the entire control period for three different human donors (n = 6/donor). For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 18). Statistical differences between colon regions (AC vs TC vs DC) are indicated by assigning different letters to the respective colon regions (p < 0.05).
| AC | TC | DC | |
|---|---|---|---|
| Acetate (mM) | 12.2a ± 2.1 | 26.0b ± 2.0 | 30.1c ± 2.3 |
| Propionate (mM) | 2.7a ± 0.7 | 8.3b ± 0.7 | 9.4c ± 0.7 |
| Butyrate (mM) | 11.3a ± 0.9 | 12.7b ± 0.7 | 12.0c ± 0.5 |
| Lactate (mM) | 0.2a ± 0.1 | 0.4b ± 0.1 | 0.3 a,b ± 0.2 |
| Branched SCFA (mM) | 1.7a ± 0.1 | 2.2b ± 0.1 | 2.5c ± 0.1 |
| Ammonium (mg/L) | 169.5a ± 17.7 | 296.5b ± 27.1 | 354.6c ± 39.0 |
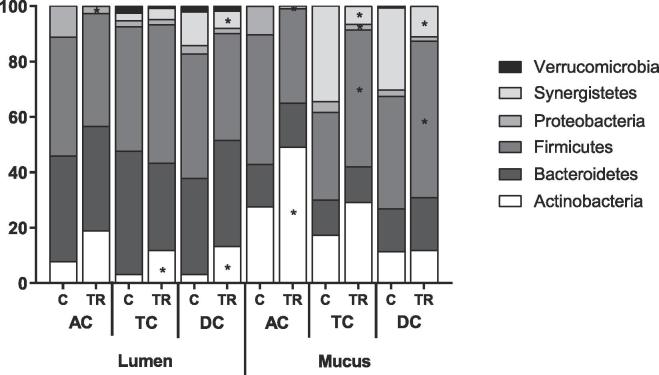
The 16S-targeted Illumina sequencing revealed that the main phyla in the microbial community across the three different donors were Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, Synergistetes and Verrucomicrobia. Families that were specifically enriched in the AC included Bifidobacteriaceae, Lactobacillaceae, Veillonellaceae and Enterobacteriaceae (Table 2). Several families within the Firmicutes phylum, e.g. Lachnospiraceae and Ruminococcaceae, specifically colonized the distal colon areas (TC and DC), which was also observed for the Rikenellaceae and Tannerellaceae family within the Bacteroidetes phylum. A similar tendency was observed for the Akkermansiaceae family, though not reaching statistical significance, mainly because donor 1 showed a much lower abundance of Akkermansiaceae (<1%) as compared to the other donors in the distal colon areas. Desulfovibrionaceae and Synergistetes specifically colonized the DC compartment. Finally, a phylum-specific colonization of the lumen versus the mucus layer was observed with higher levels of Actinobacteria in the mucosal environment as compared to the lumen, which was especially attributed to the significant mucosal enhancement of Bifidobacteriaceae in all colon compartments. The luminal environment was enriched in species from the Bacteroidetes phylum (reaching significance in the TC and DC), while significantly higher levels of Synergistetes were observed in the mucus layer.
Table 2.
Colon-region specific colonization of microbial community. Average abundance (%) of microbial families belonging to specific phyla in the luminal (L) and mucosal (M) environment of the ascending (AC), transverse (TC) and descending colon (DC) of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) at the end of the control period (n = 1/donor). For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 3). Statistical differences between colon regions (AC vs TC vs DC) are indicated by assigning different letters to the respective colon regions (small letters are used for the luminal environment, while capital letters are used for the mucosal environment). Statistical differences between the lumen and the mucus layer are indicated by their respective p-values in each of the colon regions. P-values < 0.05 are considered as statistically significant differences.
| Phylum | Family | Abundance (%) |
p-value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| L |
M |
L vs M |
||||||||
| AC | TC | DC | AC | TC | DC | AC | TC | DC | ||
| Actinobacteria | Bifidobacteriaceae | 7.8 a | 3.0 a | 3.0 a | 27.6 A | 16.6B | 10.3B | 0.006 | 0.006 | 0.024 |
| Coriobacteriaceae | 0.0 a | 0.1b | 0.1b | 0.0 A | 0.6B | 1.0B | 0.374 | 0.079 | 0.016 | |
| Bacteroidetes | Bacteroidaceae | 28.5 a | 37.6 a | 26.9 a | 13.4 A | 10.8 A | 12.8 A | 0.284 | 0.008 | 0.096 |
| Prevotellaceae | 9.2 a | 0.7 a | 0.2 a | 1.5 A,B | 0.1 A | 0.0B | 0.381 | 0.029 | 0.009 | |
| Rikenellaceae | 0.0 a | 0.1b | 0.2b | 0.0 A | 0.4B | 1.1C | – | 0.022 | 0.003 | |
| Tannerellaceae | 0.5 a | 6.2b | 7.4b | 0.2 A | 1.4B | 1.4B | 0.532 | 0.049 | 0.040 | |
| Firmicutes | Acidaminococcaceae | 0.1 a | 0.8b | 1.2 a,b | 0.0 A | 0.5B | 1.4B | 0.460 | 0.364 | 0.748 |
| Christensenellaceae | 0.0 a | 0.0 a | 0.0 a | 0.0 A | 0.0 A | 0.2B | – | 0.145 | 0.012 | |
| Clostridiaceae_1 | 0.0 a | 0.0 a | 0.0 a | 0.0 A | 0.1 A | 0.6B | 0.586 | 0.138 | 0.005 | |
| Erysipelotrichaceae | 0.0 a | 0.0 a | 0.1 a | 0.0 A | 0.1 A,B | 0.2B | – | 0.364 | 0.050 | |
| Eubacteriaceae | 0.0 a | 0.0 a | 0.1 a | 0.0 A | 0.1 A,B | 1.0B | – | 0.355 | 0.062 | |
| Lachnospiraceae | 11.0 a | 30.4b | 29.6b | 25.6 A | 24.5 A | 28.9 A | 0.117 | 0.217 | 0.889 | |
| Lactobacillaceae | 0.0 a | 0.0 a | 0.0 a | 2.2 A | 0.1B | 0.0C | 0.012 | 0.020 | 0.654 | |
| Ruminococcaceae | 0.0 a | 2.2 a | 2.2 a | 0.0 A | 1.0B | 4.6C | 0.463 | 0.543 | 0.161 | |
| Veillonellaceae | 31.9 a | 11.4b | 11.6b | 19.0 A | 5.3B | 3.4B | 0.069 | 0.094 | 0.019 | |
| Proteobacteria | Burkholderiaceae | 0.2 a,b | 0.2 a | 0.4b | 0.2 A | 0.3 A | 1.0 A | 0.903 | 0.182 | 0.143 |
| Desulfovibrionaceae | 0.0 a | 0.8b | 1.5c | 0.1 A | 2.9B | 0.8C | 0.251 | 0.004 | 0.035 | |
| Enterobacteriaceae | 10.3 a | 0.5b | 0.3b | 9.9 A | 0.3B | 0.1B | 0.924 | 0.222 | 0.009 | |
| Pseudomonadaceae | 0.7 a | 0.3 a,b | 0.2b | 0.1 A | 0.0B | 0.0B | 0.003 | 0.085 | 0.057 | |
| uncultured | 0.0 a | 0.4 a | 0.5 a | 0.0 A | 0.4 A | 0.3 A | – | 0.947 | 0.766 | |
| Synergistetes | Synergistaceae | 0.0 a | 2.8b | 12.1c | 0.0 A | 34.5B | 29.7B | 0.214 | 0.007 | 0.015 |
| Verrucomicrobia | Akkermansiaceae | 0.0 a | 2.5 a | 2.0 a | 0.0 A | 0.1 A | 0.7 A | – | 0.345 | 0.313 |
3.2. Altered microbial metabolic activity in response to synbiotic treatment
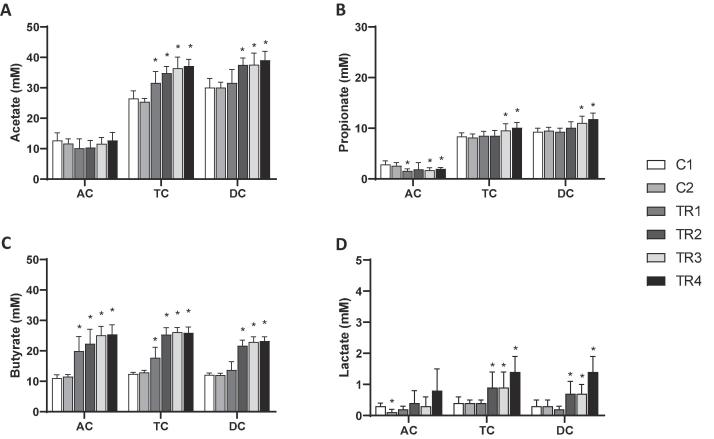
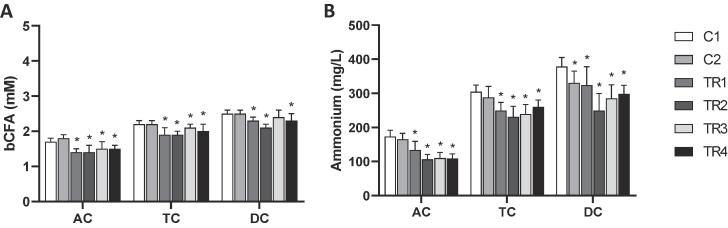
With respect to microbial SCFA production, consistent findings were made for the three different donors in response to the synbiotic treatment (Fig. S1) so that the averages of all parameters over the three donors were calculated per colon compartment for optimal visualization of donor-independent treatment effects. SCFA profiles mainly consisted of acetate, propionate and butyrate (Fig. 1) and trace amounts of branched SCFA (Fig. 2). Treatment with the synbiotic test product resulted in significantly higher acetate concentrations in the distal colon areas (TC and DC) as compared to the control period, i.e. an increase of 10.7 mM and 8.9 mM in the TC and DC respectively. As for propionate concentrations, the synbiotic test product resulted in significantly decreased levels in the AC. However, during the final two weeks of treatment, propionate levels significantly increased in the TC and DC as compared to the control period. Butyrate levels were strongly enhanced, reaching significantly higher levels in all colonic regions as compared to the control period. An average increase in butyrate concentrations of 14.4 mM in the AC, 13.6 mM in the TC and 11.1 mM in the DC was observed upon synbiotic supplementation. Despite the low levels of branched SCFA (Fig. 2), a significant reduction was detected in all colon areas upon treatment with the test product. Similar effects were observed on ammonium production (Fig. 2), with a significant average decrease of 65.3 mg/L in the AC, 44.4 mg/L in the TC and 80.5 mg/L in the DC upon synbiotic supplementation. Finally, upon treatment with the synbiotic test product lactate levels significantly increased in the distal colon areas (TC & DC) (Fig. 1). Furthermore, a trend towards increased lactate levels was observed during the final week of treatment in the AC (p = 0.061).
Fig. 1.
Effects on microbial metabolic activity. Average (±stdev) (A) acetate, (B) propionate, (C) butyrate and (D) lactate levels (mM) during the control (C1-C2; n = 3/donor) and the treatment (TR1-4; n = 3/donor) weeks in the ascending (AC), transverse (TC) and descending colon (DC) of the human gastro-intestinal tract for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented per week (n = 9). Statistically significant differences relative to the first control week are indicated with *(p < 0.05).
Fig. 2.
Effect on proteolytic markers. Average (±stdev) (A) branched SCFA (BCFA; mM) and (B) ammonium (mg/L) production during the control (C1-C2; n = 3/donor) and the treatment (TR1-4; n = 3/donor) weeks in the ascending (AC), transverse (TC) and descending colon (DC) of the human gastro-intestinal tract for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented per week (n = 9). Statistically significant differences relative to the first control week are indicated with *(p < 0.05).
3.3. Altered microbial composition in response to synbiotic treatment
Application of 16S-targeted Illumina sequencing demonstrated that the synbiotic test product increased the diversity of the gut microbiota across the three different donors in the distal colon areas (TC and DC), while a decreased diversity was observed in the AC (Table 3), reaching significance for the combined luminal and mucosal microbiota (p = 0.015 in AC, p = 0.044 in TC and p = 0.025 in DC).
Table 3.
Treatment effect of synbiotic formulation on microbial community composition at family level. Abundance (%) of microbial families level and the Reciprocal Simpson Diversity Index in the luminal and mucosal environment of the ascending (AC), transverse (TC) and descending colon (DC) of the human gastro-intestinal tract at the end of the control (C) and the treatment (TR) period upon treatment with the test product (n = 1/donor). For optimal observation of consistent effects over the different donors tested, the average of the three donors (D1/2/3) is presented (n = 3). Statistically significant differences between control and treatment (C vs TR), are indicated with *(p < 0.05).
| Phylum | Family | Average D1/2/3 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lumen |
Mucus |
||||||||||||
| AC |
TC |
DC |
AC |
TC |
DC |
||||||||
| CTRL | TR | CTRL | TR | CTRL | TR | CTRL | TR | CTRL | TR | CTRL | TR | ||
| Actinobacteria | Bifidobacteriaceae | 7.8 | 18.8 | 3.0 | 11.7* | 3.0 | 13.1* | 27.6 | 49.0* | 16.6 | 26.6 | 10.3 | 10.6 |
| Coriobacteriaceae | 0.0 | 0.0 | 0.1 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.6 | 2.2 | 1.0 | 1.1 | |
| Bacteroidetes | Bacteroidaceae | 28.5 | 1.0* | 37.6 | 16.1* | 26.9 | 21.4 | 13.4 | 1.2 | 10.8 | 9.2 | 12.8 | 14.3 |
| Prevotellaceae | 9.2 | 36.7 | 0.7 | 8.3* | 0.2 | 5.3* | 1.5 | 14.7 | 0.1 | 0.5* | 0.0 | 0.1* | |
| Rikenellaceae | 0.0 | 0.0 | 0.1 | 0.3 | 0.2 | 0.8 | 0.0 | 0.0 | 0.4 | 0.3 | 1.1 | 1.2 | |
| Tannerellaceae | 0.5 | 0.0 | 6.2 | 6.9 | 7.4 | 10.7 | 0.2 | 0.1 | 1.4 | 2.9 | 1.4 | 3.3* | |
| Firmicutes | Acidaminococcaceae | 0.1 | 0.1 | 0.8 | 1.6 | 1.2 | 1.2 | 0.0 | 0.1 | 0.5 | 0.9 | 1.4 | 1.3 |
| Bacillaceae | 0.0 | 1.0* | 0.0 | 0.2 | 0.0 | 0.2* | 0.0 | 0.4 | 0.0 | 0.2 | 0.0 | 0.2* | |
| Christensenellaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.4* | |
| Clostridiaceae_1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.1 | 0.1 | 0.6 | 0.4 | |
| Enterococcaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Erysipelotrichaceae | 0.0 | 0.0 | 0.0 | 0.5* | 0.1 | 0.5 | 0.0 | 0.0 | 0.1 | 0.6* | 0.2 | 1.0 | |
| Eubacteriaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 1.0 | 0.1 | |
| Lachnospiraceae | 11.0 | 8.0 | 30.4 | 19.4 | 29.6 | 16.3 | 25.6 | 6.1 | 24.5 | 31.0 | 28.9 | 46.0* | |
| Lactobacillaceae | 0.0 | 8.3 | 0.0 | 1.5 | 0.0 | 1.7 | 2.2 | 11.7 | 0.1 | 0.5 | 0.0 | 0.2 | |
| Ruminococcaceae | 0.0 | 0.0 | 2.2 | 16.1* | 2.2 | 10.2* | 0.0 | 0.0 | 1.0 | 8.3* | 4.6 | 3.3 | |
| Veillonellaceae | 31.9 | 23.5 | 11.4 | 10.7 | 11.6 | 8.2 | 19.0 | 15.4 | 5.3 | 7.8 | 3.4 | 3.6 | |
| Proteobacteria | Burkholderiaceae | 0.2 | 0.9 | 0.2 | 0.5* | 0.4 | 0.4 | 0.2 | 0.1 | 0.3 | 0.3 | 1.0 | 0.3 |
| Desulfovibrionaceae | 0.0 | 0.0 | 0.8 | 0.7 | 1.5 | 0.8* | 0.1 | 0.0 | 2.9 | 0.9* | 0.8 | 0.6 | |
| Enterobacteriaceae | 10.3 | 1.7* | 0.5 | 0.1* | 0.3 | 0.1* | 9.9 | 0.9* | 0.3 | 0.0 | 0.1 | 0.0* | |
| Pseudomonadaceae | 0.7 | 0.0* | 0.3 | 0.2 | 0.2 | 0.1 | 0.1 | 0.0* | 0.0 | 0.0 | 0.0 | 0.0 | |
| uncultured | 0.0 | 0.0 | 0.4 | 0.5 | 0.5 | 0.5 | 0.0 | 0.0 | 0.4 | 0.7 | 0.3 | 0.7 | |
| Synergistetes | Synergistaceae | 0.0 | 0.0 | 2.8 | 4.1 | 12.1 | 6.3* | 0.0 | 0.0 | 34.5 | 6.5* | 29.7 | 10.9* |
| Verrucomicrobia | Akkermansiaceae | 0.0 | 0.0 | 2.5 | 0.7 | 2.0 | 1.8 | 0.0 | 0.0 | 0.1 | 0.0 | 0.7 | 0.0 |
| Reciprocal Simpson Diversity Index | 5.9 | 4.1 | 12.4 | 16.2 | 12.3 | 15.9 | 6.9 | 4.6 | 7.0 | 14.2 | 8.8 | 13.5 | |
With respect to alteration of community composition, it firstly followed that the Bacillaceae family to which the probiotic strains belong increased in all colon areas, with the strongest effect being noted in the AC. At OTU level (Table 4), the stimulation of Bacillaceae was mainly related to an increase in Bacillaceae OTU 59, related to Bacillus subtilis. Further, in the AC, a significant increase in the Actinobacteria phylum (Fig. 3) was observed, which was mainly attributed to an increase in Bifidobacteriaceae at family level (Table 3). At OTU level (Table 4), the stimulation of Bifidobacteriaceae was caused by the non-significant increase of two Bifidobacterium OTUs, i.e. an OTU related to Bifidobacterium adolescentis and one related to Bifidobacterium bifidum. Additionally, a clear trend towards increased abundance of Lactobacillaceae was observed in the AC, which was attributed to an increase in an OTU related to Pediococcus acidilactici (p = 0.100 in lumen and p = 0.069 in mucus).
Table 4.
Treatment effect of synbiotic formulation on microbial community composition at OTU level. Abundance (%) at microbial OTU level in the luminal and mucosal environment of the ascending (AC), transverse (TC) and descending colon (DC) of the human gastro-intestinal tract at the end of the control (C) and the treatment (TR) period upon treatment with the test product (n = 1/donor). For optimal observation of consistent effects over the different donors tested, the average of the three donors (D1/2/3) is presented (n = 3). The 31 most abundant OTUs are presented together with the most abundant Bacillaceae OTU. Statistically significant differences between control and treatment (C vs TR), are indicated with* (p < 0.05).
| Phylum | Family | OTU | Related to | Average D1/2/3 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lumen |
Mucus |
||||||||||||||
| AC |
TC |
DC |
AC |
TC |
DC |
||||||||||
| CTRL | TR | CTRL | TR | CTRL | TR | CTRL | TR | CTRL | TR | CTRL | TR | ||||
| Actinobacteria | Bifidobacteriaceae | Otu0004 | Bifidobacterium adolescentis | 7.6 | 15.4 | 2.8 | 10.5* | 2.7 | 11.9 | 18.2 | 30.0 | 2.7 | 4.0 | 1.0 | 2.0 |
| Otu0005 | Bifidobacterium bifidum | 0.1 | 3.3 | 0.2 | 1.1 | 0.2 | 1.2* | 8.8 | 18.4 | 13.3 | 22.0 | 9.0 | 8.2 | ||
| Bacteroidetes | Bacteroidaceae | Otu0007 | Bacteroides fragilis | 8.9 | 0.0 | 10.6 | 0.4 | 7.5 | 0.4 | 4.4 | 0.1 | 2.1 | 1.4 | 3.3 | 1.5 |
| Otu0008 | Bacteroides dorei | 6.5 | 0.0 | 4.4 | 2.9 | 5.6 | 3.2 | 7.9 | 0.3 | 3.7 | 1.6 | 2.6 | 1.9 | ||
| Otu0009 | Bacteroides uniformis | 0.0 | 0.0 | 7.1 | 6.6 | 5.5 | 10.3 | 0.0 | 0.0 | 0.9 | 1.3 | 1.7 | 1.6 | ||
| Otu0015 | Bacteroides massiliensis | 6.3 | 0.8 | 2.5 | 1.9 | 1.7 | 2.1 | 0.3 | 0.7 | 0.5 | 2.4 | 0.6 | 4.5* | ||
| Otu0017 | Bacteroides intestinalis | 0.0 | 0.0 | 7.4 | 1.6 | 2.9 | 1.7 | 0.0 | 0.0 | 1.6 | 0.2 | 1.0 | 0.5 | ||
| Otu0019 | Bacteroides thetaiotaomicron | 0.0 | 0.0 | 1.7 | 0.8 | 1.4 | 1.4 | 0.0 | 0.0 | 0.9 | 1.1 | 2.0 | 2.8 | ||
| Otu0022 | Bacteroides ovatus | 4.7 | 0.1* | 2.0 | 0.6* | 1.1 | 0.6* | 0.6 | 0.1 | 0.2 | 0.2 | 0.2 | 0.2 | ||
| Prevotellaceae | Otu0006 | Prevotella copri | 9.1 | 36.6 | 0.7 | 8.3* | 0.2 | 5.3* | 1.5 | 14.5 | 0.1 | 0.5* | 0.0 | 0.1* | |
| Tannerellaceae | Otu0010 | Parabacteroides distasonis | 0.5 | 0.0 | 5.7 | 4.7 | 6.7 | 7.6 | 0.2 | 0.1* | 1.0 | 1.0 | 0.7 | 0.6 | |
| Otu0021 | Parabacteroides johnsonii | 0.0 | 0.0 | 0.2 | 1.7 | 0.3 | 2.6 | 0.0 | 0.0 | 0.4 | 1.7* | 0.5 | 2.5* | ||
| Firmicutes | Acidaminococcaceae | Otu0023 | Phascolarctobacterium faecium | 0.0 | 0.0 | 0.8 | 1.6 | 1.2 | 1.2 | 0.0 | 0.0 | 0.5 | 0.9 | 1.4 | 1.3 |
| Bacillaceae | Otu0059 | Bacillus subtilis | 0.0 | 0.9* | 0.0 | 0.2* | 0.0 | 0.2* | 0.0 | 0.3 | 0.0 | 0.1 | 0.0 | 0.1* | |
| Lachnospiraceae | Otu0003 | Clostridium clostridioforme | 10.4 | 1.0* | 14.6 | 2.9* | 14.3 | 2.4* | 23.9 | 1.5* | 11.9 | 5.2* | 9.6 | 3.5* | |
| Otu0011 | Clostridium oroticum | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 6.4 | 19.4* | ||
| Otu0012 | Blautia wexlerae | 0.3 | 0.1 | 2.5 | 3.6 | 2.0 | 2.7 | 0.8 | 0.2 | 4.4 | 6.0 | 1.3 | 2.3* | ||
| Otu0016 | Blautia faecis | 0.0 | 0.0 | 2.1 | 5.6 | 2.7 | 5.4 | 0.0 | 0.0 | 0.6 | 3.2* | 0.3 | 1.0* | ||
| Otu0020 | Ruminococcus torques | 0.0 | 0.0 | 0.4 | 0.6 | 0.8 | 0.7 | 0.0 | 0.0 | 0.4 | 1.3* | 1.4 | 6.1 | ||
| Otu0026 | Roseburia faecis | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.6 | 2.9* | 1.0 | 3.0 | ||
| Otu0028 | Lachnoclostridium sp. | 0.1 | 2.6 | 0.3 | 1.6 | 0.1 | 0.9 | 0.1 | 4.1 | 0.1 | 0.2 | 0.0 | 0.1 | ||
| Otu0029 | Lachnospiraceae bacterium | 0.0 | 0.0 | 2.0 | 0.7* | 1.9 | 0.5* | 0.0 | 0.0 | 0.7 | 0.4 | 0.3 | 0.1 | ||
| Otu0030 | Roseburia inulinivorans | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.6 | 1.0 | 1.5 | ||
| Otu0031 | Clostridium scindens | 0.0 | 0.0 | 0.0 | 0.3 | 0.2 | 0.2 | 0.0 | 0.0 | 0.3 | 1.3 | 1.0 | 2.1 | ||
| Lactobacillaceae | Otu0014 | Pediococcus acidilactici | 0.0 | 8.2 | 0.0 | 1.5 | 0.0 | 1.7 | 2.2 | 11.6 | 0.1 | 0.5 | 0.0 | 0.2 | |
| Ruminococcaceae | Otu0013 | Subdoligranulum sp. | 0.0 | 0.0 | 1.7 | 9.6 | 1.2 | 6.1* | 0.0 | 0.0 | 0.2 | 5.1* | 0.2 | 1.5* | |
| Otu0027 | Faecalibacterium prausnitzii | 0.0 | 0.0 | 0.1 | 3.5 | 0.1 | 1.9 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 0.5 | ||
| Otu0038 | Faecalibacterium prausnitzii | 0.0 | 0.0 | 0.2 | 2.4* | 0.1 | 1.4* | 0.0 | 0.0 | 0.0 | 1.0* | 0.0 | 0.1* | ||
| Veillonellaceae | Otu0001 | Megasphaera sp. | 28.3 | 21.8 | 10.5 | 10.2 | 11.0 | 7.9 | 10.2 | 13.8 | 3.6 | 6.6 | 1.7 | 2.3 | |
| Otu0025 | Veillonella ratti | 2.6 | 1.3 | 0.4 | 0.2 | 0.2 | 0.1 | 1.5 | 1.3 | 0.7 | 0.0 | 0.1 | 0.0 | ||
| Enterobacteriaceae | Otu0018 | Enterobacter sp. | 7.5 | 1.4 | 0.3 | 0.1 | 0.2 | 0.1 | 9.1 | 0.5* | 0.2 | 0.0 | 0.0 | 0.0 | |
| Synergistetes | Synergistaceae | Otu0002 | Cloacibacillus evryensis | 0.0 | 0.0 | 2.7 | 3.8 | 12.0 | 6.0* | 0.0 | 0.0 | 32.7 | 5.4* | 29.4 | 10.6* |
| Verrucomicrobia | Akkermansiaceae | Otu0024 | Akkermansia muciniphila | 0.0 | 0.0 | 2.5 | 0.7 | 2.0 | 1.8 | 0.0 | 0.0 | 0.1 | 0.0 | 0.7 | 0.0 |
Fig. 3.
Microbial community composition as assessed via 16S-targeted Illumina sequencing. Abundance (%) at microbial phylum level in the luminal and mucosal environment of the ascending (AC), transverse (TC) and descending colon (DC) of the human gastro-intestinal tract at the end of the control (C; n = 1/donor) and the treatment (TR; n = 1/donor) period upon treatment with the test product for three human donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 3). Statistically significant differences relative to the control period are indicated with *(p < 0.05).
Within the Bacteroidetes phylum, a strong increase in Prevotellaceae was observed in the AC upon supplementation of the synbiotic test product, reaching significance in the distal parts of the colon (TC and DC). The stimulation of the Prevotellaceae family was mainly at the expense of the Bacteroidaceae family, which significantly decreased in abundance in the lumen of the AC and TC. As for other bacterial groups belonging to Bacteroidetes and containing propionate-producing species, such as Rikenellaceae and Tannerellaceae, a specific stimulation was detected in the TC and DC. For instance, at OTU level, a significant increase was observed in the mucosal environment of the TC and DC for an OTU related to Parabacteroides johnsonii.
Another consistent effect upon treatment with the synbiotic test product was a decrease in several families within the Proteobacteria and Synergistetes phyla. Furthermore, it was found that treatment with the synbiotic test product stimulated a wide spectrum of groups containing potential butyrate-producing species, such as Erysipelotrichaceae, Lachnospiraceae and Ruminococcaceae, with a significant increase being observed in the abundance of an OTU related to Faecalibacterium prausnitzii.
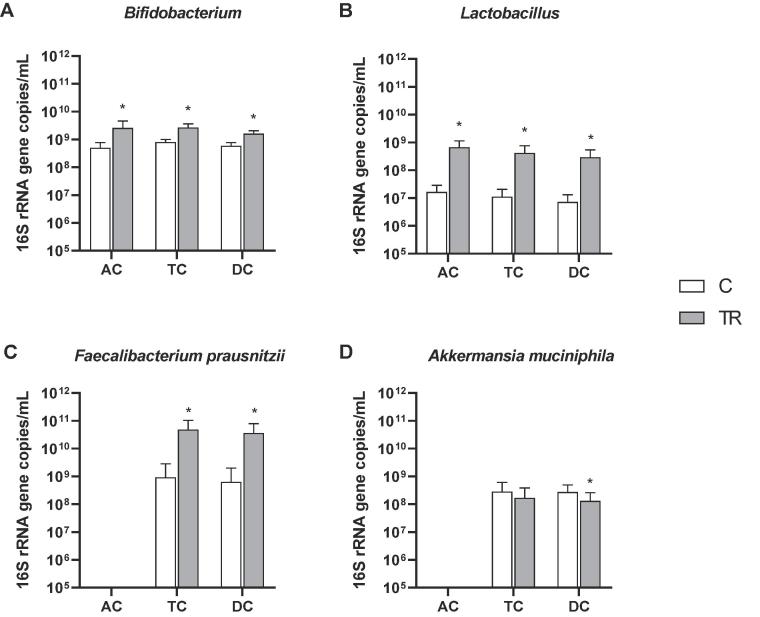
To confirm some of the aforementioned treatment effects of the synbiotic formulation on specific taxonomic groups of interest (Bifidobacterium spp., Lactobacillus spp., Faecalibacterium prausnitzii and Akkermansia muciniphila), a quantitative approach, i.e. qPCR analysis was performed (Fig. 4 for luminal environment and Fig. S2 for mucosal environment). Across the three different donors, significant increases of Bifidobacterium spp. and Lactobacillus spp. were observed in all colonic areas in both the luminal and mucosal environment, except for Bifidobacterium spp. in the mucosal environment of the AC where a trend towards increased concentrations was observed (p = 0.057). Additionally, significantly increased levels of Faecalibacterium prausnitzii were observed in the lumen of the TC and DC, while in the simulated mucus layer increased Faecalibacterium prausnitzii concentrations were detected in all colonic areas, reaching significance in the TC. As for Akkermansia muciniphila levels, a significant reduction was detected in the DC of both the luminal and mucosal environment across the three different donors.
Fig. 4.
Luminal microbial community composition as assessed via qPCR. Average (±stdev) (A) Bifidobacterium, (B) Lactobacillus, (C) Faecalibacterium prausnitzii and (D) Akkermansia muciniphila levels (16S rRNA gene copies/mL) over the entire control (C; n = 2/donor) and treatment (TR; n = 4/donor) period in the luminal environment of the ascending (AC), transverse (TC) and descending colon (DC) of the human gastro-intestinal tract for three donors tested. For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 6 for C; n = 12 for TR). Statistically significant differences between the control and treatment period are indicated with *(p < 0.05).
4. Discussion
In the present study, the effect of prolonged administration of a synbiotic formulation, containing five spore-forming Bacillus strains and a blend of FOS, GOS and XOS, on the human microbiome was assessed for three different human individuals using the validated in vitro M-SHIME® model (Van den Abbeele et al., 2012). A unique characteristic of the SHIME® model is that it allows to perform mechanistic research on an intestinal microbial community that, prior to treatment, is fully stable so that treatment effects are not obscured by natural variation in gut microbiome composition and functionality as is the case in vivo (Liu et al., 2018, Possemiers et al., 2004). By applying a novel analysis method that allowed to characterize microbiota composition at high phylogenetic resolution, i.e. 16S-targeted sequencing of the V3-V4 hypervariable regions (generating amplicons of ~425 base pairs), the current study confirmed that the M-SHIME® model maintained a diverse and representative, colon region-specific luminal and mucosal microbial community in its three consecutive colon regions. The AC was specifically enriched with members of the Bifidobacteriaceae, Lactobacillaceae, Veillonellaceae and Enterobacteriaceae, which contain several species involved in primary substrate degradation. The establishment of a saccharolytic microbial community in the AC confirms findings of Van den Abbeele et al. (2010). While the AC community was physically transferred to the distal colon (TC and DC), these regions displayed higher abundance of microbial groups with specific metabolic functions (Van den Abbeele et al., 2010), such as propionate-producing Tannerellaceae and butyrate-producing Lachnospiraceae and Ruminococcaceae. Furthermore, the TC and DC were characterized by a specific colonization of members of Akkermansiaceae and Synergistaceae. Recently, Akkermansia muciniphila, the only representative of the Akkermansiaceae family in the human gut, has been shown to degrade mucins in the distal colon while virtually being absent in the AC (Van Herreweghen et al., 2017). Synergistaceae on the other hand are known for their proteolytic fermentation, which is generally favoured in distal colon areas when sugars are depleted (Bhandari and Gupta, 2012, Liu et al., 2018). Besides the longitudinal differences in microbial community composition, a phylum-specific colonization of the lumen versus the mucus layer was observed. It was previously reported that the mucosal environment in the M-SHIME® model is specifically colonized by members of the Firmicutes phylum, resulting in enhanced butyrate levels as compared to conventional in vitro gut models (Liu et al., 2018, Van den Abbeele et al., 2013). This was confirmed in the current study given the mucosal enrichment of Lachnospiraceae (in the AC), Clostridiaceae (in the DC) and Erysipelotrichaceae (in the DC) (Liu et al., 2018). Further, also Actinobacteria were enriched in the mucosal environment of all colon regions, especially ascribed to increased levels of Bifidobacteriaceae. On the other hand, the luminal environment was characterized by higher levels of Bacteroidetes species (only significant in TC and DC), again confirming previous findings (Liu et al., 2018, Van den Abbeele et al., 2013), while Synergistetes specifically colonized the mucus layer (Liu et al., 2018). The maintenance of a highly diverse and representative colon-region specific microbiota in the SHIME® model provided an excellent platform for doing mechanistic research to evaluate the effect of the synbiotic treatment.
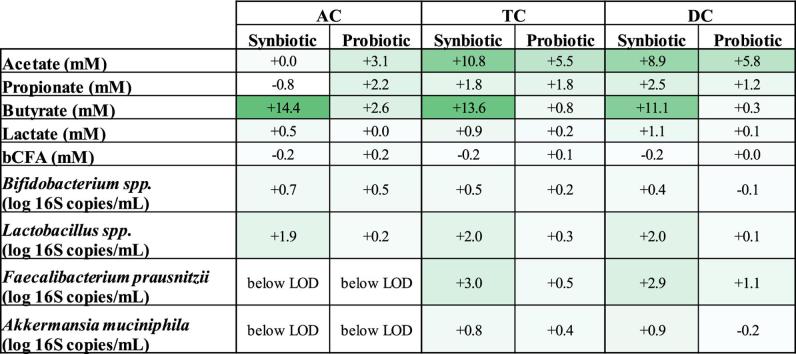
In contrast with recent observations done by our research group upon testing the repeated daily administration of the same probiotic strains in a similar in vitro study (Duysburgh et al., 2019), treatment with the synbiotic test product in the current study resulted in a consistent stronger increase of acetate levels in the TC and DC, propionate levels in the DC as well as butyrate and lactate concentrations along the entire colon (Table 5). While for the probiotic formulation a butyrogenic effect was only shown for one of the donors tested, treatment with the synbiotic test product in the current study resulted in a consistently strong increase in butyrate levels for all donors tested, resulting in an average increase that was 11.7 mM, 12.8 mM and 10.8 mM higher as compared to probiotic administration in the AC, TC and DC respectively. At microbial community level, this was mainly attributed to significant increases in Faecalibacterium prausnitzii levels in the distal colon areas, which were not observed upon probiotic supplementation (Table 5). Furthermore, in the present study a significant increase in Lactobacillus spp. was observed in the AC upon synbiotic administration, while this effect was absent when testing the probiotic blend alone (Table 5). Overall, these observations stress the fact that combining functional fibers with probiotic strains could be a useful strategy in modulating the microbial community, as they might selectively stimulate different microbial groups.
Table 5.
Comparison of probiotic and synbiotic treatment. Average increase in acetate (mM), propionate (mM), butyrate (mM), lactate (mM), branched SCFA (mM), Bifidobacterium (log 16S rRNA gene copies/mL), Lactobacillus (log 16S rRNA gene copies/mL), Faecalibacterium prausnitzii (log 16S rRNA gene copies/mL) and Akkermansia muciniphila (log 16S rRNA gene copies/mL) levels in the ascending (AC), transverse (TC) and descending colon (DC) of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) at the end of the treatment period with the probiotic and synbiotic formulation as compared to their respective control period for three different human donors (n = 3/donor). For optimal observation of consistent effects over the different donors tested, the average of the three donors is presented (n = 9). Results from the probiotic formulation were retrieved from a previous study performed by our research group (Duysburgh et al., 2019). Statistically significant differences between the probiotic and the synbiotic treatment group, are indicated with bold (p < 0.05).
| AC |
TC |
DC |
||||
|---|---|---|---|---|---|---|
| Synbiotic | Probiotic | Synbiotic | Probiotic | Synbiotic | Probiotic | |
| Acetate (mM) | +0.02 | +3.13 | +10.75 | +5.48 | +8.94 | +5.84 |
| Propionate (mM) | −0.83 | +2.16 | +1.75 | +1.75 | +2.49 | +1.23 |
| Butyrate (mM) | +14.36 | +2.62 | +13.58 | +0.80 | +11.08 | +0.33 |
| Lactate (mM) | +0.51 | −0.04 | +0.93 | +0.18 | +1.09 | +0.11 |
| Branched SCFA (mM) | −0.23 | +0.21 | −0.19 | +0.08 | −0.22 | +0.01 |
|
Bifidobacterium spp. (log 16S copies/mL) |
+0.66 | +0.49 | +0.48 | +0.16 | +0.38 | −0.07 |
|
Lactobacillus spp. (log 16S copies/mL) |
+1.89 | +0.23 | +2.04 | +0.26 | +2.02 | +0.14 |
|
Faecalibacterium prausnitzii (log 16S copies/mL) |
below LOD | below LOD | +2.96 | +0.49 | +2.85 | +1.11 |
|
Akkermansia muciniphila (log 16S copies/mL) |
below LOD | below LOD | +0.76 | +0.37 | +0.90 | −0.18 |
With respect to the synbiotic treatment, it was observed that the family to which the five spore-forming strains of the product belong to, i.e. Bacillaceae, significantly increased in the AC upon supplementation, suggesting a successful engraftment in the colonic microbiota that could have resulted in the microbial modulation that was subsequently observed. Firstly, the synbiotic treatment increased microbial diversity in the distal colon compartments (TC and DC), which is considered as an important feature for the improvement of gut health, whereas decreased diversity was observed in the AC. Prebiotic supplementation can indeed result in reduced bacterial diversity in the proximal colon mainly due to stimulation of specific microbial groups at the first site of fermentation (Zhang et al., 2015). The specific treatment effect considered a strong increase in Actinobacteria in the AC, which was mainly attributed to a mucosal increase in Bifidobacteriaceae. Bifidobacterial stimulation has extensively been reported upon supplementation of prebiotic compounds like FOS (Meyer and Stasse-Wolthuis, 2009), GOS (Depeint et al., 2008) and XOS (Makelainen et al., 2010). The strong bifidogenic effect might have led to good cross-feeding interactions with other resident microbiota resulting in increased levels of propionate and mostly butyrate (Moens et al., 2016). While acetate is a key metabolite of Bifidobacterium species, acetate was not increased in the AC given its likely efficient conversion to butyrate. Butyrate is a major energy source for the colonic epithelium and exerts several health-promoting properties such as anti-inflammatory activity, anti-cancer effects, promotion of satiety and reduction of oxidative stress (Hamer et al., 2008). In the distal colon, this butyrogenic effect was mainly associated with a significant increase in Faecalibacterium prausnitzii levels. Recently it has been shown that consumption of FOS from gold kiwifruit, as used in the present study, significantly increased Faecalibacterium prausnitzii levels in functionally constipated individuals (Blatchford et al., 2017). Faecalibacterium prausnitzii exerts strong anti-inflammatory activity in the intestinal environment which is mainly linked with the production of butyrate stimulation of regulatory T-cells (Furusawa et al., 2013, Sokol et al., 2008). Furthermore, increased Faecalibacterium prausnitzii levels have been associated with reduction of inflammatory markers (Furet et al., 2010) and reduction of endotoxemia in obese subjects (Gonzalez-Sarrias et al., 2018). Interestingly, McFarlin et al. reported that the same probiotic formulation as the one that was used during the current study decreased post-prandial metabolic endotoxemia in human subjects (McFarlin et al., 2017), thereby suggesting a possible role of microbial intervention in tackling obesity-related disorders.
Besides a stimulation of butyrate, also a significant increase in acetate, lactate and propionate levels was observed in the distal colon regions upon synbiotic supplementation over the three donors tested. At community level, this was linked with increased abundance of an OTU related to Prevotella copri (as part of the Prevotellaceae family) (Hayashi et al., 2007) and one related to Parabacteroides johnsonii (as part of the Tannerellaceae family) (Sakamoto et al., 2007), which both produce acetate and succinate as their major metabolic end-products, with succinate being able to be further converted to propionate by succinate-converting, propionate producing micro-organisms such as Bacteroides and Veillonella species (Hosseini et al., 2011). Indeed, in the present study, increased abundance of an OTU related to Bacteroides massiliensis was observed in the mucosal DC, which might have contributed to the increased propionate levels that were observed towards the end of the treatment period.
From this study, it can be concluded that the synbiotic formulation, containing a mixture of five spore-forming Bacillus strains and a prebiotic blend of FOS, GOS and XOS, consistently affected microbial activity and composition in the human gastrointestinal tract in vitro, with profound effects being observed on colonic butyrate production. Moreover, when compared with a previous study investigating the impact of the probiotic strains on colonic functionality, the generated data suggest a synergistic effect on the gut microbiome for the synbiotic test product. Given the fact that the probiotic formulation has already been shown to impact metabolic endotoxemia in human individuals, it might be interesting to further investigate the efficacy of the synbiotic formulation in tackling metabolic disorders.
Acknowledgments
Acknowledgement
The work was financially supported by Microbiome Labs.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
Kiran Krishnan and Thomas F. Bayne are employees of Microbiome Labs.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpx.2019.100021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bent S.J., Forney L.J. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2008;2:689–695. doi: 10.1038/ismej.2008.44. [DOI] [PubMed] [Google Scholar]
- Bhandari, V., Gupta, R., 2012. Molecular signatures for the phylum Synergistetes and some of its subclades. [DOI] [PubMed]
- Blatchford P., Stoklosinski H., Eady S., Wallace A., Butts C., Gearry R., Gibson G., Ansell J. Consumption of kiwifruit capsules increases Faecalibacterium prausnitzii abundance in functionally constipated individuals: a randomised controlled human trial. J. Nutr. Sci. 2017;6 doi: 10.1017/jns.2017.52. e52–e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon N., Top E.M., Verstraete W., Siciliano S.D. Bioaugmentation as a tool to protect the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl. Environ. Microbiol. 2003;69:1511–1520. doi: 10.1128/AEM.69.3.1511-1520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartman S.T., La Ragione R.M., Woodward M.J. Bacillus subtilis spores germinate in the chicken gastrointestinal tract. Appl. Environ. Microbiol. 2008;74:5254–5258. doi: 10.1128/AEM.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casula G., Cutting S.M. Bacillus probiotics: spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 2002;68:2344–2352. doi: 10.1128/AEM.68.5.2344-2352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M.C., Derrien M., Isolauri E., de Vos W.M., Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weirdt R., Possemiers S., Vermeulen G., Moerdijk-Poortvliet T.C., Boschker H.T., Verstraete W., Van de Wiele T. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol. Ecol. 2010;74:601–611. doi: 10.1111/j.1574-6941.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- Depeint F., Tzortzis G., Vulevic J., I'Anson K., Gibson G.R. Prebiotic evaluation of a novel galactooligosaccharide mixture produced by the enzymatic activity of Bifidobacterium bifidum NCIMB 41171, in healthy humans: a randomized, double-blind, crossover, placebo-controlled intervention study. Am. J. Clin. Nutr. 2008;87:785–791. doi: 10.1093/ajcn/87.3.785. [DOI] [PubMed] [Google Scholar]
- Duysburgh, C., Van den Abbeele, P., Krishnan, K., Bayne, T.F., Marzorati, M., 2019. A mix of spore-forming probiotic Bacillus strains exerts inter-individual effects on microbial metabolism and composition in the human distal colon. Under review.
- Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., Guiot Y., Derrien M., Muccioli G.G., Delzenne N.M., de Vos W.M., Cani P.D. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. PNAS. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farid R., Ahanchian H., Jabbari F., Moghiman T. Effect of a new synbiotic mixture on atopic dermatitis in children: a randomized-controlled trial. Iran. J. Pediatr. 2011;21:225–230. [PMC free article] [PubMed] [Google Scholar]
- Fujimori S., Gudis K., Mitsui K., Seo T., Yonezawa M., Tanaka S., Tatsuguchi A., Sakamoto C. A randomized controlled trial on the efficacy of synbiotic versus probiotic or prebiotic treatment to improve the quality of life in patients with ulcerative colitis. Nutrition (Burbank, Los Angeles County, Calif.) 2009;25:520–525. doi: 10.1016/j.nut.2008.11.017. [DOI] [PubMed] [Google Scholar]
- Furet J.-P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., Doré J., Corthier G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009;68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x. [DOI] [PubMed] [Google Scholar]
- Furet J.P., Kong L.C., Tap J., Poitou C., Basdevant A., Bouillot J.L., Mariat D., Corthier G., Dore J., Henegar C., Rizkalla S., Clement K. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N.N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J.M., Topping D.L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., Salminen S.J., Scott K., Stanton C., Swanson K.S., Cani P.D., Verbeke K., Reid G. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sarrias A., Romo-Vaquero M., Garcia-Villalba R., Cortes-Martin A., Selma M.V., Espin J.C. The endotoxemia marker lipopolysaccharide-binding protein is reduced in overweight-obese subjects consuming pomegranate extract by modulating the gut microbiota: a randomized clinical trial. Mol. Nutr. Food Res. 2018;62 doi: 10.1002/mnfr.201800160. [DOI] [PubMed] [Google Scholar]
- Hamer H.M., Jonkers D., Venema K., Vanhoutvin S., Troost F.J., Brummer R.J. Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Shibata K., Sakamoto M., Tomita S., Benno Y. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2007;57:941–946. doi: 10.1099/ijs.0.64778-0. [DOI] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., Calder P.C., Sanders M.E. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- Hong H.A., Duc le H., Cutting S.M. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 2005;29:813–835. doi: 10.1016/j.femsre.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Hosseini E., Grootaert C., Verstraete W., Van de Wiele T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 2011;69:245–258. doi: 10.1111/j.1753-4887.2011.00388.x. [DOI] [PubMed] [Google Scholar]
- Ipar N., Aydogdu S.D., Yildirim G.K., Inal M., Gies I., Vandenplas Y., Dinleyici E.C. Effects of synbiotic on anthropometry, lipid profile and oxidative stress in obese children. Beneficial Microb. 2015;6:775–782. doi: 10.3920/BM2015.0011. [DOI] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glockner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucl. Acids Res. 2013;41 doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Firrman J., Tanes C., Bittinger K., Thomas-Gahring A., Wu G.D., Van den Abbeele P., Tomasula P.M. Establishing a mucosal gut microbial community in vitro using an artificial simulator. PLoS One. 2018;13 doi: 10.1371/journal.pone.0197692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makelainen H., Forssten S., Saarinen M., Stowell J., Rautonen N., Ouwehand A.C. Xylo-oligosaccharides enhance the growth of bifidobacteria and Bifidobacterium lactis in a simulated colon model. Beneficial Microb. 2010;1:81–91. doi: 10.3920/BM2009.0025. [DOI] [PubMed] [Google Scholar]
- McFarlin B.K., Henning A.L., Bowman E.M., Gary M.A., Carbajal K.M. Oral spore-based probiotic supplementation was associated with reduced incidence of post-prandial dietary endotoxin, triglycerides, and disease risk biomarkers. World J. Gastrointest. Pathophysiol. 2017;8:117–126. doi: 10.4291/wjgp.v8.i3.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D., Stasse-Wolthuis M. The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur. J. Clin. Nutr. 2009;63:1277–1289. doi: 10.1038/ejcn.2009.64. [DOI] [PubMed] [Google Scholar]
- Moens F., Weckx S., De Vuyst L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 2016;231:76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Molly K., Vande Woestyne M., Verstraete W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- Pandey K.R., Naik S.R., Vakil B.V. Probiotics, prebiotics and synbiotics- a review. J. Food Sci. Technol. 2015;52:7577–7587. doi: 10.1007/s13197-015-1921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemiers S., Verthe K., Uyttendaele S., Verstraete W. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 2004;49:495–507. doi: 10.1016/j.femsec.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Rhee K.J., Sethupathi P., Driks A., Lanning D.K., Knight K.L. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J. Immunol. (Baltimore, Md. 1950) 2004;172:1118–1124. doi: 10.4049/jimmunol.172.2.1118. [DOI] [PubMed] [Google Scholar]
- Rinttilä T., Kassinen A., Malinen E., Krogius L., Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- Safavi M., Farajian S., Kelishadi R., Mirlohi M., Hashemipour M. The effects of synbiotic supplementation on some cardio-metabolic risk factors in overweight and obese children: a randomized triple-masked controlled trial. Int. J. Food Sci. Nutr. 2013;64:687–693. doi: 10.3109/09637486.2013.775224. [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Kitahara M., Benno Y. Parabacteroides johnsonii sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2007;57:293–296. doi: 10.1099/ijs.0.64588-0. [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeir J., de Vrese M. Probiotics, prebiotics, and synbiotics—approaching a definition. Am. J. Clin. Nutr. 2001;73:361s–364s. doi: 10.1093/ajcn/73.2.361s. [DOI] [PubMed] [Google Scholar]
- Shakeri H., Hadaegh H., Abedi F., Tajabadi-Ebrahimi M., Mazroii N., Ghandi Y., Asemi Z. Consumption of synbiotic bread decreases triacylglycerol and VLDL levels while increasing HDL levels in serum from patients with type-2 diabetes. Lipids. 2014;49:695–701. doi: 10.1007/s11745-014-3901-z. [DOI] [PubMed] [Google Scholar]
- J. Funct. Foods. 2016;24:549–557. [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L.G., Gratadoux J.J., Blugeon S., Bridonneau C., Furet J.P., Corthier G., Grangette C., Vasquez N., Pochart P., Trugnan G., Thomas G., Blottiere H.M., Dore J., Marteau P., Seksik P., Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. PNAS. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Seksik P., Furet J.P., Firmesse O., Nion-Larmurier I., Beaugerie L., Cosnes J., Corthier G., Marteau P., Dore J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Sumi H., Yatagai C., Wada H., Yoshida E., Maruyama M. [Effect of Bacillus natto-fermented product (BIOZYME) on blood alcohol, aldehyde concentrations after whisky drinking in human volunteers, and acute toxicity of acetaldehyde in mice]. Arukoru kenkyu to yakubutsu izon = Japan. J. Alcohol Stud. Drug Dependence. 1995;30:69–79. [PubMed] [Google Scholar]
- Tam N.K., Uyen N.Q., Hong H.A., Duc le H., Hoa T.T., Serra C.R., Henriques A.O., Cutting S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006;188:2692–2700. doi: 10.1128/JB.188.7.2692-2700.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wiele T., Van den Abbeele P., Ossieur W., Possemiers S., Marzorati M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) In: Verhoeckx K., Cotter P., López-Expósito I., Kleiveland C., Lea T., Mackie A., Requena T., Swiatecka D., Wichers H., editors. The Impact of Food Bioactives on Health: in vitro and ex vivo models. Springer International Publishing; Cham: 2015. pp. 305–317. [PubMed] [Google Scholar]
- Van den Abbeele P., Belzer C., Goossens M., Kleerebezem M., De Vos W.M., Thas O., De Weirdt R., Kerckhof F.M., Van de Wiele T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P., Grootaert C., Marzorati M., Possemiers S., Verstraete W., Gerard P., Rabot S., Bruneau A., El Aidy S., Derrien M., Zoetendal E., Kleerebezem M., Smidt H., Van de Wiele T. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 2010;76:5237–5246. doi: 10.1128/AEM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P., Roos S., Eeckhaut V., MacKenzie D.A., Derde M., Verstraete W., Marzorati M., Possemiers S., Vanhoecke B., Van Immerseel F., Van de Wiele T. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 2012;5:106–115. doi: 10.1111/j.1751-7915.2011.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herreweghen F., Van den Abbeele P., De Mulder T., De Weirdt R., Geirnaert A., Hernandez-Sanabria E., Vilchez-Vargas R., Jauregui R., Pieper D.H., Belzer C., De Vos W.M., Van de Wiele T. In vitro colonisation of the distal colon by Akkermansia muciniphila is largely mucin and pH dependent. Beneficial Microb. 2017;8:81–96. doi: 10.3920/BM2016.0013. [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Yin A., Li H., Wang R., Wu G., Shen J., Zhang M., Wang L., Hou Y., Ouyang H., Zhang Y., Zheng Y., Wang J., Lv X., Wang Y., Zhang F., Zeng B., Li W., Yan F., Zhao Y., Pang X., Zhang X., Fu H., Chen F., Zhao N., Hamaker B.R., Bridgewater L.C., Weinkove D., Clement K., Dore J., Holmes E., Xiao H., Zhao G., Yang S., Bork P., Nicholson J.K., Wei H., Tang H., Zhang X., Zhao L. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine. 2015;2:968–984. doi: 10.1016/j.ebiom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.