Abstract
Nociceptors located in the trigeminal ganglion (TG) and DRG are the primary sensors of damaging or potentially damaging stimuli for the head and body, respectively, and are key drivers of chronic pain states. While nociceptors in these two tissues show a high degree of functional similarity, there are important differences in their development lineages, their functional connections to the CNS, and recent genome-wide analyses of gene expression suggest that they possess some unique genomic signatures. Here, we used translating ribosome affinity purification to comprehensively characterize and compare mRNA translation in Scn10a-positive nociceptors in the TG and DRG of male and female mice. This unbiased method independently confirms several findings of differences between TG and DRG nociceptors described in the literature but also suggests preferential utilization of key signaling pathways. Most prominently, we provide evidence that translational efficiency in mechanistic target of rapamycin (mTOR)-related genes is higher in the TG compared with DRG, whereas several genes associated with the negative regulator of mTOR, AMP-activated protein kinase, have higher translational efficiency in DRG nociceptors. Using capsaicin as a sensitizing stimulus, we show that behavioral responses are greater in the TG region and this effect is completely reversible with mTOR inhibition. These findings have implications for the relative capacity of these nociceptors to be sensitized upon injury. Together, our data provide a comprehensive, comparative view of transcriptome and translatome activity in TG and DRG nociceptors that enhances our understanding of nociceptor biology.
SIGNIFICANCE STATEMENT The DRG and trigeminal ganglion (TG) provide sensory information from the body and head, respectively. Nociceptors in these tissues are critical first neurons in the pain pathway. Injury to peripheral neurons in these tissues can cause chronic pain. Interestingly, clinical and preclinical findings support the conclusion that injury to TG neurons is more likely to cause chronic pain and chronic pain in the TG area is more intense and more difficult to treat. We used translating ribosome affinity purification technology to gain new insight into potential differences in the translatomes of DRG and TG neurons. Our findings demonstrate previously unrecognized differences between TG and DRG nociceptors that provide new insight into how injury may differentially drive plasticity states in nociceptors in these two tissues.
Keywords: DRG, mTOR, neuropathic pain, TG, TRAP
Introduction
Mechanical, thermal, and chemical peripheral stimuli are detected by the pseudo-unipolar sensory neurons of the DRG and the trigeminal ganglion (TG) (Devor, 1999; Woolf and Ma, 2007; Dubin and Patapoutian, 2010). Neurons in the DRG transmit signals from the limbs and body, including much of the viscera, to the CNS through the dorsal horn of the spinal cord. TG neurons relay sensory information from the head and face through a region of the dorsal brainstem known as the trigeminal nucleus caudalis. Although TG and DRG neurons express similar markers and are often considered as very similar, there are differences in their cellular populations (Price and Flores, 2007). The tissues also have distinct embryonic origins with important functional consequences (Durham and Garrett, 2010). Finally, neurons in these ganglia innervate distinct targets in the periphery (e.g., the teeth and dura mater for the TG) and in the CNS. An excellent example of this differential innervation in the CNS is the discovery of a subset of TG nociceptors that bypass the traditional second-order relay in the nucleus caudalis projecting directly to the parabrachial nucleus (Rodriguez et al., 2017). These findings suggest distinct molecular signatures of DRG and TG neurons that may be important for understanding sensory neurobiology from these different regions of an organism.
Advances in next-generation sequencing have allowed the characterization of DRG and TG tissues at the genome-wide level using RNA sequencing (RNA-seq) (Manteniotis et al., 2013; Reynders et al., 2015; Gong et al., 2016; Hu et al., 2016; Kogelman et al., 2017). These studies provide significant insight into genes that are differentially expressed between these tissues, including differences between species (Manteniotis et al., 2013; Flegel et al., 2015; Kogelman et al., 2017). However, these studies lack cell-type specificity and fail to capture translational efficiency. Cell specificity is a key advantage for single-cell transcriptomic methods (Usoskin et al., 2015; Hu et al., 2016) and other cellular enrichment protocols (Isensee et al., 2014; Thakur et al., 2014; Lopes et al., 2017) that have now been applied to the DRG and/or TG. However, only one direct comparison has thus far been made between TG and DRG transcriptomes using neuronal enrichment followed by RNA-seq (Lopes et al., 2017). Examining ribosome-bound RNA is advantageous because there is strong evidence that transcriptional and translational efficiencies are decoupled in most cells (Fortelny et al., 2017). Methods that sequence ribosome-bound RNAs give more accurate predictions of cellular proteomes (Heiman et al., 2008; Ingolia, 2016). Two techniques have emerged in this area. The first, ribosome footprint profiling, comprehensively and quantitatively provides a snapshot of translation activity at single codon resolution through deep sequencing of ribosome-protected mRNA fragments from cells or tissues (Ingolia, 2016). This technique, which has recently been applied to the DRG (Uttam et al., 2018), does not allow insight into cell-type-specific translational profiling. A second technique is translating ribosome affinity purification (TRAP), which relies on genetic tagging of ribosomal proteins for cell-specific pulldown of translating ribosomes bound to mRNAs for RNA-seq (Doyle et al., 2008; Heiman et al., 2008, 2014). This technique lacks the single codon resolution of ribosome footprint profiling but allows for precise assessment of cellular translatomes in vitro and in vivo.
Here we used the TRAP technology using the Nav1.8Cre mouse (Stirling et al., 2005) to achieve sensory neuron-specific ribosome tagging with enrichment in the nociceptor population. We then compared TG and DRG nociceptor translatomes and quantified mRNAs that are differentially expressed at the transcriptional and/or translational level. Interestingly, we found that translational activity of mechanistic target of rapamycin (mTOR)-related genes is higher in the TG compared with DRG. Given the key role that this signaling pathway plays in rapid sensitization of nociceptors (Khoutorsky and Price, 2018), this result is intriguing because activation of nociceptors in the facial region produces greater sensitization and perceived pain in human subjects (Schmidt et al., 2015, 2016), an effect that our experiments also demonstrate in mice. Therefore, our work pinpoints important signaling differences between DRG and TG nociceptors that have direct functional consequences on the susceptibility of these nociceptors to rapid sensitization.
Materials and Methods
Transgenic animals: Nav1.8cre/Rosa26fsTRAP mice.
All animal procedures were approved by the Institutional Animal Care and Use Committee of University of Texas at Dallas.
Rosa26fsTRAP mice were purchased from The Jackson Laboratory (stock #022367). Transgenic mice expressing Cre recombinase under the control of the Scn10a (Nav1.8) promoter were obtained initially from Professor John Wood (University College London) but are commercially available from Infrafrontier (EMMA ID: 04582). The initial characterization of these mice demonstrated that the introduction of the Cre recombinase in heterozygous animals does not affect pain behavior, and their DRG neurons have normal electrophysiological properties (Stirling et al., 2005). Nav1.8cre mice on a C57BL/6J genetic background were maintained and bred at the University of Texas at Dallas. Upon arrival, Rosa26fsTRAP mice were crossed to Nav1.8cre to generate the Nav1.8-TRAP mice that express a fused EGFP-L10a protein in Nav1.8-expressing neurons. All experiments were performed using male and female littermates 8–12 weeks old. Mice were group housed (4 maximum) in nonenvironmentally enriched cages with food and water ad libitum on a 12 h light-dark cycle. Room temperature was maintained at 21 ± 2°C.
TRAP.
Nav1.8-TRAP male and female mice were decapitated and DRG and TG rapidly dissected in ice-cold dissection buffer (1× HBSS; Invitrogen, 14065006), 2.5 mm HEPES, 35 mm glucose, 4 mm NaHCO3, 100 μg/ml cycloheximide, 0.001V 2 mg/ml emetine). DRGs or TGs were transferred to ice-cold polysome buffer (20 mm HEPES, 12 mm MgCl2, 150 mm KCl, 0.5 mm DTT, 100 μg/ml cycloheximide, 20 μg/ml emetine, 40 U/ml SUPERase IN, Promega, 1 μl DNase, and protease inhibitor) and homogenized using a Dounce homogenizer. Samples were centrifuged at 3000 × g for 10 min to prepare postnuclear fraction (S1). Then, 1% NP-40 and 30 mm 1,2-dihexanoyl-sn-glycero-3-phosphocholine were added to the S1 fraction and then centrifuged at 15,000 × g for 15 min to generate a postmitochondrial fraction (S20). A 200 μl sample of S20 was removed for use as Input, and 800 μl of S20 was incubated with protein G-coated Dynabeads (Invitrogen) bound to 50 μg of anti-GFP antibodies (HtzGFP-19F7 and HtzGFP-19C8, Memorial Sloan Kettering Centre) for 3 h at 4°C with end-over-end mixing. Anti-GFP beads were washed with high salt buffer (20 mm HEPES, 5 mm MgCl2, 350 mm KCl, 1% NP-40, 0.5 mm DTT, and 100 μg/ml cycloheximide), and RNA was eluted from all samples using a Direct-zol kit (Zymo Research) according to the manufacturer's instructions. RNA yield was quantified using a Nanodrop system (Thermo Fisher Scientific), and RNA quality was determined by fragment analyzer (Advanced Analytical Technologies).
Library generation and sequencing.
Libraries were generated from 100 ng to 1 μg of total RNA using Quantseq 3′ mRNA-Seq library kit (Lexogen) with RiboCop rRNA depletion kit (Lexogen) treatment according to the manufacturer's protocols. The endpoint PCR amplification cycle number for each sample was determined by qPCR assay with PCR Add-on kit for Illumina (Lexogen). The cycle number was selected when the fluorescence value reached 33% of the maximum for each sample. Purified libraries were quantified by Qubit (Invitrogen), and the average size was determined by fragment analyzer (Advanced Analytical Technologies) with high-sensitivity next-generation sequencing fragment analysis kit. Libraries were then sequenced on an Illumina NextSeq500 Sequencer using 50 bp single-end reads.
Sequence files generated by the Illumina NextSeq500 Sequencer were downloaded from BaseSpace. An initial quality check using FastQC 0.11.5 (Babraham Bioinformatics) was done on the sequencing files, and then trimming was performed on the server with the FASTQ Toolkit. Sequences were trimmed with optimized parameters (13 bases from 3′ end, 17 bases from 5′ end, and any poly-adenine longer than 2 bases from the 3′ side). Trimming parameters were optimized based on FastQC results and mapping rate, as well as manually checking high reads or abundant chromosomal regions with IGV 2.3.80. The trimmed sequencing samples were then processed using TopHat 2.1.1 (with Bowtie 2.2.9) and mapped to the mouse reference genome (NCBI reference assembly GRCm38.p4) and reference transcriptome (Gencode vM10) generating files in .bam format. Processed .bam files were then quantified for each gene using Cufflinks 2.2.1 with gencode.vM10 genome annotation. Because reads only mapped to the 3′UTR of the gene, read counts were not normalized by length by using the Cufflinks option − no-length-correction. Relative abundance for the ith gene was determined by calculating TPM (transcripts per million) values as follows:
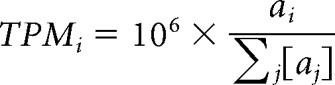 |
where aj is the Cufflinks reported relative abundance. Finally, TPM values were normalized to upper decile for each biological replicate, and udTPM (upper decile TPM) were used for analysis (Glusman et al., 2013). This was done to provide uniform processing for samples with varying sequencing depth and because of varying number of genes in the transcriptome and translatome samples.
Behavioral procedures.
Female C57BL/6J mice were injected subdermally with capsaicin (0.1 μm) into either cheek or hindpaw in a volume of 10 μl with Hamilton syringe and 30G needle. For cheek injections, mice cheeks were shaved 3 d before injections. AZD8055 (mTORC1 inhibitor) or vehicle was administered intraperitoneally (10 mg/kg) 2 h before capsaicin injections into the cheek. AZD8055 was dissolved in DMSO (50 mg/ml) and further diluted in 30% (w/v) cyclodextrin to make up the correct dose for each animal. Vehicle consisted of 10% DMSO and 30% w/v cyclodextrin. Baseline videos were recorded for 15 min for each mouse. After cheek or hindpaw injections, experimental videos were recorded for 60 min. The recording setup consisted of one camera in front and one in the back. The sum of facially directed behaviors with the forepaws following injection of capsaicin into the whisker pad as well as the number of hindpaw directed behaviors for the hindpaw were scored and classified as nocifensive behaviors.
The Mouse Grimace Scale was used to quantify affective aspects of pain in mice (Langford et al., 2010). We scored the changes in the facial expressions (using the facial action coding system) at baseline and then 15 and 30 min after intraplantar or facial injection of capsaicin.
qRT-PCR.
Lumbar DRGs and TGs were isolated from 4 male mice per genotype and flash-frozen on dry ice and stored at −80°C until ready to be processed. Tissues were homogenized using a pestle, and total RNA was extracted using RNAqueous Total RNA Isolation kits (Thermo Fisher Scientific). RNA was subsequently treated with TURBO DNase (Thermo Fisher Scientific) according to the manufacturer's instructions. RNA concentration was measured on a NanoDrop 2000 (Thermo Fisher Scientific). cDNA was synthesized using iScript Reverse Transcriptase (Bio-Rad). qRT-PCR was done using a Applied Biosystems Lightcycler 7500 Real-Time PCR system using iTaq Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions with 3 technical replicates per biological replicate (averages of the technical replicates per biological replicate are reported) using primers pairs: Gapdh forward 5′-GACAACTTTGGCATTGTGGA-3′ and Gapdh reverse 5′-CATCATACTTGGCAGGTTTCTC-3′, Rraga forward 5′-ACGTCCGATTCTTGGGGAAC-3′ and Rraga reverse 5′-TACGGAAGATGTTGTCCCGC-3′, Fth forward 5′-GCACTGCACTTGGAAAAGAGT-3′ and Fth reverse 5′-ACGTGGTCACCCAGTTCTTT-3′. Primers were made by Integrated DNA Technologies.
Primer efficiency curves were determined by diluting total RNA of DRG and TG samples with 6 points of 1:5 serial dilutions. RNA dilutions were then converted to cDNA, and standard curves were determined for DRG and TG with each primer set separately. Concentrations resulting in multiple products or incorrect product size via melt-curve analysis (derivative reporter vs temperature) were omitted. Efficiencies for each primer set for DRGs and TGs were calculated using the Applied Biosystems 7500 software version 2.3. Total RNA (115 ng) used in experiments fell within primer standard curves with efficiencies between 85% and 110%. Data were analyzed as 2−ΔΔCt and normalized as shown in Results.
Antibodies.
The peripherin antibody used for immunohistochemistry were obtained from Sigma-Aldrich. Isolectin B4 (IB4) conjugated to AlexaFluor-568 and secondary AlexaFluor antibodies were purchased from Invitrogen. Calcitonin gene-related peptide (CGRP) antibody was purchased from Peninsula Laboratories. RagA and Akt1s1 (also known as PRAS40) antibodies were from Cell Signaling Technology. Antibodies for TRAP (HtzGFP-19F7 and HtzGFP-19C8) were obtained from Sloan Memorial Kettering Centre, after establishing Material Transfer Agreements with the laboratory of Prof. Nathaniel Heintz (Rockefeller University).
Immunohistochemistry.
Animals were anesthetized with isoflurane (4%) and killed by decapitation, and tissues were flash-frozen in OCT on dry ice. Sections of TG (20 μm) were mounted onto SuperFrost Plus slides (Thermo Fisher Scientific) and fixed in ice-cold 10% formalin in 1× PBS for 45 min, and then subsequently washed 3 times for 5 min each in 1× PBS. Slides were then transferred to a solution for permeabilization made of 1× PBS with 0.2% Triton X-100 (Sigma-Aldrich). After 30 min, slides were washed 3 times for 5 min each in 1× PBS. Tissues were blocked for at least 2 h in 1× PBS and 10% heat-inactivated normal goat serum. TG or DRG slices were stained with peripherin, CGRP, and IB4 conjugated to AlexaFluor-568. Immunoreactivity was visualized following 1 h incubation with goat anti-rabbit, goat anti-mouse, and goat anti-guinea pig AlexaFluor antibodies at room temperature. All immunohistochemical images are representations of samples taken from 3 animals per genotype. Images were taken using an Olympus FluoView 1200 confocal microscope. Analysis of images was done using ImageJ Version 1.48 for Apple OSX (National Institutes of Health).
Western blotting.
Male and female mice were used for all Western blotting experiments and were killed by decapitation while under anesthesia and tissues (DRG or TG) were flash frozen on dry ice. Frozen tissues were homogenized in lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, pH 8.0, and 1% Triton X-100) containing protease and phosphatase inhibitors (Sigma-Aldrich), and homogenized using a pestle. A total of 15 μg of protein was boiled for 5 min in loading dye and then loaded into each well and separated by a 10%–12% SDS-PAGE gel. Proteins were transferred to a 0.45 μm PVDF membrane (Millipore) at 25 V overnight at 4°C. Subsequently, membranes were blocked with 5% nonfat dry milk in 1× Tris buffer solution containing Tween 20 (TTBS) for 3 h. Membranes were washed in 1× TTBS 3 times for 5 min each, then incubated with primary antibody overnight at 4°C. The following day, membranes were washed 3 times in 1× TTBS for 5 min each, then incubated with the corresponding secondary antibody at room temperature for 1 h. Membranes were then washed with 1× TTBS 5 times for 5 min each. Signals were detected using Immobilon Western Chemiluminescent HRP substrate (Millipore). Bands were visualized using film (Kodak) or with a Bio-Rad ChemiDoc Touch. Membranes were stripped using Restore Western Blot Stripping buffer (Thermo Fisher Scientific) and reprobed with another antibody. Analysis was performed using Image Lab (Bio-Rad).
Statistics.
All data are presented as mean ± SEM. All analysis was done using GraphPad Prism 6 version 6.0 for Mac OS X. Single comparisons were performed using Student's t test or one-way ANOVA if multiple groups were compared. For behavioral experiments, two-way ANOVA (time × treatment) was used to measure effects across time between different groups. If significant effects were found by ANOVA, post hoc analyses were performed. Multiple comparisons between groups/within groups were performed using Sidak's correction. Statistical results can be found in the figure legends.
Statistics for RNA sequencing.
Differential expression analysis was performed using MATLAB scripts. TPM values were normalized to their 90th percentile to generate udTPMs, and the probability density function of the udTPM was used to set the threshold value for further analysis. Genes showing consistent expression above the set threshold across biological replicates were then used to generate lists of differentially expressed genes. Standard t test was first performed assuming unequal variances between experimental groups generating p values for each gene as follows. A q value for the ith test was then calculated using Benjamini–Hochberg correction for multiple comparisons as follows:
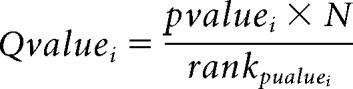 |
where N is the number of tests.
Finally, the cumulative density function of the fold change was plotted and used to set the fold change for the input and TRAP fraction for both DRG and TG datasets. Gene set enrichment analyses were performed with Enrichr (Kuleshov et al., 2016) using the Gene Ontology molecular function 2015 term, the biological process 2015 term, and the Reactome 2015 libraries.
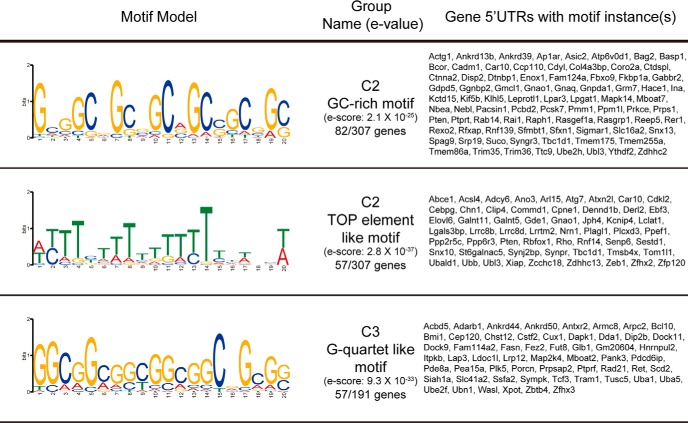
For motif finding, 5′-UTR sequences of corresponding genes were obtained from gencode.vM10 (mouse genome assembly GRCm38), with all transcript isoforms kept for analysis. As most 5′-UTRs of different isoforms from the same gene share partial/whole sequences with each other, when a 5′-UTR sequence was fully shared by another longer 5′-UTR isoform of the same gene, the shorter version was removed to prevent genes with a large amount of isoforms being overrepresented in the motif analysis. All 5′-UTR sequences remaining after filtering were then passed through MEME Suite 5.0.2 for motif discovery, with the following parameters: all motifs are within 10–20 bp length range, only found on the provided strand, and appear in at least 10% of the genes provided. Motifs appearing in >30% of the genes with significant E value are shown in the text.
Results
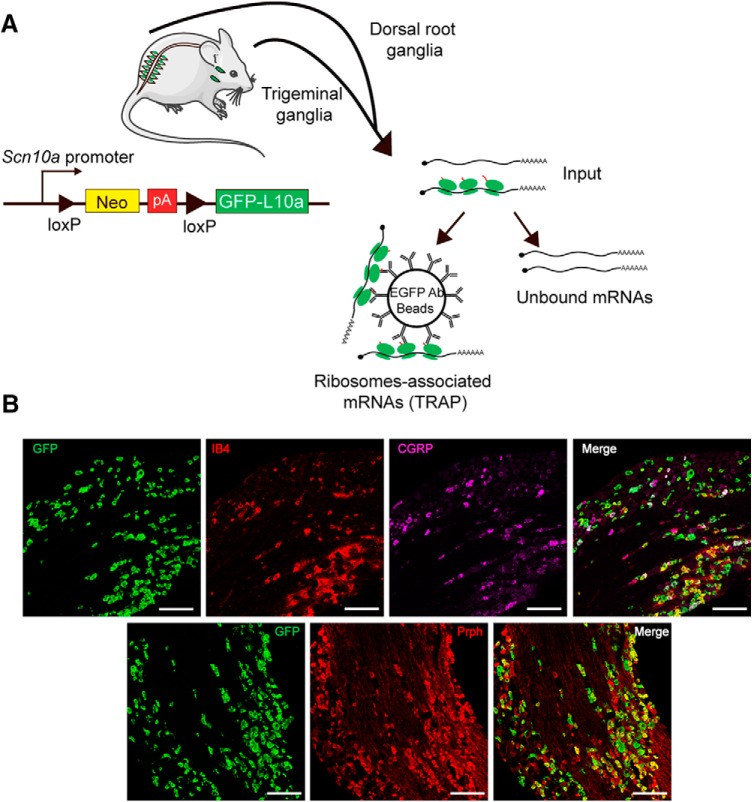
To generate nociceptor-TRAP mice, Nav1.8cre animals were crossed with Rosa26fs-TRAP (Zhou et al., 2013) to express the eGFP fused to the ribosomal L10a protein in Nav1.8+ neurons. This approach generates Nav1.8-TRAP neurons in both the DRG and TG. While the specificity of our approach was recently shown in the DRG (Megat et al., 2019), we characterized expression of the transgene in the TG (Fig. 1A). We found that eGFP-L10a-positive neurons primarily colocalized with small-diameter peripherin-positive neurons and that extensive overlap was found with both CGRP immunoreactivity and with IB4 staining (Fig. 1B). These findings demonstrate that this technique labels an equivalent subset of neurons in the DRG and TG of mice.
Figure 1.
TRAP-seq strategy and expression in TG. A, Schematic representation of TRAP-seq approach showing isolation of translating ribosomes with immunoprecipitation using anti-GFP-coated beads. B, Immunostaining of CGRP, IB4, and peripherin (Prph) on TG sections from Nav1.8-TRAP mice (GFP). Scale bar, 100 μm.
Having confirmed that the Nav1.8-TRAP approach yields robust expression in nociceptors in the TG, we set out to conduct TRAP sequencing to compare nociceptor translatomes in the DRG and TG. To successfully isolate ribosome-associated mRNAs from Nav1.8-TRAP cells, we determined that TGs from 4 animals were required for a single biological replicate. This number matches the number of DRGs needed for TRAP sequencing. To make comparisons between the TG and DRG, we generated TRAP sequencing from the TG that was then compared with our previously generated DRG dataset (GSE 113941). We sequenced the total mRNA input from all biological replicates and mRNAs associated with translating ribosomes in the Nav1.8 subset of TG neurons, equivalently to what was done from DRG (Megat et al., 2019). This approach allowed us to make comparisons between the whole tissue transcriptional and Nav1.8+ neuron translational landscapes between DRG and TG.
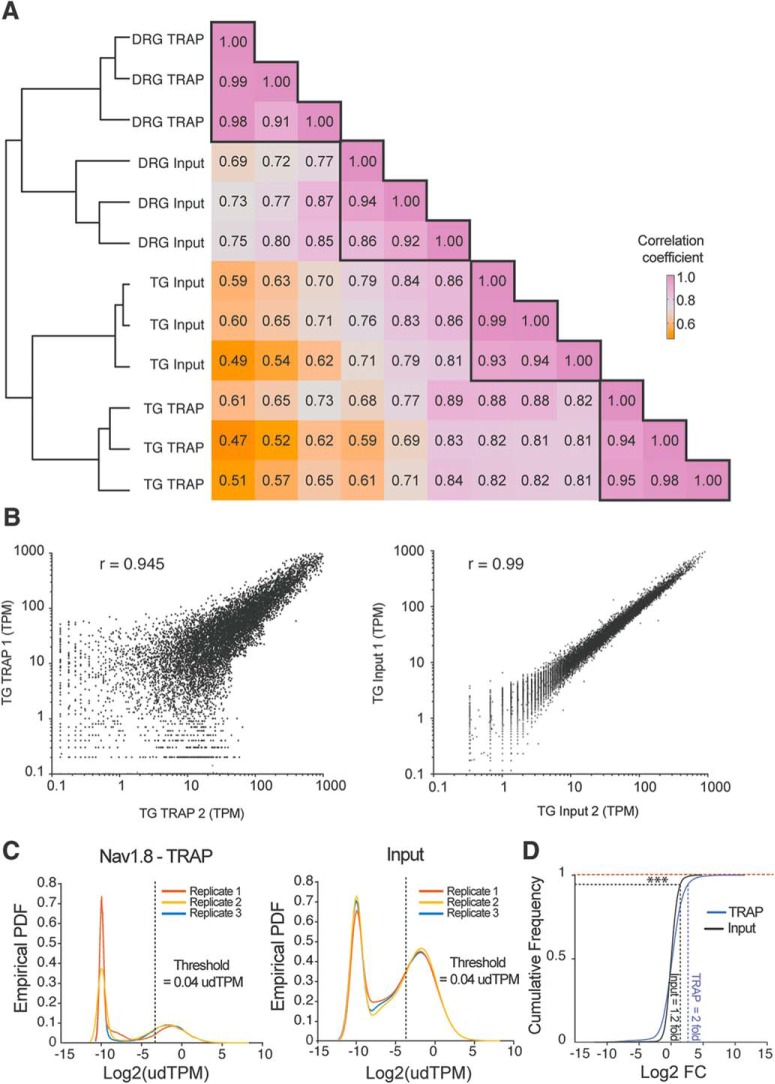
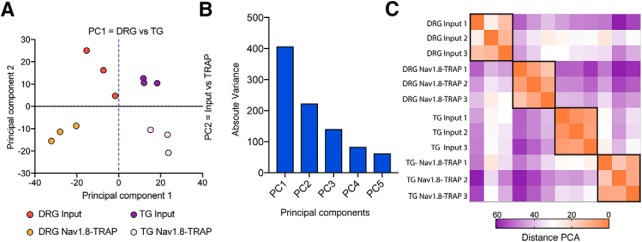
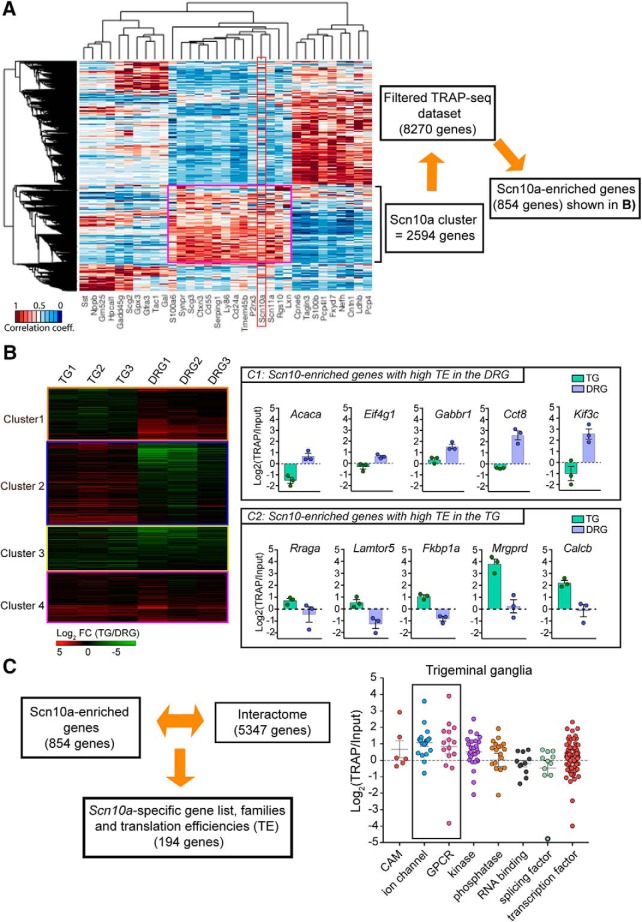
The first dimension of the clustering analysis identified clear differences between TG and DRG as well as distinctions within each subcluster comprised of the input (transcriptome) and TRAP (translatome) RNA sequencing (Fig. 2A). We observed strong correlation coefficients between biological replicates demonstrating low variability in the experimental protocol (Fig. 2B). Gene expression values (TPMs) were normalized to the 90th percentile for each biological replicate, and the empirical probability density function of the normalized expression level (upper decile (ud)TPM) was plotted for the input and TRAP fractions (Fig. 2C). The probability density function identified 2 peaks, and the inflection point was used to set the threshold expression values according to the sequencing depth (Fig. 2C). After further filtering, based on consistent expression among biological replicates, we included a total of 7358 genes in the final analysis to make comparisons between the DRG and TG transcriptomes and Nav1.8-TRAP translatomes. Finally, we plotted the cumulative frequency distribution as a function of the log twofold change for each of these 7358 genes in TG and DRG biological replicates, and the 95th percentile was used to set the threshold fold change values for the input and TRAP fractions (Fig. 2D). Principal component (PC) analysis indicated that PC1 distinguished between TG and DRG, whereas PC2 detected a difference between input and Nav1.8-TRAP, suggesting a clear transcriptional and translational signature for both of these tissues (Fig. 3A). Detailed analysis of the variances for each PC clearly showed that the first 2 PCs (PC1 = difference between DRG and TG transcriptomes; and PC2 = difference between the DRG and TG Nav1.8-TRAP translatomes) explained the majority of the variance seen in the dataset (Fig. 3B). Further clustering analysis confirmed the findings of the PC analysis (Fig. 3C).
Figure 2.
DRG and TG TRAP-seq shows high correlation between biological replicates and similar sequencing depth. A, Heatmap of the correlation coefficient and cluster analysis showing clear separation between DRG and TG as well as in between TRAP-seq and bulk RNA-seq from each tissue. B, Scatter plot of input and TRAP-seq shows high correlation between biological replicates for each approach. C, Empirical probability density function (PDF) of the TPM for all genes in analysis shows a similar distribution between replicates, which are each shown as a different color, for TRAP-seq and input. D, Cumulative distribution of the fold change (FC) in input and TRAP-seq shows higher FCs in TRAP-seq samples. Kolgomorov-Smirnov test, ***p < 0.001.
Figure 3.
PC analysis shows a clear difference between transcriptomes and translatomes in TG and DRG. A, PC analysis shows that differences between TG and DRGs whole tissue transcriptomes represent the first PC, whereas differences between transcriptome and translatome are the second PC. B, Absolute variances for each PC show that PC1 and PC2 provide the majority of variation in the entire datasets. C, Heatmap of the absolute PC distances showing 4 distinct clusters, each of which is defined by whole transcriptome (input) versus TRAP-seq and the tissue.
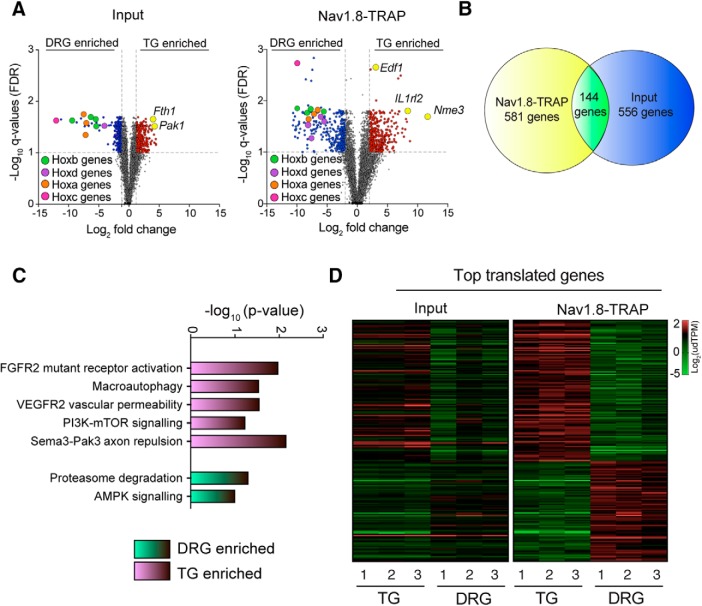
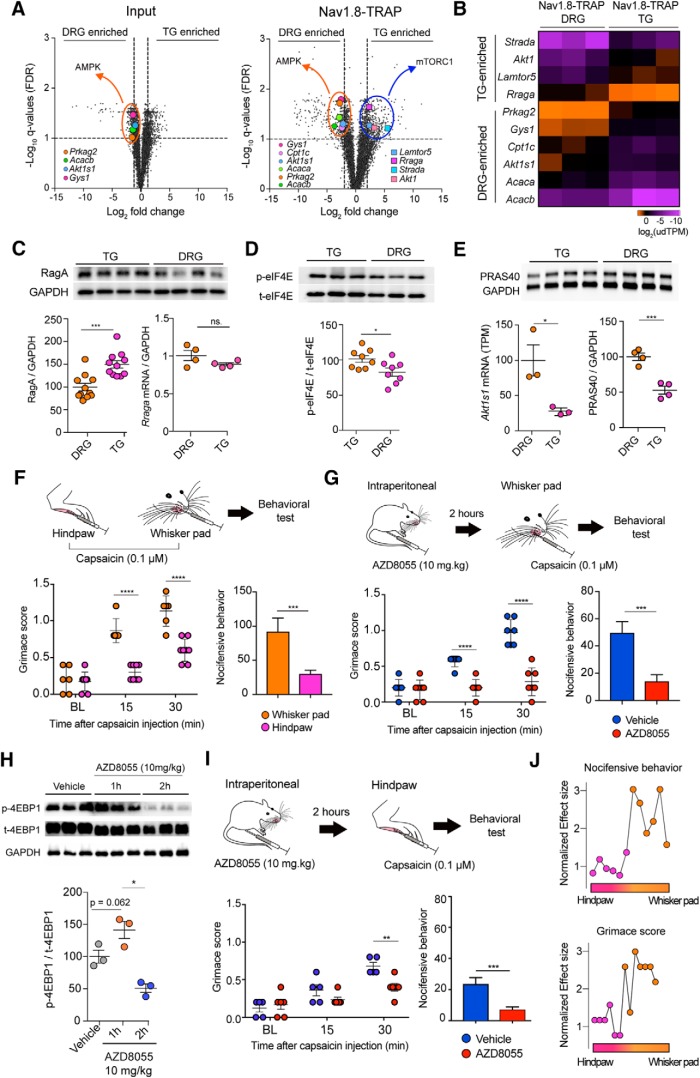
Analysis of the input transcriptome data between TG and DRG revealed that 379 genes were significantly enriched in the TG and 315 in the DRG (Fig. 4A; Tables 1, 2). Among these 315 genes in the DRG, we observed enrichment of the Hox family transcription factors (Fig. 4A). These genes are well-known regulators of rostral to caudal segmental development, so enrichment in DRG is expected given the rostral–caudal extent of the DRG (Kammermeier and Reichert, 2001). Among the 379 genes enriched in the TG, we found particularly high expression and enrichment of Fth1 and Pak1 (Fig. 4A). Analysis of the Nav1.8-TRAP dataset revealed 372 genes enriched in the TG and 348 in the DRG (Fig. 4A; Tables 3, 4). Consistent with the transcriptome results, the Hox genes showed a highly enriched translational profile in the DRG (Fig. 4A). Among the top mRNAs highly associated with ribosomes in the TG Nav1.8-TRAP dataset, we found Nme3, Il1rl2, and Edf1(Fig. 4A). None of these 3 genes has been associated with a specific TG function previously, although the Il1rl2 gene encodes a receptor for interleukin 1β (IL1β), which activates TG nociceptors through a mechanism that has previously been attributed to IL1β Type 1 receptors (Takeda et al., 2008). GO term analysis of the differentially expressed genes in the Nav1.8-TRAP datasets revealed an enrichment in specific pathways, including VEGFR, FGFR, as well as the PI3K-mTOR pathway (Fig. 4B). Interestingly, we observed an enrichment in AMP-activated protein kinase (AMPK)-related genes in the DRG-TRAP dataset (Fig. 4B). This finding is intriguing because the AMPK pathway is a negative regulator of PI3K-mTOR signaling (Hardie, 2014, 2015) and suggests shifting in the balance between these two signaling pathways between the DRG and TG.
Figure 4.
Transcriptomic and translatomic differences between the TG and DRG of mice. A, B, Volcano plots showing genes that are enriched in the DRG or TG in the whole tissue transcriptome (input) or in the TRAP-seq sample (Nav1.8-TRAP) with genes highlighted in the text labeled (yellow dots). C, GO term analysis of the TRAP-seq-enriched mRNAs in DRG or TG using EnrichR (adjusted p value < 0.05) shows an enrichment in AMPK-related genes in the DRGs, whereas mTOR-related genes are highly translated in the TG. D, Heatmaps showing the expression level of enriched mRNAs (input) and enriched translated mRNAs (Nav1.8 TRAP) in both tissues showing discordance between the transcriptome and translatome mRNA levels.
Table 1.
Genes upregulated in the TG input
| Genes | Log2 fold change | p | q | Genes | Log2 fold change | p | q |
|---|---|---|---|---|---|---|---|
| 1700037C18Rik | 1.847 | 0.006 | 0.058 | Mrpl36 | 1.645 | 0.024 | 0.100 |
| 6430548M08Rik | 1.249 | 0.000 | 0.026 | Mrpl44 | 1.331 | 0.020 | 0.093 |
| 9130401M01Rik | 1.884 | 0.023 | 0.098 | Mrpl46 | 1.476 | 0.005 | 0.052 |
| Aard | 2.059 | 0.001 | 0.030 | Mrps11 | 1.718 | 0.003 | 0.042 |
| Abca2 | 1.917 | 0.000 | 0.026 | Mrps14 | 1.614 | 0.004 | 0.050 |
| Abhd6 | 1.493 | 0.001 | 0.031 | Mrps23 | 1.332 | 0.001 | 0.031 |
| Adarb1 | 1.426 | 0.011 | 0.072 | Mrps36 | 1.262 | 0.002 | 0.034 |
| Adck2 | 1.969 | 0.004 | 0.049 | Mt2 | 3.017 | 0.023 | 0.097 |
| Ak5 | 1.564 | 0.004 | 0.050 | Mt3 | 2.988 | 0.011 | 0.071 |
| Alkbh3 | 2.585 | 0.002 | 0.039 | Mtap | 1.600 | 0.017 | 0.086 |
| Amdhd2 | 1.234 | 0.001 | 0.030 | Mtfp1 | 1.528 | 0.001 | 0.034 |
| Anapc13 | 1.952 | 0.003 | 0.046 | Mtmr4 | 1.402 | 0.005 | 0.052 |
| Ap4s1 | 1.506 | 0.006 | 0.057 | Mxd3 | 1.297 | 0.020 | 0.092 |
| Apbb1 | 3.191 | 0.005 | 0.054 | Mxra8 | 2.100 | 0.004 | 0.051 |
| Apip | 1.677 | 0.023 | 0.099 | Myl12a | 2.141 | 0.000 | 0.027 |
| Apmap | 1.356 | 0.007 | 0.061 | Mylk | 1.285 | 0.019 | 0.090 |
| Apod | 3.523 | 0.001 | 0.029 | Naa38 | 3.315 | 0.019 | 0.090 |
| Apoe | 1.621 | 0.018 | 0.088 | Nap1l2 | 1.759 | 0.002 | 0.035 |
| Arfip2 | 1.328 | 0.000 | 0.026 | Ndp | 1.418 | 0.005 | 0.053 |
| Arhgef3 | 1.405 | 0.014 | 0.079 | Ndufa12 | 1.436 | 0.012 | 0.073 |
| Arhgef4 | 1.255 | 0.005 | 0.055 | Ndufa13 | 1.921 | 0.024 | 0.100 |
| Arih2 | 1.638 | 0.008 | 0.066 | Ndufb2 | 3.404 | 0.005 | 0.053 |
| Armc5 | 1.203 | 0.020 | 0.093 | Necab3 | 1.663 | 0.007 | 0.062 |
| Arpc5l | 1.248 | 0.021 | 0.095 | Nefh | 1.820 | 0.001 | 0.031 |
| Atp5c1 | 2.858 | 0.012 | 0.074 | Nif3l1 | 1.999 | 0.018 | 0.087 |
| Atp5d | 2.923 | 0.004 | 0.052 | Nme3 | 3.589 | 0.008 | 0.063 |
| Atp5j | 2.373 | 0.011 | 0.070 | Nnat | 1.368 | 0.023 | 0.099 |
| Atp5sl | 1.335 | 0.005 | 0.053 | Nr2f6 | 1.334 | 0.016 | 0.084 |
| Atxn7l3 | 1.365 | 0.015 | 0.080 | Nsg1 | 2.505 | 0.003 | 0.041 |
| Avpi1 | 1.810 | 0.015 | 0.082 | Nsmaf | 1.511 | 0.002 | 0.037 |
| B930041F14Rik | 2.887 | 0.004 | 0.051 | Nubp2 | 2.798 | 0.000 | 0.026 |
| Bad | 1.797 | 0.005 | 0.052 | Nudt1 | 1.395 | 0.017 | 0.086 |
| Bet1l | 1.486 | 0.014 | 0.079 | Nudt13 | 1.358 | 0.007 | 0.062 |
| Bod1 | 1.529 | 0.013 | 0.077 | Odc1 | 2.129 | 0.005 | 0.053 |
| Cacng5 | 2.058 | 0.008 | 0.063 | Otud3 | 2.021 | 0.002 | 0.037 |
| Calb2 | 3.253 | 0.005 | 0.053 | P2rx6 | 1.581 | 0.009 | 0.068 |
| Calu | 1.213 | 0.001 | 0.031 | Pacs2 | 1.388 | 0.000 | 0.029 |
| Camkk1 | 1.514 | 0.013 | 0.076 | Pak1 | 3.982 | 0.001 | 0.030 |
| Casp3 | 1.506 | 0.002 | 0.035 | Pard6a | 1.806 | 0.011 | 0.072 |
| Cbx7 | 1.324 | 0.015 | 0.081 | Pced1a | 1.460 | 0.018 | 0.088 |
| Ccdc12 | 1.344 | 0.018 | 0.087 | Pcp4l1 | 1.512 | 0.000 | 0.020 |
| Ccdc124 | 1.505 | 0.015 | 0.082 | Pdia4 | 1.410 | 0.002 | 0.037 |
| Ccdc63 | 3.251 | 0.018 | 0.088 | Pdlim2 | 2.857 | 0.001 | 0.030 |
| Cd81 | 1.413 | 0.005 | 0.055 | Pex11b | 2.281 | 0.017 | 0.086 |
| Cda | 1.665 | 0.003 | 0.044 | Pgbd5 | 1.688 | 0.001 | 0.031 |
| Cdc37 | 1.442 | 0.014 | 0.079 | Pin1 | 1.948 | 0.013 | 0.076 |
| Cdk5r1 | 1.482 | 0.005 | 0.055 | Pkdcc | 1.610 | 0.003 | 0.045 |
| Cdpf1 | 1.216 | 0.014 | 0.079 | Pkm | 1.408 | 0.014 | 0.080 |
| Cdr2l | 1.629 | 0.015 | 0.082 | Pla2g16 | 2.793 | 0.000 | 0.022 |
| Cela1 | 1.982 | 0.014 | 0.080 | Plcd4 | 1.204 | 0.015 | 0.082 |
| Cenpf | 3.061 | 0.020 | 0.091 | Plekha4 | 1.487 | 0.001 | 0.030 |
| Cep19 | 1.321 | 0.005 | 0.053 | Plk5 | 2.167 | 0.007 | 0.063 |
| Cgrrf1 | 2.813 | 0.002 | 0.035 | Pllp | 1.905 | 0.002 | 0.040 |
| Chchd1 | 2.920 | 0.001 | 0.029 | Plpp1 | 2.148 | 0.003 | 0.046 |
| Chchd3 | 1.514 | 0.018 | 0.088 | Plxdc1 | 1.659 | 0.003 | 0.046 |
| Chga | 1.707 | 0.007 | 0.063 | Pnpla2 | 1.296 | 0.013 | 0.078 |
| Chgb | 1.656 | 0.002 | 0.039 | Polr2b | 1.215 | 0.021 | 0.095 |
| Chmp6 | 2.654 | 0.003 | 0.045 | Polr2l | 4.483 | 0.010 | 0.070 |
| Chpf2 | 1.312 | 0.015 | 0.082 | Pon2 | 1.207 | 0.002 | 0.040 |
| Chrac1 | 1.481 | 0.002 | 0.035 | Ppa2 | 1.552 | 0.005 | 0.052 |
| Ckmt1 | 1.257 | 0.020 | 0.093 | Ppdpf | 1.387 | 0.023 | 0.097 |
| Clcn7 | 1.490 | 0.005 | 0.053 | Ppfia4 | 2.311 | 0.001 | 0.030 |
| Clec2l | 2.448 | 0.002 | 0.036 | Ppm1f | 1.386 | 0.011 | 0.071 |
| Clu | 1.283 | 0.000 | 0.029 | Ppp1r16b | 1.798 | 0.023 | 0.097 |
| Clybl | 1.665 | 0.015 | 0.081 | Ppp2r4 | 1.920 | 0.003 | 0.044 |
| Cnnm4 | 1.420 | 0.022 | 0.097 | Prorsd1 | 1.630 | 0.018 | 0.088 |
| Cnp | 2.467 | 0.000 | 0.024 | Prpsap1 | 1.397 | 0.001 | 0.031 |
| Cnpy3 | 1.844 | 0.008 | 0.063 | Prss12 | 2.286 | 0.000 | 0.029 |
| Cnst | 1.991 | 0.017 | 0.085 | Prx | 1.489 | 0.001 | 0.033 |
| Cops3 | 1.363 | 0.008 | 0.064 | Psmb11 | 2.692 | 0.021 | 0.094 |
| Coq10a | 2.084 | 0.010 | 0.068 | Psmb7 | 2.699 | 0.003 | 0.045 |
| Cotl1 | 1.523 | 0.002 | 0.035 | Ptcd2 | 1.699 | 0.016 | 0.085 |
| Cox6b1 | 1.564 | 0.018 | 0.088 | Ptgds | 1.888 | 0.008 | 0.063 |
| Cox7a2l | 1.430 | 0.005 | 0.053 | Rab11fip5 | 1.791 | 0.008 | 0.063 |
| Cplx1 | 1.869 | 0.008 | 0.065 | Rab35 | 1.527 | 0.008 | 0.063 |
| Crip1 | 1.529 | 0.014 | 0.079 | Rab3ip | 1.739 | 0.005 | 0.053 |
| Crispld2 | 1.532 | 0.006 | 0.056 | Rad54l | 1.412 | 0.018 | 0.088 |
| Cspg5 | 1.427 | 0.010 | 0.069 | Rarres1 | 1.432 | 0.001 | 0.031 |
| Csrp2 | 2.150 | 0.001 | 0.030 | Rcor2 | 1.946 | 0.006 | 0.056 |
| Cst3 | 1.798 | 0.004 | 0.048 | Rep15 | 3.803 | 0.006 | 0.059 |
| Ctif | 1.444 | 0.023 | 0.099 | Rhbdd2 | 1.301 | 0.002 | 0.035 |
| Ctnnbl1 | 1.636 | 0.019 | 0.089 | Rhot2 | 1.367 | 0.013 | 0.076 |
| Ctsf | 1.591 | 0.000 | 0.029 | Rimklb | 1.410 | 0.021 | 0.094 |
| Cyb5a | 1.289 | 0.009 | 0.068 | Rnaseh2c | 2.276 | 0.010 | 0.069 |
| Cyc1 | 3.399 | 0.002 | 0.034 | Rnf114 | 1.743 | 0.001 | 0.029 |
| Dbi | 1.474 | 0.015 | 0.081 | Rnf121 | 1.390 | 0.005 | 0.053 |
| Dexi | 2.255 | 0.000 | 0.026 | Rnf157 | 1.611 | 0.005 | 0.055 |
| Dffa | 1.337 | 0.011 | 0.072 | Rom1 | 2.221 | 0.000 | 0.032 |
| Dhdh | 4.211 | 0.008 | 0.064 | Rpl10a | 2.066 | 0.008 | 0.063 |
| Dlg2 | 1.409 | 0.004 | 0.048 | Rprm | 1.661 | 0.004 | 0.048 |
| Dnajb9 | 1.633 | 0.001 | 0.030 | Rps27 | 2.271 | 0.023 | 0.099 |
| Dnajc11 | 1.379 | 0.004 | 0.050 | S100a4 | 1.233 | 0.003 | 0.045 |
| Dnal4 | 1.470 | 0.019 | 0.090 | Sac3d1 | 3.758 | 0.002 | 0.037 |
| Dpm3 | 2.497 | 0.012 | 0.074 | Sap18 | 1.448 | 0.024 | 0.100 |
| Dpp9 | 1.459 | 0.007 | 0.061 | Sat1 | 1.390 | 0.005 | 0.055 |
| Eaf1 | 1.246 | 0.002 | 0.035 | Scg5 | 1.738 | 0.011 | 0.072 |
| Edf1 | 2.374 | 0.017 | 0.086 | Scn4b | 1.595 | 0.005 | 0.052 |
| Efcc1 | 4.477 | 0.009 | 0.068 | Scrn1 | 1.936 | 0.001 | 0.030 |
| Egln2 | 1.685 | 0.001 | 0.031 | Scx | 2.374 | 0.004 | 0.049 |
| Eif2b2 | 2.535 | 0.001 | 0.031 | Scyl3 | 3.634 | 0.010 | 0.068 |
| Eif3l | 2.072 | 0.012 | 0.075 | Sec13 | 2.615 | 0.017 | 0.086 |
| Elp3 | 1.598 | 0.001 | 0.031 | Selm | 2.482 | 0.009 | 0.067 |
| Eme1 | 1.587 | 0.009 | 0.068 | Sepp1 | 2.699 | 0.002 | 0.035 |
| Eme2 | 1.501 | 0.006 | 0.056 | Sfxn5 | 1.714 | 0.003 | 0.046 |
| Endod1 | 1.402 | 0.002 | 0.034 | Sh3bgr | 1.391 | 0.024 | 0.100 |
| Enho | 1.447 | 0.001 | 0.031 | Sh3gl2 | 1.278 | 0.010 | 0.069 |
| Eno2 | 1.548 | 0.020 | 0.092 | Sh3rf1 | 1.571 | 0.002 | 0.038 |
| Eny2 | 1.328 | 0.009 | 0.067 | Shd | 1.737 | 0.002 | 0.038 |
| Epn3 | 1.200 | 0.014 | 0.079 | Sirt2 | 1.287 | 0.004 | 0.050 |
| Esrrg | 1.705 | 0.007 | 0.063 | Slc22a17 | 4.451 | 0.005 | 0.054 |
| Etl4 | 1.537 | 0.005 | 0.052 | Slc25a25 | 1.508 | 0.015 | 0.082 |
| Fabp3 | 1.275 | 0.022 | 0.096 | Slc25a43 | 3.074 | 0.021 | 0.095 |
| Fabp7 | 3.165 | 0.002 | 0.038 | Slc25a5 | 3.786 | 0.002 | 0.036 |
| Faim2 | 1.328 | 0.002 | 0.034 | Slc38a10 | 1.399 | 0.017 | 0.087 |
| Fam160b2 | 1.237 | 0.015 | 0.083 | Slc4a2 | 1.322 | 0.014 | 0.079 |
| Fam162a | 2.032 | 0.005 | 0.054 | Slc6a8 | 1.844 | 0.008 | 0.063 |
| Fam19a5 | 2.083 | 0.016 | 0.084 | Slc9a3r1 | 2.002 | 0.004 | 0.051 |
| Fam57b | 2.035 | 0.005 | 0.054 | Slco2b1 | 2.443 | 0.004 | 0.051 |
| Fars2 | 1.969 | 0.002 | 0.038 | Smim1 | 1.230 | 0.009 | 0.066 |
| Fbxl12 | 1.201 | 0.006 | 0.057 | Smim4 | 1.838 | 0.002 | 0.035 |
| Fbxo27 | 2.483 | 0.001 | 0.031 | Smoc2 | 1.302 | 0.004 | 0.049 |
| Fbxo44 | 1.807 | 0.004 | 0.048 | Smox | 2.826 | 0.001 | 0.030 |
| Fchsd1 | 1.429 | 0.005 | 0.053 | Smpx | 1.427 | 0.001 | 0.034 |
| Fdx1l | 1.385 | 0.018 | 0.088 | Sncb | 1.711 | 0.007 | 0.060 |
| Fhdc1 | 1.306 | 0.000 | 0.026 | Snn | 2.308 | 0.012 | 0.074 |
| Fkbp2 | 1.980 | 0.015 | 0.083 | Snx22 | 2.722 | 0.009 | 0.068 |
| Fkbp4 | 1.407 | 0.005 | 0.055 | Sphkap | 2.242 | 0.001 | 0.030 |
| Fth1 | 4.168 | 0.000 | 0.022 | Sptb | 1.567 | 0.020 | 0.093 |
| Fuca1 | 1.308 | 0.013 | 0.076 | Srm | 1.623 | 0.012 | 0.075 |
| Gatb | 1.370 | 0.008 | 0.064 | Stard3 | 1.798 | 0.002 | 0.039 |
| Glb1l2 | 2.686 | 0.001 | 0.031 | Stk32c | 1.467 | 0.022 | 0.096 |
| Gle1 | 1.391 | 0.020 | 0.091 | Stmn4 | 2.074 | 0.000 | 0.028 |
| Glyr1 | 1.263 | 0.014 | 0.079 | Stxbp6 | 1.486 | 0.024 | 0.100 |
| Gps1 | 2.939 | 0.008 | 0.065 | Suclg1 | 2.146 | 0.008 | 0.064 |
| Gpx1 | 1.719 | 0.023 | 0.097 | Supt4a | 3.247 | 0.001 | 0.031 |
| Grk6 | 1.513 | 0.007 | 0.061 | Suv420h1 | 2.426 | 0.019 | 0.090 |
| Gtf2h4 | 4.152 | 0.003 | 0.045 | Syn2 | 1.312 | 0.001 | 0.030 |
| Gtf2i | 1.710 | 0.003 | 0.045 | Syne4 | 2.723 | 0.002 | 0.037 |
| Gtf2ird1 | 2.143 | 0.010 | 0.069 | Sys1 | 1.933 | 0.003 | 0.046 |
| Haghl | 1.514 | 0.008 | 0.063 | Taf6l | 1.539 | 0.006 | 0.056 |
| Hapln4 | 1.571 | 0.012 | 0.075 | Tango2 | 1.243 | 0.008 | 0.065 |
| Harbi1 | 1.772 | 0.011 | 0.070 | Tecr | 1.980 | 0.024 | 0.100 |
| Haus8 | 3.145 | 0.010 | 0.069 | Tfb1m | 1.304 | 0.012 | 0.073 |
| Hax1 | 1.463 | 0.007 | 0.063 | Thap11 | 1.324 | 0.004 | 0.049 |
| Hebp2 | 1.567 | 0.001 | 0.033 | Tifab | 1.602 | 0.018 | 0.088 |
| Hhatl | 2.106 | 0.003 | 0.045 | Timm9 | 1.715 | 0.009 | 0.066 |
| Hid1 | 1.773 | 0.013 | 0.077 | Tmco1 | 1.240 | 0.014 | 0.079 |
| Hist3h2ba | 1.307 | 0.005 | 0.052 | Tmem101 | 1.649 | 0.020 | 0.092 |
| Hlcs | 1.772 | 0.012 | 0.075 | Tmem126a | 1.893 | 0.001 | 0.030 |
| Homer3 | 2.804 | 0.001 | 0.031 | Tmem132c | 2.205 | 0.007 | 0.059 |
| Hpca | 1.469 | 0.003 | 0.046 | Tmem14c | 1.461 | 0.022 | 0.096 |
| Hs3st1 | 1.520 | 0.012 | 0.075 | Tmem18 | 1.588 | 0.002 | 0.040 |
| Hsdl2 | 1.375 | 0.005 | 0.053 | Tmem201 | 1.336 | 0.001 | 0.030 |
| Htra1 | 2.177 | 0.007 | 0.063 | Tmem203 | 1.637 | 0.018 | 0.087 |
| Hunk | 3.047 | 0.007 | 0.061 | Tmem229b | 1.572 | 0.002 | 0.038 |
| Iba57 | 1.357 | 0.014 | 0.080 | Tmem242 | 1.386 | 0.018 | 0.088 |
| Id3 | 2.405 | 0.005 | 0.053 | Tmem25 | 2.039 | 0.001 | 0.030 |
| Idh2 | 1.732 | 0.002 | 0.036 | Tmem258 | 3.156 | 0.013 | 0.076 |
| Idh3b | 1.848 | 0.007 | 0.062 | Tmem60 | 1.268 | 0.001 | 0.031 |
| Imp3 | 2.481 | 0.007 | 0.060 | Tnfrsf1a | 1.468 | 0.005 | 0.055 |
| Impdh2 | 1.694 | 0.004 | 0.051 | Tpbgl | 2.734 | 0.008 | 0.064 |
| Inpp5j | 2.890 | 0.006 | 0.058 | Trak1 | 1.435 | 0.005 | 0.053 |
| Itih5 | 1.235 | 0.006 | 0.058 | Trappc3 | 2.247 | 0.020 | 0.093 |
| Itm2c | 1.551 | 0.002 | 0.035 | Trf | 2.565 | 0.002 | 0.037 |
| Jam3 | 2.138 | 0.000 | 0.030 | Trp53rka | 1.316 | 0.001 | 0.030 |
| Kat2a | 1.756 | 0.009 | 0.067 | Tspan3 | 1.262 | 0.012 | 0.074 |
| Kcnq4 | 2.581 | 0.001 | 0.030 | Ttc9b | 1.870 | 0.011 | 0.071 |
| Kctd15 | 2.542 | 0.016 | 0.084 | Txnl4b | 1.574 | 0.003 | 0.044 |
| Krt10 | 1.758 | 0.008 | 0.064 | Tyr | 2.829 | 0.023 | 0.097 |
| Lancl1 | 1.907 | 0.000 | 0.029 | Tyro3 | 2.928 | 0.019 | 0.089 |
| Laptm4b | 1.441 | 0.001 | 0.031 | U2af1l4 | 1.981 | 0.002 | 0.035 |
| Ldhb | 1.497 | 0.014 | 0.080 | Ube2v1 | 1.615 | 0.006 | 0.058 |
| Letm1 | 1.305 | 0.015 | 0.081 | Ubl5 | 1.577 | 0.021 | 0.093 |
| Lgi3 | 1.359 | 0.006 | 0.055 | Ufsp1 | 2.295 | 0.001 | 0.034 |
| Limd1 | 1.389 | 0.000 | 0.028 | Ulk1 | 1.439 | 0.008 | 0.064 |
| Lrp1 | 1.242 | 0.001 | 0.033 | Uqcc2 | 2.331 | 0.006 | 0.058 |
| Lyz2 | 3.211 | 0.002 | 0.040 | Uqcc3 | 1.376 | 0.005 | 0.054 |
| Lztr1 | 1.525 | 0.018 | 0.088 | Uqcrh | 1.253 | 0.016 | 0.084 |
| Maged2 | 1.849 | 0.012 | 0.074 | Vasp | 1.204 | 0.024 | 0.100 |
| Map1lc3b | 1.800 | 0.001 | 0.030 | Vim | 1.384 | 0.007 | 0.060 |
| Mark4 | 2.680 | 0.002 | 0.040 | Vwa7 | 1.728 | 0.001 | 0.033 |
| Mars | 1.517 | 0.003 | 0.046 | Wbp1 | 1.987 | 0.009 | 0.068 |
| Mat2a | 1.570 | 0.001 | 0.030 | Wfs1 | 2.149 | 0.001 | 0.031 |
| Meis2 | 1.776 | 0.004 | 0.051 | Wwox | 1.523 | 0.023 | 0.098 |
| Mgat5 | 1.417 | 0.001 | 0.032 | Yif1a | 1.236 | 0.006 | 0.059 |
| Mgst3 | 3.432 | 0.001 | 0.035 | Zfand2b | 2.929 | 0.005 | 0.054 |
| Mief1 | 1.479 | 0.013 | 0.078 | Zfp180 | 1.595 | 0.014 | 0.080 |
| Mmd2 | 2.595 | 0.003 | 0.044 | Zfp335 | 1.645 | 0.017 | 0.085 |
| Mobp | 1.996 | 0.015 | 0.082 | Zfp771 | 1.604 | 0.023 | 0.097 |
| Mpc2 | 1.877 | 0.004 | 0.052 |
Table 2.
Genes upregulated in the DRG input
| Genes | Log2 fold change | p | q | Genes | Log2 fold change | p | q |
|---|---|---|---|---|---|---|---|
| 1700019D03Rik | −1.367 | 0.001 | 0.030 | Mpped2 | −2.004 | 0.000 | 0.026 |
| 9330159F19Rik | −1.337 | 0.001 | 0.031 | Mpv17l2 | −1.509 | 0.006 | 0.055 |
| 9330182L06Rik | −1.668 | 0.006 | 0.056 | Mt-Co3 | −1.553 | 0.007 | 0.063 |
| Abca5 | −1.456 | 0.008 | 0.063 | Myc | −1.925 | 0.001 | 0.030 |
| Acacb | −1.267 | 0.010 | 0.070 | Myh1 | −4.915 | 0.002 | 0.037 |
| Acbd5 | −1.220 | 0.018 | 0.088 | Myl1 | −7.949 | 0.000 | 0.022 |
| Acpp | −1.662 | 0.000 | 0.022 | Myo1b | −1.242 | 0.004 | 0.049 |
| Acsl4 | −1.505 | 0.000 | 0.025 | Myom1 | −2.336 | 0.002 | 0.038 |
| Acta1 | −6.161 | 0.000 | 0.027 | Myt1l | −1.439 | 0.005 | 0.055 |
| Actn1 | −1.467 | 0.002 | 0.036 | Nectin1 | −1.609 | 0.010 | 0.069 |
| Adcyap1 | −1.814 | 0.003 | 0.045 | Nedd4l | −1.349 | 0.001 | 0.029 |
| Adgrd1 | −1.531 | 0.006 | 0.057 | Nek1 | −1.475 | 0.017 | 0.086 |
| Adgrf5 | −1.678 | 0.004 | 0.049 | Nfya | −1.752 | 0.018 | 0.087 |
| Adk | −2.250 | 0.000 | 0.026 | Nhs | −1.233 | 0.001 | 0.031 |
| Agtr1a | −2.244 | 0.002 | 0.038 | Nktr | −1.403 | 0.008 | 0.063 |
| Ammecr1 | −1.266 | 0.012 | 0.074 | Noct | −1.573 | 0.015 | 0.082 |
| Ank3 | −1.585 | 0.000 | 0.029 | Nptx2 | −1.297 | 0.002 | 0.040 |
| Ankrd6 | −1.453 | 0.003 | 0.043 | Nptxr | −1.481 | 0.001 | 0.031 |
| Ano3 | −1.205 | 0.003 | 0.044 | Npy2r | −2.372 | 0.007 | 0.061 |
| Arfgef2 | −1.305 | 0.000 | 0.026 | Nras | −1.633 | 0.012 | 0.075 |
| Arhgap23 | −1.415 | 0.012 | 0.074 | Nrip1 | −1.304 | 0.000 | 0.027 |
| Arhgap26 | −2.827 | 0.000 | 0.026 | Nrxn3 | −1.701 | 0.008 | 0.064 |
| Arrdc3 | −1.213 | 0.023 | 0.099 | Nt5e | −2.145 | 0.001 | 0.034 |
| Ass1 | −1.757 | 0.002 | 0.040 | Nup88 | −1.244 | 0.023 | 0.098 |
| Astn1 | −1.500 | 0.007 | 0.061 | Ocrl | −1.219 | 0.004 | 0.049 |
| Atp2b4 | −2.126 | 0.001 | 0.030 | Ormdl1 | −1.558 | 0.004 | 0.048 |
| Auts2 | −1.836 | 0.000 | 0.027 | Osbpl3 | −1.696 | 0.000 | 0.026 |
| B630005N14Rik | −1.380 | 0.017 | 0.086 | Pabpc1 | −1.654 | 0.020 | 0.093 |
| Bdnf | −1.450 | 0.003 | 0.042 | Pabpn1 | −1.450 | 0.009 | 0.067 |
| Bnc2 | −1.401 | 0.004 | 0.050 | Palm2 | −2.056 | 0.005 | 0.055 |
| Brms1l | −1.357 | 0.012 | 0.073 | Palmd | −1.212 | 0.000 | 0.026 |
| Cacna2d1 | −1.978 | 0.000 | 0.039 | Pam | −1.491 | 0.006 | 0.057 |
| Camk2a | −1.912 | 0.001 | 0.031 | Panx1 | −1.364 | 0.002 | 0.040 |
| Camk2d | −1.608 | 0.011 | 0.071 | Paqr3 | −1.230 | 0.006 | 0.058 |
| Camta1 | −1.640 | 0.013 | 0.077 | Pde10a | −1.289 | 0.001 | 0.030 |
| Capn1 | −1.429 | 0.003 | 0.042 | Pde11a | −1.938 | 0.000 | 0.021 |
| Car8 | −1.627 | 0.011 | 0.072 | Pdlim1 | −1.524 | 0.005 | 0.052 |
| Casz1 | −1.381 | 0.004 | 0.048 | Pfkp | −1.296 | 0.004 | 0.052 |
| Ccdc141 | −1.908 | 0.002 | 0.035 | Pfn1 | −2.156 | 0.001 | 0.030 |
| Cct8 | −1.364 | 0.009 | 0.068 | Pgm2l1 | −1.494 | 0.000 | 0.029 |
| Cd274 | −1.797 | 0.005 | 0.052 | Phip | −1.266 | 0.004 | 0.050 |
| Cd2ap | −1.457 | 0.012 | 0.074 | Pitpnc1 | −1.209 | 0.000 | 0.026 |
| Cd44 | −1.559 | 0.001 | 0.030 | Pitpnm2 | −1.297 | 0.015 | 0.080 |
| Cd47 | −1.588 | 0.000 | 0.020 | Pkia | −1.817 | 0.003 | 0.042 |
| Cd55 | −2.309 | 0.001 | 0.031 | Plcb3 | −1.560 | 0.000 | 0.025 |
| Cdc14b | −1.372 | 0.000 | 0.025 | Plekha6 | −1.243 | 0.008 | 0.064 |
| Celf4 | −1.240 | 0.005 | 0.052 | Plvap | −1.298 | 0.018 | 0.087 |
| Celf6 | −1.775 | 0.013 | 0.077 | Plxnc1 | −1.740 | 0.005 | 0.053 |
| Cep170 | −1.468 | 0.006 | 0.058 | Plxnd1 | −1.345 | 0.002 | 0.035 |
| Cfap157 | −1.378 | 0.002 | 0.040 | Polr2a | −1.357 | 0.001 | 0.031 |
| Chml | −1.782 | 0.012 | 0.074 | Pou1f1 | −2.038 | 0.019 | 0.089 |
| Chpt1 | −1.408 | 0.010 | 0.068 | Ppef1 | −1.257 | 0.004 | 0.049 |
| Ciapin1 | −1.263 | 0.020 | 0.093 | Ppp1r12a | −1.564 | 0.001 | 0.030 |
| Ckm | −7.336 | 0.001 | 0.031 | Ppp3ca | −1.498 | 0.000 | 0.021 |
| Clgn | −1.231 | 0.009 | 0.067 | Ppp6c | −1.218 | 0.010 | 0.068 |
| Clip2 | −2.335 | 0.003 | 0.046 | Prdm8 | −2.355 | 0.001 | 0.030 |
| Cmip | −1.580 | 0.002 | 0.037 | Prg2 | −3.717 | 0.001 | 0.030 |
| Cnot1 | −1.720 | 0.000 | 0.025 | Prkag2 | −1.331 | 0.022 | 0.097 |
| Cnot4 | −1.280 | 0.004 | 0.048 | Prkar2b | −1.651 | 0.000 | 0.025 |
| Cntrl | −1.361 | 0.004 | 0.050 | Prkca | −2.159 | 0.000 | 0.025 |
| Cpeb1 | −1.200 | 0.005 | 0.053 | Ptgdr | −1.577 | 0.002 | 0.035 |
| Cpne2 | −1.790 | 0.018 | 0.087 | Ptger1 | −1.387 | 0.008 | 0.063 |
| Cpsf7 | −1.260 | 0.010 | 0.069 | Ptms | −1.523 | 0.000 | 0.026 |
| Csrnp3 | −1.215 | 0.000 | 0.025 | Ptprt | −2.355 | 0.001 | 0.029 |
| Ctsl | −1.450 | 0.002 | 0.034 | Ptrf | −1.688 | 0.002 | 0.040 |
| Ddx3x | −1.330 | 0.001 | 0.030 | Pum1 | −1.405 | 0.008 | 0.065 |
| Deptor | −1.652 | 0.001 | 0.033 | Pura | −1.301 | 0.006 | 0.056 |
| Dgkh | −1.341 | 0.005 | 0.055 | Purb | −1.209 | 0.021 | 0.095 |
| Dgkz | −1.775 | 0.000 | 0.024 | Pygl | −1.338 | 0.001 | 0.031 |
| Disp2 | −1.423 | 0.010 | 0.069 | Rab27b | −1.526 | 0.016 | 0.083 |
| Dpp10 | −1.486 | 0.000 | 0.021 | Rab39b | −1.515 | 0.003 | 0.045 |
| Dpp6 | −1.293 | 0.002 | 0.039 | Rab3c | −1.439 | 0.013 | 0.075 |
| Ebf3 | −1.919 | 0.002 | 0.035 | Rabgap1l | −1.317 | 0.003 | 0.044 |
| Eif4e3 | −1.421 | 0.001 | 0.032 | Raph1 | −1.327 | 0.007 | 0.060 |
| Etnk1 | −1.896 | 0.001 | 0.030 | Rasgrp1 | −1.547 | 0.001 | 0.031 |
| F2rl2 | −1.621 | 0.012 | 0.075 | Rbms1 | −1.558 | 0.002 | 0.034 |
| Fabp4 | −5.383 | 0.000 | 0.026 | Reps2 | −1.310 | 0.003 | 0.041 |
| Fam102b | −1.588 | 0.000 | 0.025 | Rgmb | −1.207 | 0.015 | 0.082 |
| Fam122b | −2.137 | 0.004 | 0.050 | Rgs17 | −2.168 | 0.000 | 0.026 |
| Fam179b | −1.540 | 0.003 | 0.041 | Rnf144a | −1.423 | 0.000 | 0.029 |
| Fam214b | −1.405 | 0.001 | 0.031 | Robo2 | −1.321 | 0.001 | 0.030 |
| Fam222b | −1.668 | 0.007 | 0.062 | Rps11 | −1.411 | 0.005 | 0.055 |
| Filip1 | −1.223 | 0.001 | 0.030 | Rspo2 | −1.809 | 0.000 | 0.026 |
| Fsd2 | −2.160 | 0.012 | 0.073 | Runx1 | −1.204 | 0.006 | 0.055 |
| Gal | −2.045 | 0.002 | 0.035 | Ryr1 | −3.459 | 0.010 | 0.069 |
| Ghr | −2.439 | 0.004 | 0.049 | S100a11 | −2.366 | 0.007 | 0.063 |
| Gm17305 | −1.767 | 0.003 | 0.046 | S100a8 | −3.282 | 0.001 | 0.029 |
| Gm42417 | −4.801 | 0.000 | 0.029 | S100a9 | −4.220 | 0.000 | 0.021 |
| Gmfb | −1.259 | 0.004 | 0.049 | Safb | −1.574 | 0.011 | 0.070 |
| Gna14 | −1.468 | 0.021 | 0.095 | Samsn1 | −2.440 | 0.001 | 0.030 |
| Gnai3 | −1.748 | 0.011 | 0.072 | Scd1 | −1.809 | 0.001 | 0.034 |
| Gnao1 | −1.462 | 0.001 | 0.032 | Scg2 | −1.809 | 0.009 | 0.066 |
| Gnaq | −1.531 | 0.002 | 0.034 | Scg3 | −1.743 | 0.002 | 0.038 |
| Gnb4 | −1.236 | 0.008 | 0.064 | Scn9a | −1.579 | 0.000 | 0.027 |
| Gp1bb | −1.271 | 0.020 | 0.093 | Scyl2 | −1.375 | 0.001 | 0.031 |
| Grip1 | −1.423 | 0.013 | 0.076 | Sdcbp | −1.233 | 0.001 | 0.030 |
| Grm7 | −1.784 | 0.001 | 0.033 | Sema4b | −1.437 | 0.002 | 0.039 |
| H2-K1 | −1.611 | 0.019 | 0.091 | Sepw1 | −1.253 | 0.013 | 0.077 |
| Hace1 | −1.481 | 0.008 | 0.065 | Slc16a3 | −2.036 | 0.001 | 0.029 |
| Hba-a2 | −1.762 | 0.004 | 0.052 | Slc27a3 | −1.610 | 0.008 | 0.065 |
| Hbb-bt | −1.668 | 0.004 | 0.052 | Slc35a5 | −1.304 | 0.000 | 0.038 |
| Hcn3 | −1.929 | 0.003 | 0.045 | Slc37a1 | −1.336 | 0.016 | 0.085 |
| Hgf | −1.762 | 0.003 | 0.044 | Slc39a6 | −1.502 | 0.003 | 0.041 |
| Hmbox1 | −1.450 | 0.011 | 0.072 | Slc51a | −1.622 | 0.002 | 0.035 |
| Hmgcl | −1.314 | 0.013 | 0.077 | Slc5a3 | −1.415 | 0.014 | 0.080 |
| Hoxa10 | −7.016 | 0.000 | 0.020 | Slc9a6 | −1.278 | 0.002 | 0.039 |
| Hoxa7 | −7.044 | 0.000 | 0.045 | Smim10l1 | −1.212 | 0.002 | 0.036 |
| Hoxa9 | −6.931 | 0.000 | 0.027 | Smim5 | −1.524 | 0.015 | 0.080 |
| Hoxb2 | −5.272 | 0.001 | 0.029 | Socs2 | −1.430 | 0.001 | 0.031 |
| Hoxb4 | −6.121 | 0.000 | 0.026 | Sorl1 | −1.403 | 0.002 | 0.034 |
| Hoxb5 | −6.619 | 0.000 | 0.020 | Spred2 | −1.203 | 0.008 | 0.064 |
| Hoxb9 | −9.266 | 0.000 | 0.024 | Spryd7 | −1.357 | 0.010 | 0.068 |
| Hoxc6 | −11.922 | 0.000 | 0.024 | Srek1 | −1.309 | 0.009 | 0.067 |
| Hoxd10 | −11.238 | 0.000 | 0.023 | Ssbp3 | −1.358 | 0.008 | 0.064 |
| Hoxd4 | −2.142 | 0.003 | 0.046 | St8sia3 | −1.270 | 0.007 | 0.060 |
| Hoxd8 | −1.603 | 0.004 | 0.048 | Stag2 | −1.202 | 0.005 | 0.053 |
| Hoxd9 | −3.929 | 0.001 | 0.030 | Sycp3 | −1.430 | 0.019 | 0.089 |
| Hs6st2 | −2.118 | 0.004 | 0.050 | Synpr | −1.887 | 0.001 | 0.029 |
| Hsp90ab1 | −1.392 | 0.006 | 0.056 | Syt1 | −1.331 | 0.000 | 0.021 |
| Idh1 | −2.275 | 0.000 | 0.047 | Syt4 | −2.148 | 0.004 | 0.052 |
| Idi1 | −1.751 | 0.022 | 0.097 | Syt7 | −1.673 | 0.001 | 0.029 |
| Ids | −1.571 | 0.009 | 0.067 | Syt9 | −1.899 | 0.000 | 0.026 |
| Il10rb | −1.208 | 0.020 | 0.093 | Tac1 | −1.726 | 0.000 | 0.028 |
| Il6st | −1.838 | 0.001 | 0.030 | Taf1 | −1.263 | 0.024 | 0.100 |
| Impad1 | −1.490 | 0.002 | 0.035 | Taok1 | −1.691 | 0.005 | 0.055 |
| Ina | −2.171 | 0.000 | 0.028 | Tlx3 | −1.349 | 0.001 | 0.030 |
| Irf2 | −1.226 | 0.000 | 0.029 | Tmem158 | −1.431 | 0.003 | 0.045 |
| Izumo4 | −1.366 | 0.009 | 0.068 | Tmem164 | −1.228 | 0.008 | 0.063 |
| Kcna4 | −1.408 | 0.010 | 0.069 | Tmem185b | −1.358 | 0.019 | 0.088 |
| Kcnab1 | −1.296 | 0.003 | 0.046 | Tmem200a | −1.785 | 0.003 | 0.041 |
| Kcnb2 | −2.196 | 0.001 | 0.032 | Tmem233 | −1.233 | 0.019 | 0.089 |
| Kcnt1 | −1.215 | 0.017 | 0.086 | Tmem255a | −1.367 | 0.001 | 0.033 |
| Kdelc2 | −1.279 | 0.010 | 0.070 | Tmem56 | −1.466 | 0.009 | 0.068 |
| Kdm7a | −1.460 | 0.009 | 0.068 | Tmtc2 | −1.914 | 0.015 | 0.082 |
| Kif5b | −1.301 | 0.008 | 0.064 | Tmx3 | −1.230 | 0.008 | 0.064 |
| Klf7 | −1.346 | 0.000 | 0.026 | Tnnc2 | −6.667 | 0.001 | 0.030 |
| Larp1 | −1.374 | 0.006 | 0.056 | Tnnt3 | −7.079 | 0.000 | 0.028 |
| Lbh | −1.629 | 0.003 | 0.042 | Top2a | −1.930 | 0.008 | 0.064 |
| Lcor | −1.431 | 0.008 | 0.063 | Tra2a | −1.451 | 0.015 | 0.081 |
| Ldb2 | −1.834 | 0.001 | 0.031 | Trp53bp1 | −1.257 | 0.004 | 0.049 |
| Ldlr | −1.708 | 0.002 | 0.034 | Trpc3 | −1.616 | 0.012 | 0.073 |
| Lifr | −1.742 | 0.005 | 0.052 | Trpv1 | −2.016 | 0.008 | 0.065 |
| Lonrf1 | −1.453 | 0.002 | 0.035 | Ubb | −1.257 | 0.022 | 0.097 |
| Lox | −2.537 | 0.018 | 0.087 | Ubqln2 | −1.407 | 0.004 | 0.050 |
| Lpar3 | −1.721 | 0.003 | 0.046 | Ugcg | −1.398 | 0.007 | 0.060 |
| Lrfn1 | −1.409 | 0.000 | 0.022 | Unc13c | −1.455 | 0.002 | 0.037 |
| Lrrc8b | −1.402 | 0.001 | 0.030 | Usp17la | −1.946 | 0.011 | 0.072 |
| Lrrtm2 | −1.389 | 0.018 | 0.088 | Usp9x | −1.500 | 0.011 | 0.072 |
| Ly86 | −2.557 | 0.009 | 0.068 | Vwa5a | −1.352 | 0.002 | 0.040 |
| Magi3 | −1.665 | 0.001 | 0.030 | Wasf1 | −1.501 | 0.000 | 0.020 |
| Mal2 | −2.994 | 0.000 | 0.029 | Wfdc2 | −1.246 | 0.019 | 0.090 |
| Mb | −6.746 | 0.000 | 0.027 | Xirp2 | −8.656 | 0.000 | 0.026 |
| Mbnl1 | −1.392 | 0.018 | 0.088 | Yod1 | −1.273 | 0.009 | 0.067 |
| Mbnl2 | −1.795 | 0.000 | 0.029 | Zbtb44 | −1.315 | 0.015 | 0.080 |
| Mcoln1 | −1.305 | 0.010 | 0.070 | Zdhhc13 | −1.497 | 0.000 | 0.028 |
| Mfsd7b | −1.521 | 0.007 | 0.062 | Zeb2 | −1.453 | 0.011 | 0.070 |
| Mon1a | −1.310 | 0.000 | 0.029 | Zfhx2 | −1.542 | 0.016 | 0.084 |
| Zfhx3 | −1.317 | 0.007 | 0.060 |
Table 3.
Genes upregulated in the TG Nav1.8-TRAP dataset
| Genes | Log2 fold change | p | q | Genes | Log2 fold change | p | q |
|---|---|---|---|---|---|---|---|
| 0610009B22Rik | 3.295 | 0.001 | 0.031 | Nap1l5 | 2.348 | 0.001 | 0.029 |
| 1110032A03Rik | 2.542 | 0.007 | 0.059 | Nbea | 2.525 | 0.001 | 0.028 |
| 1110065P20Rik | 5.685 | 0.001 | 0.032 | Nbl1 | 2.738 | 0.015 | 0.085 |
| 1700037C18Rik | 2.913 | 0.010 | 0.072 | Ndel1 | 3.132 | 0.000 | 0.010 |
| 2010107E04Rik | 2.008 | 0.000 | 0.016 | Ndufa1 | 2.495 | 0.002 | 0.034 |
| 2210013O21Rik | 2.570 | 0.006 | 0.056 | Ndufb2 | 3.065 | 0.002 | 0.034 |
| 2700094K13Rik | 3.894 | 0.004 | 0.049 | Ndufb5 | 2.214 | 0.001 | 0.027 |
| 9430016H08Rik | 3.239 | 0.016 | 0.089 | Ndufb9 | 2.915 | 0.000 | 0.017 |
| Aard | 7.736 | 0.005 | 0.050 | Ndufs2 | 2.315 | 0.003 | 0.043 |
| Acp1 | 2.522 | 0.004 | 0.049 | Ngfr | 2.166 | 0.011 | 0.075 |
| Adam9 | 4.085 | 0.004 | 0.045 | Nme3 | 11.466 | 0.000 | 0.020 |
| Adra2c | 5.673 | 0.001 | 0.025 | Nrn1l | 4.040 | 0.009 | 0.068 |
| Akirin1 | 2.298 | 0.009 | 0.068 | Nsun3 | 3.157 | 0.018 | 0.093 |
| Akt1 | 3.554 | 0.006 | 0.058 | Nt5m | 3.252 | 0.017 | 0.090 |
| Alkbh3 | 7.497 | 0.002 | 0.035 | Nubp2 | 5.238 | 0.007 | 0.063 |
| Amacr | 5.927 | 0.013 | 0.082 | Nudt10 | 4.615 | 0.010 | 0.072 |
| Anapc13 | 3.962 | 0.015 | 0.087 | Nudt11 | 3.346 | 0.018 | 0.093 |
| Ankrd24 | 2.576 | 0.012 | 0.078 | Nudt7 | 4.780 | 0.002 | 0.037 |
| Anxa5 | 2.142 | 0.004 | 0.047 | Nup37 | 4.108 | 0.000 | 0.017 |
| Apip | 7.421 | 0.003 | 0.042 | Ost4 | 2.585 | 0.004 | 0.050 |
| Apln | 4.200 | 0.009 | 0.069 | Ostf1 | 2.376 | 0.003 | 0.042 |
| Arl5a | 2.178 | 0.002 | 0.038 | Pacsin1 | 2.217 | 0.016 | 0.089 |
| Arl6 | 4.831 | 0.003 | 0.042 | Pak1 | 5.331 | 0.006 | 0.056 |
| Arl8a | 2.045 | 0.008 | 0.066 | Pak3 | 3.228 | 0.009 | 0.069 |
| Armc1 | 2.080 | 0.016 | 0.089 | Parp3 | 2.863 | 0.010 | 0.072 |
| Arpc1b | 3.965 | 0.000 | 0.020 | Pcbd2 | 3.529 | 0.002 | 0.037 |
| Arpc5l | 2.223 | 0.002 | 0.039 | Pcolce2 | 3.567 | 0.003 | 0.042 |
| Asna1 | 3.806 | 0.010 | 0.073 | Pcp4l1 | 3.151 | 0.004 | 0.045 |
| Atg4c | 4.484 | 0.001 | 0.028 | Pcsk1n | 2.681 | 0.010 | 0.072 |
| Atox1 | 2.116 | 0.010 | 0.072 | Pcsk7 | 4.142 | 0.005 | 0.050 |
| Atp5c1 | 2.674 | 0.003 | 0.040 | Pdcd6 | 2.770 | 0.009 | 0.067 |
| Atp5d | 2.552 | 0.006 | 0.058 | Pde6d | 5.181 | 0.004 | 0.050 |
| Atp5g1 | 2.613 | 0.003 | 0.040 | Pdlim2 | 3.067 | 0.001 | 0.027 |
| Atp5s | 6.255 | 0.000 | 0.018 | Pdzd9 | 2.834 | 0.005 | 0.050 |
| Atp6v0d1 | 3.072 | 0.000 | 0.018 | Pex11b | 7.187 | 0.000 | 0.003 |
| Avpi1 | 2.049 | 0.005 | 0.053 | Pfdn1 | 2.368 | 0.013 | 0.082 |
| Banf1 | 2.411 | 0.001 | 0.028 | Pfkm | 2.196 | 0.011 | 0.074 |
| Bbs9 | 4.103 | 0.021 | 0.099 | Phospho2 | 2.332 | 0.013 | 0.082 |
| Bloc1s1 | 2.178 | 0.015 | 0.085 | Phpt1 | 2.748 | 0.007 | 0.063 |
| Bloc1s3 | 4.164 | 0.003 | 0.042 | Pigh | 2.609 | 0.011 | 0.075 |
| Btbd2 | 2.239 | 0.002 | 0.036 | Pin1 | 3.518 | 0.001 | 0.031 |
| C77080 | 3.683 | 0.000 | 0.016 | Pla2g16 | 2.041 | 0.017 | 0.091 |
| Cacng7 | 2.455 | 0.005 | 0.054 | Plekhb1 | 3.646 | 0.014 | 0.083 |
| Calca | 2.290 | 0.001 | 0.027 | Plpp1 | 4.280 | 0.017 | 0.091 |
| Calm2 | 2.036 | 0.014 | 0.085 | Pole3 | 3.127 | 0.020 | 0.097 |
| Camk2g | 2.396 | 0.003 | 0.044 | Polr2d | 6.787 | 0.000 | 0.004 |
| Cbln1 | 6.228 | 0.004 | 0.047 | Polr2j | 6.438 | 0.001 | 0.027 |
| Ccdc88a | 2.029 | 0.007 | 0.059 | Polr2l | 2.072 | 0.011 | 0.076 |
| Cdc123 | 2.146 | 0.020 | 0.098 | Polr2m | 2.330 | 0.005 | 0.050 |
| Cdk2ap1 | 4.137 | 0.003 | 0.040 | Pomgnt1 | 3.461 | 0.020 | 0.099 |
| Cdkn1b | 2.713 | 0.001 | 0.024 | Ppm1j | 2.163 | 0.009 | 0.067 |
| Cdr2l | 2.794 | 0.007 | 0.061 | Ppp2r5c | 2.205 | 0.014 | 0.084 |
| Cebpzos | 3.122 | 0.004 | 0.049 | Praf2 | 2.266 | 0.019 | 0.096 |
| Cela1 | 4.351 | 0.004 | 0.049 | Prkcd | 2.054 | 0.003 | 0.040 |
| Cenpq | 4.971 | 0.002 | 0.037 | Prkcdbp | 6.444 | 0.002 | 0.035 |
| Cfap69 | 3.913 | 0.012 | 0.077 | Prkrir | 2.325 | 0.008 | 0.064 |
| Cfl1 | 2.008 | 0.001 | 0.032 | Prorsd1 | 5.144 | 0.008 | 0.065 |
| Chchd1 | 3.792 | 0.000 | 0.017 | Psma1 | 2.561 | 0.017 | 0.091 |
| Chchd10 | 3.927 | 0.011 | 0.074 | Psmb11 | 2.717 | 0.000 | 0.021 |
| Chchd4 | 2.146 | 0.003 | 0.040 | Psmc3ip | 6.150 | 0.007 | 0.061 |
| Chd3os | 2.704 | 0.003 | 0.040 | Qars | 2.763 | 0.012 | 0.079 |
| Chmp6 | 2.299 | 0.021 | 0.099 | Rab10 | 2.106 | 0.010 | 0.071 |
| Chodl | 5.030 | 0.016 | 0.089 | Rab15 | 3.719 | 0.013 | 0.080 |
| Chp1 | 4.425 | 0.000 | 0.017 | Rab1a | 4.034 | 0.008 | 0.064 |
| Clmp | 2.253 | 0.019 | 0.094 | Rab28 | 2.269 | 0.001 | 0.032 |
| Cnih2 | 4.508 | 0.007 | 0.061 | Rab33a | 3.270 | 0.015 | 0.085 |
| Cnrip1 | 2.211 | 0.008 | 0.066 | Rab35 | 2.067 | 0.012 | 0.078 |
| Commd1 | 3.042 | 0.003 | 0.045 | Rab4b | 2.707 | 0.008 | 0.065 |
| Commd4 | 2.980 | 0.019 | 0.094 | Rad54l | 5.057 | 0.001 | 0.026 |
| Coq3 | 3.088 | 0.011 | 0.075 | Rad9b | 2.382 | 0.012 | 0.079 |
| Cox6b1 | 2.708 | 0.000 | 0.023 | Rho | 4.096 | 0.003 | 0.040 |
| Cox7a2 | 2.299 | 0.006 | 0.056 | Rhog | 4.929 | 0.012 | 0.078 |
| Cox7b | 4.295 | 0.000 | 0.017 | Rnase4 | 2.128 | 0.018 | 0.092 |
| Cox7c | 2.255 | 0.001 | 0.028 | Rnf114 | 3.644 | 0.013 | 0.082 |
| Cox8a | 2.576 | 0.000 | 0.021 | Rnf215 | 4.125 | 0.006 | 0.058 |
| Crlf2 | 3.905 | 0.011 | 0.075 | Rnf7 | 2.072 | 0.013 | 0.081 |
| Crtc2 | 5.091 | 0.001 | 0.023 | Romo1 | 2.456 | 0.000 | 0.021 |
| Crtc3 | 3.703 | 0.018 | 0.092 | Rpl10 | 3.076 | 0.013 | 0.080 |
| Ctxn3 | 2.686 | 0.000 | 0.023 | Rpl28 | 2.167 | 0.019 | 0.095 |
| Cyb5a | 2.330 | 0.012 | 0.079 | Rpl29 | 5.333 | 0.021 | 0.099 |
| Cyc1 | 3.989 | 0.013 | 0.080 | Rpl35 | 3.039 | 0.003 | 0.040 |
| Cystm1 | 3.446 | 0.002 | 0.038 | Rpl37 | 2.967 | 0.003 | 0.042 |
| Dad1 | 4.088 | 0.006 | 0.056 | Rpl39 | 3.241 | 0.001 | 0.027 |
| Dalrd3 | 7.942 | 0.002 | 0.036 | Rps23 | 2.219 | 0.020 | 0.097 |
| Dda1 | 2.393 | 0.017 | 0.091 | Rps29 | 2.912 | 0.002 | 0.037 |
| Dlg2 | 4.233 | 0.010 | 0.072 | Rpusd1 | 3.617 | 0.008 | 0.064 |
| Dnajc12 | 3.352 | 0.016 | 0.088 | Rraga | 2.131 | 0.000 | 0.023 |
| Dnal4 | 4.100 | 0.012 | 0.076 | Rtn4r | 2.805 | 0.001 | 0.028 |
| Dpep2 | 4.510 | 0.001 | 0.026 | Rxrg | 4.463 | 0.004 | 0.049 |
| Dpm3 | 3.461 | 0.019 | 0.095 | Sac3d1 | 4.162 | 0.009 | 0.070 |
| Dzank1 | 3.857 | 0.011 | 0.074 | Sap18 | 3.653 | 0.019 | 0.095 |
| Edf1 | 2.907 | 0.000 | 0.002 | Sdhb | 4.145 | 0.013 | 0.082 |
| Eef1a1 | 2.126 | 0.016 | 0.088 | Sdhd | 2.350 | 0.008 | 0.066 |
| Efcc1 | 5.725 | 0.001 | 0.032 | Sec13 | 5.366 | 0.002 | 0.037 |
| Eif4a3 | 2.748 | 0.008 | 0.066 | Sec23ip | 2.817 | 0.014 | 0.083 |
| Emb | 2.251 | 0.000 | 0.017 | Sep15 | 2.455 | 0.009 | 0.068 |
| Enox1 | 3.300 | 0.017 | 0.090 | Sepp1 | 2.412 | 0.013 | 0.081 |
| Epm2a | 2.756 | 0.003 | 0.041 | Serp2 | 3.709 | 0.004 | 0.046 |
| Esyt1 | 4.806 | 0.004 | 0.045 | Serping1 | 4.031 | 0.007 | 0.060 |
| Exd2 | 5.752 | 0.000 | 0.016 | Sh2d3c | 3.339 | 0.019 | 0.095 |
| Exosc6 | 4.476 | 0.005 | 0.053 | Sh3bgrl | 2.659 | 0.004 | 0.049 |
| Fabp7 | 3.901 | 0.001 | 0.024 | Sh3bgrl3 | 2.229 | 0.001 | 0.032 |
| Fam105a | 3.610 | 0.013 | 0.082 | Shd | 8.734 | 0.007 | 0.061 |
| Fam188a | 2.065 | 0.011 | 0.075 | Shisa5 | 2.629 | 0.011 | 0.074 |
| Fam58b | 3.355 | 0.002 | 0.036 | Sirt2 | 3.344 | 0.020 | 0.098 |
| Fam89a | 3.170 | 0.010 | 0.072 | Sirt3 | 3.145 | 0.017 | 0.090 |
| Far2 | 3.913 | 0.006 | 0.056 | Sla2 | 2.598 | 0.002 | 0.037 |
| Farp1 | 6.925 | 0.000 | 0.016 | Slc22a17 | 3.985 | 0.018 | 0.092 |
| Fbxl16 | 2.675 | 0.007 | 0.060 | Slc24a2 | 2.209 | 0.001 | 0.028 |
| Fbxo2 | 3.859 | 0.007 | 0.063 | Slc25a3 | 3.404 | 0.006 | 0.058 |
| Fbxo27 | 8.071 | 0.000 | 0.023 | Slc25a43 | 2.557 | 0.019 | 0.095 |
| Fgf9 | 4.057 | 0.007 | 0.063 | Slc35d2 | 6.734 | 0.000 | 0.018 |
| Fgfr2 | 6.744 | 0.003 | 0.043 | Slc3a2 | 3.247 | 0.001 | 0.025 |
| Fhl1 | 2.487 | 0.001 | 0.024 | Slc45a4 | 2.344 | 0.009 | 0.068 |
| Fkbp2 | 2.064 | 0.008 | 0.065 | Slc46a3 | 5.451 | 0.003 | 0.040 |
| Fsd1 | 2.330 | 0.010 | 0.072 | Slc6a15 | 3.041 | 0.014 | 0.084 |
| Fth1 | 3.259 | 0.001 | 0.023 | Slco2b1 | 5.287 | 0.004 | 0.047 |
| Fxyd6 | 2.042 | 0.006 | 0.056 | Slitrk1 | 3.031 | 0.018 | 0.094 |
| Gabrg1 | 3.806 | 0.000 | 0.018 | Smdt1 | 2.724 | 0.000 | 0.019 |
| Galnt18 | 2.412 | 0.013 | 0.080 | Smim12 | 2.038 | 0.000 | 0.021 |
| Gatad1 | 2.512 | 0.004 | 0.047 | Smim8 | 2.292 | 0.003 | 0.042 |
| Gipc1 | 3.901 | 0.018 | 0.093 | Snapc5 | 2.360 | 0.016 | 0.088 |
| Glb1l2 | 4.152 | 0.008 | 0.066 | Snn | 3.703 | 0.015 | 0.085 |
| Gm15440 | 5.964 | 0.012 | 0.079 | Snrpb | 2.974 | 0.002 | 0.037 |
| Gm5113 | 6.649 | 0.001 | 0.032 | Snrpn | 3.974 | 0.002 | 0.037 |
| Gmnn | 4.878 | 0.001 | 0.026 | Snx3 | 2.491 | 0.002 | 0.037 |
| Gng5 | 4.458 | 0.000 | 0.018 | Spcs1 | 2.441 | 0.021 | 0.099 |
| Gng8 | 4.700 | 0.006 | 0.057 | Spon1 | 3.949 | 0.021 | 0.099 |
| Golga1 | 3.883 | 0.016 | 0.089 | Srp14 | 2.998 | 0.000 | 0.021 |
| Gpr35 | 3.338 | 0.017 | 0.091 | Ssr4 | 3.347 | 0.011 | 0.075 |
| Gramd1b | 2.251 | 0.000 | 0.020 | Stau2 | 2.158 | 0.008 | 0.066 |
| Grpel2 | 3.678 | 0.018 | 0.092 | Strada | 5.449 | 0.008 | 0.064 |
| Gtf2i | 3.014 | 0.011 | 0.075 | Stx2 | 3.170 | 0.000 | 0.017 |
| H2-T23 | 4.183 | 0.010 | 0.070 | Suclg1 | 2.738 | 0.010 | 0.073 |
| H2afz | 2.207 | 0.003 | 0.042 | Supt4a | 3.527 | 0.001 | 0.032 |
| Hist3h2ba | 2.569 | 0.016 | 0.089 | Tagln3 | 3.422 | 0.004 | 0.047 |
| Hsbp1 | 2.142 | 0.016 | 0.089 | Tbc1d14 | 2.192 | 0.009 | 0.070 |
| Idh3b | 3.457 | 0.001 | 0.025 | Tfb1m | 3.745 | 0.013 | 0.082 |
| Ift122 | 3.334 | 0.011 | 0.074 | Thoc7 | 2.488 | 0.008 | 0.066 |
| Ift20 | 2.144 | 0.021 | 0.099 | Tifab | 5.602 | 0.004 | 0.045 |
| Ift43 | 6.053 | 0.018 | 0.094 | Timm8b | 4.092 | 0.007 | 0.061 |
| Igsf21 | 4.754 | 0.014 | 0.084 | Tjap1 | 3.096 | 0.021 | 0.099 |
| Il1rl2 | 8.506 | 0.000 | 0.016 | Tmem106b | 2.653 | 0.011 | 0.076 |
| Imp3 | 2.343 | 0.001 | 0.032 | Tmem199 | 4.071 | 0.014 | 0.084 |
| Inpp5j | 4.481 | 0.008 | 0.064 | Tmem216 | 8.212 | 0.001 | 0.026 |
| Isca2 | 2.096 | 0.011 | 0.074 | Tmem230 | 2.058 | 0.006 | 0.058 |
| Iscu | 3.142 | 0.006 | 0.058 | Tmem258 | 5.441 | 0.000 | 0.017 |
| Kcnip1 | 5.122 | 0.001 | 0.032 | Tmem53 | 3.911 | 0.009 | 0.069 |
| Kcnip4 | 2.177 | 0.002 | 0.038 | Tmem62 | 4.157 | 0.014 | 0.083 |
| Kctd8 | 2.176 | 0.006 | 0.055 | Tnfrsf1a | 5.108 | 0.008 | 0.065 |
| Klf8 | 4.565 | 0.004 | 0.047 | Tnfsfm13 | 5.814 | 0.015 | 0.085 |
| Lamtor5 | 2.577 | 0.005 | 0.055 | Tomm7 | 2.169 | 0.000 | 0.017 |
| Lcmt1 | 2.326 | 0.006 | 0.056 | Tpd52l1 | 4.373 | 0.009 | 0.068 |
| Lgals3bp | 6.428 | 0.008 | 0.064 | Tpgs2 | 3.087 | 0.004 | 0.049 |
| Limk1 | 2.887 | 0.002 | 0.034 | Tppp3 | 2.673 | 0.000 | 0.015 |
| Lipa | 2.672 | 0.005 | 0.054 | Trappc1 | 3.056 | 0.019 | 0.095 |
| Lix1 | 2.853 | 0.002 | 0.035 | Trdmt1 | 5.930 | 0.000 | 0.021 |
| Lrrc8a | 2.077 | 0.000 | 0.018 | Triap1 | 2.123 | 0.008 | 0.064 |
| Ltbr | 3.603 | 0.007 | 0.063 | Trim12a | 2.989 | 0.011 | 0.075 |
| Lxn | 2.922 | 0.001 | 0.026 | Trim9 | 3.505 | 0.014 | 0.084 |
| Lyrm2 | 4.541 | 0.000 | 0.023 | Trnp1 | 2.255 | 0.003 | 0.040 |
| Lyrm5 | 2.559 | 0.019 | 0.095 | Tspan17 | 5.060 | 0.008 | 0.066 |
| M6pr | 2.171 | 0.007 | 0.062 | Tspan3 | 2.352 | 0.003 | 0.040 |
| Manbal | 2.620 | 0.009 | 0.069 | Tspan7 | 2.270 | 0.001 | 0.027 |
| Map1lc3b | 2.978 | 0.001 | 0.028 | Tub | 6.954 | 0.000 | 0.016 |
| Mapkap1 | 2.645 | 0.010 | 0.070 | Tubb4b | 2.426 | 0.002 | 0.037 |
| Mapkapk5 | 2.476 | 0.004 | 0.045 | Tyro3 | 5.176 | 0.000 | 0.023 |
| Mast2 | 4.666 | 0.004 | 0.049 | Ubald1 | 3.325 | 0.003 | 0.040 |
| Mbd2 | 3.693 | 0.012 | 0.078 | Ubl5 | 2.981 | 0.006 | 0.058 |
| Mblac2 | 3.198 | 0.007 | 0.063 | Ulk1 | 2.369 | 0.009 | 0.067 |
| Mboat7 | 4.459 | 0.005 | 0.055 | Uqcrb | 3.669 | 0.001 | 0.025 |
| Mgst3 | 5.093 | 0.004 | 0.046 | Uqcrfs1 | 2.994 | 0.005 | 0.052 |
| Mid2 | 6.101 | 0.001 | 0.028 | Usmg5 | 3.313 | 0.001 | 0.027 |
| Minpp1 | 4.786 | 0.004 | 0.047 | Vkorc1l1 | 3.000 | 0.010 | 0.072 |
| Mipol1 | 3.394 | 0.015 | 0.086 | Vta1 | 2.955 | 0.012 | 0.076 |
| Mlf2 | 2.377 | 0.000 | 0.018 | Vwc2l | 3.597 | 0.010 | 0.071 |
| Mob3b | 3.803 | 0.020 | 0.099 | Wbp1 | 4.041 | 0.019 | 0.095 |
| Mrpl18 | 2.783 | 0.000 | 0.023 | Wbp2 | 2.195 | 0.000 | 0.002 |
| Mrpl27 | 2.877 | 0.013 | 0.081 | Wdr59 | 3.560 | 0.004 | 0.049 |
| Mrps14 | 4.314 | 0.004 | 0.049 | Wfs1 | 4.090 | 0.009 | 0.067 |
| Mrps36 | 4.605 | 0.006 | 0.058 | Wisp1 | 5.770 | 0.008 | 0.064 |
| Msra | 3.657 | 0.013 | 0.082 | Ypel3 | 2.511 | 0.003 | 0.042 |
| mt-Nd2 | 2.268 | 0.007 | 0.063 | Ywhaq | 2.191 | 0.001 | 0.026 |
| Mxd3 | 2.495 | 0.007 | 0.060 | Zdhhc6 | 4.565 | 0.008 | 0.064 |
| Mzt1 | 2.130 | 0.000 | 0.021 | Zfp932 | 3.625 | 0.020 | 0.097 |
| Naa38 | 4.005 | 0.018 | 0.093 | Zfp944 | 6.108 | 0.006 | 0.056 |
Table 4.
Genes upregulated in the DRG Nav1.8-TRAP dataset
| Genes | Log2 fold change | p | q | Genes | Log2 fold change | p | q |
|---|---|---|---|---|---|---|---|
| 2810417H13Rik | −7.030 | 0.002 | 0.036 | Mif | −2.343 | 0.012 | 0.078 |
| A430078G23Rik | −4.575 | 0.005 | 0.055 | Mmp15 | −2.137 | 0.005 | 0.055 |
| Abca6 | −10.621 | 0.000 | 0.017 | Mon1a | −2.894 | 0.002 | 0.037 |
| Abhd17c | −2.949 | 0.000 | 0.017 | Mrpl14 | −3.067 | 0.020 | 0.099 |
| Abt1 | −2.637 | 0.008 | 0.065 | Mrpl37 | −2.040 | 0.003 | 0.044 |
| Acaca | −2.654 | 0.002 | 0.037 | Mrto4 | −3.186 | 0.006 | 0.056 |
| Acacb | −3.534 | 0.005 | 0.055 | Msn | −2.617 | 0.003 | 0.040 |
| Acot6 | −7.991 | 0.001 | 0.029 | Mt-Co3 | −6.804 | 0.001 | 0.026 |
| Acta1 | −5.881 | 0.006 | 0.057 | Mterf4 | −2.725 | 0.009 | 0.068 |
| Actb | −2.404 | 0.000 | 0.016 | Mvd | −2.888 | 0.002 | 0.035 |
| Adap1 | −2.420 | 0.010 | 0.073 | Myc | −6.466 | 0.001 | 0.030 |
| Adgrb3 | −4.063 | 0.019 | 0.095 | Myh1 | −10.133 | 0.002 | 0.035 |
| Adra2a | −8.313 | 0.000 | 0.017 | Myh9 | −2.359 | 0.004 | 0.046 |
| Akap5 | −8.506 | 0.000 | 0.017 | Myo10 | −4.730 | 0.006 | 0.056 |
| Akr7a5 | −2.071 | 0.013 | 0.081 | Myoc | −3.234 | 0.015 | 0.085 |
| Akt1s1 | −2.002 | 0.004 | 0.050 | Nat9 | −3.387 | 0.012 | 0.078 |
| Anapc5 | −2.274 | 0.001 | 0.023 | Ndfip1 | −4.237 | 0.010 | 0.071 |
| Ankrd13d | −2.118 | 0.006 | 0.058 | Ndst3 | −4.583 | 0.007 | 0.062 |
| Anp32b | −2.272 | 0.003 | 0.040 | Ndufa3 | −2.237 | 0.017 | 0.090 |
| Ap1s1 | −2.586 | 0.003 | 0.040 | Ndufs5 | −2.374 | 0.018 | 0.092 |
| Ap2s1 | −2.371 | 0.012 | 0.078 | Ndufs6 | −3.439 | 0.000 | 0.018 |
| Aqr | −2.439 | 0.019 | 0.094 | Nes | −5.213 | 0.001 | 0.030 |
| Arhgap26 | −3.451 | 0.000 | 0.017 | Neurl4 | −2.326 | 0.005 | 0.052 |
| Arhgap39 | −2.113 | 0.006 | 0.058 | Nfkbil1 | −4.943 | 0.001 | 0.027 |
| Arhgef11 | −2.432 | 0.016 | 0.088 | Nfya | −4.346 | 0.018 | 0.093 |
| Ascc2 | −10.550 | 0.001 | 0.024 | Noct | −2.815 | 0.013 | 0.081 |
| Astn2 | −8.207 | 0.001 | 0.027 | Nol10 | −2.001 | 0.001 | 0.024 |
| Atf6b | −2.528 | 0.010 | 0.072 | Nop16 | −2.605 | 0.001 | 0.032 |
| Atg2a | −2.559 | 0.001 | 0.028 | Nptx1 | −2.018 | 0.005 | 0.055 |
| Atm | −2.175 | 0.017 | 0.090 | Npy1r | −3.647 | 0.007 | 0.061 |
| Atp2b4 | −2.641 | 0.000 | 0.017 | Nsun2 | −2.052 | 0.002 | 0.037 |
| Atp6v0a1 | −3.155 | 0.001 | 0.032 | Nts | −6.880 | 0.003 | 0.040 |
| B3gnt8 | −2.129 | 0.015 | 0.085 | Nuak1 | −2.011 | 0.003 | 0.040 |
| Bach1 | −4.987 | 0.002 | 0.035 | Nup155 | −5.434 | 0.000 | 0.021 |
| Baz1b | −2.451 | 0.016 | 0.089 | Nup88 | −2.178 | 0.005 | 0.050 |
| C130074G19Rik | −6.277 | 0.001 | 0.027 | Nyap1 | −2.210 | 0.003 | 0.045 |
| Cadm4 | −2.525 | 0.005 | 0.050 | Obfc1 | −5.064 | 0.003 | 0.041 |
| Capn1 | −2.714 | 0.001 | 0.028 | Obox3 | −3.209 | 0.002 | 0.035 |
| Cast | −2.300 | 0.000 | 0.017 | Ogfr | −3.157 | 0.004 | 0.047 |
| Ccdc130 | −5.043 | 0.003 | 0.040 | P2rx3 | −2.594 | 0.015 | 0.086 |
| Ccdc3 | −2.703 | 0.021 | 0.099 | Pabpn1 | −2.231 | 0.008 | 0.065 |
| Cct5 | −2.433 | 0.002 | 0.037 | Palm3 | −2.239 | 0.010 | 0.072 |
| Cct7 | −2.291 | 0.004 | 0.045 | Panx1 | −2.446 | 0.006 | 0.056 |
| Cct8 | −4.490 | 0.002 | 0.034 | Pcbp2 | −5.332 | 0.003 | 0.041 |
| Cdc26 | −2.164 | 0.002 | 0.039 | Pcdh11x | −7.446 | 0.000 | 0.003 |
| Cdh1 | −2.230 | 0.004 | 0.049 | Pex6 | −2.364 | 0.005 | 0.055 |
| Cdk11b | −3.476 | 0.004 | 0.047 | Pfdn2 | −2.988 | 0.004 | 0.045 |
| Cep85l | −9.647 | 0.001 | 0.026 | Pfkp | −2.031 | 0.001 | 0.032 |
| Cetn2 | −2.177 | 0.010 | 0.072 | Pfn1 | −2.932 | 0.000 | 0.020 |
| Ckb | −2.670 | 0.002 | 0.035 | Pgls | −2.444 | 0.009 | 0.068 |
| Ckm | −8.225 | 0.006 | 0.056 | Phldb2 | −8.699 | 0.004 | 0.046 |
| Clgn | −2.261 | 0.000 | 0.017 | Pih1d1 | −2.509 | 0.002 | 0.035 |
| Clint1 | −2.048 | 0.009 | 0.067 | Plcb3 | −3.864 | 0.000 | 0.018 |
| Clip2 | −4.567 | 0.000 | 0.015 | Plec | −3.238 | 0.000 | 0.017 |
| Cln6 | −3.765 | 0.001 | 0.025 | Plekhm1 | −4.182 | 0.005 | 0.053 |
| Col1a1 | −2.067 | 0.011 | 0.074 | Pnpo | −3.077 | 0.008 | 0.066 |
| Col5a2 | −7.121 | 0.021 | 0.100 | Poll | −4.050 | 0.002 | 0.035 |
| Col5a3 | −5.712 | 0.005 | 0.050 | Polr2 h | −3.280 | 0.011 | 0.075 |
| Col8a1 | −7.657 | 0.002 | 0.037 | Pop1 | −7.207 | 0.000 | 0.001 |
| Cops6 | −2.017 | 0.002 | 0.037 | Prcc | −2.257 | 0.002 | 0.033 |
| Coq7 | −2.086 | 0.015 | 0.085 | Prg2 | −10.331 | 0.000 | 0.020 |
| Cpe | −2.053 | 0.002 | 0.034 | Prkag2 | −2.591 | 0.000 | 0.019 |
| Cpt1c | −2.190 | 0.006 | 0.059 | Prrx1 | −5.132 | 0.004 | 0.047 |
| Cpxm1 | −3.325 | 0.005 | 0.052 | Psmb3 | −2.915 | 0.002 | 0.036 |
| Csgalnact1 | −9.636 | 0.001 | 0.028 | Psmb5 | −2.213 | 0.002 | 0.036 |
| Csnk2a2 | −2.122 | 0.006 | 0.059 | Psmc1 | −2.405 | 0.000 | 0.016 |
| Csrnp1 | −10.669 | 0.001 | 0.030 | Psmd13 | −2.670 | 0.003 | 0.044 |
| Ctu2 | −3.003 | 0.014 | 0.084 | Psmd2 | −2.182 | 0.001 | 0.027 |
| Cul7 | −3.621 | 0.007 | 0.062 | Psmd4 | −2.809 | 0.003 | 0.044 |
| Cwc25 | −2.930 | 0.004 | 0.045 | Ptger1 | −3.477 | 0.018 | 0.094 |
| Dapk2 | −2.224 | 0.014 | 0.084 | Ptms | −3.871 | 0.002 | 0.037 |
| Ddx56 | −2.158 | 0.004 | 0.045 | Ptpn23 | −7.273 | 0.002 | 0.036 |
| Dgkz | −2.686 | 0.003 | 0.040 | Ptprb | −7.624 | 0.000 | 0.004 |
| Disp1 | −2.321 | 0.015 | 0.085 | Ptrf | −4.042 | 0.005 | 0.053 |
| Dlg3 | −3.824 | 0.008 | 0.064 | Pycr2 | −2.066 | 0.015 | 0.085 |
| Dnaja2 | −2.167 | 0.001 | 0.028 | Pygl | −2.181 | 0.014 | 0.083 |
| Dnm1 | −2.469 | 0.009 | 0.068 | Pygo2 | −2.586 | 0.007 | 0.063 |
| Dph2 | −6.419 | 0.000 | 0.017 | Qsox1 | −2.054 | 0.003 | 0.040 |
| Dph7 | −6.003 | 0.000 | 0.014 | R3hdm4 | −2.054 | 0.013 | 0.082 |
| Dpp7 | −2.645 | 0.013 | 0.080 | Rack1 | −2.006 | 0.005 | 0.055 |
| Dpp8 | −2.446 | 0.003 | 0.042 | Rae1 | −2.346 | 0.014 | 0.084 |
| Dpy19l4 | −6.024 | 0.001 | 0.028 | Rara | −4.005 | 0.005 | 0.053 |
| Dpysl5 | −2.386 | 0.016 | 0.088 | Rbm14 | −6.125 | 0.001 | 0.032 |
| Drg2 | −2.986 | 0.011 | 0.076 | Rbm6 | −2.191 | 0.007 | 0.061 |
| Ebpl | −2.024 | 0.006 | 0.056 | Rcc2 | −2.150 | 0.002 | 0.037 |
| Edc4 | −3.342 | 0.003 | 0.040 | Reps1 | −4.535 | 0.000 | 0.022 |
| Ehmt2 | −2.944 | 0.008 | 0.066 | Rexo4 | −3.169 | 0.015 | 0.087 |
| Eif2b4 | −2.226 | 0.009 | 0.068 | Rfc2 | −2.711 | 0.005 | 0.054 |
| Eif2b5 | −2.424 | 0.002 | 0.033 | Riok1 | −2.211 | 0.017 | 0.090 |
| Eif3j1 | −2.155 | 0.018 | 0.092 | Rnf122 | −2.237 | 0.000 | 0.018 |
| Eif3m | −2.731 | 0.006 | 0.057 | Rnf2 | −2.259 | 0.015 | 0.086 |
| Eif5b | −2.048 | 0.008 | 0.066 | Rpia | −2.151 | 0.017 | 0.090 |
| Emc1 | −2.455 | 0.004 | 0.047 | Rpl24 | −3.600 | 0.003 | 0.040 |
| Eps8 | −7.159 | 0.003 | 0.044 | Rplp2 | −2.166 | 0.003 | 0.040 |
| Fabp4 | −7.365 | 0.000 | 0.021 | Rrp1 | −2.684 | 0.001 | 0.025 |
| Fam195b | −2.538 | 0.003 | 0.043 | Rrp7a | −2.173 | 0.010 | 0.070 |
| Fam21 | −2.569 | 0.001 | 0.024 | Rsph9 | −3.818 | 0.009 | 0.067 |
| Fam65b | −7.543 | 0.003 | 0.044 | Rsrc1 | −2.223 | 0.005 | 0.054 |
| Fbn1 | −5.976 | 0.004 | 0.046 | Ryr1 | −9.068 | 0.021 | 0.099 |
| Fh1 | −2.129 | 0.006 | 0.056 | S100a8 | −7.163 | 0.001 | 0.025 |
| Fhl3 | −3.273 | 0.014 | 0.084 | S100a9 | −6.621 | 0.002 | 0.033 |
| Fkbp10 | −8.824 | 0.001 | 0.027 | Sae1 | −2.758 | 0.008 | 0.064 |
| Fkbp14 | −2.261 | 0.005 | 0.053 | Sart3 | −2.174 | 0.001 | 0.024 |
| Fnbp4 | −4.246 | 0.012 | 0.078 | Sass6 | −2.773 | 0.015 | 0.087 |
| Frg1 | −2.094 | 0.001 | 0.030 | Scn2a1 | −4.227 | 0.008 | 0.066 |
| Ftsj3 | −2.299 | 0.004 | 0.048 | Sdad1 | −2.183 | 0.014 | 0.084 |
| Gab2 | −2.044 | 0.004 | 0.047 | Sdk2 | −4.549 | 0.013 | 0.081 |
| Gdap1l1 | −2.493 | 0.011 | 0.074 | Sdsl | −4.243 | 0.017 | 0.092 |
| Gfod2 | −2.657 | 0.010 | 0.071 | Selenbp1 | −8.748 | 0.001 | 0.027 |
| Gga1 | −2.391 | 0.004 | 0.049 | Senp1 | −5.212 | 0.018 | 0.092 |
| Gm21967 | −3.888 | 0.010 | 0.073 | Sept6 | −2.159 | 0.006 | 0.058 |
| Gm42417 | −9.870 | 0.005 | 0.053 | Serpina11 | −3.644 | 0.014 | 0.085 |
| Golga2 | −2.272 | 0.003 | 0.045 | Sertad1 | −2.157 | 0.006 | 0.059 |
| Golga7b | −2.577 | 0.004 | 0.047 | Sfrp5 | −2.379 | 0.001 | 0.028 |
| Gp1bb | −2.047 | 0.001 | 0.030 | Sin3b | −2.401 | 0.002 | 0.032 |
| Gpr179 | −8.139 | 0.001 | 0.028 | Slc16a3 | −10.302 | 0.005 | 0.050 |
| Gpx4 | −2.608 | 0.009 | 0.069 | Slc25a24 | −8.902 | 0.000 | 0.017 |
| Grcc10 | −2.915 | 0.003 | 0.040 | Slc27a4 | −2.479 | 0.020 | 0.097 |
| Gsn | −2.133 | 0.015 | 0.085 | Slc39a6 | −2.296 | 0.002 | 0.036 |
| Gtf2f2 | −9.634 | 0.003 | 0.042 | Slc43a1 | −4.289 | 0.009 | 0.068 |
| Gys1 | −2.353 | 0.000 | 0.016 | Slc51a | −6.814 | 0.000 | 0.015 |
| H2afy2 | −3.646 | 0.001 | 0.026 | Slc7a3 | −2.557 | 0.018 | 0.093 |
| Hba-a2 | −3.246 | 0.010 | 0.072 | Slc7a5 | −3.490 | 0.000 | 0.020 |
| Hddc2 | −2.355 | 0.005 | 0.055 | Slfn2 | −8.252 | 0.014 | 0.084 |
| Hdgfrp2 | −2.558 | 0.001 | 0.028 | Smarcd2 | −3.411 | 0.018 | 0.092 |
| Hgf | −3.196 | 0.008 | 0.065 | Smarce1 | −2.804 | 0.016 | 0.089 |
| Hmgcl | −2.512 | 0.006 | 0.058 | Snf8 | −2.088 | 0.006 | 0.056 |
| Hnrnpf | −2.067 | 0.005 | 0.053 | Snrnp200 | −2.004 | 0.018 | 0.093 |
| Hnrnpk | −2.677 | 0.000 | 0.016 | Snw1 | −2.689 | 0.000 | 0.016 |
| Hnrnpm | −2.317 | 0.006 | 0.057 | Snx9 | −7.202 | 0.001 | 0.029 |
| Hoxa10 | −7.037 | 0.000 | 0.018 | Sox8 | −8.131 | 0.000 | 0.012 |
| Hoxa7 | −6.596 | 0.000 | 0.018 | Sppl3 | −2.328 | 0.012 | 0.079 |
| Hoxa9 | −6.957 | 0.000 | 0.023 | Spred3 | −4.439 | 0.003 | 0.040 |
| Hoxb2 | −5.943 | 0.000 | 0.019 | Srcap | −3.676 | 0.007 | 0.060 |
| Hoxb4 | −8.161 | 0.000 | 0.017 | Ssb | −2.140 | 0.005 | 0.053 |
| Hoxb5 | −7.221 | 0.000 | 0.017 | Supt16 | −2.360 | 0.000 | 0.018 |
| Hoxb9 | −9.174 | 0.000 | 0.016 | Syp | −2.076 | 0.015 | 0.087 |
| Hoxc6 | −9.853 | 0.000 | 0.002 | Taf4b | −10.301 | 0.002 | 0.037 |
| Hoxd10 | −7.982 | 0.001 | 0.030 | Tango6 | −10.199 | 0.002 | 0.037 |
| Hoxd4 | −4.810 | 0.001 | 0.026 | Tarbp2 | −3.521 | 0.003 | 0.040 |
| Hoxd8 | −5.752 | 0.000 | 0.020 | Tbc1d10b | −2.056 | 0.011 | 0.074 |
| Hoxd9 | −7.597 | 0.005 | 0.053 | Thap4 | −6.562 | 0.000 | 0.017 |
| Hps4 | −5.021 | 0.003 | 0.042 | Timm50 | −2.701 | 0.011 | 0.074 |
| Hsp90ab1 | −2.714 | 0.000 | 0.018 | Tkt | −3.256 | 0.010 | 0.072 |
| Hspa9 | −2.531 | 0.000 | 0.023 | Tlx3 | −2.618 | 0.002 | 0.035 |
| Ier5l | −5.059 | 0.006 | 0.057 | Tmem101 | −2.205 | 0.015 | 0.087 |
| Il11ra1 | −2.334 | 0.012 | 0.078 | Tmem201 | −5.403 | 0.000 | 0.020 |
| Ino80 | −4.042 | 0.006 | 0.057 | Tmem205 | −4.532 | 0.001 | 0.026 |
| Irf5 | −8.535 | 0.000 | 0.004 | Tnnc2 | −9.833 | 0.001 | 0.030 |
| Irgq | −2.836 | 0.010 | 0.072 | Tnnt3 | −7.545 | 0.002 | 0.038 |
| Kdm1a | −2.263 | 0.006 | 0.058 | Tnpo2 | −2.492 | 0.007 | 0.063 |
| Kdm4b | −8.283 | 0.010 | 0.073 | Tnrc18 | −2.271 | 0.005 | 0.053 |
| Kif3a | −2.029 | 0.014 | 0.084 | Top2a | −7.193 | 0.019 | 0.095 |
| Kif3c | −4.039 | 0.002 | 0.035 | Ttbk1 | −2.591 | 0.007 | 0.061 |
| Klf2 | −6.811 | 0.001 | 0.025 | Tubb3 | −2.963 | 0.000 | 0.015 |
| Lcmt2 | −4.928 | 0.003 | 0.040 | Txnrd2 | −2.946 | 0.008 | 0.066 |
| Ldlr | −2.419 | 0.001 | 0.024 | Ubc | −2.316 | 0.002 | 0.037 |
| Leo1 | −2.433 | 0.008 | 0.065 | Ube2s | −2.379 | 0.009 | 0.069 |
| Lig1 | −2.253 | 0.002 | 0.034 | Uchl1 | −3.022 | 0.009 | 0.069 |
| Loxl1 | −6.618 | 0.001 | 0.030 | Uck2 | −2.829 | 0.006 | 0.056 |
| Lrrc17 | −3.531 | 0.008 | 0.064 | Upf3b | −3.615 | 0.007 | 0.060 |
| Lrrfip1 | −3.487 | 0.005 | 0.050 | Urgcp | −4.220 | 0.001 | 0.029 |
| Lta4 h | −2.258 | 0.003 | 0.042 | Usp17la | −7.689 | 0.000 | 0.016 |
| Ltbp3 | −2.457 | 0.007 | 0.062 | Utp18 | −4.338 | 0.009 | 0.068 |
| Man2b2 | −8.690 | 0.000 | 0.018 | Vezt | −2.143 | 0.015 | 0.087 |
| Map1a | −2.487 | 0.002 | 0.035 | Vps33a | −2.623 | 0.008 | 0.066 |
| Mb | −9.449 | 0.002 | 0.035 | Vps8 | −3.249 | 0.000 | 0.018 |
| Mboat1 | −5.624 | 0.003 | 0.041 | Xirp2 | −8.750 | 0.002 | 0.038 |
| Mcoln1 | −2.973 | 0.012 | 0.079 | Yipf1 | −2.497 | 0.001 | 0.025 |
| Mdfic | −10.177 | 0.001 | 0.032 | Zfp212 | −2.281 | 0.013 | 0.081 |
| Med16 | −2.428 | 0.018 | 0.092 | Zfp292 | −2.238 | 0.015 | 0.086 |
| Med29 | −2.799 | 0.008 | 0.064 | Zfp30 | −6.842 | 0.002 | 0.035 |
| Mfap4 | −4.773 | 0.003 | 0.045 | Zfp384 | −4.615 | 0.004 | 0.049 |
| Mgat4c | −5.486 | 0.002 | 0.035 | Zfp428 | −2.044 | 0.003 | 0.045 |
Next, we evaluated correlation between differentially transcribed and translated mRNAs between the TG and DRG. To do this, we plotted the 379 mRNAs with higher transcript levels in TG and 315 with higher levels in the DRG. We plotted these against TPMs from the Nav1.8-TRAP datasets from both tissues. We did the same thing for the 372 Nav1.8-TRAP enriched mRNAs from TG and 348 from DRG and compared these with TPMs from input RNA sequencing (Fig. 4C). We observed that only 144 genes were shared between these datasets, suggesting that transcriptional and translational regulation is decoupled in these tissues, at least for the most highly enriched genes. This finding is consistent with genome-wide experiments showing that transcription and translation are decoupled for many, if not most, mRNAs (Liu et al., 2016).
We then sought to validate some specific findings from whole transcriptome or Nav1.8-TRAP sequencing data obtained from the comparison between TG and DRG. Analysis of the differentially expressed genes between TG and DRG showed that Fth1 is highly enriched in the TG (Fig. 5A). We used qRT-PCR on mRNA prepared from both tissues to validate that there is a significant enrichment of Fth1 mRNA in the TG by this method (Fig. 5B). Comparisons of the TG and DRG transcriptome showed that multiple genes among the AMPK pathway were enriched in the DRG, such as Prkag2, Acacb, Akt1s1, and Gys (Fig. 6A). Interestingly, these same mRNAs were among the 144 that were regulated at the translational level as well (Fig. 6A), but there were also a number of additional mRNAs involved in the AMPK pathway that were only found in the translatome dataset, including Cpt1c and Acaca. In stark contrast, we observed an enrichment of mRNAs in the translatome in the TG that are associated with the PI3K-mTORC1 pathway, including Strada, Lamtor5, Akt1, and Rraga (Fig. 6A,B). As mentioned previously, this predicts a higher level of mTOR activity in TG than in the DRG nociceptors. To begin to address this prediction, we examined steady-state protein levels for selected targets between DRG and TG. We chose to focus on RragA, which encodes the RagA protein, because it is a critical activator of mTORC1 activity that links mTORC1 to amino acid and glucose signaling at the interface with lysosomes (Efeyan et al., 2013, 2014). Consistent with transcriptome data, we observed no differences in the level of Rraga mRNA between TG and DRG, but we did detect a significant increase in protein level in the TG versus the DRG (Fig. 6C). We have previously shown that Rraga mRNA translation is finely controlled by the activity of Mnk1 and correlated with the level of eIF4E phosphorylation. Here, we also detected a higher level of eIF4E phosphorylation in the TG compared with DRG (Fig. 6D), suggesting that TG nociceptors may display higher translational activity via this pathway than their DRG counterparts (Megat et al., 2019). We also focused on Akt1s1, which encodes the PRAS40 protein, because this is a negative regulator of mTORC1 activity with actions that are inversely related to RagA (Wiza et al., 2012; Chong, 2016). In the DRG, we observed that the level of the ribosome-associated Akt1s1 mRNAs was higher in the DRG compared with TG, and this was validated by increased PRAS40 protein in DRG (Fig. 6E).
Figure 5.
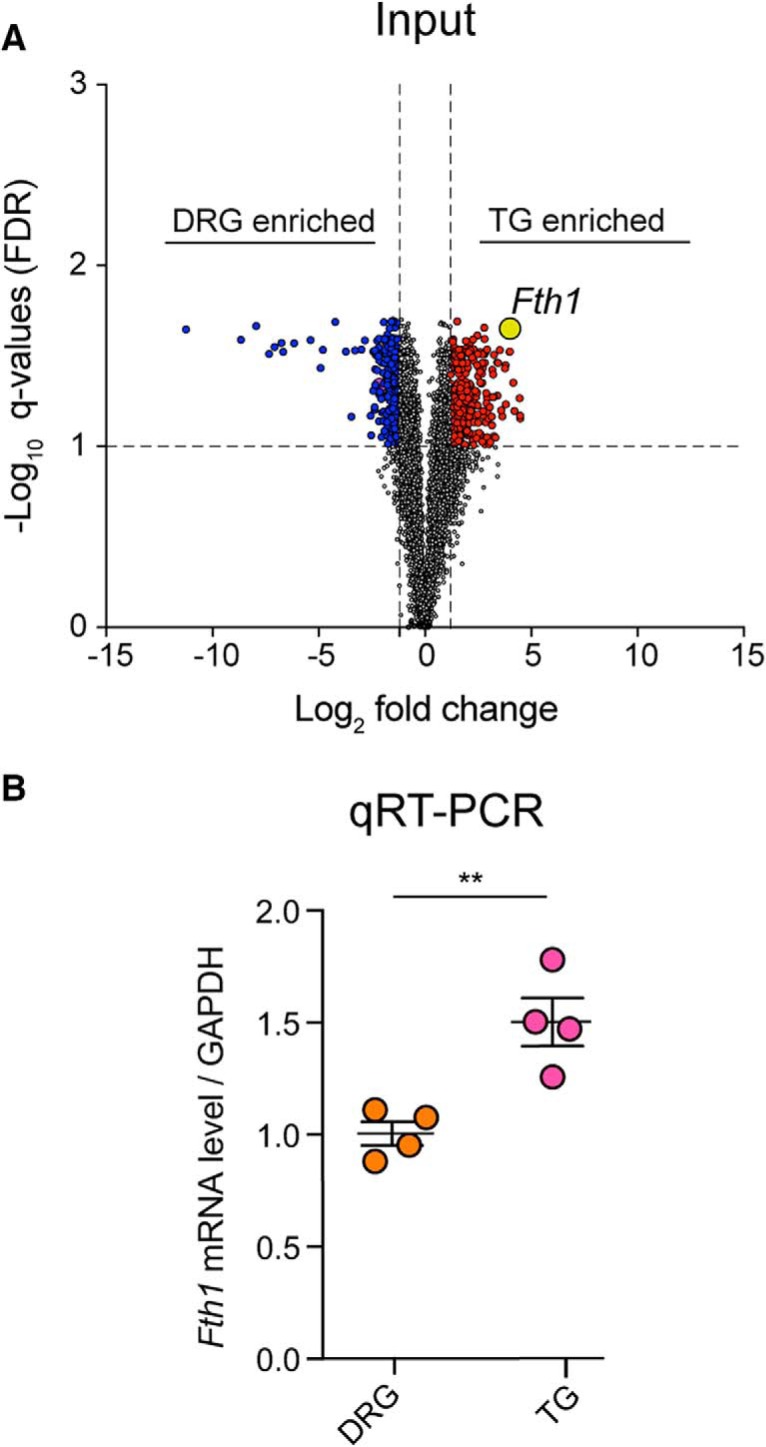
RNA-seq analysis reveals Fth1 as differentially expressed in the TG and validated through qRT-PCR. A, Volcano plot shows Fth1 (yellow dot) as being significantly enriched in TG versus DRGs. B, qRT-PCR shows a 50% increase in Fth1 mRNA expression in TG. (paired t test, t = 4,15; df = 6; **p = 0.0048).
Figure 6.
TRAP-seq analysis reveals that AMPK- and mTORC1-related genes are differential expressed and/or translated in the DRG and TG, respectively. A, Volcano plot showing an enrichment in AMPK-related genes in the input DRG sample, including Prkag2, Akt1s1, Gys1, Acacb, as well as in TRAP-seq (including Prkag2, Akt1s1, Gys1, Acacb, Acaca, and Cpt1c). In converse, mTORC1-related genes are enriched in TG, such as Strada, Rraga, Akt, and Lamtor5. B, Heatmap shows increase translation of AMPK and mTORC1 genes in the DRGs and TG, respectively. C, Immunoblotting shows an upregulation of RagA protein inTG (RagA: DRG = 100 ± 8.39,T = 149.8 ± 8.03, *p = 0.0003, n = 11), whereas Rraga mRNA measured by qRT-PCR was not different between DRGs and TG (Rraga: DRG = 1.120 ± 0.075, TG = 1.01 ± 0.024, p = 0.152, n = 4). D, Immunoblotting shows a lower level of eIF4E phosphorylation in the DRG compared with TG (p-eIF4E: TG = 100.5 ± 4.28, DRG = 79.98 ± 7.13, *p = 0.0404). E, The negative regulator of mTORC1, PRAS40 (Akt1s1) mRNA, and TE was significantly increased in DRGs and confirmed by an increase in protein level (Akt1s1: DRG = 100 ± 5.28, TG = 52.62 ± 5.48, ***p = 0.008, n = 4). F, Nocifensive behavior and grimace score after injection of capsaicin (0.1 μm) into the whisker pad or the hindpaw. Capsaicin induces a more intense affective response when injected into the whisker pad compared with the hindpaw as shown by the mouse grimace score at 15 and 30 min (two-way ANOVA: F(2,24) = 22.98, ****p < 0.0001, post hoc Sidak ****p < 0.0001 at 15 and 30 min). Likewise, nocifensive behavior is more pronounced when capsaicin was injected into the whisker pad compared with the hindpaw (F(1,12) = 11.62, **p < 0.0052, post hoc Sidak ***p = 0.002 at 60 min after capsaicin). G, Pretreatment with an mTORC1 inhibitor (AZD8055, 10 mg/kg) blocked capsaicin-induced nocifensive behavior in the whisker pad (F(2,24) = 13.93, ****p < 0.0001, post hoc Sidak ***p = 0.002 at 60 min after capsaicin) and affective pain (F(2,24) = 21.62, ****p < 0.0001, post hoc Sidak ****p < 0.0001 at 15 and 30 min). H, Intraperitoneal injection of AZD8055 (10 mg/kg) decreased the level of p-4EBP1 at 2 h (one-way ANOVA: F(2,6) = 19.15, **p = 0.0025, post hoc Dunnett: Veh vs 2 h, *p = 0.027) in the TG. I, AZD8055 inhibited capsaicin-induced grimace at 30 min (F(2,27) = 4.52, *p = 0.02, post hoc Sidak **p = 0.0034) and nocifensive behavior (F(1,9) = 17.45, **p < 0.0024, post hoc Sidak ***p < 0.001 at 60 min after capsaicin) when injected into the hindpaw. J, For each group of animals, the difference between the vehicle- and AZD8055-treated values was calculated and plotted for the nocifensive behavior and mouse grimace score. We observed a significantly larger effect size of AZD8055 in nocifensive behavior (unpaired t test, t = 3.52, df = 11, **p = 0.0048) and grimacing (unpaired t test, t = 5.54, df = 11, ***p = 0.0002) when capsaicin was injected in the whisker pad. ns, not significant.
Collectively, the results described above suggest that the balance of mTORC1 signaling through the lysosome is shifted toward activation in the TG compared with the DRG, which could influence nociceptive responses in the facial area compared with areas innervated by the DRG. To test this hypothesis, we gave injections of a low dose of capsaicin (0.1 μm), a TRPV1 agonist, into the hindpaw and the whisker pad (facial area). We observed a significantly more pronounced spontaneous pain response following facial capsaicin compared with the hindpaw (Fig. 6F). Also, the intensity/number of the nocifensive behavior was significantly larger following injection of capsaicin into the cheek, again suggesting that nociceptive stimuli trigger larger behavioral responses when administered in the facial area (Fig. 6F). We next sought to investigate whether capsaicin-induced nocifensive behavior was dependent on mTORC1 activity in the TG region. We treated animals with an mTORC1 inhibitor (AZD8055, 10 mg/kg) 2 h before injection of capsaicin into the whisker pad. We observed that the mTORC1 inhibitor significantly attenuated grimace responses and nocifensive behaviors (Fig. 6G), and this behavioral change correlated with a significant decrease in the level of p-4EBP1 (Fig. 6H), a downstream target of mTORC1. While we also observed that mTORC1 inhibition significantly attenuated grimace responses and nocifensive behavior induced by a plantar injection of capsaicin (Fig. 6I), the effect size was significantly smaller compared with capsaicin into the whisker pad (Fig. 6J). Previous clinical findings reported that repetitive noxious heat stimulation, which also acts via TRPV1, creates greater sensitization in the TG region in people (Schmidt et al., 2015). Our findings parallel these observations and support a model wherein enhanced mTORC1 signaling in TG nociceptors is a cause of this enhanced sensitization.
Combining the datasets described above with single-cell RNA sequencing from existing data sources (Usoskin et al., 2015; Hu et al., 2016) allowed us to infer translation efficiencies (TEs) for all mRNAs translated in Nav1.8 neurons. First, we used the most discriminative genes in each cell type cluster (Hu et al., 2016) and calculated the correlation coefficients with all the protein coding genes in our Nav.8-TRAP sequencing datasets. Then, we plotted the heatmap of the correlation coefficient, and we observed a clear cluster of genes highly correlated with Scn10a (Fig. 7A). The Scn10a cluster (2594 genes) was compared with our TRAP-filtered dataset (7358 genes), which generated a list of 854 Scn10a-enriched genes (Fig. 7A). We then looked at the expression level of the Scn10a-enriched genes and calculated the TEs (the ratio of the TRAP and Input values) for each gene in TG and DRG datasets. Cluster 1 (C1) identified the Scn10a-enriched genes showing high TEs in the DRG (Fig. 7B; Fig. 7-1). Among them, we again found Acaca that codes for the protein ACC (acetyl-CoA carboxylase 1), a downstream target of AMPK (Hardie, 2014). Cluster 2 (C2) identified genes showing high TEs in the TG, such as Lamtor5, Rraga, and Fkbp1a, all important regulators of the mTORC1 pathway (Fig. 7B; Fig. 7-1). This cluster also identified the CGRPβ mRNA Calcb and the MrgprD receptor mRNA. Cluster (C3) contained genes with low TEs in both TG and DRG, and cluster 4 (C4) identifies genes with high TE in TG and DRG (Fig. 7-1). Finally, we examined functional gene families (e.g., ion channels, GPCRs and kinases) for any systematic differences in TEs for mRNAs expressed in Nav1.8+ nociceptors in the TG. Interestingly, we observed that ion channels and GPCRs tend to show higher TEs compared with other gene families, such as kinases or transcription factors (Fig. 7C; Fig. 7-2), a finding that is consistent with observations in DRG Nav1.8-expressing neurons (Megat et al., 2019).
Figure 7.
TE analysis for Scn10a-enriched genes in TG- and DRG-TRAP-seq shows differential TEs between tissues. A, Heatmap showing the correlation coefficient of the protein coding genes with the most discriminative expression between cell populations based on the DRG single-cell dataset published previously (Usoskin et al., 2015; Hu et al., 2016) with Nav1.8 (Scn10a) highlighted. A cluster of 2547 genes was identified as highly enriched in the Scn10a-positive neuronal population. Those 2547 genes were then merged to the TRAP-seq filtered dataset (∼8000 genes) to identify a group of 854 mRNAs that were highly enriched in the single-cell population that also expressed Scn10a and not found in other cell populations. B, Heatmap of the TE for the 854 mRNAs shows 4 separate clusters. C1 identifies mRNAs with high TEs in the DRG but lower in TG. C2 shows genes with high TE in the TG and low TEs in the DRG. C3 identifies mRNAs with low TEs in both tissues. C4 identifies mRNAs with high TEs in both tissues. C, Calculation of TE efficiencies for gene families in the TG shows higher TEs for mRNAs coding for ion channels and GPCRs compared with splicing and transcription factors. Figure 7-1 shows estimated TEs for all genes shown in clusters in Figure 7A. Figure 7-2 shows estimated TEs by gene family.
Estimated TE for all genes in clusters shown in Fig 7A. The Table shows estimated TEs in the DRG and TG for each cluster shown in Fig 7A. Download Figure 7-1, DOCX file (106.5KB, docx)
Estimated TE for all genes by gene family. The Table shows estimated TEs in the DRG and TG for each of the gene families mentioned in the text. Download Figure 7-2, DOCX file (41.1KB, docx)
Finally, we used MEME Suite (Bailey et al., 2015) to search for motifs in the 5′ UTRs of mRNAs in clusters 1–4 described above. We only considered motifs that were found in >30% of genes in each of the clusters. In C1, we did not identify any enriched motifs; however, in C2, we identified 2 motifs in mRNAs of 5′-UTRs for genes with increased TE in the TG versus the DRG (Fig. 8). One of these was a GC-rich motif found in 82 of 307 mRNAs, and another was a terminal oligo pyrimidine tract motif found in 57 of 307 mRNAs. The latter motif is interesting because it is consistent with the finding that mTORC1 genes are more translated in the TG because terminal oligo pyrimidine tract element containing mRNAs show increased TE when mTORC1 activity is high (Thoreen et al., 2012). In the C3 cluster, which contains mRNAs with low TEs in both TG and DRG, we found a G quadruplex motif (57 of 193 mRNAs) (Fig. 8) that is likely a target for eIF4A-mediated translation control (Wolfe et al., 2014), suggesting that eIF4A activity might be low under normal conditions in TG and DRG neurons. We did not find any enriched motifs in C4.
Figure 8.
mRNA motifs enriched in 5′-UTRs from clusters of genes that show altered TEs between TG and DRG. Two motifs were found in cluster C2 (higher TE in TG than in DRG) and 1 motif was found in cluster C3 (low TE in both TG and DRG). Genes with motifs found in their 5′-UTRs are shown to the right of the corresponding motifs.
Discussion
Our work uses the TRAP technology to highlight differences in the translatomes of Nav1.8+ neurons in the DRG and TG. Although there are many consistencies between these tissues, as would be expected by the similar function of Nav1.8+ neurons in the DRG and TG, there are some striking differences that may have important functional implications. Prominent among these are higher levels of protein synthesis for many regulators of the mTORC1 pathway in the TG and higher protein synthesis for members of the AMPK signaling pathway in the DRG. mTORC1 is a well-known downstream target of the AMPK kinase (Alers et al., 2012). It has been documented that, under energy-low conditions, increases in AMPK activity inhibit mTORC1, resulting in decreased overall protein synthesis and promotion of autophagy mechanisms (Schmidt et al., 2016). Because these signaling pathways regulate one another, this suggests that mRNAs that are regulated by the mTORC1 pathway are likely to have higher translational efficiencies in the TG than in the DRG. Previous psychophysical studies in humans have shown that painful stimulation of the TG area causes greater sensitization than stimulation of DRG-innervated regions (Schmidt et al., 2015, 2016). These studies have also demonstrated a lack of habitation in the TG region with repeated painful thermal stimulation (Schmidt et al., 2015). It is now well established that the mTORC1 signaling pathway plays a key role in controlling nociceptor excitability and sensitization (Melemedjian et al., 2010; Moy et al., 2017; Khoutorsky and Price, 2018), and this sensitization is strongly attenuated by activation of the AMPK pathway (Melemedjian et al., 2011; Burton et al., 2017). Our findings are in line with somatotopic differences in response to painful stimulation and a higher propensity to sensitization in TG nociceptors. While this might be explained by the biological relevance of the head and facial area for vital functions, our data show that differences in basal mTORC1 activity between TG and DRG nociceptors could drive differences in magnitude of sensitization following injury. However, it is also important to note that recently discovered anatomical differences between central projections of DRG and TG neurons may also mediate these differences (Rodriguez et al., 2017), in particular as they relate to enhanced fear and anxiety from painful stimulation of the TG region (Schmidt et al., 2016).
A recent paper examined differences in mRNA expression on FACS-sorted TG and DRG neurons from mice, demonstrating that >99% of mRNA showed consistent expression between TG and DRG neurons (Lopes et al., 2017). These authors only identified 24 mRNAs with differential expression, but these included Hox genes, as we also found, and an arginine vasopressin receptor (Lopes et al., 2017). Many other differentially expressed genes they attributed to non-neuronal cell types. We found >300 differentially expressed genes in the whole tissue transcriptome of the DRG versus the TG, and many of these mRNAs may be attributable to non-neuronal cells because we did not sort cells for whole transcriptome. This likely explains the major discrepancies between transcriptomes in these two papers. However, major differences in translatome findings cannot be attributable to non-neuronal cells because the approach we use is specific to Nav1.8-expressing neurons, most of which are nociceptors. Our work also identifies potential differences in translation regulation signaling between the DRG and TG that provides a plausible explanation for these difference in the translatome. This is especially important considering that the mTOR (Patursky-Polischuk et al., 2009; Thoreen et al., 2012) and AMPK (Dowling et al., 2007) pathways have distinct effects on TE of specific subsets of mRNAs.
There are limitations to our approach. Primary among these is that our TE estimates could only be applied to a subset of genes that have been identified as highly enriched in the Nav1.8+ population of neurons by single-cell RNA sequencing. Future efforts may use cell-sorting techniques (Thakur et al., 2014; Lopes et al., 2017) for transcriptome generation in combination with TRAP sequencing to make estimates of the TE across the active genome of Nav1.8+ population of cells. A technical shortcoming of this potential approach is that tissue homogenization and cellular dissociation protocols that are needed to sort cells for transcriptomic analysis cause induction of classes of genes, including molecular chaperones and immediate early genes that can bias transcriptomes and distort TE estimates (van den Brink et al., 2017). A second limitation is that, while our data are suggestive of important differences in mTORC1 and AMPK signaling between these two tissues that may regulate susceptibility to nociceptor sensitization, we have not shown this directly with behavioral or electrophysiological evidence. However, the notion of that TG nociceptors are more intensely sensitized by noxious stimuli is supported by preclinical models and human psychophysical data (Schmidt et al., 2015, 2016). For example, it has recently been demonstrated that injury to TG nerves induces a grimacing effect in both rats and mice (Akintola et al., 2017). This is in stark contrast to effects of injury to the sciatic nerve where grimacing effects are not observed (Langford et al., 2010). These findings suggest that injury to TG nerves induces a stronger ongoing pain phenotype in both of these rodent species. Additional work is needed to clarify whether this is driven by the mTOR signaling axis in the TG.
The results presented here add to a growing body of literature that there are important differences between the DRG and TG that are likely relevant for understanding pain disorders that originate from these regions. These include differential developmental origins (Zou et al., 2004), differential expression of neuronal subtype markers (Price and Flores, 2007), and altered response to injury, such as sympathetic sprouting into the DRG in response to injury (Chung et al., 1996; Chien et al., 2005; Xie et al., 2007, 2015), which does not occur in the TG (Bongenhielm et al., 1999). Our use of the TRAP technique to define the translatomes of Nav1.8+ neurons in DRG and TG points to a host of newly discovered differences between these two tissues and generates a new resource that can be mined to gain addition insight.
Footnotes
This work was supported by National Institutes of Health Grant R01NS065926 to T.J.P., Grant R01NS098826 to T.J.P. and G.D., and Grant R01NS100788 to Z.T.C., University of Texas STARS program to T.J.P. and G.D., and a postdoctoral CONACYT fellowship program to P.B.-I. Raw RNA sequencing data are available through GEO: GSE 113941. Transgenic mice are available through The Jackson Laboratory. All raw data and code are available upon request.
The authors declare no competing financial interests.
References
- Akintola T, Raver C, Studlack P, Uddin O, Masri R, Keller A (2017) The grimace scale reliably assesses chronic pain in a rodent model of trigeminal neuropathic pain. Neurobiol Pain 2:13–17. 10.1016/j.ynpai.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B (2012) Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol 32:2–11. 10.1128/mcb.06159-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME Suite. Nucleic Acids Res 43:W39–W49. 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongenhielm U, Boissonade FM, Westermark A, Robinson PP, Fried K (1999) Sympathetic nerve sprouting fails to occur in the trigeminal ganglion after peripheral nerve injury in the rat. Pain 82:283–288. 10.1016/S0304-3959(99)00064-0 [DOI] [PubMed] [Google Scholar]
- Burton MD, Tillu DV, Mazhar K, Mejia GL, Asiedu MN, Inyang K, Hughes T, Lian B, Dussor G, Price TJ (2017) Pharmacological activation of AMPK inhibits incision-evoked mechanical hypersensitivity and the development of hyperalgesic priming in mice. Neuroscience 359:119–129. 10.1016/j.neuroscience.2017.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien SQ, Li C, Li H, Xie W, Pablo CS, Zhang JM (2005) Sympathetic fiber sprouting in chronically compressed dorsal root ganglia without peripheral axotomy. J Neuropathic Pain Symptom Palliation 1:19–23. 10.3109/J426v01n01_05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong ZZ. (2016) Targeting PRAS40 for multiple diseases. Drug Discov Today 21:1222–1231. 10.1016/j.drudis.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Chung K, Lee BH, Yoon YW, Chung JM (1996) Sympathetic sprouting in the dorsal root ganglia of the injured peripheral nerve in a rat neuropathic pain model. J Comp Neurol 376:241–252. [DOI] [PubMed] [Google Scholar]
- Devor M. (1999) Unexplained peculiarities of the dorsal root ganglion. Pain Suppl 6:S27–S35. 10.1016/S0304-3959(99)00135-9 [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N (2007) Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res 67:10804–10812. 10.1158/0008-5472.CAN-07-2310 [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135:749–762. 10.1016/j.cell.2008.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, Patapoutian A (2010) Nociceptors: the sensors of the pain pathway. J Clin Invest 120:3760–3772. 10.1172/JCI42843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham PL, Garrett FG (2010) Development of functional units within trigeminal ganglia correlates with increased expression of proteins involved in neuron-glia interactions. Neuron Glia Biol 6:171–181. 10.1017/S1740925X10000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM (2013) Regulation of mTORC1 by the rag GTPases is necessary for neonatal autophagy and survival. Nature 493:679–683. 10.1038/nature11745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Schweitzer LD, Bilate AM, Chang S, Kirak O, Lamming DW, Sabatini DM (2014) RagA, but not RagB, is essential for embryonic development and adult mice. Dev Cell 29:321–329. 10.1016/j.devcel.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel C, Schöbel N, Altmüller J, Becker C, Tannapfel A, Hatt H, Gisselmann G (2015) RNA-Seq analysis of human trigeminal and dorsal root ganglia with a focus on chemoreceptors. PLoS One 10:e0128951. 10.1371/journal.pone.0128951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortelny N, Overall CM, Pavlidis P, Freue GV (2017) Can we predict protein from mRNA levels? Nature 547:E19–E20. 10.1038/nature22293 [DOI] [PubMed] [Google Scholar]
- Glusman G, Caballero J, Robinson M, Kutlu B, Hood L (2013) Optimal scaling of digital transcriptomes. PLoS One 8:e77885. 10.1371/journal.pone.0077885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Wu J, Zhou S, Wang Y, Qin J, Yu B, Gu X, Yao C (2016) Global analysis of transcriptome in dorsal root ganglia following peripheral nerve injury in rats. Biochem Biophys Res Commun 478:206–212. 10.1016/j.bbrc.2016.07.067 [DOI] [PubMed] [Google Scholar]
- Hardie DG. (2014) AMPK-sensing energy while talking to other signaling pathways. Cell Metab 20:939–952. 10.1016/j.cmet.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. (2015) AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol 33:1–7. 10.1016/j.ceb.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N (2008) A translational profiling approach for the molecular characterization of CNS cell types. Cell 135:738–748. 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N (2014) Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat Protoc 9:1282–1291. 10.1038/nprot.2014.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Huang K, Hu Y, Du G, Xue Z, Zhu X, Fan G (2016) Single-cell RNA-seq reveals distinct injury responses in different types of DRG sensory neurons. Sci Rep 6:31851. 10.1038/srep31851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. (2016) Ribosome footprint profiling of translation throughout the genome. Cell 165:22–33. 10.1016/j.cell.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee J, Wenzel C, Buschow R, Weissmann R, Kuss AW, Hucho T (2014) Subgroup-elimination transcriptomics identifies signaling proteins that define subclasses of TRPV1-positive neurons and a novel paracrine circuit. PLoS One 9:e115731. 10.1371/journal.pone.0115731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier L, Reichert H (2001) Common developmental genetic mechanisms for patterning invertebrate and vertebrate brains. Brain Res Bull 55:675–682. 10.1016/S0361-9230(01)00559-7 [DOI] [PubMed] [Google Scholar]
- Khoutorsky A, Price TJ (2018) Translational control mechanisms in persistent pain. Trends Neurosci 41:100–114. 10.1016/j.tins.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogelman LJ, Christensen RE, Pedersen SH, Bertalan M, Hansen TF, Jansen-Olesen I, Olesen J (2017) Whole transcriptome expression of trigeminal ganglia compared with dorsal root ganglia in rattus norvegicus. Neuroscience 350:169–179. 10.1016/j.neuroscience.2017.03.027 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford DJ, Bailey AL, Chanda ML, Clarke SE, Drummond TE, Echols S, Glick S, Ingrao J, Klassen-Ross T, Lacroix-Fralish ML, Matsumiya L, Sorge RE, Sotocinal SG, Tabaka JM, Wong D, van den Maagdenberg AM, Ferrari MD, Craig KD, Mogil JS (2010) Coding of facial expressions of pain in the laboratory mouse. Nat Methods 7:447–449. 10.1038/nmeth.1455 [DOI] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Lopes DM, Denk F, McMahon SB (2017) The molecular fingerprint of dorsal root and trigeminal ganglion neurons. Front Mol Neurosci 10:304. 10.3389/fnmol.2017.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteniotis S, Lehmann R, Flegel C, Vogel F, Hofreuter A, Schreiner BS, Altmüller J, Becker C, Schöbel N, Hatt H, Gisselmann G (2013) Comprehensive RNA-seq expression analysis of sensory ganglia with a focus on ion channels and GPCRs in trigeminal ganglia. PLoS One 8:e79523. 10.1371/journal.pone.0079523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megat S, Ray PR, Moy JK, Lou TF, Barragán-Iglesias P, Li Y, Pradhan G, Wanghzou A, Ahmad A, Burton MD, North RY, Dougherty PM, Khoutorsky A, Sonenberg N, Webster KR, Dussor G, Campbell ZT, Price TJ (2019) Nociceptor translational profiling reveals the RagA-mTORC1 network as a critical generator of neuropathic pain. J Neurosci 39:393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Asiedu MN, Tillu DV, Sanoja R, Yan J, Lark A, Khoutorsky A, Johnson J, Peebles KA, Lepow T, Sonenberg N, Dussor G, Price TJ (2011) Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol Pain 7:70. 10.1186/1744-8069-7-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Asiedu MN, Tillu DV, Peebles KA, Yan J, Ertz N, Dussor GO, Price TJ (2010) IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J Neurosci 30:15113–15123. 10.1523/JNEUROSCI.3947-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy JK, Khoutorsky A, Asiedu MN, Black BJ, Kuhn JL, Barragán-Iglesias P, Megat S, Burton MD, Burgos-Vega CC, Melemedjian OK, Boitano S, Vagner J, Gkogkas CG, Pancrazio JJ, Mogil JS, Dussor G, Sonenberg N, Price TJ (2017) The MNK-eIF4E signaling axis contributes to injury-induced nociceptive plasticity and the development of chronic pain. J Neurosci 37:7481–7499. 10.1523/JNEUROSCI.0220-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J, Rüegg MA, Hall MN, Meyuhas O (2009) The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol 29:640–649. 10.1128/MCB.00980-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TJ, Flores CM (2007) Critical evaluation of the colocalization between calcitonin gene-related peptide, substance P, transient receptor potential vanilloid subfamily type 1 immunoreactivities, and isolectin B4 binding in primary afferent neurons of the rat and mouse. J Pain 8:263–272. 10.1016/j.jpain.2006.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders A, Mantilleri A, Malapert P, Rialle S, Nidelet S, Laffray S, Beurrier C, Bourinet E, Moqrich A (2015) Transcriptional profiling of cutaneous MRGPRD free nerve endings and C-LTMRs. Cell Rep 10:1007–1019. 10.1016/j.celrep.2015.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E, Sakurai K, Xu J, Chen Y, Toda K, Zhao S, Han BX, Ryu D, Yin H, Liedtke W, Wang F (2017) A craniofacial-specific monosynaptic circuit enables heightened affective pain. Nat Neurosci 20:1734–1743. 10.1038/s41593-017-0012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Schunke O, Forkmann K, Bingel U (2015) Enhanced short-term sensitization of facial compared with limb heat pain. J Pain 16:781–790. 10.1016/j.jpain.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Schmidt K, Forkmann K, Sinke C, Gratz M, Bitz A, Bingel U (2016) The differential effect of trigeminal vs. peripheral pain stimulation on visual processing and memory encoding is influenced by pain-related fear. Neuroimage 134:386–395. 10.1016/j.neuroimage.2016.03.026 [DOI] [PubMed] [Google Scholar]
- Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH, Nassar MA (2005) Nociceptor-specific gene deletion using heterozygous NaV1.8-cre recombinase mice. Pain 113:27–36. 10.1016/j.pain.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Takeda M, Takahashi M, Matsumoto S (2008) Contribution of activated interleukin receptors in trigeminal ganglion neurons to hyperalgesia via satellite glial interleukin-1beta paracrine mechanism. Brain Behav Immun 22:1016–1023. 10.1016/j.bbi.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Thakur M, Crow M, Richards N, Davey GI, Levine E, Kelleher JH, Agley CC, Denk F, Harridge SD, McMahon SB (2014) Defining the nociceptor transcriptome. Front Mol Neurosci 7:87. 10.3389/fnmol.2014.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485:109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P (2015) Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18:145–153. 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]
- Uttam S, Wong C, Amorim IS, Jafarnejad SM, Tansley SN, Yang J, Prager-Khoutorsky M, Mogil JS, Gkogkas CG, Khoutorsky A (2018) Translational profiling of dorsal root ganglia and spinal cord in a mouse model of neuropathic pain. Neurobiol Pain 4:35–44. 10.1016/j.ynpai.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, Robin C, van Oudenaarden A (2017) Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods 14:935–936. 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- Wiza C, Nascimento EB, Ouwens DM (2012) Role of PRAS40 in Akt and mTOR signaling in health and disease. Am J Physiol Endocrinol Metab 302:E1453–E1460. 10.1152/ajpendo.00660.2011 [DOI] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, Schatz JH, Rodrigo CM, Zhao C, Rondou P, de Stanchina E, Teruya-Feldstein J, Kelliher MA, Speleman F, Porco JA Jr, Pelletier J, et al. (2014) RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513:65–70. 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf CJ, Ma Q (2007) Nociceptors-noxious stimulus detectors. Neuron 55:353–364. 10.1016/j.neuron.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Xie W, Strong JA, Li H, Zhang JM (2007) Sympathetic sprouting near sensory neurons after nerve injury occurs preferentially on spontaneously active cells and is reduced by early nerve block. J Neurophysiol 97:492–502. 10.1152/jn.00899.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Strong JA, Zhang JM (2015) Local knockdown of the NaV1.6 sodium channel reduces pain behaviors, sensory neuron excitability, and sympathetic sprouting in rat models of neuropathic pain. Neuroscience 291:317–330. 10.1016/j.neuroscience.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Zhang Y, Ma Q, Gu F, Day DS, He A, Zhou B, Li J, Stevens SM, Romo D, Pu WT (2013) Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci U S A 110:15395–15400. 10.1073/pnas.1304124110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Fritzsch B, Xu PX (2004) Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development 131:5561–5572. 10.1242/dev.01437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Estimated TE for all genes in clusters shown in Fig 7A. The Table shows estimated TEs in the DRG and TG for each cluster shown in Fig 7A. Download Figure 7-1, DOCX file (106.5KB, docx)
Estimated TE for all genes by gene family. The Table shows estimated TEs in the DRG and TG for each of the gene families mentioned in the text. Download Figure 7-2, DOCX file (41.1KB, docx)