Abstract
We describe the protocol development and optimization of asymmetric flow field-flow fractionation (AF4) technology for separating and characterizing extracellular nanoparticles (ENPs), particularly small extracellular vesicles, known as exosomes, and even smaller novel nanoparticles, known as exomeres. This technique fractionates ENPs based on hydrodynamic sizes and demonstrates a unique capability to separate nanoparticles with sizes ranging from a few nanometers to undefined level of micrometers. ENPs are resolved by two perpendicular flows, channel flow and cross flow, in a flat thin channel with a semi-permissive bottom wall membrane. The AF4 separation method offers several advantages over other isolation methods for ENP analysis, including being label-free, gentle, rapid (< 1 hour), and highly reproducible, and providing efficient recovery of analytes. Most importantly, in contrast to other available techniques, AF4 can separate ENPs at high resolution (1 nm) and provide a large dynamic range of size-based separation. In conjunction with real-time monitors, such as ultraviolet absorbance and dynamic light scattering, and an array of post-separation characterizations, AF4 facilitates the successful separation of distinct subsets of exosomes and the identification of exomeres. Though the whole procedure of cell culture and ENP isolation from the conditioned media by ultracentrifugation can take approximately three days, the AF4 fractionation step takes only one hour to perform. Users of this technology will require expertise in the working principle of AF4 to operate and customize protocol applications. AF4 can contribute to the development of high-quality, exosome- and exomere-based molecular diagnostics and therapeutics.
Keywords: asymmetric-flow field-flow fractionation (AF4), Exosomes, exomeres, extracellular vesicles
INTRODUCTION
Cells secrete a wide variety of soluble factors and extracellular vesicles (EVs) to mediate intercellular communication (locally and systemically) under both physiological and pathological conditions, including cancers1–3. EVs are heterogeneous and comprise various subclasses, including exosomes, which are extracellular membrane vesicles of endosomal origin and whose size usually falls within the range of 50 nm to 150 nm, and microvesicles, which are large vesicles shed directly by budding from the cellular plasma membrane and whose size commonly falls within the range of 150 nm to 500 nm or even larger to >10μm. Cancer cells shed atypically large vesicles, known as large oncosomes (0.5 μm to 10 μm) which result from alterations in specific signaling pathways (e.g. Ras Homolog Family Member A/Rho-associated protein kinase (RhoA/Rock) signaling)4–6. Extensive research has shown that functional molecules, including proteins, genetic material, metabolites and lipids, are selectively recruited and packaged into EVs and horizontally transferred to recipient cells, thereby acting as vehicles of intercellular communication7–10. In addition, through our recent work, we have identified a novel population of non-membranous nanoparticles termed ‘exomeres’ (~35 nm), which are the predominant extracellular nanoparticles (ENPs) (including both exomeres and EVs) secreted by most types of cells11.
Although our knowledge of the biology, function and translational potential of ENPs is rapidly expanding, the heterogeneous nature of these nanoparticles and technical limitations in efficiently separating ENP subpopulations have hindered the characterization of their molecular composition and dissection of the pathways involved in their biogenesis. Nonetheless, it is evident that the differences in size, surface molecules and chemistry, and mechanical properties among ENP subpopulations all collectively contribute to their biodistribution and systemic functions in vivo. Intensive translational research efforts have focused on identifying potential ENP-based diagnostic/prognostic biomarkers as well as on developing ENP-based therapeutic strategies12–18. More advanced characterization of molecular signatures associated with each subset of ENPs will facilitate these areas of research.
To date, various strategies have been developed in an attempt to isolate pure EV subpopulations, especially exosomes, including differential ultracentrifugation (dUC), immuno-affinity capture (IAC), ultrafiltration (UF) and size-exclusion chromatography (SEC), polymer-based precipitation, and microfluidics19–24. However, the purity, yield and integrity of isolated exosomes as well as the labor- and time-efficiency remain to be improved. Asymmetric flow field-flow fractionation (AF4) exhibits unique capabilities to separate nanoparticles and has been widely utilized to characterize nanoparticles and polymeric nanoparticles in the pharmaceutical industry as well as biological macromolecules, protein complexes and viruses25, 26. In this protocol, we provide a detailed description of the method development and optimization of AF4 for characterizing and separating exomeres and exosome subpopulations used in our previously published work11.
Overview of the procedure and development of the AF4 protocol
The technique of field-flow fraction (FFF) was first developed by Giddings in 196627 and further evolved into a class of flexible analytical fractionating techniques with unique capabilities to separate analytes ranging a few nanometers up to about 100 μm in size with high resolution25. The separating device is usually a flat, thin channel with height varying between 50 μm and 500 μm. A parabolic laminar channel flow is generated that carries the samples forward from the inlet to the outlet (see Figure 1). Perpendicular to the forward channel flow, an external physical field is applied to the channel to drive the accumulation of samples at the bottom wall. However, the counteracting Brownian motion of the molecules results in their diffusion toward the channel’s interior. Therefore, the equilibrium of these two driving forces resolves the samples at different layers within the channel relative to the bottom wall and elute them out at varying time points. The more elevated the samples are from the bottom wall, the faster the flow rate of the laminae in which the samples reside. As a result, particles at higher positions will be eluted out of the channel earlier as compared to less elevated particles. Different types of external fields, such as gravitational field by centrifugation, temperature difference, electric field and cross-flow, have been applied to separate samples based on different inherent biophysical properties28–32.
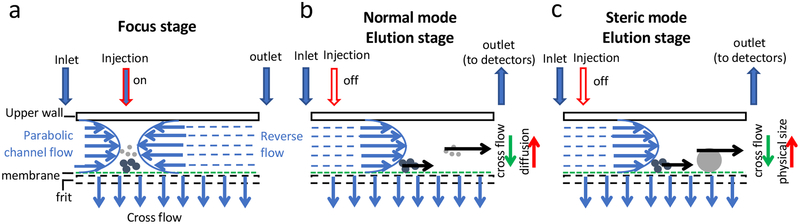
Figure 1.
Schematic illustration of the AF4 working principle. (a-c) Shown are the side views of the AF4 channel, whose height is usually several hundreds of μm. The part size shown in the figure is for illustration only and not drawn to scale. (a) shows that in the Focus stage, two flows in opposing directions are pumped into the channel from the inlet and outlet ports and balanced near the injection port. Samples are injected during the Focus stage and focused in a thin band by the two opposing flows. Particles reach level heights related to their diffusion coefficients. (b) shows that in the Elution stage of the normal mode, a single direction of channel flow from the inlet to the outlet is applied and particles with small hydrodynamic size and high diffusion coefficient are eluted at an early time point, whereas particles with large hydrodynamic size and low diffusion coefficient elute later. (c) shows that when the physical size of a particle is too large to be considered as a point mass compared to the channel height, it elutes in the Steric mode. In contrast to the Normal model as shown in (b), large particles elute earlier than the smaller ones. The fine blue arrows indicate the channel flow (horizontal ones) and cross flow (vertical ones) as marked in the figure, respectively; The fine black arrows illustrate the channel flow carrying the ENPs through the channel during the elution step (the length of the arrows represents the magnitude of the flow rate); the blue block arrows represent the Inlet and Outlet flows as marked in the figure; and the red block arrows depict the Injection flow (the filled arrow indicates the Injection flow is ON, and the empty arrow indicates the Injection flow is OFF). All the arrow heads point to the flow direction. The green fine arrows indicate the direction of particle movement caused by the crossflow; the red fine arrows in (b) and (c) indicate the direction of particle movement due to their diffusion (b) or physical position of the particles in the channel due to their physical sizes (c), respectively.
AF4 was then developed on the basis of flow FFF by Granger, Wahlund and Giddings in the 1980s33, 34. In AF4, the bottom wall is replaced with a semi-permissive membrane with a specific cutoff size, allowing the penetration of solvent and small molecules below the cutoff size but retaining sample components larger than the cutoff size. The cross-flow, perpendicular to the forward channel flow, is the driving force for sample accumulation at the bottom wall (the membrane, in this case). The separation of samples is solely determined by the differences in their inherent diffusion coefficients. The adjustable cross-flow makes AF4 a powerful fractionating technique with great flexibility to accommodate samples with a large size range. Also, due to the compatibility with both organic and aqueous buffers, AF4 has been widely utilized to characterize biological and non-biological analytes, such as proteins, plasmids, polysaccharides, lipoproteins, virus and virus-like particles, liposomes and various polymers35–40.
However, to date, AF4 has been rarely tested for EV analysis. Although a few studies use AF4 to analyze exosomes, these studies are limited by their small sample size and a lack of thorough characterization of the biophysical and molecular properties of exosome fractions required for method optimization41–44. The purpose of our work is to develop the AF4 method and optimize the instrumental parameters used to separate and characterize exosome subpopulations and exomeres. Our protocol can also serve as a reference for further modifications for other types of ENP analysis.
As the volume limit of sample injection is determined by the injection loop size, (which is 100 μL in our instrumental setting), and considering the complexity of the sample composition, and the desired resolution (to separate exosome subpopulations, in our case), we chose ENPs derived from cultured cells and prepared them using dUC. This method enriched for exosomes as the input samples. In brief, conditioned media from cultured murine melanoma cell line B16-F10 was collected and subjected to a sequential centrifugation at 500 × g for 10 min to remove cells and cell debris, at 12,000 × g for 20 min to remove large EVs, such as microparticles and large oncosomes, and finally at 100,000 × g to pellet ENPs that are still heterogeneous in particle size but enriched for exosomes and exomeres. The pelleted samples were washed once and resuspended in PBS, ready for AF4 fractionation. Real-time detectors for ultraviolet (UV) absorbance and quasi-elastic (dynamic) light scattering (QELS) were installed for real time determination of the fractionated particle concentration and hydrodynamic size, respectively. Post-separation characterization, including dynamic light scattering (DLS) measurement in batch mode, nanosight tracking analysis (NTA), transmission electron microscopy, and other biophysical/biochemical analyses, were also employed to validate the fractionation quality and further characterize the separated ENPs, as described previously11. However, these post-separation characterization methods are not detailed in this protocol.
Through our pilot studies, we evaluated the influence of several key parameters of AF4 that were shown to be critical for high-resolution separation of distinct exosome subsets. These included cross-flow, channel height, sample focusing, type of membrane, and the amount of loaded sample. These factors collectively determine fractionation quality, and changing one parameter usually affects the influence of other factors on resolution power. Testing different combinations of these factors, however, can be expensive, time-consuming, and labor-intensive, and thus can be impractical. Understanding the working principles of AF4 and determining the complexity of the analyzed samples (e.g. prior examination by electron microscopy, NTA, and/or western blotting analysis of markers of potential constituents) will be useful to guide the method development process.
Cross-flow
According to the AF4 theory, cross-flow is the driving force counteracting the Brownian motion of particles to resolve particles with different hydrodynamic sizes at different channel-flow laminae at steady state. Thus, cross-flow is a defining factor in AF4 fractionation quality. To determine the optimal cross-flow for exosome fractionation, various cross-flow settings were evaluated. Exosome fractionation profiles (fractograms of UV absorbance and DLS) were devised from representative cross-flow settings, as shown in Figure 2. Specifically, linear gradients of cross-flow with different starting flow rates (at 0.3, 0.5 and 1.0 mL/min) and slopes (i.e., how fast the cross-flow drops to 0 mL/min; tested conditions: a decrease in flow rate from 0.5 to 0 mL/min within 15, 30 and 45 min) were examined.
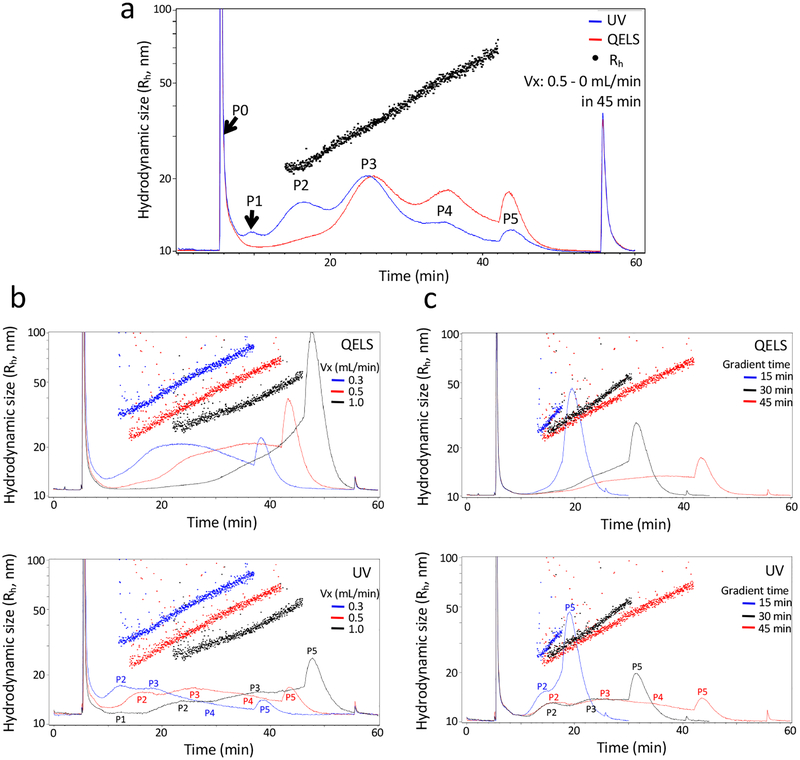
Figure 2.
Influence of cross-flow on AF4 fractionation. (a) Shown is a representative AF4 fractionation profile of B16-F10 sEVs collected by applying a linear cross-flow gradient with an initial flow rate at 0.5 mL/min within 45 min (Peaks are marked as P0-P5; UV (red line), QELS (blue line), Rh (black dots)), or (b) with an initial flow rate at 0.3 mL/min (blue line), 0.5 mL/min (red line) or 1.0 mL/min (black line) and dropping to 0 mL/min over 45 min, or (c) with an initial flow rate at 0.5 mL/min and dropping to 0 mL/min over 15 min (blue), 30 min (black) or 45 min (red). Top, QELS (quasi-elastic (dynamic) light scattering) at 100°; bottom, UV absorbance at 280nm. The other AF4 parameters are: channel flow rate, 1.0 mL/min; channel height, 490 μm; sample focus time, 2 min; membrane, regenerated cellulose (RC), input amount, 40 μg. QELS, quasi-elastic (dynamic) light scattering; Vx, cross-flow rate.
As shown in Figure 2a, a linear separation of the small extracellular vesicles (sEV) mixture was achieved when the cross-flow decreased from 0.5 to 0 mL/min within 45 min. In this graph, the hydrodynamic radius (black dots, Y axis) was plotted along the time course (X axis). The red (QELS) and blue (UV) lines show the total intensity of dynamic light scattering (DLS) and UV absorbance (indicating the protein concentration and abundance of particles) at each time point, respectively. Both QELS and UV were shown in a relative scale to illustrate the relative abundance of each subpopulation in the same sample and therefore the axes were not shown. Based on the properties of DLS data collected at each time point, Astra 6 deduced the corresponding hydrodynamic size of particles resolved at that time point and plotted along the Y axis (black dots). Under this specific AF4 setup, three major peaks (P2, P3 and P4) were observed. These peaks represented the exomeres and two exosome subsets (i.e., small exosomes [Exo-S] and large exosomes [Exo-L]), respectively, as reported in our previous work11. Proteomic characterization of these sEV subsets have revealed the enrichment of markers of endosomal origin, especially in the Exo-S subpopulation, indicating the enrichment of bona-fide exosomes in this subset separated via AF411. Whereas Exo-L has characteristics of both exosomes and EVs potentially larger than 150nm, suggesting that they may represent non-canonical exosomes or probably sEVs of different subcellular origins11. Among the other peaks, P0 is the void peak, resulting from flow disturbance when switching from the focus/injection mode to the elution mode. P1 is a very minor peak, generated by the concomitant elution of the void peak and species that were smaller than exomeres. Depending on the ENP preparation, P1 was sometimes barely detected. P5 was generated due to loss of control on flow rate when it decreased below ~0.08 mL/min and all retained sample components (larger microparticles and/or aggregates of small particles) were eluted out.
To elucidate, AF4 is highly reproducible and yields almost identical fractograms of both UV and QELS if the same sample is analyzed repeatedly under identical conditions11. However, we have observed that factors, such as cell culture conditions, nutrient availability, and passage number of cell lines, may influence the biogenesis and release of ENPs from particular cell lines11. One consequence is that the relative abundance of different subpopulations of ENPs may vary, reflected by the peak size of AF4 fractograms. The difference in the AF4 fractograms observed between Figure 2a and 2b–c when the same AF4 method (Vx 0.5 mL/min, gradient time 45 min) was utilized for fractionation is mainly due to the difference between two independent biological samples. Furthermore, peaks identified on the AF4-UV charts of samples resolved under diverse conditions examined below (data demonstrated in Figures 2–6, Astra 6 data files are provided at https://figshare.com/s/6f22aede51fb279a3f81.) in this protocol, as compared to Figure 2a, were labeled as peaks P2 to P5 in a similar manner. However, the quality/resolution of matching peaks between samples resolved under varying conditions can be divergent.
As shown in Figure 2b, when the initial cross-flow rate was increased to 1.0 mL/min, no additional shoulder peaks were observed to separate further from peaks P2-P4, indicating the uniformity of these three populations of particles. A delay in the elution of all three peaks was observed. Moreover, a much higher P5 peak was observed and this is due to insufficient time for elution of large particles, including Exo-L, in the given time and based on the channel size. In contrast, when the initial cross-flow rate was set to 0.3 mL/min, the samples eluted much earlier, indicating that this flow rate was not fast enough to retain the sample constituents inside the channel and resolve them efficiently. Therefore, an initial cross-flow rate of 0.5 mL/min was used throughout the procedure.
Next, we evaluated the impact of different slopes of the cross-flow gradient on separation quality. A linear decrease of the cross-flow rate from 0.5 to 0 mL/min within a time span of 15, 30 and 45 min were compared. Clearly, the peaks became narrower and the separation quality was compromised when shorter time spans were used (Figure 2c). When a time span of 15 min was used, samples were incompletely resolved and only one peak was partially eluted prior to elution of peak P5, as shown clearly in Fig 2c, bottom chart (blue line and dots). The maximum hydrodynamic diameter of resolved ENPs is less than 80 nm. In addition, a larger P5 peak was observed when a shorter time span was used, indicating insufficient time for separation and elution of large particles. The resolution was improved when the gradient time span was increased, as we observed the increase of the size range of separated ENPs, the smaller peak P5, and more peaks of fractionated samples resolved. When longer time spans were used, the peaks broadened but with improved separation quality. This setting is desired when high-purity particles in discrete fractions need to be recovered for further post-separation characterization. However, when longer time spans are used, other practical issues, such as the dilution of samples and the sensitivity limit of real-time detectors for accurate measurement, have to be taken into account. Based on this consideration, and given the successful separation of distinct subsets of exosomes and exomeres, we chose the linear gradient of the cross-flow decreasing from 0.5 to 0 mL/min within 45 min for our study and did not test longer time spans.
Channel height
Based on the working principle of AF4, the channel’s geometry, including its width, height and shape, is critical for fractionation quality. The short channel utilized in our study is a product of Wyatt Technology (Santa Barbara, USA), which has a trapezoidal geometry45, 46 with a tip-to-tip length of 152 mm and a linear decrease of the channel width from 21.5 mm (close to the injection port and about 12 mm away from the inlet tip) to 3 mm. With the shape and width already optimized and fixed, the height (i.e., the thickness of the channel, determined by the spacer used between the upper wall and the bottom accumulation membrane) is the only parameter available for further optimization. A series of spacers with different thicknesses (190, 250, 350 and 490 μm) are provided by the manufacturer. The channel height affects the parabolic laminar flow rate profile and thus the separation resolution. It also affects the channel capacity, with a thicker channel allowing for analysis of larger sample amounts. Since we needed to recover enough sample for downstream analysis, the loading capacity is an important factor for our fractionation and so we only considered employing spacers with a thickness of 350 μm and 490 μm. As shown in Figure 3, the channel with the 350-μm spacer eluted samples earlier but with narrower peaks and a reduced separation resolution compared to the channel with the 490-μm spacer. Therefore, the 490-μm spacer was chosen for our work.
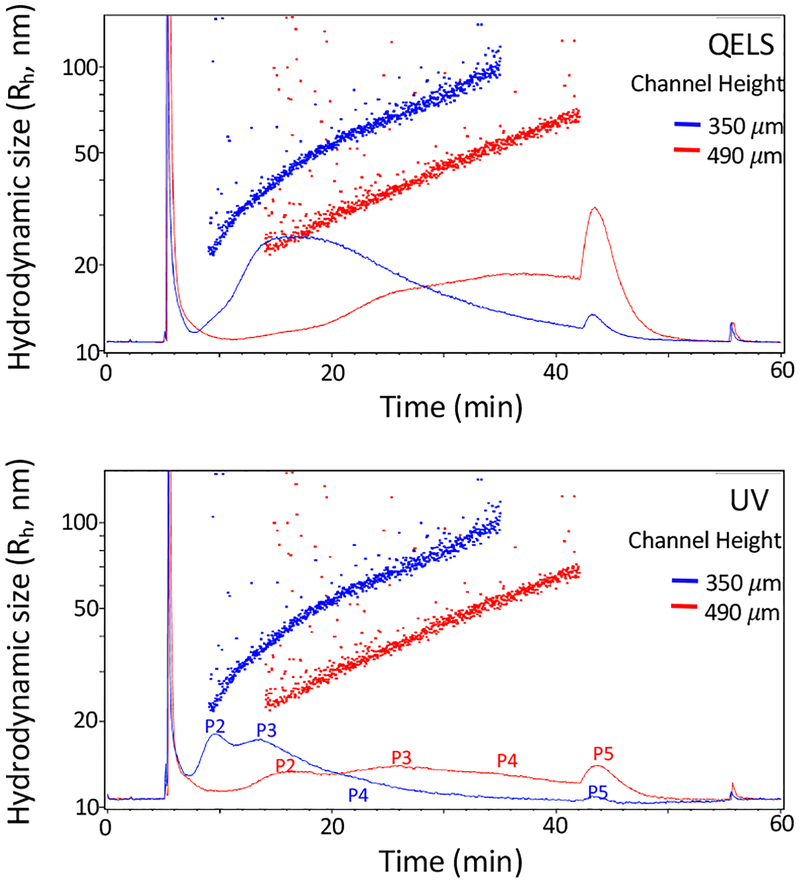
Figure 3.
Effect of the channel height upon AF4 fractionation. Shown are AF4 fractionation profiles of B16-F10 sEVs collected using a channel with a spacer of 350 μm (blue) and 490 μm (red). Top, QELS at 100°; bottom, UV absorbance at 280nm. The other AF4 parameters are: channel flow rate, 1.0 mL/min; a linear gradient of cross-flow decreasing from 0.5 mL/min to 0 mL/min over 45 min; sample focus time, 2 min; membrane, regenerated cellulose (RC), input amount, 40 μg.
Focusing
A 100-μL sample loop was used in our instrument for sample loading. It is a significant portion of the total channel capacity, which usually ranges from 200 μL to 1000 μL. Once injected into the channel, the sample would spread throughout the channel and lead to insufficient fractionation. To avoid this, a flow opposing the channel forward flow was introduced from the outlet and, together with the channel flow, focused the sample into a narrow band close to the injection port (i.e., focus mode). First, the focus flow was established and then the samples were injected in the focus mode and given enough time to reach steady-state equilibrium before elution. The focusing flow rate and focusing time determine focusing efficiency. Here, we fixed the focusing flow rate at 0.5 mL/min, the same as the initial cross-flow rate for elution, and then tested different time periods (2, 5, and 10 min) for focusing efficiency. As shown in Figure 4, different focusing times did not significantly affect peak shape or resolution power. Moreover, before exomere elution occurred, the fractograms of both UV and DLS reached similar baselines. Notably, we observed that the P5 signal intensity increased as focusing time increased, suggesting potential particle aggregation caused by extensive focusing. Therefore, we chose a focusing time of 2 min for our study.
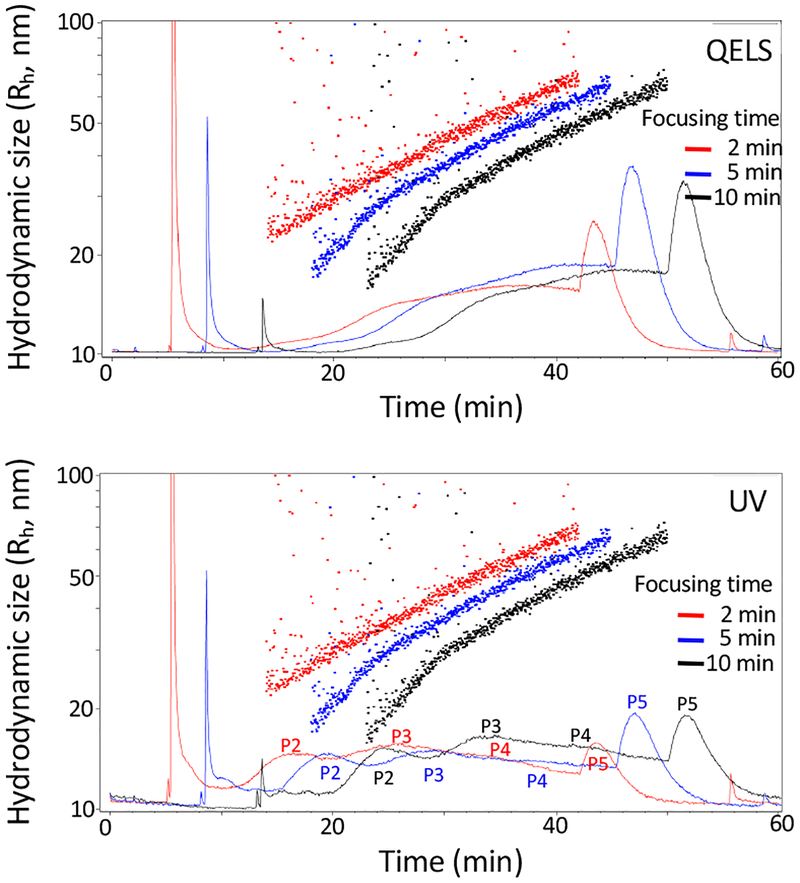
Figure 4.
Effect of the focus time upon AF4 fractionation. Shown are AF4 fractionation profiles of B16-F10 sEVs collected using a sample focus time of 2 min (red), 5 min (blue) or 10 min (black). Top, QELS at 100°; bottom, UV absorbance at 280nm. The other AF4 parameters are: channel flow rate, 1.0 mL/min; a linear gradient of cross-flow decreasing from 0.5 mL/min to 0 mL/min over 45 min; channel height, 490 μm; membrane, regenerated cellulose (RC), input amount, 40 μg.
Membrane choice
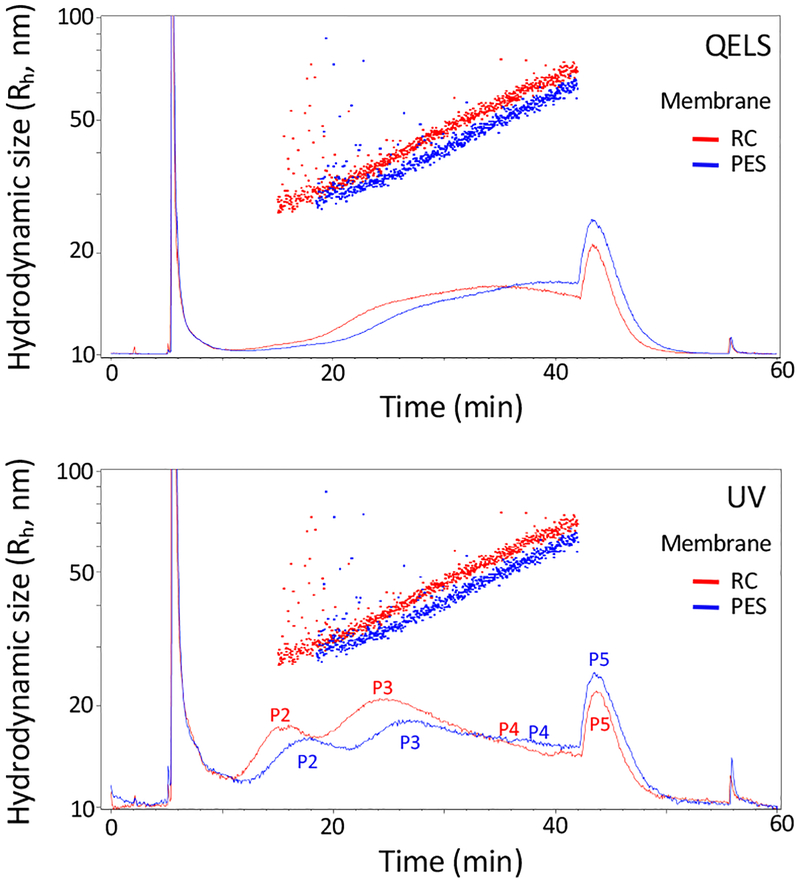
Since the sample fractionation is performed close to the membrane, in addition to the pore size (should be below the size of the smallest solute to fractionate in your sample) of the membrane, the compatibility of the membrane material with the samples also needs to be considered. For example, the sample may bind to the membrane non-specifically. Two different types of membranes that are commonly used for biological material concentration or filtration, regenerated cellulose (RC) and polyethersulfone (PES), were tested for exosome fractionation. While keeping other AF4 parameters exactly the same, we observed a delay of sample elution and broader peaks in the channel with PES compared with RC, suggesting potential non-specific interactions between samples and the membrane (Figure 5). Therefore, the RC membrane was selected for our studies.
Figure 5.
Comparison of the AF4 performance for separating EVs using different membranes: regenerated cellulose (RC, red) versus poly(ether)sulfone (PES, blue). B16-F10 sEVs were analyzed using the following AF4 parameters: channel flow rate, 1.0 mL/min; a linear gradient of cross-flow decreasing from 0.5 mL/min to 0 mL/min over 45 min; channel height, 490 μm; sample focus time, 2 min; input amount, 40 μg. Top, QELS at 100°; bottom, UV absorbance at 280nm.
Amount of input sample
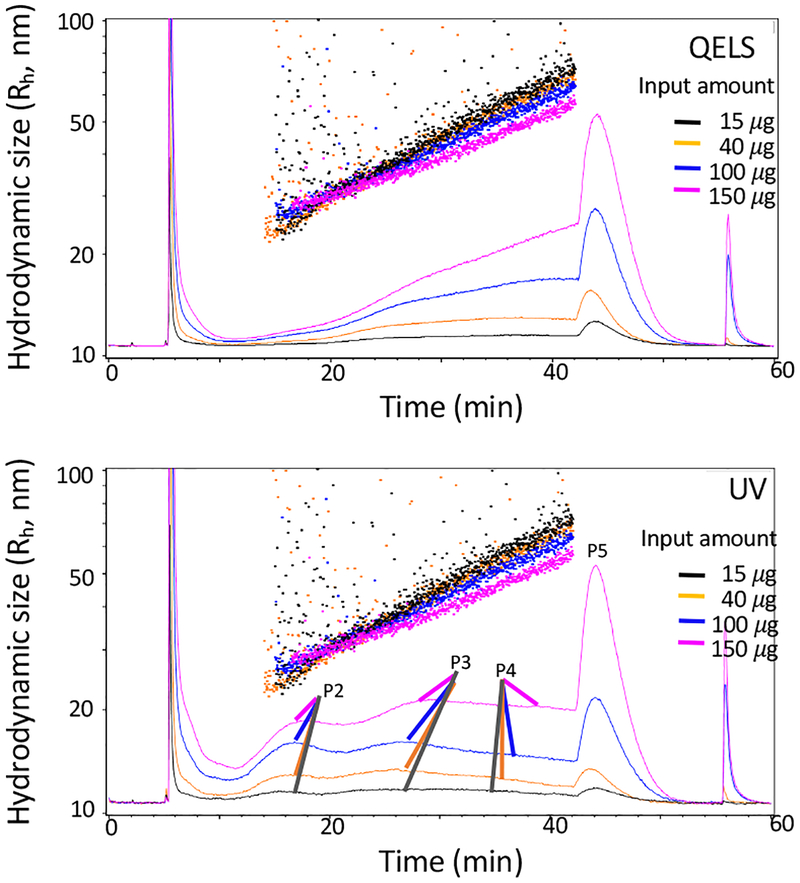
Once we determined the key fractionation parameters, we then examined the loading capacity of AF4. The minimal amount of material required for AF4 is determined primarily by the sensitivity limit of the real-time detectors, such as DLS and UV monitors. The signal/noise ratio must be adequate for accurate data collection and interpretation. The maximal amount of material is determined by the required resolution of fractionation, which depends on the purpose of the experiment and the complexity of the sample to be analyzed. To efficiently separate exomeres and the two exosome subsets that we reported previously11 from sEVs prepared using dUC, we tested different amounts of B16-F10-derived sEV input samples ranging from 15 μg to 165 μg. As shown in Figure 6, 15 μg was the lower limit of material for this analysis, as we started to detect a high level of noise, especially at the low end of hydrodynamic size. Inputs of 40 μg and 100 μg yielded almost identical fractionation profiles and hydrodynamic size determinations, indicating comparable fractionation resolution and robust signal detection. However, when the amount of input increased to 165 μg, the elution of all peaks was delayed significantly, resulting in incomplete elution of Exo-L. Bleed-through of each particle population to the adjacent populations increased (and thus poorer separation occurred), as indicated by the increased signal intensity at the valleys between peaks. Therefore, an input ranging from 40 μg to 100 μg was used for this study.
Figure 6.
Examination of the sample (B16-F10 sEVs) loading capacity for AF4 analysis. Shown are AF4 fractionation profiles of B16-F10 sEVs with an input of 15μg (black), 40 μg (orange); 100 μg (blue), or 150 μg (purple). Top, QELS at 100°; bottom, UV absorbance at 280nm. The other AF4 parameters are: channel flow rate, 1.0 mL/min; a linear gradient of cross-flow decreasing from 0.5 mL/min to 0 mL/min over 45 min; channel height, 490 μm; sample focus time, 2 min; membrane, regenerated cellulose (RC).
Applications of the method
As illustrated in the above assessments and in our previous publication11, AF4 technology provides unique capabilities to separate nanoparticles with high resolution within a large size range and we show AF4 can separate distinct exosome subpopulations and exomeres. Our findings exhibit the potential of AF4 methodology in identifying other distinct EV subpopulations. Coupled with real-time monitoring (e.g., multi-angle light scattering (MALS), DLS, UV absorbance and fluorescent detection) and post-separation analyses (e.g., microscopy, mass spectrometry of proteins, lipids, glycans and metabolites, and DNA and RNA sequencing), AF4 can yield valuable data on ENP analytes, including particle morphology and size, relative abundance, molecular composition, and other biophysical and biochemical properties. AF4 can help researchers decipher the complexities and heterogeneity of ENPs that cannot be well addressed with other existing techniques.
Our AF4 protocol describes the fractionation of exomeres and exosome subsets from sEVs isolated from the conditioned media of B16-F10 cells and a panel of more than 20 different cancer cell lines and 5 normal cell lines (please refer to Supplementary Table 1 in Ref. 11 for the detailed list of cell lines examined; and unpublished data [Haiying Zhang & David Lyden] on normal mouse and human mammary epithelial cell lines CommaD (mouse), HMEC124 (human), and HMEC240L (human)). The characterization of exomeres, Exo-S and Exo-L isolated from five cancer cell lines (i.e. B16-F10, 4T1, Pan02, MDA-MB-4175, and AsPC-1) were reported previously (Ref. 11). The AF4 fractionation analysis of sEVs isolated from additional cell lines also indicate the detection and separation of exomeres, ExoS and Exo-L as reported in our previous work11. This AF4 method can be used to fractionate and characterize sEVs isolated from an array of bodily fluids (including blood plasma or serum, lymphatic fluid, bone marrow plasma, cerebrospinal fluid, urine, saliva, bronchoalveolar lavage, milk and amniotic fluid), given their similar particle compositional complexity. One point to emphasize is that AF4 fractionation of sEVs isolated from bodily fluids usually requires prior stratification of the samples to remove confounding substances (e.g. lipoproteins in plasma) with similar sizes of sEVs. Since all cells are capable of shedding EVs, our protocol can be employed to study the EV biology of any organism.
In addition to use for biological discovery, our protocol can also be modified for use in the field of quality control in exosome-based pharmaceutical production. Exosomes have become attractive therapeutic delivery vehicles for treating cancer and other types of diseases47. AF4 coupled with sensitive molecular assays can serve as an improved analytic tool to evaluate purity, drug loading efficiency, and the integrity of the exosome product by detecting debris or aggregates.
Last but not least, this protocol can serve as a reference to further develop and optimize methods for fractionating and characterizing other types of ENPs. Some unique advantages of AF4 are its high resolution and large size range of fractionation. Furthermore, different conditions, such as cross-flow setting and focus time, can be easily tested by simply programing the settings into the software, with minimal handling of the channel. Besides their use in analyzing exosomes and other sEVs, fractionation protocols for large EVs, such as larger microparticles and oncosomes, can be further developed. Specific caution should be taken when fractionating large particles since they may be too large to elute in the normal mode (when the particle is small and considered as point-mass compared to the channel height) but in the steric model instead (Figure 1). Moreover, other fields, such as an electric field, can also be applied to AF4 to stratify particles based on additional biophysical properties other than size, allowing even broader application of AF4 technology.
Taken together, the separation and characterization of distinct EV subpopulations by AF4 are critical to advancing our knowledge of the biology of EVs and their functional roles in physiological and pathological conditions. By profiling the molecular cargo of EVs, we can identify signature proteins, lipids, glycans and genes as well as specific signaling pathways associated with disease progression, facilitating the identification of potential diagnostic/prognostic biomarkers, including those related to cancers. Such knowledge will also provide a rationale for developing ENP-based therapies in clinical trials.
Comparison with other methods
A multitude of technologies, in addition to AF4, have been developed to isolate pure exosomes and other EV subpopulations. The most commonly used technique makes use of dUC and separates the particles based on their hydrodynamic size and density. Successive centrifugation at different centrifugal forces eliminates dead cells and cellular debris (500 × g, 10 min), large oncosomes and apoptotic bodies (2000~3000 × g, 20 min) and larger microparticles (10,000~12,000 × g, 20 min), and subsequently pellets sEVs (100,000 × g, 70 min)19,48,49. Centrifuge rotor type, centrifugal force and centrifugation time are key factors influencing the product yield and purity of this method. Its performance also varies depending on the cell types studied51. dUC can process large volumes and high amounts of sample, but the purity of the material recovered is poor. It can only roughly partition particles into groups, such as large vesicles, microparticles, and sEVs (enriched for exomeres and exosomes), with expected heterogeneity within each group and contamination for other groups19, 24. The high centrifugal force may also cause sample aggregation. With our protocol, we took advantage of dUC by first stratifying and concentrating the sEV population and then analyzing particles at much higher resolution to further fractionate exomeres and exosome subsets.
Density gradient floatation (DGF) is often used to further purify sEVs first isolated using dUC. In DGF, EVs are overlaid upon a gradient of increasing dilutions of a viscous solution (sucrose or iodixanol are commonly used) and, upon centrifugation, they migrate to the equilibrium density determined by the EV’s size, shape and density. DGF is often used to remove non-membranous particles from EVs and has also been employed in several studies to address exosome heterogeneity12,50,51. The major drawbacks of DGF, when compared to the performance of AF4, include its time-consuming preparations, lack of automation, operator-dependent reproducibility and low yield19, 24. Long periods of incubation with high sucrose concentrations can also damage EV integrity, necessitating additional washing steps for its removal (personal observation). In contrast, AF4 is rapid, fully automated, highly reproducible, robust, and compatible with many buffer choices that mimic physiological conditions. Resolution and size range in EV fractionation is far superior with AF4 than with DGF24.
SEC, a gentle means of nanoparticle fractionation, has been extensively used for protein and protein complex analysis in biochemical and biophysical studies. Recently, it has been adopted to fractionate EVs52–55. In SEC, particles are separated in a column filled with porous polymer beads (stationary phase) based on their size and shape. Smaller-sized particles with a globular shape can penetrate the porous beads more readily, taking a longer route and more time to elute, whereas the larger particles are excluded from penetrating the pores and subsequently elute more rapidly. The elution of particles with abnormal shapes is more complicated due to its potential steric interference with particle traveling through the pores. Compared to other technologies, SEC has a resolution most similar to that of AF4. Still, AF4 demonstrates superior resolution over a much wider size range25. SEC resolution drops when particles are close to or larger than the upper limits of pore size. Furthermore, SEC is not as flexible as AF4 in changing separation parameters and its size range of separation is fixed for a given column with a specific solitary phase. Moreover, AF4 contains a hollow channel with only a membrane at the accumulation wall but, unlike SEC, requires no stationary phase. This stationary phase in SEC generates shear stress and renders a much larger surface area than AF4 for nonspecific binding of analytes. Similar to AF4 methods, the input sample loading volume for SEC must be restricted and there is an upper limit for the sample capacity to compromise a balance between sufficient yield and exemplary fractionation quality. Sample stratification by dUC and concentration methodologies prior to separation greatly facilitate the separation power of SEC.
UF allows for straightforward isolation of EV populations based on their size by filtering the sample through a series of semipermeable membranes with defined pore sizes (i.e., as reflected by molecular weight cutoffs)56, 57. Smaller particles below the cutoff size can penetrate through the pores while larger ones are retained. UF provides a crude separation of EVs due to limitations of membrane pore size availability. Most EVs are not rigid spheres but rather flexible particles and can transfigure to pass through the pores, especially when pressure is applied. Another concern is the uniformity of membrane pore size, which is critical for separation purity. Though the separation power of UF is inferior to AF4, UF can serve as a means to pre-stratify and concentrate input samples for further analysis by AF4.
Distinguishable from methods which separate EVs mainly by their size, IAC relies on the antigenic recognition of EV surface molecules (primarily proteins). IAC is highly selective, fast and flexible to scale for either preparation or analytic purpose. This separation principle has been adapted for different formats of analysis and preparation, including precipitation using immunomagnetic beads, flow analysis, detection by microarray, microscopy, western or ELISA assay, and microfluidic separation19,22,23,58. The inherent limitation of IAC is that knowledge about the surface antigen is a prerequisite. The other concern is that the IAC antigen may be represented in multiple subpopulations of EVs with divergent sizes and/or origins. Thus, the application of AF4 may further necessitate EV separation based on size. EVs captured by IAC are ideal for molecular content characterization but not for further functional studies due to inefficient removal of the capturing antibody, which may interfere with the functional assay or targeting and uptake by recipient cells. In contrast, AF4 is label-free and enables these functional analyses feasible.
Level of expertise needed to implement the protocol
The AF4 instrument is commercially available (e.g., Wyatt Technology) and the manufacturer can perform the initial set-up. The method development for specific sample analysis, routine maintenance of instruments, and troubleshooting require a good understanding of the working principles of AF4 and installed detectors, training for handing the AF4 channel and detectors, and being familiar with software used for AF4 operation and data collection. Previous experience with chromatography and/or microfluidics is helpful in mastering the AF4 application. However, once the AF4 fractionation method training has been achieved, only minimal skills, such as familiarity with software interface and proper instructions, are necessary to complete the fractionation process since nearly all the steps are automatic and programmed.
Limitations
One inherent limitation of AF4 is that it fractionates samples based on their size. As a consequence, particles with the same hydrodynamic size but with different morphologies, surface molecules and other biophysical properties cannot be separated from each other via AF4 alone. However, other fields, such as an electric field, can also be applied in conjunction with AF4 to provide further separation according to additional characteristics such as particle surface charge. Special consideration is also required when developing a protocol for large particles whose sizes are too large to be considered as point-mass compared to the channel height. These large particles will elute in the steric mode rather than the normal mode, as illustrated in Figure 1.
A second inherent drawback is that AF4 can accommodate only small amounts of sample (e.g., 40 μg to 100 μg in our case), which is often not efficient for large-scale preparations in more detailed assessments of nanoparticle properties. The sample instead can be divided into multiple fractionation analyses for improved characterization of specific nanoparticle subsets. A third limitation of AF4 is that due to the loading capacity limitation, the input sample requires UC preparation or other means to first stratify and concentrate the analytes (i.e. sEVs in our study) prior to fractionation.
Furthermore, it has to be pointed out that no single formula can be universally applied for analysis of different types of samples. The fractionation method and key parameters discussed above in the protocol development section have to be developed and optimized based on the complexity (i.e. size and abundance of each component) of the sample of interest. In certain cases, different running methods and instrument settings may have to be combined sequentially to efficiently separate different components within a complex sample.
Experimental design
Preparation of sEVs from cell culture.
With the aim to separate distinct cellular nanoparticles, such as exomeres and exosome subsets, we decided to study sEVs isolated using dUC as the input samples for AF4 due to its capability to process large volume of sample and short time preparation. Alternative methods, such as DGF, UF and SEC, can also be used for sEV input sampling. EVs captured by IAC can be applied, as well, if the antibody can be removed from the EVs.
This protocol has been developed and optimized using sEVs derived from cell culture model systems. Conditioned media is sequentially spun to remove cells, cell debris, and large EVs and finally pellet down the sEVs. We have previously reported that fresh versus frozen sEV samples do not markedly differ in AF4 profiles11, indicating that the structural integrity of EVs is well preserved during the freeze-thaw process. Of note, the culture conditions, such as growing cells in hypoxic conditions, and the passage of cells can influence EV production and composition (i.e. the percent of each particle type in a sample)11. Thus, these changes in ENP composition may require modifications of the AF4 methods for further optimization to achieve the desired separation quality.
As stated in the “Application of the method” section, this protocol could also be applied to sEVs prepared from other resources, such as bodily fluids, including plasma, in conjugation with other means of prior stratification of the samples. AF4 parameters, such as the cross-flow gradient, can be further adjusted to meet the specific requirements of particular samples (for example, existence of additional types of ENPs). However, for certain sample types, the EV composition is more complicated than that derived from conditioned media of cell cultures. For example, the presence of lipoprotein particles in blood plasma may interfere with the separation of exosomes due to their partial overlap in size. In this case, other means (such as immuno-affinity capture) of prior removal of lipoproteins from the plasma sample is desired before their further fractionation via AF4.
AF4 fractionation of sEVs and real-time data collection and analysis.
The AF4 method for separation of exomeres and exosome subsets (i.e., Exo-S and Exo-L) from cell culture-derived sEVs is illustrated in Figure 7 and Table 1. Based on the complexity of the EV samples and the goal of each specific study, this running method can be further adjusted, as described in the “Overview of the procedure and development of the protocol.”
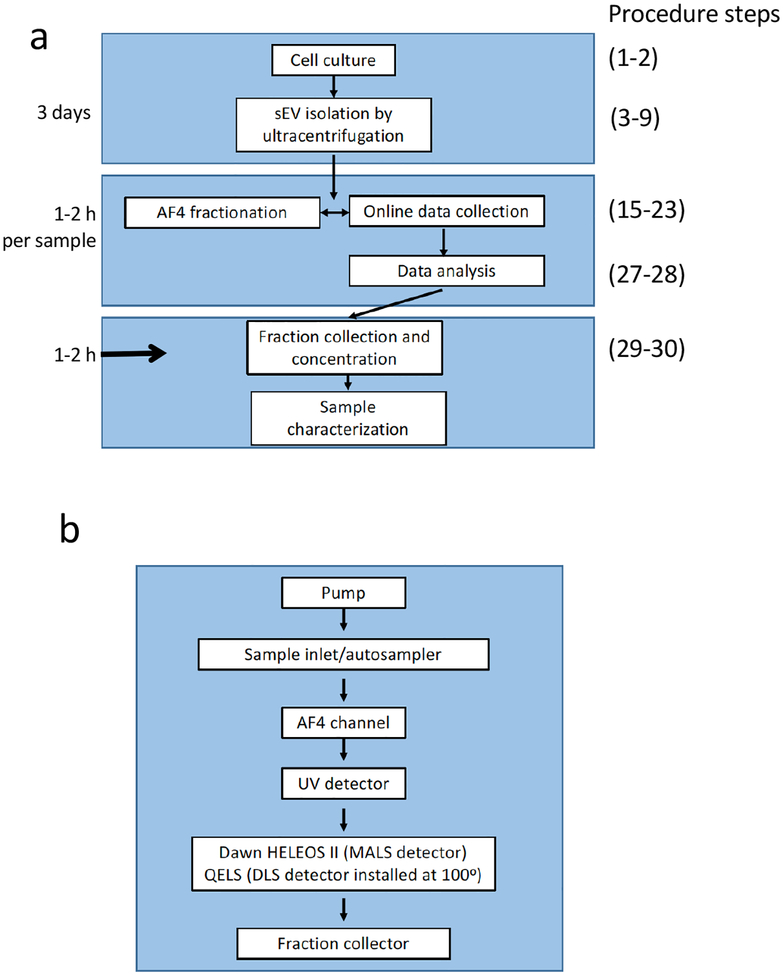
Figure 7.
Schematic illustration of the overall procedure, the instrument flow route, and operative methods for AF4 of ENPs. (a) The overview of experimental design for cell culture-derived ENP isolation and AF4 fractionation. (b) Illustration of the AF4 flow route and arrangement of inline real-time detectors.
Table 1.
AF4 operative method programmed for ENP fractionation.
| Start Time (min) |
End Time (min) |
Duration (min) |
Mode | Vx Sta rt (mL/min) |
Vx End (mL/min) |
|---|---|---|---|---|---|
| 0 | 2 | 2 | Elution | 0.5 | 0.5 |
| 2 | 3 | 1 | Focus | ||
| 3 | 5 | 2 | Focus + Injection | ||
| 5 | 50 | 45 | Elution | 0.5 | 0 |
| 50 | 55 | 5 | Elution | 0 | 0 |
| 55 | 60 | 5 | Elution + Injection | 0 | 0 |
Forward channel flow (Detector flow) 1.0 mL/min
Focus flow 0.5mL/min
Injection flow 0.2 mL/min
Focusing (%) 30
For real-time monitoring and analysis of the fractionation of particles, several real-time detectors are usually installed in place immediately after the AF4 channel. Our laboratory has the DAWN HELEOS-II (MALS detector) with QELS (DLS detector) installed at the detector 12 (100°) position (Wyatt Technology) and the Agilent 1260 Infinity Multiple Wavelength Detector (set at 280 nm for UV absorbance detection) in place. We mainly use the DLS measurement to determine the hydrodynamic size of the fractionated particles in real time. The primary data from a DLS measurement is the autocorrelation function, which plots the average overall changes in the scattered light intensity of molecules with time (see example in Figure 8). The exponential decay rate of the autocorrelation function determines the translational diffusion coefficient (Dt) of molecules in solution based on the following equation:
Based on the Stokes-Einstein relation (below), we can further deduce an effective hydrodynamic radius (Rh) from Dt.
The assumption for this calculation is that the solute (EV, in our case) is a sphere undergoing Brownian motion. Rh is the radius of a sphere with the same translational diffusion coefficient as the analyzed solute. Rh depends only on the physical size of the solute and its size-related behavior, such as diffusion and viscosity, but is not affected by its density or molecular weight. The measurement range of 0.5 nm to 1000 nm radius makes DLS an effective tool to measure the size of sEVs.
Figure 8.
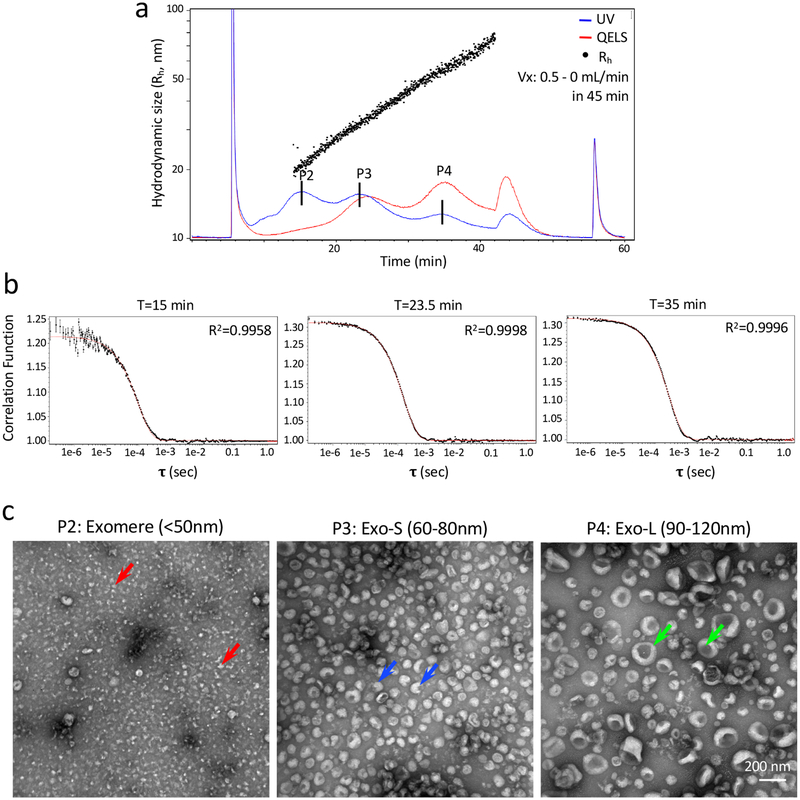
Representative AF4 fractionation analysis of B16-F10 sEVs. Shown are representative AF4 fractionation profile of B16-F10 sEVs (a) and autocorrelation functions at specific time points (b). (c) TEM imaging analysis of combined fractions for peaks P2 (exomere), P3 (Exo-S), and P4 (Exo-L) (The methods for sample preparation and TEM imaging analysis were described in Zhang H et al. Nat Cell Biol 20, 332–343 (2018) (Ref 11). Scale bar, 200 nm. Colored arrows point to representative particles in each subpopulation. TEM images are adapted with permission from Zhang et al. (ref 11), Springer Nature.
Combining AF4 with real-time DLS measurements is critical for accurate size determination. In a polydisperse sample, DLS measurement yields an average Rh and the specific information on each compositional species in a given sample is missing. However, fractionation results in the separation of solutes with different sizes and each fraction contains only a very small admixture of different Rh particles. Thus, fractionation allows the size of each species to be more accurately measured. For such monodisperse samples, the resulting autocorrelation functions are single exponentials, which are simple to interpret. Fitting the data to a single exponential function is performed in the Astra software using the Cumulants model59. By examining the ideal fitting to a single exponential, one can further evaluate the separation quality.
One requirement for accurate DLS measurement of Rh is that the sample concentration must be sufficient so that the sample scatters at least three times more light than the solvent to obtain an acceptable signal/noise ratio. In particular, small molecules, such as exomeres, scatter less light and require even higher concentrations of analyte to optimize results.
Besides DLS, static light scattering (SLS) detected by MALS measures the radius of gyration (Rg). Rg is defined as the mass averaged distance of each point in a molecule from its gravity center and is generally different from Rh. Comparing Rg to Rh can further reveal the compactness of a solute (i.e., empty versus filled particles). In general, the MALS detector is more sensitive than DLS monitoring and thus it will be of specific use when only little amount of material available for analysis.
The UV detector is used as part of our instrumentation for real-time concentration measurements. The intensity of UV absorbance can provide us an approximation of the relative abundance of different species in the sample, despite not having defined extinction coefficients for different species in the EV sample mixture. The peaks of UV absorbance are useful in guiding the choice of combining fractions of similar particles. However, we also often conduct the bicinchoninic acid assay (BCA assay) and NTA after fraction collection for quantification purposes. Once pure EVs are obtained and further characterization can be performed, the extinction coefficients of each species for improved interpretation of the UV absorbance data to concentration can then be determined.
One key consideration for real-time detectors is that it must have exceptional sensitivity due to the limited amount of material passing through the detector at each single time point. Besides the detectors mentioned above, other sensors, such as differential refractive index (dRI) and fluorescence detectors (FLDs), are often included as standard parts of the instrumentation for a variety of macromolecular characterization techniques. dRI, considered a universal concentration detector, is accurate and versatile in all types of solvents and independent of chromophores or fluorophores. FLDs are useful if autofluorescent molecules or artificial fluorescent labeling are present in specific subsets of EVs. It should be noted that, with additional detectors assembled inline, the fractionated particles take a longer path and more time to reach the fraction collector. As a result, diffusion of molecules will lead to broadening of peaks, dilution of fractionated samples and reduction in separation resolution. Therefore, only detectors considered essential for the real-time monitoring should be installed.
Fraction collection, concentrations and characterization.
AF4 fractions can be collected automatically or manually for downstream post-separation characterization. We have installed the Agilent Fraction Collector (1260 series) to automatically collect fractions into 96-well plates, but similar fraction collectors can be utilized for accurate and reproducible fraction collections. Fractions can be collected either by volume or over time, and fractions of particles with the same identity based on real-time and post-separation characterization can be further pooled together for downstream analysis. For example, in our previous work to identify exomeres and distinct subsets of exosomes11, we first examined representative fractions across the whole time course of fractionation by real-time DLS and post-separation transmission electron microscopy analysis and then pooled the fractions of particles with similar size and morphology together for further characterization. This step was also guided by the peaks of UV absorbance (indicating the most abundant fraction of each type of particles). To validate that the pooled fractions are relatively pure and not contaminated significantly by other types of adjacent particles, only fractions centered around the peaks were pooled together. Depending on the resolution of the fractionation, this fraction combination step can be empirically determined. Due to the different composition of EV subpopulations in a given sample, occasionally the UV peaks are not identifiable and thus the fraction combination will rely more on other properties, such as size and morphology. For instance certain cell lines secreted Exo-S and/or Exo-L at very low levels compared to exomeres, resulting in no corresponding peaks observed in UV and/or QELS/DLS fractograms.
The individual or combined fractions can be directly utilized for downstream analysis or subjected to a concentration step before further characterization. We usually concentrate the collected fractions using the Amicon Ultra-series of centrifugal filter units with Ultracel-30 (30 KDa cutoff) membrane (Millipore). Other alternative means of concentration, such as tangential filtration centrifugation, direct UF, UC, or IAC, can be applied depending on the need for the downstream analysis. A variety of analyses can be performed on fractionated EVs, including but not limited to: BCA assay, NTA, atomic force microscopy, electron microscopy, mass spectrometry of molecular contents (e.g., proteins, lipids, glycan, and metabolites), western blotting or the enzyme-linked immunosorbent assay, sequencing of genetic material (DNA and RNAs), DLS measurement in batch mode, and zeta potential measurement. The functional roles of the fractionated EV subpopulations can be further investigated in vitro or in vivo.
MATERIALS
REAGENTS
B16-F10 cell line (ATCC, CRL-6475) ! CAUTION Cell lines should be regularly checked to ensure that they are authentic and free of mycoplasma contamination.
DMEM (VWR, Catalog No. 45000–304)
Premium Grade Fetal Bovine Serum (FBS) (VWR, Catalog No. 97068–085)
L-Glutamine, 100x (Corning, Catalog No. 25–005-CI)
Penicillin-Streptomycin Solution, 50X (Corning, Catalog No. 30–001-CI)
Sterile PBS (VWR, Catalog No. 45000–446)
TrypLE (Thermo Fisher Scientific, Catalog No. 12604–039P)
ATCC Universal Mycoplasma Detection Kit (ATCC, Catalog # 30–1012K)
Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Catalog No. 23225)
ATCC Universal Mycoplasma Detection kit (ATCC, Catalog No. 30–1012K)
Bovine serum albumin (BSA) (Sigma, Catalog No. A1900)
Water filtered using the Milli Q system (Millipore)
Ethanol (Sigma, Catalog No. 459828) ! CAUTION This chemical is flammable and should be stored in a cabinet for flammable materials.
Contrad 70 (Decon Labs, Inc Catalog No. 1003)
Sodium Dodecyl Sulfate (SDS) (Omnipur, Catalog No. 7910) ! CAUTION This chemical is corrosive and toxic, and can cause severe skin and eye irritation. Wear protective gloves, mask, eyeshield, faceshield, and protective clothing to handle.
Nitric Acid (Fisher Scientific, Catalog No. 7697–37-2) ! CAUTION This chemical is highly corrosive and can cause severe eye and skin burns, severe respiratory and digestive tract burns. Wear proper protective equipment (gloves, eyeshield, faceshield, clothing, respirators) and handle it in a chemical hood. The waste should be treated as a hazardous waste following state and local hazardous waste regulations.
EQUIPMENT
Labconco Purifier Class II biosafety cabinet
Tissue culture incubator
EVOS FL Microscope (Thermo Fisher Scientific)
Optima XPN-100 Ultracentrifuge (Beckman Coulter, Catalog No. A94469)
Type 45 Ti Rotor, Fixed-Angle, Titanium (Beckman Coulter, Catalog No. 41103909) (The k factor of this rotor is 133 at maximum speed, and 299 at 100,000 × g speed)
Table-top Heraeus Multifuge x3R Centrifuge Series (Thermo Fisher Scientific, Catalog No. 75004501)
Microcentrifuge (Eppendorf, 5424R)
AccuScan GO UV/Vis Microplate Spectrophotometer (Fisher Scientific, Catalog No. 14–377-579)
Milli Q system (Millipore)
4 °C Refrigerator (Thermo Fisher Scientific)
−20 °C freezer (Thermo Fisher Scientific)
−80 °C freezer (Thermo Fisher Scientific)
37 °C Incubator (Thermo Fisher Scientific)
Autoclave (Tuttnauer)
Dishwasher (Steelco)
Consumables
150×25mm tissue culture dish with Grid (VWR, Catalog No. 25383–103)
5/10/25mL Serological pipettes (VWR, Catalog No. 82050–478/82050–482/82051–182)
Disposable Tips (Denville, Catalog No. P1096-FR/P1121/P1122/P1126)
500 ml Supor machV PES Filter Units (VWR, Catalog No. 73520–984)
15mL/ 50mL conical tubes (VWR, Catalog No. 82050–276/ 82050–346)
1.7 ml Microcentrifuge Tubes (VWR, Catalog No. 53550–698)
96-well plate (VWR, Catalog No. 62406–081)
Blue screw caps (Agilent, Catalog No. 5182–0717)
Screw cap vials (Agilent, Catalog No. 5182–0714)
vial insert, 250ul pulled point glass (Agilent, Catalog No. 5183–2085)
96-well plate, 1.0ml, polypropylene (Agilent, Catalog No.8010–0534)
Sealing tape, clear polyolefin (Thermo Fisher Scientific, Catalog No. 232701)
Ultracentrifuge tube (Beckman Coulter, Catalog No. 355628)
Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-30 membrane (Millipore, Catalog No. UFC903024)
Amicon Ultra-4 Centrifugal Filter Unit with Ultracel-30 membrane (Millipore, Catalog No. UFC803024)
Millipore Reg. Cellulose membrane 10KD SC (Wyatt technology, Catalog No. 4057)
Nadir Polyethersulfone membrane 10KD SC (Wyatt technology, Catalog No. 1903)
Inline filter membrane 0.1μm (Wyatt Technology, Catalog No.1871)
Dry wipes (Kimtech, Catalog No. 7552)
2L glass bottles (VWR, Catalog No.10754–822)
Hemocytometer (Weber Scientific, Catalog 3048–12)
AF4 instrument parts
Agilent 1260 Infinity Analytical- and Preparative-scale Fraction Collectors (G1364C)
Agilent 1290 Thermostat (G1330B)
Agilent 1260 Infinity Standard Autosampler (G1329B)
Agilent 1260 Infinity Multiple Wavelength Detector (G1365D)
Agilent 1260 Infinity Isocratic Pump (G1310B)
GASTORR TG-14 HPLC vacuum degasser
Wyatt Technology DAWN HELEOS-II with QELS installed at detector 12 (100°)
Wyatt Technology Eclipse AF4
Wyatt Technology short channel
Computer installed with Chemstation and Astra 6 software
Software
Chemstation (Agilent Technologies) integrated with the Eclipse module (Wyatt Technology) to operate the AF4 flow
Astra 6 (Wyatt Technology) for MALS, DLS and UV data acquisition and analysis
REAGENT SETUP
B16-F10 cell culture medium 500 mL of DMEM is supplemented with 10% (vol/vol) FBS (exosome-depleted), 50 units/mL Penicillin, 50 μg/mL Streptomycin, and 2mM L-Glutamine, and stored at 4 °C for up to a month.
-
Exosome-depleted FBS FBS is depleted of exosomes by ultracentrifugation at 100,000 × g, 10 °C for 90 min and sterilized with a 0.22 μm filter unit. Aliquots of exosome-depleted FBS can be stored at −20°C for long term.
! CAUTION To avoid contamination, the rotor needs to be first sterilized by wiping with 70% Ethanol and all tubes for ultracentrifugation should be autoclaved. The entire FBS handling process should be carried out in a Biological Safety Cabinet for tissue culture.
CRITICAL In our recent work, we have reported that negligible amount of ENPs can be contributed by the blank media supplemented with FBS that has been previously spun at 100,000 × g for 90 min to deplete ENPs11. However, researchers should judge based on their specific study goals and decide what type of media to use in their work, such as serum-free media, serum-free media supplemented with specific growth factors, or media supplemented with FBS depleted of ENPs by ultracentrifugation for longer durations (e.g. overnight). Consideration should be taken into account that the biogenesis, secretion and content of ENPs derived from cultured cells might be affected when different medium is used for culture.
20% (vol/vol) Ethanol Milli Q filtered water is used to make the dilution of Ethanol. The final solution is made freshly and filtered with a 0.22μm filter unit.
0.5 mg/mL BSA solution Dissolve BSA powder in PBS at a concentration of 0.5 mg/mL and store aliquot at −20 °C for long term.
1% (vol/vol) Contrad 70 Dilute Contrad 70 with Milli Q water to a final concentration of 1% (vol/vol), and store at room temperature (RT, ~22 °C) for long term.
10% (wt/vol) SDS Dissolve SDS powder in Milli Q water at a concentration of 10% (wt/vol) (i.e.10 g per 100 mL) and store at RT for long term. ! CAUTION This chemical is corrosive and toxic, and can cause severe skin and eye irritation. Wear protective gloves, mask, eyeshield, faceshield, and protective clothing to handle.
10% (vol/vol) Nitric Acid Dilute nitric acid with Milli Q water to a final concentration of 10% (vol/vol), and store at RT for long term. ! CAUTION This chemical is highly corrosive and can cause severe eye and skin burns, severe respiratory and digestive tract burns. Wear proper protective equipment (gloves, eyeshield, faceshield, clothing, respirators) and handle it in a chemical hood. The waste should be treated as a hazardous waste following state and local hazardous waste regulations.
EQUIPMENT SETUP
All the parts of the AF4 instrument should be installed, configured, calibrated and certified by the manufactures (Agilent and Wyatt Technology) before utilization.
PROCEDURE
Preparation of sEVs from the conditioned media of cell culture. TIMING ~3 days
CRITICAL: B16-F10 murine melanoma cell line is used as a model system in this protocol. A schematic flow diagram summarizing the key steps of the entire procedure and the flow route of AF4 is shown in Figure 7.
-
1
Seed ~2.25×106 B16-F10 cells per P150 tissue culture plate in 25mL of the DMEM complete medium supplemented with exosome-depleted FBS, and seed a total of 12 plates.
CAUTION Cell lines should be regularly checked to ensure that they are authentic and free of mycoplasma contamination. We conduct monthly examination using the ATCC Universal Mycoplasma Detection Kit (ATCC, Catalog # 30–1012K) to ensure our cells are free of mycoplasma contamination.
CAUTION Follow sterile procedure at this step.
CRITICAL STEP The passage number of the B16-F10 cells influences sEV composition, reflected by the changes in the relative abundance of different subsets of sEVs (7). So, avoid comparing experimental data using B16-F10 cells with a big difference (e.g. a difference of 8 passages as reported in Ref. 11) in their passage numbers.
-
2
Keep cells in a humidified tissue culture incubator for 72 h under standard conditions (5%CO2, 37 °C). The cell culture should just reach confluence without cell death and any abnormal phenotypical changes.
CRITICAL STEP Cells should be allowed to reach confluence to get highest sEV yield, but no cell death and stressed phenotype (e.g. cells demonstrate a needle-like morphology or spread like a large square) should be apparent by the harvesting time to ensure the purity of the sEVs.
CAUTION Follow sterile procedure at this step.
-
3
Collect the conditioned media from all 12 plates of B16-F10 cells into 50 mL conical tubes and centrifuge at 500 × g at 10°C for 10 min in a table-top centrifuge.
PAUSE POINT Alternatively, the supernatant can be spun at 3000 × g at 10 °C for 20 min in the table-top centrifuge, transferred to new tubes and placed at −80 °C for long term storage. Though the overall structural integrity of ENPs are maintained after the freezing/thawing steps, how this freezing/storage step may influence the functionality of ENPs has not been well examined in the field. Researchers should decide how to proceed through the whole protocol at the listed pause points based on their specific study design. However, avoid freezing-thawing or keeping samples on ice for a long period when feasible.
-
4
Transfer the supernatant to ultracentrifuge tubes (6 × 50 mL/tube) and centrifuge at 12,000 × g at 10°C for 20min in Type 45-Ti ultracentrifuge rotor (pre-chilled at 4°C).
CRITICAL STEP For ultracentrifugation, the opposing pair of tubes across the center of rotation need to be balanced with each other. Do not load more than 50 mL per tube to avoid sample spilling. The rotors should be kept at 4 °C when not in use.
-
5
Transfer the supernatant to new ultracentrifuge tubes and centrifuge at 100,000 × g at 10°C for 70min in the same rotor.
CRITICAL STEP At the end of each ultracentrifugation step, make sure the supernatant is transferred immediately to avoid the loosening of the pellet and either contaminating the supernatant or losing the pelleted samples.
-
6
Discard the supernatant and resuspend the pellets in 1 mL of ice-cold PBS gently. Avoid introducing air bubbles. Combine all the sample into one ultracentrifuge tube and bring the final volume to 50 mL with ice-cold PBS.
-
7
Centrifuge at 100,000 × g at 10 °C for another 70 min. Resuspend the final pellet in ~0.5 mL of PBS and transfer to a 1.7 mL microcentrifuge tube on ice (the sEV sample for AF4 fraction in the next section).
CRITICAL STEP The pellet sometimes may be hard to break and difficult to resuspend into a homogeneous suspension. If so, the samples can be kept on ice for another 15 to 30 min or an extra volume of PBS can be added to the sample. Then gently pipette up and down to resuspend the samples and transfer to a 1.7 mL microcentrifuge tube for quantification. Avoid introducing air bubbles.
-
8
Quantify the sEV yield by taking a 10 μL aliquot and measuring the protein concentration using the Pierce BCA Protein Assay Kit. Follow the manufacture’s instruction to mix the samples or BSA standards provided by the kit with the reagents in a 96-well plate and incubate at 37 °C for 30 min. Read the absorbance at 562 nm using the AccuScan GO UV/Vis Microplate Spectrophotometer and calculate the concentration of the samples based on the BSA standards included. Meanwhile, keep the rest of the sample on ice for the same day or next day AF4 fractionation, or freeze at −80°C for long term storage.
PAUSE POINT The samples can be frozen at −80°C for long term storage.
?TROUBLE SHOOTING
Assemble the AF4 channel TIMING ~1 h
-
9
Before assembling the AF4 channel, select the membrane type and cutoff size (see discussion in “Overview of the procedure and development of the AF4 protocol: membrane choice”. we use RC membrane with 10 KDa cutoff size) and the spacer with desired thickness (i.e. the channel height, we use 490μm) first.
-
10
Rinse all parts of the AF4 channel (i.e. the top and bottom plates, spacer, membrane, O- ring, and frit) with Milli Q water and assemble in the order of the top plate, spacer, membrane and bottom plates with the frit/O-ring in place). Bolt the parts together using a torque wrench with 5 Nm and 7.5 Nm torques applied sequentially.
CRITICAL STEP Since the sample specimen is positioned in the channel laminae very close to the membrane, it is critical that the membrane should be smooth and unruffled. Wear gloves and do not bend the membrane during the assembly procedure.
CRITICAL STEP Since ethanol can cause the “membrane swelling” phenomena and reduce the effective channel height for fractionation, avoid exposing the membrane to ethanol.
CRITICAL STEP It is critical that the channel is tightly sealed and its height is precise and even across the whole channel. To assemble the AF4 channel, a metered wrench such as a torque wrench should be used to apply force precisely. A good practice is to tighten the two bolts in the center first and then the ones at the corners in diagonal order.
-
11
Connect the tubing to the inlet, crossflow and injection ports but with the outlet port unconnected. Program the flow rate settings and operate using ChemStation to start to run water at a channel flow rate of 1 mL/min through the system for at least 30 min and let air in the channel run out from the outlet port. Then connect the tubing from the outlet to the detectors. No air bubbles should be observed in the channel.
PAUSE POINT For short term storage, the instrument can be operated in the Night Rinse mode with a constant channel flow of 0.2 mL/min overnight, up to a few days.
CRITICAL STEP Do not leave the system in still aqueous solvents for a long period. For long term storage, dissemble the membrane from the channel and maintain the system in 20% ethanol.
CRITICAL STEP Keep the tubing from the channel to the detectors and the fraction collector as short as possible to reduce the peak broadening and sample dilution effects and to avoid decreases in separation resolution.
?TROUBLE SHOOTING
Equilibrate and coat the membrane with BSA TIMING 2 ~ 3 h
-
12
Change the aqueous solvent to PBS, and keep running at a channel flow of 1 mL/min for at least 30 min to 1 h.
PAUSE POINT The instrument can be operated in the Night Rinse mode overnight, with a constant channel flow of 0.2 mL/min.
CRITICAL STEP If the system has been maintained in ethanol or isopropanol, it should be flushed completely with water first before switching to PBS. Mixing PBS with alcohol will cause salt precipitation.
?TROUBLE SHOOTING
-
13
[Optional step: This step is only needed when a new membrane is installed.] Load 30 to 40 μg of BSA (0.5 mg/mL) onto AF4 and run the sample using the same AF4 method as for sEV fractionation (following steps 14–24). Exceptions are there is no need to collect fractions for BSA coating; and for the second elution step illustrated in Table 1, use a constant crossflow of 3 mL/min for 15 min for BSA instead. Repeat, by running BSA 1–2 more times.
PAUSE POINT The instrument can be operated in the Night Rinse mode overnight, up to a few days after membrane coating.
CRITICAL STEP The purpose of this step is to block non-specific binding of the samples to the membrane. The sample to be analyzed, if extra sample is available, can be used for this blocking step, too. This step is only needed when a new membrane is installed.
CRITICAL STEP At the end of the day, after all samples have been processed, turn on the COMET of the DAWN HELEOS II detector for about 30 min to clean the flow cell.
?TROUBLE SHOOTING
AF4 fractionation of sEVs TIMING 1–2h per sample
-
14
To initialize the instrument, first open Chemstation and load the AF4 running method as described in Table 1 (The running method is programmed, edited and saved in the “Method” module of Chemstation).
-
15
Set both thermostats for the autosampler and the fraction collector at 4 °C; turn on the UV lamp for the MWD detector (280 nm) at least 30 min before sample analysis; turn on the laser for DAWN HELEOS II (664 nm).
-
16
Turn on the fraction collector and choose the collection mode (either based on volume or time interval); install 96-well plates for fraction collection (Ensure plates are installed according to the configuration of the fraction collector). We collect fractions based on time intervals of 0.5 min, so two plates are needed to collect the fractions from one sample.
CRITICAL STEP All operations should be done using the ChemStation software except for switching on the laser of DAWN HELEOS II using the instrument’s front control panel.
-
17
Open Astra 6 and start a new experiment file for data collection.
For Configuration, select “PBS, aqueous” as the system solvent; specify UV wavelength at 280 nm” and enable “Band Broadening” option; for HELEOS, enable “Band Broadening” and “Temperature Control” options; and for QELS, select “Use QELS dithering”. For Procedure: specify the time interval for MALS data collection at 1 sec and QELS interval at 2 sec; set the duration for data collection at 60 min; select “Trigger on Auto-Inject”;
Click the “Run” button and the data collection will automatically start once triggered by the signal from the autosampler.
-
18
Prepare the AF4 input samples by adjusting the concentration of the sEVs isolated in Step 8 to 1 μg/μL with PBS. Spin at 12,000 × g at 4 °C for 5 min to remove insoluble aggregates right before loading onto AF4.
CRITICAL STEP Pre-spinning of the sample before loading onto AF4 is critical to avoid analyzing artifacts of aggregates formed during the high-speed ultracentrifugation.
?TROUBLE SHOOTING
-
19
Transfer the supernatant into a pre-chilled screw cap glass vial. Use a 250 μL pulled point glass vial insert if the total volume of the sample is small, and put it onto the autosampler platform at the designated position from which the autosampler is set to pick up the sample automatically.
-
20
In the Autosampler module of Chemstation, specify the sample volume to analyze (40 to 100 μL; i.e. 40 to 100 μg at 1 μg/μL), and then click the “Single Sample” to start the fractionation, real time data collection (MALS, DLS, and UV absorption), and fraction collection (by time slice of 0.5 min, fraction volume will be 0.5 mL/fraction since the forward channel flow (i.e. the detector flow) rate is set at 1 mL/min).
CRITICAL STEP Our pilot study has determined a range from 40 to 100 μg of B16-F10 sEVs is suitable for the current AF4 running method we have developed. This will need to be further adjusted for specific samples due to their composition complexity following discussion in the “Overview of the procedure and development of the AF4 protocol: Amount of input sample”.
CRITICAL STEP It is critical to avoid getting air bubbles into the system. Make sure that no air bubbles are trapped in the sample vial and have a larger volume of sample in the vial than the volume to be analyzed.
?TROUBLE SHOOTING
-
21
During the running of the sample, check the real flow rates in the panel of “Wyatt Eclipse Status” and make sure they are close enough (within the range certified by the manufacture) to the set flow rates.
CRITICAL STEP If the real flow rates are quite different from the set ones, something is wrong with the flow control and repairing/maintenance by the manufacturer is needed to ensure the fractionation quality.
CRITICAL STEP It is critical to keep the channel flow rate (the detector flow) constant during the fractionation. Changes in the flow rate can cause artifact signal detection by the monitors.
-
22
Once the fractionation is finished, take the 96-well plates out of the fraction collector and seal them using adhesive tape. Keep the plates on ice or at 4 °C for the next procedure (see steps 28–33).
-
23
Click “Reset the fraction collector” so that the starting position for fraction collection is reset to its original position. Otherwise, the instrument will resume the fraction collection of the next sample from the last fraction position of the previous sample.)
-
24Perform real-time data analysis after each run:
- In Astra 6 select the experiment to be analyzed and adjust the baselines: we usually set up the baseline for the MALS signal collected from t7he LS 11 (90°) detector first, and then apply it to all the other detectors. Make sure to check individual detectors to ensure the baselines are set up correctly.
- Select peak regions to analyze. Either single or multiple regions can be selected to analyze simultaneously.
- Examine the fractionation quality by checking the fitting of the autocorrelation function at representative fractions to a single exponential model. The closer of R2 to 1, the better purity of the separation.
-
Open a new window of EASI Graph, and plot Hydrodynamic Radius (Rh), QELS (DLS) and UV signals versus time. The hydrodynamic radius (Rh) of particles is deduced solely from DLS signal using equations described above in Experimental design. The Hydrodynamic Radius (Rh) plot displays the size of particles eluted at each time point. The UV signal intensity can reveal the relative abundance of particles with different sizes. Based on these plots (and together with potential post-separation characterizations following steps 28–33), one can judge the AF4 separation quality and the sample composition (i.e. the relative abundance of particles with different sizes). Other types of analysis can be plotted as well by choosing different axes to display in EASI Graph according to the need.CRITICAL STEP Besides real-time UV detection, other means of quantification such as BCA assay and NTA analysis can be used to measure the concentration of the fractions (see steps 28–33).CRITICAL STEP The sample concentration should be high enough to scatter enough light for accurate Rh determination, especially for small size particles as they scatter much less light.?TROUBLE SHOOTING
-
25
To fractionate more of the same biological sample or a new sample, install new 96-well fraction collection plates and start fractioning the next sample by repeating steps 17–24.
CRITICAL STEP If multiple samples need to be analyzed but the collection of separated fractions is not required, a “Sequence” (a series of methods and samples programmed to be run sequentially) can be run instead of the method for a single sample. Users can refer to the manual from the manufacturer for details.
-
26
Run a blank control of the running buffer (i.e. PBS or other types of running buffer used to analyze the samples) using the same AF4 running method for the samples on the same day by following steps 17–24. This blank control can help evaluate the instrument performance such as background noise level and identify systemic problems than may influence sample analysis.
CRITICAL STEP ENPs isolated from the equal volume of blank media can be run as a control in parallel to those isolated from the conditioned medial of specific cell lines. This will yield the information on the potential contamination of ENPs present in the medium in the final ENP product.
-
27
After all samples have been analyzed for the day, turn on the COMET of the DAWN HELEOS II for ~30 min and switch off the laser, UV lamp, thermostats and fraction collector. Change the aqueous solvent to water and run in the Night Rinse mode overnight or up to a few days. For long term storage, dissemble the membrane from the channel, run ~50–100 mL of 20% ethanol through the system, and then maintain the system in 20% ethanol.
Fraction collection and concentration for post-separation characterization. TIMING 1–2h
-
28
Depending on the characterization to be conducted, examine the individual fractions directly or pool fractions with similar properties (especially from the same peak region) for further concentration (see step 29) before examination. If a high amount of material is required for the downstream characterization, repeat multiple runs of the same sEV sample and combine similar fractions from each run.
-
29
Concentrate individual fraction or pooled fractions of sEVs using Millipore centrifugal filter columns with Ultracel-30 membrane (30 kDa cutoff).
First, pre-rinse the filter columns by adding 5 mL (for Ultra-4 filter column) or 15 mL (for Ultra-15 filter column) of ice-cold PBS followed by spinning at 3,700 × g at 4 °C for 5 min. Discard the flow-through and liquid remaining in the top filter columns.
-
30
Transfer pooled fractionated samples into the top filter column and spin at 3,700 × g at 4 °C for 7–8 min. Retain the concentrated samples in the top filter columns and discard the flow-through buffer which collects at the bottom of the collection tubes.
-
31
Repeat step 30 until each sample is concentrated to the desired volume (e.g. <100 μL for Ultra-4 filter column and <200 μL for Ultra-15 filter column). For each sample, the same filter column can be repeatedly loaded and spun to concentrate the sample.
-
32
Transfer the concentrated samples to 1.7 mL microcentrifuge tubes on ice. Record the volume and take an aliquot (5 to 10 μL) for BCA measurements to determine the protein concentration.
-
33
Store the concentrated samples on ice for short-term storage (up to 2~3 days) or frozen at −80°C for long term storage. Downstream molecular characterizations (see discussion in Experimental design) and functional study can be followed up on these concentrated fractionated samples.
CRITICAL STEP For an unknown sample that is analyzed using AF4 for the first time, we check the morphology of representative individual fractions by TEM first before pooling fractions together for further analysis.
CRITICAL STEP It is possible that particles with same hydrodynamic size but different morphology are eluted together from AF4. Other means to separate these particles based on their distinct biophysical/biochemical properties (such as density, surface molecule expression, and charge) should be explored in combination with AF4 for further fractionation.
CRITICAL STEP We usually pool fractions together based on the hydrodynamic size, morphology and purity of representative fractions. If baseline separation of two adjacent, distinct populations of particles is not achieved, avoid collecting those fractions in the “valley” between the peaks of two populations for further characterization.
?TROUBLE SHOOTING
TROUBLESHOOTING
See Table 2 for Troubleshooting guidance.
Table 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 8 | Low sEV yield | Low cell confluence by the CM harvest time | Seed a higher number of cells per plate or a bigger number of plates, or use a longer cell cutlure time |
| Lost the sEV pellet of 100,000x g ultracentrifugation | Remove the supernatant immediately from the sEV pellet of 100,000x g ultracentrifugation | ||
| Abnormally high sEV yield | Contamination due to inefficient washing | Resuspend the pellet from Step 6 completely and use a large volume of PBS to wash in Step 7 | |
| Too much carryover of media in Step 6 | Invert the tubes from Step 6 on paper towel to drain the leftover of media or suck it off using the vacuum system before washing with PBS | ||
| Contamination from the pellet of 12,000x g ultracentrifugation | Transfer the supernatant immediately to new tubes from the pellet of 12,000x g ultracentrifugation in Steps 4–5 | ||
| 11 | Leaking channel and/or tubing connection | The AF4 channel was not assembled properly | Reassemble the channel following Steps 10–11. Make sure the channel is assembled using the proper and precise force with a torque wrench |
| The O-ring is damaged or not placed properly | Change to a new O-ring if it is damaged; install it evenly and smoothly in the groove along the frit on the bottom plate of the channel | ||
| The screw thread is damaged or worn out | Replace with new screws, or use a piece of Teflon (polytetrafluoroethylene (PTFE) film) tape to help seal the thread | ||
| 12 | high LS background noise | Particle contaminants in the system | Change the inline filter for mobile phase solution |
| Thoroughly rinse the membrane with Milli Q water before assembling; equilibrate the membrane in the AF4 channel with PBS overnight; flush the channel thoroughly before connecting to detectors. | |||
| 13, 20 | No signal or much lower signal than expected for the sample | Membrane installed inproperly | Reassemble the channel and make sure the smooth side of the membrane facing the inside of the channel |
| Lost sample due to leaking channel/connection | See above troubleshooting for the Step 11 “Leaking channel and/or tubing connection” | ||
| Lost sample due to damaged membrane | Replace with new membrane | ||
| Inefficient focus | Use a colored sample such as blue Dextran to test the focus efficiency. A narrow band of the sample should be located close to the inject port. If not, increase the focus time post the injection step | ||
| 13, 20 | High noise | Contaminant present in the mobile phase solution | Filter the buffer before use and use an inline filter (0.1 μm, and change it routinely, about once a month) |
| If compatible with downstream analysis, include sodium azide in the mobile phase solution | |||
| Particle contaminants in the system | Use COMMET after running samples to clean the MALS flow channel | ||
| Flush the channel and the system thoroughly with a large volume of filter water; | |||
| If flushing with water does not solve the problem, clean the detectors by running and incubating in 10% SDS, 1% Contrad 70, or 10% Nitric acid for 30 min up to overnight, then thoroughly rinse with filtered water | |||
| 13, 20 | Baseline drifting | Particle contaminants in the system | See above troubleshooting for the Step 13, 20 “High noise” |
| Collect AF4 profile of PBS blank control right before or after the AF4 fractionation of the samples of interest, and then use the PBS control profile to perform baseline subtraction from the profile of the samples. | |||
| Laser performance quality decreased | Replace with new laser | ||
| 13, 20 | Sharp jump in signal intensity | Unstable voltage | Use the power supply that provides stable voltage and current |
| Air bubble introduced into the system | Degas the buffer before use and/or use an online degaser; make sure there is no air bubble in the sample vial and have an excess of sample to inject | ||
| 13, 20 | Sample elutes too early or too late than expected | Aberrant flow rate | Compare the real flow rate versus the set flow rate shown in the panel of “Wyatt Eclipse Status”. If the difference is big, repair by the manufacture is required. |
| 18 | Too large pellet | Insufficient resuspension of sEV pellet in Step 8. | Repeat pipetting up and down gently to resuspend the pellet; use a larger volume of PBS to resuspend the pelleted sEVs. To get more accurate loading of the sample, conduct the BCA assay upon the supernatant from the brief spin at 12,000 xg in Step 18. |
| 20 | A shift toward late elution | Membrane used for extended period and bound non-specifically with contaminant | Replace with a new membrane |
| 20 | The signal not reaching the baseline level by the end of AF4 fractionation | Not enough elution time | Use a longer cross flow gradient time and/or a longer time for the last two steps (Elution, Elution + Inject) of the AF4 running method |
| 24 | Curling-up tail of the Rh plot at the small Rh end | Inaccurate Rh determination from the DLS measurement due to insuffient amount of the sample, especially for particles of small size | Increase the amount of the sample to analyze |
| Insufficient separation of the particles, especially at the beginning of AF4 fractionation | Use other means such as EM and NTA analysis to validate the purity | ||
| Increase the Focus time after the injection step | |||
| Increase the initial cross flow rate and a longer time span of fractionation to allow better separation | |||
| 24 | Scattered Rh plot | Inaccurate Rh determination from the DLS measurement due to insuffient amount of the sample, especially for particles of small size | Increase the amount of the sample to analyze |
| High background noise | See above troubleshooting for the Step 13, 20 “High noise” | ||
| 24 | Too big P5-corresponding peak | Not enough elution time | Use a longer cross flow gradient time |
| 33 | No sample recovered | Lost sample due to damaged membrane of the filter unit | Save the flow through and apply to new filter unit for concentration |
TIMING
Step 1–8, cell culture and isolation of sEVs: ~3 days.
Step 9–11, the AF4 channel assembling: ~1 h.
Step 12–13, Equilibration and coating the membrane with BSA: 2 ~ 3 h.
(Step 13 are only needed when a new membrane is installed.)
Step 14–27, AF4 fractionation of sEVs: 1–2 h per sample.
Step 28–33, fraction collection and concentration for post-separation characterization: 1–2 h.
ANTICIPATED RESULTS
We have provided a detailed protocol for optimal sEV preparation and fractionation via AF4. We have highlighted and discussed the key steps for successful of AF4 separation: (i) the preparation of sEVs from conditioned media of cell culture; (ii) development and optimization of the AF4 running methods; (iii) real-time data analysis and fraction collection for post-separation characterization.
Pre-stratification of the sEVs using methods such as UC is critical to reduce the complexity of the samples to be analyzed in their particle composition. This allows enough material for each subpopulation of sEVs present in the samples to be analyzed by a single run of AF4. Otherwise, a series of AF4 methods for best separation of particles within different size ranges have to be adapted. Another key factor for successful AF4 analysis and fractionation is the amount of input samples loaded onto the AF4 system. Overloading the system will result in poor resolution and inefficient separation of nanoparticles; whereas loading too little a sample will lead to poor signal detection and inaccurate data deduction, as shown in Figure 6.
Five major parameters for AF4 running method optimization have been discussed, including cross flow, channel height, focus time, loading amount, and membrane type (see Figures 2–5). A representative AF4 fractionation profile of B16-F10 derived sEVs is shown in Figure 8. Based on the method described here, three major subpopulations of sEVs are identified (Figure 8a, i.e. exomeres, Exo-S and Exo-L, corresponding to peaks P2, P3 and P4, respectively). The autocorrelation function is a key factor to determine the purity of each fraction (Figure 8b). The separated particles can be further recovered and usually need further concentration for a variety of post-separation analyses, such as TEM, NTA, BCA assay, biophysical/biochemical property characterization, molecular composition determination, and functional studies. Shown in Figure 8c is TEM imaging analysis of combined fractions for B16-F10 exomeres, Exo-S and Exo-L, revealing the distinct morphology of each sEV subset. As a reference, an estimation has been calculated using three independent experiments for the final yield of each subpopulation from two representative cell lines (B16-F10 and AsPC-1) (Supplementary Figure 1). Resulted from an input of 100 μg sEVs, the yield of exomeres, Exo-S and Exo-L from B16-F10 are 2.7 μg, 10.7 μg and 5.2 μg, respectively; and 8.4 μg, 4.5 μg and 1.7 μg from AsPC-1, respectively. The concentration of an individual fraction immediately post AF4 fractionation is too dilute to be measured accurately using the BCA assay. However, a rough estimation by back calculation yields the following average concentration for each subpopulation: 0.36 μg/mL, 0.88 μg/mL and 0.66 μg/mL of exomeres, Exo-S and Exo-L from B16-F10, respectively; and 2.8 μg/mL, 0.76 μg/mL and 0.27 μg/mL of exomeres, Exo-S and Exo-L from AsPC-1, respectively. Due to the loss of samples during the concentration step, the original concentration of an individual fraction should be slightly higher. One point to make is that the yield of each subpopulation depends on the specific biological sample (for example, different cell lines and cell culture conditions) and sample preparation. The stringency for combining fractions for each subpopulation and the recovery rate of the concentration step have to be considered as well.
Supplementary Material
Supplementary Figure 1. Estimation of the yield recovered for eoxmeres, Exo-S and Exo-L derived from B16-F10 and AsPC-1. Shown in (a) and (b) are the yield recovered for exomere, Exo-S and Exo-L derived from B16-F10 and AsPC-1 (100μg of sEV input for AF4), respectively. (c) and (d) show the average concentration of unconcentrated fraction post AF4 fractionation for exomere, Exo-S and Exo-L derived from B16-F10 and AsPC-1, respectively. Data are presented as mean ± s.e.m. and three independent experiments for each cell type are analyzed.
ACKNOWLEDGEMENT
We are grateful for the great AF4 technical support from Wyatt Technology. We thank our colleagues Irina Matei and Candia Kenific for comments on this protocol. Our study was supported by National Cancer Institute (U01-CA169538, D.L.), National Institutes of Health (R01-CA169416, D.L.; R01-CA218513, D.L. and H.Z.), United States Department of Defense (W81XWH-13-1-0249, D.L.), W81XWH-13-1-0427 (D.L.), Sohn Conference Foundation (H.Z.), the Children’s Cancer and Blood Foundation (D.L.), The Manning Foundation (D.L.), The Hartwell Foundation (D.L.), The Nancy C. and Daniel P. Paduano Foundation (D.L.), The Starr Cancer Consortium (D.L. and H.Z.), Pediatric Oncology Experimental Therapeutic Investigator Consortium (POETIC, D.L.), James Paduano Foundation (D.L.), National Institutes of Health/WCM CTSC (NIH/NCATS (UL1TR00457) (H.Z.); NIH/NCATS (UL1TR002384) (D.L. and H.Z.)). Thompson Family Foundation (D.L.), and Malcolm Hewitt Wiener Foundation (D.L.)
Footnotes
COMPETING INTERESTS
The authors have no competing financial interests.
DATA AVAILABILITY STATEMENT
All Astra 6 data files used for producing the plots presented in figures have been deposited at https://figshare.com/s/6f22aede51fb279a3f81.
REFERENES
- 1.Théry C, Zitvogel L & Amigorena S Exosomes: composition, biogenesis and function. Nat Rev Immunol 2, 569–579 (2002). [DOI] [PubMed] [Google Scholar]
- 2.EL Andaloussi S, Mäger I, Breakefield XO & Wood MJ Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 12, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Raposo G & Stoorvogel W Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Vizio D et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res 69, 5601–5609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morello M et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle 12, 3526–3536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minciacchi VR et al. MYC Mediates Large Oncosome-Induced Fibroblast Reprogramming in Prostate Cancer. Cancer Res 77, 2306–2317 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Balaj L et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2, 180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi DS, Kim DK, Kim YK & Gho YS Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 13, 1554–1571 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Thakur BK et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 24, 766–769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tetta C, Ghigo E, Silengo L, Deregibus MC & Camussi G Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine 44, 11–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 20, 332–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aalberts M et al. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol Reprod 86, 82 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G & Bonnerot C Exosomal-like vesicles are present in human blood plasma. Int Immunol 17, 879–887 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Huebner AR et al. Exosomes in urine biomarker discovery. Adv Exp Med Biol 845, 43–58 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Ogawa Y et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull 34, 13–23 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Admyre C et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol 179, 1969–1978 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Navabi H et al. Preparation of human ovarian cancer ascites-derived exosomes for a clinical trial. Blood Cells Mol Dis 35, 149–152 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Street JM et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med 10, 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Théry C, Amigorena S, Raposo G & Clayton A Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3.22 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Merchant ML et al. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin Appl 4, 84–96 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Lässer C, Eldh M & Lötvall J Isolation and characterization of RNA-containing exosomes. J Vis Exp, e3037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 10, 505–511 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jørgensen M et al. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J Extracell Vesicles 2, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tauro BJ et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Fraunhofer W & Winter G The use of asymmetrical flow field-flow fractionation in pharmaceutics and biopharmaceutics. Eur J Pharm Biopharm 58, 369–383 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Yohannes G, Jussila M, Hartonen K & Riekkola ML Asymmetrical flow field-flow fractionation technique for separation and characterization of biopolymers and bioparticles. J Chromatogr A 1218, 4104–4116 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Giddings CJ A New Concept Based on a Coupling of Concentration and Flow Nonuniformities. Separation Science 1, 123–125 (1966). [Google Scholar]
- 28.Berg HC, Purcell EM & Stewart WW A Method for Separating according to Mass a Mixture of Macromolecules or Small Particles Suspended in a Fluid, 3. Experiments in a Centrifugal Fluid. Proc. Natl. Acad. Sci. USA, 58, 1818–1828 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang FJF, Myers MN & Giddings JC Programmed sedimentation field-flow fractionation. Analytical Chemistry 46, 1924–1930 (1974). [Google Scholar]
- 30.Giddings JC, Martin M & Myers MN High-speed polymer separations by thermal field-flow fractionation. Journal of Chromatography A 158, 419–435 (1978). [Google Scholar]
- 31.Caldwell KD & Gao YS Electrical field-flow fractionation in particle separation. 1. Monodisperse standards. Anal Chem 65, 1764–1772 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Giddings JC, Yang FJ & Myers MN Flow-field-flow fractionation: a versatile new separation method. Science 193, 1244–1245 (1976). [DOI] [PubMed] [Google Scholar]
- 33.Granger J, Dodds J, Leclerc D & Midoux N Flow and diffusion of particles in a channel with one porous wall: Polarization chromatography. Chemical Engineering Science 41, 3119–3128 (1986). [Google Scholar]
- 34.Wahlund KG & Giddings JC Properties of an asymmetrical flow field-flow fractionation channel having one permeable wall. Anal Chem 59, 1332–1339 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Litzén A & Wahlund KG Improved separation speed and efficiency for proteins, nucleic acids and viruses in asymmetrical flow field flow fractionation. J Chromatogr 476, 413–421 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Wahlund KG & Litzén A Application of an asymmetrical flow field-flow fractionation channel to the separation and characterization of proteins, plasmids, plasmid fragments, polysaccharides and unicellular algae. J Chromatogr 461, 73–87 (1989). [DOI] [PubMed] [Google Scholar]
- 37.Yohannes G et al. Miniaturization of asymmetrical flow field-flow fractionation and application to studies on lipoprotein aggregation and fusion. Anal Biochem 354, 255–265 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Wittgren B & Wahlund K-G Fast molecular mass and size characterization of polysaccharides using asymmetrical flow field-flow fractionation-multiangle light scattering. Journal of Chromatography A 760, 205–218 (1997). [Google Scholar]
- 39.Wei Z et al. Biophysical characterization of influenza virus subpopulations using field flow fractionation and multiangle light scattering: correlation of particle counts, size distribution and infectivity. J Virol Methods 144, 122–132 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Chuan YP, Fan YY, Lua L & Middelberg AP Quantitative analysis of virus-like particle size and distribution by field-flow fractionation. Biotechnol Bioeng 99, 1425–1433 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Oh S et al. Miniaturized asymmetrical flow field-flow fractionation: application to biological vesicles. J Sep Sci 30, 1082–1087 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Sitar S et al. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal Chem 87, 9225–9233 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Petersen KE et al. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal Bioanal Chem 406, 7855–7866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashby J et al. Distribution profiling of circulating microRNAs in serum. Anal Chem 86, 9343–9349 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Cölfen H & Antonietti M Field-flow fractionation techniques for polymer and colloid analysis. Adv. Polym. Sci, 150, 67–187 (2000). [Google Scholar]
- 46.Litzen A & Wahlund CG Zone broadening and dilution in rectangular and trapezoidal asymmetrical flow field-flow fractionation channels. Analytical Chemistry 63, 1001–1007 (1991). [Google Scholar]
- 47.Batrakova EV & Kim MS Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release 219, 396–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeppesen DK et al. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J Extracell Vesicles 3, 25011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cvjetkovic A, Lötvall J & Lässer C The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bobrie A, Colombo M, Krumeich S, Raposo G & Théry C Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles 1, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willms E et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep 6, 22519, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mol EA, Goumans MJ, Doevendans PA, Sluijter JPG & Vader P Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 13, 2061–2065 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Nordin JZ et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 11, 879–883 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Böing AN et al. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willis GR, Kourembanas S & Mitsialis SA Toward Exosome-Based Therapeutics: Isolation, Heterogeneity, and Fit-for-Purpose Potency. Front Cardiovasc Med 4, 63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu R, Greening DW, Rai A, Ji H & Simpson RJ Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87, 11–25 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Xu R, Simpson RJ & Greening DW A Protocol for Isolation and Proteomic Characterization of Distinct Extracellular Vesicle Subtypes by Sequential Centrifugal Ultrafiltration. Methods Mol Biol 1545, 91–116 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Ko J et al. miRNA Profiling of Magnetic Nanopore-Isolated Extracellular Vesicles for the Diagnosis of Pancreatic Cancer. Cancer Res. 78, 3688–3697 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Wyatt Technology Corporation. DYNAMICS User’s Guide. Version 7.0 (M1400 Rev. I) Appendix A-2 (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Estimation of the yield recovered for eoxmeres, Exo-S and Exo-L derived from B16-F10 and AsPC-1. Shown in (a) and (b) are the yield recovered for exomere, Exo-S and Exo-L derived from B16-F10 and AsPC-1 (100μg of sEV input for AF4), respectively. (c) and (d) show the average concentration of unconcentrated fraction post AF4 fractionation for exomere, Exo-S and Exo-L derived from B16-F10 and AsPC-1, respectively. Data are presented as mean ± s.e.m. and three independent experiments for each cell type are analyzed.