Abstract
Pregnenolone and dehydroepiandrosterone (DHEA) are hydroxysteroids that serve as biosynthetic precursors for steroid hormones in human body. SULT2B1b has been reported to be critically involved in the sulfation of pregnenolone and DHEA, particularly in the sex steroid-responsive tissues. The current study was designed to investigate the impact of the genetic polymorphisms of SULT2B1 on the sulfation of DHEA and pregnenolone by SULT2B1b allozymes. Ten SULT2B1b allozymes previously prepared were shown to exhibit differential sulfating activities toward DHEA and pregnenolone in comparison to the wild-type enzyme. Kinetic studies revealed further significant changes in their substrate-binding affinity and catalytic activity toward DHEA and pregnenolone. Taken together, these results indicated clearly a profound effect of SULT2B1 genetic polymorphisms on the sulfating activity of SULT2B1b allozymes toward DHEA and pregnenolone, which may have implications in inter-individual variations in the homeostasis of these two important steroid precursors.
Keywords: SULT, SULT2B1b, single nucleotide polymorphisms, DHEA, pregnenolone
1. Introduction
Dehydroepiandrosterone (DHEA), the main biosynthetic precursor of sex steroids (Schiffer et al., 2018), is synthesized mainly in the adrenal glands and, to a lower extent, in ovaries and testes (Labrie, 2010). In the adrenal glands, cholesterol is first transformed to pregnenolone under the action of cytochrome P450scc (Schiffer et al., 2018). The conversion of cholesterol to pregnenolone is the rate limiting step in steroid hormone biosynthesis pathway (Neunzig and Bernhardt, 2014). Pregnenolone then serves as a precursor for glucocorticoids, mineralocorticoids, and DHEA (Neunzig and Bernhardt, 2014). In peripheral tissues, the adrenal secreted DHEA acts as a precursor for estrogen and androgen hormones through a process called intracrinology (Schiffer et al., 2018; Neunzig and Bernhardt, 2014), which involves the intracellular formation, inactivation, and action of sex steroids (Labrie, 2010). Sulfation of DHEA forming DHEA-S limits the amount of DHEA that is available for the biosynthesis of androgen hormones (Noordam et al., 2009). In the body, the sulfation of DHEA and pregnenolone has been shown to be mediated by the cytosolic sulfotransferase (SULT) enzymes, particularly SULT2A1, SULT2B1a, and SULT2B1b (Falany and Rohn-Glowacki, 2013). It is noted that in addition to serving as steroid hormones precursors, DHEA, pregnenolone, and their sulfated metabolites synthesized independently in the nervous system are considered neurosteroids that act as neuromodulators (Vallee et al., 2001).
The cytosolic sulfotransferases (SULTs) are a group of phase II conjugation enzymes that are involved in the homeostasis and detoxification of numerous exogenous and xenobiotic compounds (Falany, 1997). The SULTs catalyze the transfer of a sulfonate group from 3’-phosphoadenosine 5’-phosphosulfate (PAPS) to the hydroxyl or amino group of acceptor compounds, leading to their increased hydrophilicity and facilitated urinary and biliary excretion from the body (Falany and Roth, 1993; Weinshilboum and Otterness, 1994). In humans, there are 13 distinct SULTs that are classified into four gene families, designated SULT1, SULT2, SULT4, and SULT6 (Glatt et al., 2001; Freimuth et al., 2004). SULT2 family is previously known as the hydroxysteroid sulfotransferase family and consists of three isoforms: SULT2A1 (previously called a DHEA sulfotransferase), SULT2B1a (a pregnenolone sulfotransferase), and SULT2B1b (a cholesterol sulfotransferase) (Falany and Rohn-Glowacki, 2013). SULT2B1a and SULT2B1b isoforms are coded by the same gene, designated SULT2B1, and are generated as a result of alternative initiation and splicing, leading to the formation of the two isoforms with distinct N-terminal regions (Her et al., 1998). Compared with other known SULT enzymes, the two SULT2B1 isoforms carry a unique carboxy-terminal extension of about 53 amino acid residues (He and Falany, 2006). A previous study using SULT2B1b demonstrated that this carboxy-terminal extension influenced the thermostability, kinetic properties, subcellular localization, and immunogenicity, as well as posttranslational modification by phosphorylation (He and Falany, 2006). While the three SULT2 isoforms display overlapping substrate specificity toward different hydroxysteroids such as DHEA and pregnenolone, they exhibit tissue-specific distribution (Falany et al., 2006; Geese and Raftogianis, 2001; Otterness and Weinshilboum, 1994; Thomae et al., 2002). For example, SULT2A1 is expressed mainly in the liver, adrenal glands, and intestine (Otterness and Weinshilboum, 1994; Thomae et al., 2002), whereas SULT2B1b is highly expressed in the placenta, prostate, breast, endometrium, ovary, uterus, small intestine, colon, lung, platelet, brain, and skin (Falany et al., 2006; Geese and Raftogianis, 2001). In contrast, no expression of SULT2B1a protein was detected in any of the tissues examined (Falany and Rohn-Glowacki, 2013). SULT2B1b thus is more likely the main enzyme responsible for the sulfation of DHEA and pregnenolone in steroid-responsive tissues as well as in the brain. SULT2B1 genetic polymorphisms have been reported (Hyland et al., 2013, Levesque et al., 2014, Mostaghel 2013, Yang et al., 2013, Hu et al., 2015, Vickman et al., 2016, Chen et al., 2016; Ji et al., 2007). It is an interesting question whether SULT2B1 missense coding SNPs (cSNPs) may influence the sulfating activity of the resulting SULT2B1b allozymes and thus affect steroid-related physiology and pathology in different individuals.
In this study, ten SULT2B1b allozymes, previously prepared by site-directed mutagenesis in conjunction with bacterial expression using the pGEX gene fusion system, were examined for sulfating activities toward DHEA and pregnenolone. Kinetic parameters of SULT2B1b allozymes in mediating the sulfation of DHEA and pregnenolone were determined to delineate their differential substrate affinity and catalytic activity toward DHEA and pregnenolone in comparison to the wild-type enzyme.
2. Materials and Methods
2.1. Materials.
Pregnenolone, DHEA, adenosine 5′-triphosphate (ATP), dimethyl sulfoxide (DMSO), N-2-hydroxylpiperazine-N′-2-ethanesulfonic acid (HEPES), dithiothreitol (DTT), and Trizma base, were products of Sigma-Aldrich (St. Louis, MO, USA). PrimeSTAR® Max DNA polymerase was a product of Takara Bio (Mountain View, CA, USA). Oligonucleotide primers were synthesized by Eurofins Genomics (Louisville, KY, USA). Cellulose TLC plates and Ultrafree-MC 5000 NMWL filter units were from EMD Millipore (Billerica, MA, USA). Ecolume liquid scintillation cocktail was a product of MP Biomedicals, LLC. (Irvine, CA, USA). Carrier-free sodium [35S]sulfate was from American Radiolabeled Chemicals (St. Louis, MO, USA). PAP[35S] was synthesized using recombinant human bifunctional PAPS synthase based on a previously established procedure (Yanagisawa et al., 1998). All other chemicals were of the highest grade commercially available.
2.2. Preparation of human SULT2B1 allozymes.
As described previously, SNP databases located at the U.S. National Center for Biotechnology Information (NCBI), the Ensembl Variation database, and the Universal Protein Resource (UniProt), were systematically searched. Ten SULT2B1 coding SNPs (cSNPs) were selected based on the location and chemical nature of the amino acid variations in coded SULT2B1b allozymes (Alherz et al., 2018). The designated names and SNP ID numbers of these 10 cSNPs are: SULT2B1bPro69Ala reference SNP (rs777924668), SULT2B1b-Gly72Val (rs746398875), SULT2B1b-Thr73Met (rs527454384), SULT2B1b-Arg147His (rs777140014), SULT2B1b-Asp191Asn (rs16982158), SULT2B1b-Arg230His (rs16982169), SULT2B1b-Ser244Thr (rs765224593), SULT2B1b-Arg274Gln (rs762765702), SULT2B1b-Gly276Val (rs774212320), and SULT2B1b-Pro345Leu (rs17842463) (Table 1). The corresponding cDNAs were generated by site-directed mutagenesis, and the recombinant SULT2B1b allozymes were expressed using pGEX-4T-2 prokaryotic expression vectors and affinity purified and cleaved from the Glutathione S-transferases fusion proteins as described previously (Alherz et al., 2018).
Table 1.
Allele frequencies of human SULT2B1b enzyme
| SNP ID | SULT2B1b Allozyme | Allele Frequency | Reference |
|---|---|---|---|
| rs777924668 | SULT2B1b-Pro69Ala | 0.00001 | NCBI |
| rs746398875 | SULT2B1b-Gly72Val | 0.00001 | NCBI |
| rs527454384 | SULT2B1b-Thr73Met | 0.00002 | NCBI |
| rs777140014 | SULT2B1b-Arg147His | 0.00001 | NCBI |
| rs16982158 | SULT2B1b-Asp191Asn | 0.008 AA | Ji et al., 2007 |
| rs16982169 | SULT2B1b-Arg230His | 0.008 AA | Ji et al., 2007 |
| rs765224593 | SULT2B1b-Ser244Thr | 0.00006 | NCBI |
| rs762765702 | SULT2B1b-Arg274Gln | 0.0001 | NCBI |
| rs774212320 | SULT2B1b-Gly276Val | 0.0000 | NCBI |
| rs17842463 | SULT2B1b-Pro345Leu | 0.025 CA | Ji et al., 2007 |
AA, African American population. CA, Caucasian American population.
2.3. Sulfotransferase assay.
To quantify the sulfating activity of the recombinant SULT2B1b allozymes, PAP[35S] was used as the sulfate donor. The standard assay mixture, with a final volume of 20 μL, contained 50 mM HEPES buffer (pH 7.4), 1 mM DTT, 14 μM PAP[35S] (14.4 Ci/mmol), 0.5 μg of wild-type or SULT2B1b allozyme, and DHEA or pregnenolone (dissolved in DMSO at 10 times the final concentration in the assay mixture) as a substrate. The final concentration of DMSO in the assay mixture was thus 10% (volume/volume). A control with DMSO alone was installed in parallel. The reaction was performed for 10 min at 37°C and stopped by incubating the reaction mixture for 3 min at 100°C. To analyze the production of [35S]sulfated DHEA or pregnenolone, 1 μL of the final reaction mixture was spotted on a cellulose TLC plate, followed by TLC using a solvent system containing n-butanol: isopropanol: formic acid: water in a ratio of 3:1:1:1 (by volume). The [35S]sulfated DHEA or pregnenolone spot was located by autoradiography, cut out from the TLC plate, and eluted with 0.5 ml H2O. The [35S]radioactivity of the eluate was quantified using a liquid scintillation counter as described previously (Hui and Liu, 2015). To determine the kinetic parameters of individual SULT2B1b allozymes, varying concentrations of DHEA or pregnenolone were used, based on the same procedure described above. To determine the Km for PAPS, varying concentrations of PAPS (ranging 0.1 to 50 μM), with 50 μM of DHEA or 10 μM of pregnenolone as substrate, were tested.
2.4. Statistical analysis.
GraphPad Prism® v 6.0 software was used for calculating the kinetic constants, Km, Vmax, Kcat, and Kcat/Km based on Michaelis-Menten kinetics using non-linear regression. To determine statistical differences between the wild-type SULT2B1b and individual SULT2B1b allozymes, one-way ANOVA was used for inter-group comparison, followed by Dunnett’s test, with p-value < 0.05 considered being statistically significant.
2.5. Rotamer analysis of single point mutations.
Side-chain conformation of a mutated amino acid residue was simulated using the Dunbrack backbone-dependent rotamer library (Dunbrack, 2002). The structure of SULT2B1b, resolved with pregnenolone and PAP, (Protein Data Bank code: 1Q20), was referred to as the wild-type. Hydrophobic and hydrogen-binding interactions of the point-mutated amino acid residue with other residues, pregnenolone, or PAP, were also simulated by Find Clashes/Contacts tool in USCF Chimera software (Pettersen et al., 2004).
2.6. Substrate-binding simulation and molecular dynamics simulation analysis.
The molecular simulations for the docking of PAPS and pregnenolone into the substrate-binding sites were carried out with the crystal structure of SULT2B1b in complex with PAP and pregnenolone (1Q20) as a template. PAP in the complex was removed and PAPS was docked into the PAPS-binding site of SULT2B1b structure with amino acid substitution for each allozyme using AutoDock Vina (Trott and Olson, 2010). Pregnenolone in the complex was removed and docked into the substrate-binding site of SULT2B1b structure with substitution for each allozyme using AutoDock Vina. Molecular dynamics simulation of individual SULT2B1 allozymes with PAP and pregnenolone was performed using MD/Ensemble analysis tool in conjunction with Amber parameter of USCF Chimera software (Pettersen et al, 2004).
3. Results
3.1. Characterization of the DHEA-sulfating activity of purified human SULT2B1b allozymes.
The sulfating activity of the recombinant SULT2B1b allozymes toward DHEA was examined. Three of the ten tested SULT2B1b allozymes (SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val) showed no detectable activity, whereas the other seven SULT2B1b allozymes displayed differential sulfating activity toward DHEA (Fig. 1). Among these seven SULT2B1b allozymes, SULT2B1b-Arg274Gln showed the greatest decrease in DHEA-sulfating activity, with a 27-fold reduction compared with the SULT2B1b-wt. Of the other six allozymes, four (SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, SULT2B1b-Ser244Thr, and SULT2B1b-Pro345Leu) displayed a considerable decrease (1.7-fold or greater) in DHEA-sulfating activity, while the other two (SULT2B1b-Pro69Ala and SULT2B1b-Thr73Met) exhibited a much greater (more than 8-fold) decrease in DHEA-sulfating activity compared with the wild-type enzyme.
Fig. 1. Specific activity of the human SULT2B1b allozymes toward DHEA.
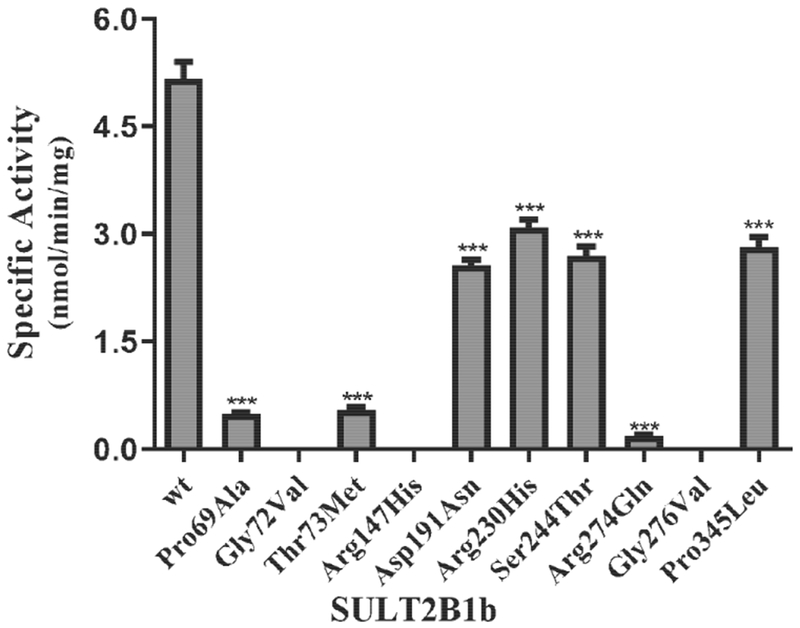
Concentration of DHEA used in the enzymatic assays was 50 μM. Data shown represent mean ± standard deviation derived from three separate determinations. One-way ANOVA was performed followed by Dunnett’s post hoc analysis. *** Statistical significant p<0.001 from SULT2B1b-wt. Three allozymes, SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val, showed no detectable activity.
To investigate further the effects of genetic polymorphisms on the DHEA-sulfating activity of SULT2B1b allozymes, kinetic experiments were performed using varying concentrations of DHEA as a substrate. The sulfation of DHEA appeared to follow the Michaelis-Menten kinetics (cf. Fig 2). The determined kinetic constants, Km, Vmax, Kcat, and Kcat/Km, for the wild-type and SULT2B1b allozymes are compiled in Table 2. Of the seven SULT2B1b allozymes examined, two (SULT2B1b-Thr73Met and SULT2B1b-Arg274Gln) showed dramatic increases in Km value (at least 5 times) compared with SULT2B1b-wt, indicating that the amino acid changes resulted in decreased DHEA binding affinity. All tested SULT2B1b allozymes exhibited lower catalytic activity (Vmax) compared with the wild-type enzyme. Among them, SULT2B1b-Asp191Asn displayed the smallest decrease (a 12.5% reduction) in the catalytic activity, while SULT2B1b-Pro345Leu and SULT2B1b-Arg230His showed more than 37% decrease in Vmax compared with the wild-type enzyme. In contrast, the other four allozymes exhibited much greater reduction. SULT2B1b-Ser244Thr and SULT2B1b-Thr73Met showed a more than 50% decrease in Vmax, whereas SULT2B1b-Pro69Ala and SULT2B1b-Arg274Gln exhibited a more than 87% decrease in Vmax, compared with SULT2B1b-wt. Consequently, the catalytic efficiency as reflected by kcat/km was significantly lower for all seven SULT2B1b allozymes. Four of them, SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, SULT2B1b-Ser244Thr, and SULT2B1b-Pro345Leu, showed more than 24% decrease compared with the wild-type (SULT2B1b-wt), while the other three (SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, and SULT2B1b-Arg274Gln) showed more dramatic reduction (up to 90%) compared with the wild-type enzyme.
Fig. 2. Kinetic analysis of the sulfation of DHEA by human wild-type SULT2B1b.
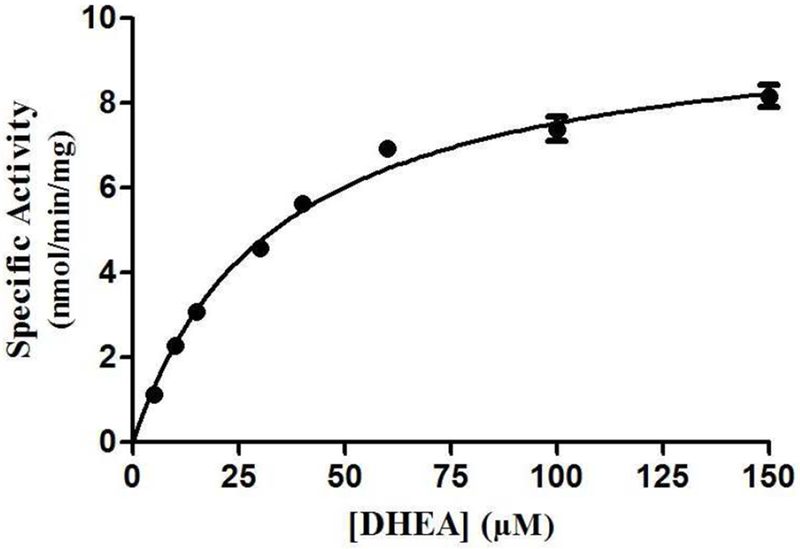
The figure shows the saturation curve analysis of the sulfation of DHEA. The fitting curve was generated based on Michaelis-Menten kinetics. Data shown represent calculated mean ± standard deviation derived from three separate experiments.
Table 2.
Kinetic constants of the human SULT2B1b allozymes in catalyzing the sulfation of DHEA
| Enzyme | DHEA | PAPS | |||
|---|---|---|---|---|---|
| Km (μM) | Vmax (nmol/min/mg) | Kcat(s−1) ×10−3 | Kcat/Km (s−1 M−1) | Km (μM) | |
| SULT2B1b-wt | 36 ± 3 | 8 ± 0.2 | 6 ± 0.1 | 167 ± 11 | 1.2 ± 0.2 |
| SULT2B1b-Pro69Ala | 62 ± 6 | 1 ± 0.0*** | 1 ± 0.0*** | 16 ± 1*** | 2.6 ± 0.6 |
| SULT2B1b-Gly72Val | N.D. | N.D. | N.D. | N.D. | N.D. |
| SULT2B1b-Thr73Met | 222 ± 24*** | 3 ± 01*** | 2 ± 0.1*** | 9 ± 1*** | 0.8 ± 0.1 |
| SULT2B1b-Arg147His | N.D. | N.D. | N.D. | N.D. | N.D. |
| SULT2B1b-Asp191Asn | 62 ± 8 | 7 ± 0.3*** | 5 ± 0.2*** | 81 ± 7*** | 0.5 ± 0.0 |
| SULT2B1b-Arg230His | 35 ± 4 | 5 ± 0.2*** | 3 ± 01*** | 86 ± 7*** | 1.0 ± 0.1 |
| SULT2B1b-Ser244Thr | 24 ± 4 | 4 ± 0.2*** | 3 ± 01*** | 127 ± 17*** | 0.3 ± 0.1 |
| SULT2B1b-Arg274Gln | 190 ± 25*** | 1 ± 0.0*** | 1 ± 0.0*** | 5 ± 1*** | 24.2 ± 4*** |
| SULT2B1b-Gly276Val | N.D. | N.D. | N.D. | N.D. | N.D. |
| SULT2B 1b-Pro345Leu | 38 ± 3 | 5 ± 0.3*** | 3 ± 0.2*** | 79 ± 1*** | 1.3 ± 0.1 |
Data shown represent mean ± SD derived from 3 independent experiments.
Statistical significance from SULT2B1b-wt (***p-value<0.0001) using one-way ANOVA followed by Dunnett’s post hoc analysis.
N.D. no detectable activity.
To investigate the binding affinity of the allozymes with the co-substrate (PAPS), the Km values for PAPS of SULT2B1b allozymes were determined with DHEA as substrate. Of all SULT2B1b allozymes tested, only SULT2B1b-Arg274Gln showed a significantly higher (with a 20-fold increase) Km value compared to the wild-type SULT2B1b.
3.2. Characterization of the pregnenolone-sulfating activity of purified human SULT2B1b allozymes.
In addition to DHEA sulfation, previous studies have shown that SULT2B1b can also sulfate pregnenolone (Falany and Rohn-Glowacki, 2013). In an initial experiment, the pregnenolone-sulfating activity of SULT2B1b allozymes was examined using 10 μM of pregnenolone as substrate. Of the ten SULT2B1b allozymes analyzed, three (SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val) showed no detectable sulfating activity. The other seven SULT2B1b allozymes exhibited differential and significantly lower sulfating activity toward pregnenolone (Fig. 3). Compared with SULT2B1b-wt, four of them (SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, SULT2B1b-Ser244Thr, and SULT2B1b-Pro345Leu) displayed a more than 1.2-fold decrease in pregnenolone-sulfating activity, while two (SULT2B1b-Pro69Ala and SULT2B1b-Thr73Met) showed a much greater decrease (by more than 5-fold) in pregnenolone-sulfating activity. Notably, SULT2B1b-Arg274Gln displayed the lowest (being greater than 35-fold lower) pregnenolone-sulfating activity compared with the wild-type enzyme.
Fig. 3. Specific activity of the human SULT2B1b allozymes toward pregnenolone.
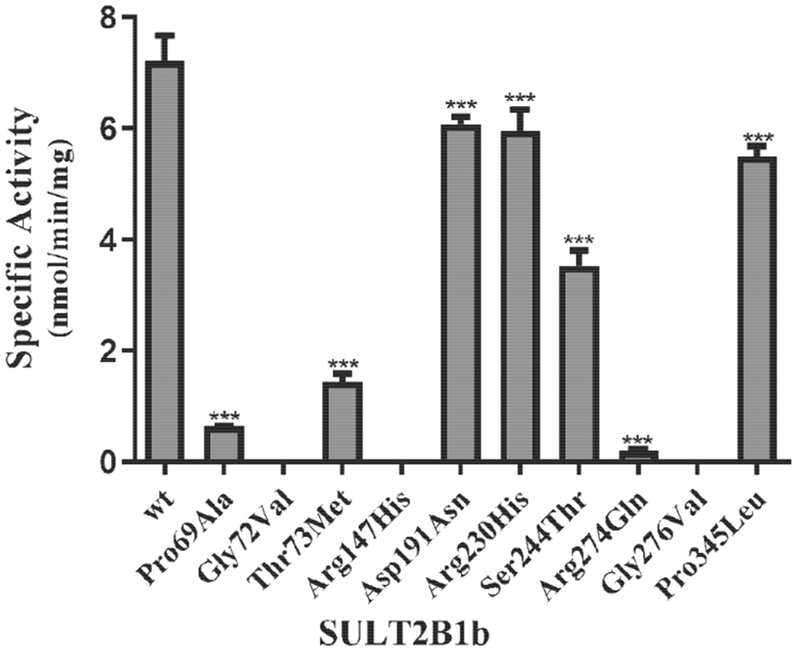
Concentration of pregnenolone used in the enzymatic assays was 10 μM. Data shown represent mean ± standard deviation derived from three separate determinations. One-way ANOVA was performed followed by Dunnett’s post hoc analysis. *** Statistical significant p<0.001 from SULT2B1b-wt. Three allozymes, SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val, showed no detectable activity.
To analyze further the effect of genetic polymorphisms on the pregnenolone-sulfating activity of SULT2B1b allozymes, kinetic experiments were performed using varying concentrations of pregnenolone as substrates. As shown in Fig. 4, pregnenolone sulfation appeared to follow the Michaelis-Menten kinetics. Kinetic constants determined for the wild-type and SULT2B1b allozymes are compiled in Table 3. Of the seven SULT2B1b allozymes analyzed, SULT2B1b-Thr73Met and SULT2B1b-Arg274Gln displayed 1.6 and 2.8-fold increase in Km value, respectively. All seven SULT2B1b allozymes examined showed a significant reduction in Vmax compared with that of SULT2B1b-wt. SULT2B1b-Pro69Ala and SULT2B1b-Arg274Gln exhibited the greatest reduction (a more than 91% decrease) in the catalytic activity toward pregnenolone compared with SULT2B1b-wt. Of the other five SULT2B1b allozymes, three (SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, and SULT2B1b-Pro345Leu) displayed a greater than 16% decrease in Vmax, while two (SULT2B1b-Thr73Met and SULT2B1b-Ser244Thr) showed a more than 58% decrease in Vmax compared with the wild-type. Based on these results, four (SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, SULT2B1b-Ser244Thr, and SULT2B1b-Arg274Gln) showed a significant reduction in the catalytic efficiency (kcat/km).Three (SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, and SULT2B1b-Arg274Gln) showed a more than 79% decrease in the catalytic efficiency compared with the wild-type (SULT2B1b-wt), while SULT2B1b-Ser244Thr showed a smaller decrease (40%) in catalytic efficiency as compared with the wild-type. Km values for PAPS of the SULT2B1b allozymes in mediating the sulfation of pregnenolone were also determined. As shown in Table 3, of the seven SULT2B1b allozymes, two (SULT2B1b-Pro69Ala and SULT2B1b-Arg274Gln) showed higher (2.8 and 18-fold, respectively) Km values, compared to SULT2B1b-wt.
Fig. 4. Kinetic analysis of the sulfation of pregnenolone by human wild-type SULT2B1b.
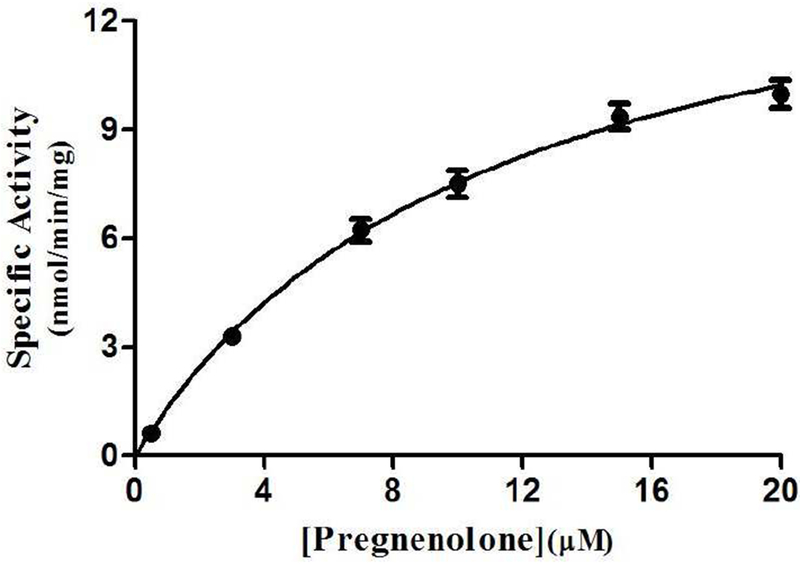
The figure shows the saturation curve analysis of the sulfation of pregnenolone. The fitting curve was generated based on Michaelis-Menten kinetics. Data shown represent calculated mean ± standard deviation derived from three separate experiments.
Table 3.
Kinetic constants of the human SULT2B1b allozymes in catalyzing the sulfation of pregnenolone
| Enzyme | Pregnenolone | PAPS | |||
|---|---|---|---|---|---|
| Km (μM) | Vmax (nmol/min/mg) | Kcat (s−1) ×10−3 | Kcat/Km (s−1 M−1) | Km (μM) | |
| SULT2B1b-wt | 8 ± 1 | 12 ± 0.7 | 8 ± 0.5 | 1005 ± 64 | 1.3 ± 0.2 |
| SULT2B 1b-Pro69Ala | 5 ± 1 | 1 ± 0.0*** | 1 ± 0.0*** | 206 ± 26*** | 3.7 ± 0.5** |
| SULT2B1b-Gly72Val | N.D. | N.D. | N.D. | N.D. | N.D. |
| SULT2B 1b-Thr73Met | 13 ± 2** | 3 ± 0.2*** | 2 ± 0.1*** | 156 ± 16*** | 1.0 ± 0.1 |
| SULT2B1b-Arg147His | N.D. | N.D. | N.D. | N.D. | N.D. |
| SULT2B 1b-Asp 191Asn | 6 ± 1 | 8 ± 0.5*** | 6 ± 0.3*** | 1013 ± 100 | 0.7 ± 0.0 |
| SULT2B 1b-Arg230His | 7 ± 1 | 10 ± 0.5*** | 7 ± 0.3** | 1010 ± 102 | 1.3 ± 0.1 |
| SULT2B 1b-Ser244Thr | 5 ± 1 | 5 ± 0.7*** | 3 ± 0.5*** | 603 ± 21*** | 0.6 ± 0.1 |
| SULT2B 1b-Arg274Gln | 22 ± 2*** | 1 ± 0.1*** | 1 ± 0.1*** | 45 ± 4*** | 23.6 ± 2*** |
| SULT2B1b-Gly276Val | N.D. | N.D. | N.D. | N.D. | N.D. |
| SULT2B 1b-Pro345Leu | 8 ± 1 | 10 ± 0.6*** | 7 ± 0.5** | 877 ± 27 | 1.4 ± 0.1 |
Data shown represent mean ± SD derived from 3 independent experiments.
Statistical significance from SULT2B1b-wt (*p-value<0.05, **p-value<0.001, ***p-value<0.0001) using one-way ANOVA followed by Dunnett’s post hoc analysis.
N.D. no detectable activity.
3.3. Structure simulation and analysis of the single amino acid mutations.
Effects of the amino acid substitution on the interaction with pregnenolone and PAP were investigated using simulated structures of SULT2B1b allozymes with corresponding amino acid substitutions (Fig. 6). Among the nine substitutions, seven substitutions (Pro69Ala, Gly72Val, Thr73Met, Arg147His, Ser244Thr, Arg174Gln, Gly276Val) were found to affect the interaction with PAP. In particular, Thr73Met was found to have altered interaction with pregnenolone as well as PAP. In contrast, Asp191Asn and Arg230His appeared to have no direct effect on the interaction with pregnenolone and PAP. Further simulation analyses were performed to predict the binding energy of SULT2B1b allozymes with pregnenolone and PAPS (Fig. 7). The docking simulation analyses suggested that substitution of Arg274 and Gly276 to Gln and Val, respectively, reduced the binding affinity with PAPS. In addition, molecular dynamics simulations were carried out to gain insight into the dynamic effects of individual amino acid substitutions on the overall structure and interaction with substrates (Fig. 8). Representative structures of most clustered group in the total 1000 structures were aligned with that of the wild-type. Most of the SULT2B1b allozyme structures were found to be similar with that of the wild-type (data not shown). The representative conformations of Pro69Ala, Thr73Met, and Arg147His are shown in Fig. 8. Interestingly, conformation of segment-2 of Arg147His appeared clearly different, which suggested that substitution of Arg147 by His may affect the interaction with PAPS. Detail observations for the simulation analyses are further elaborated in the Discussion section.
Fig. 6. Hydrophobic interaction and hydrogen bonding analyses of the SULT2B1b cSNPs.
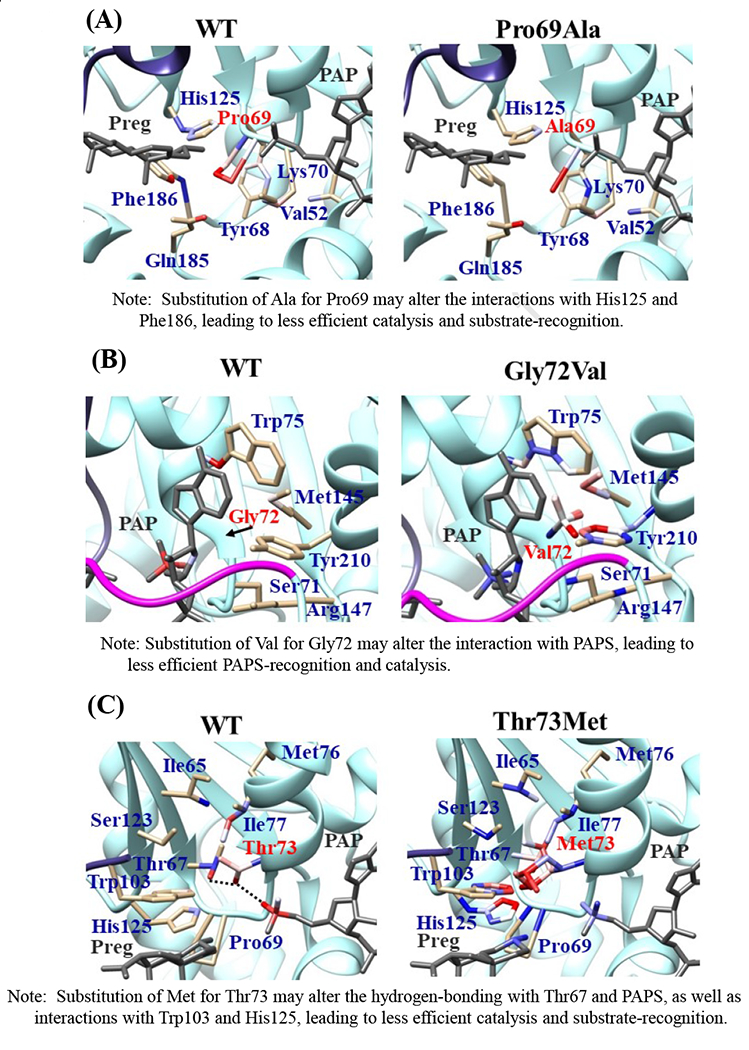
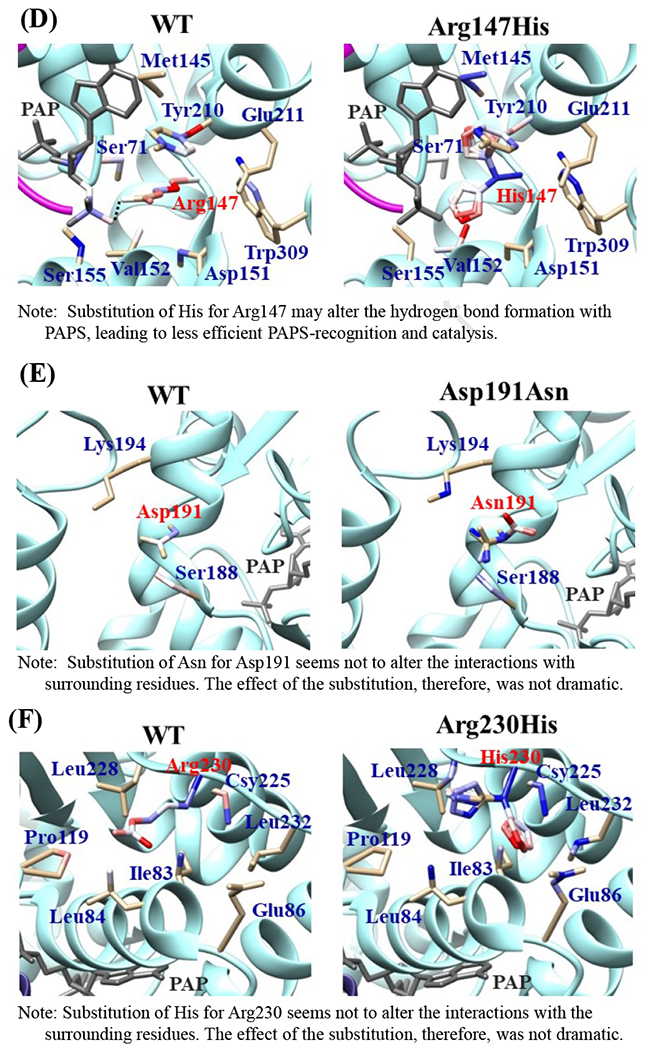
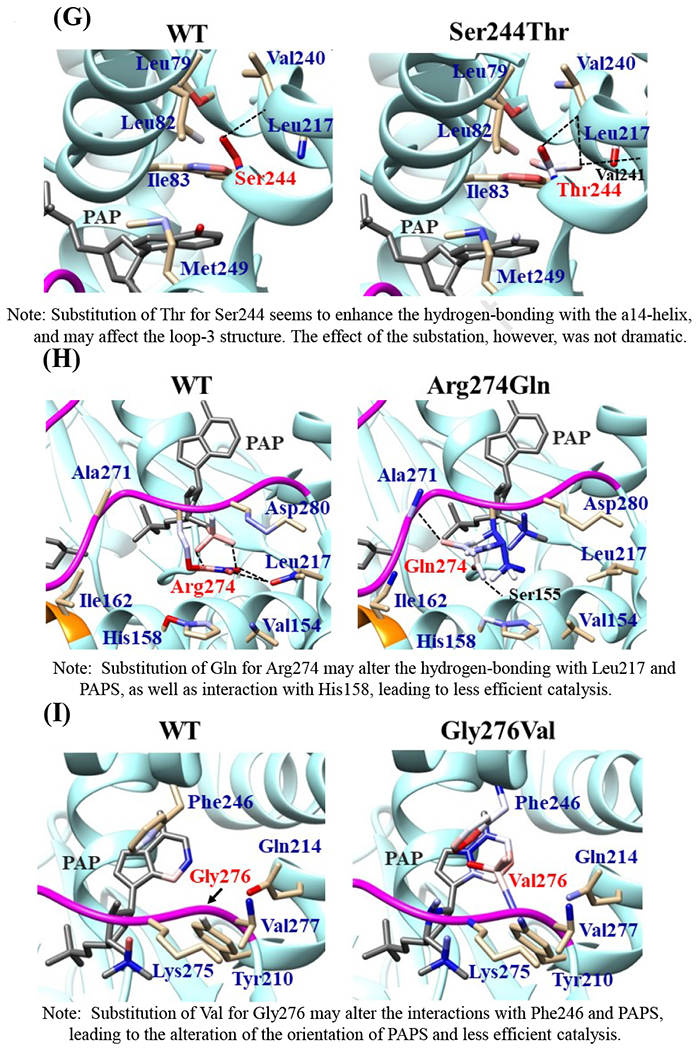
Atoms interacting with Pro69 (A), Gly72 (B), Thr73 (C), Arg147 (D), Asp191 (E), Arg230 (F), Ser244 (G), Arg274 (H), and Gly276 (I) are colored by the blue-white-red gradient (left panels). Estimated interaction formed with Ala69 in Pro69Ala (A), Val72 in Gly72Val (B), Met73 in Thr73Met (C), His147 in Arg147His (D), Asn191 in Asp191Asn (E), His230 in Arg230His (F), Thr244 in Ser244Thr (G), and Gln274 in Arg274Gln (H), Val276 in Gly276Val (I) are colored by the blue-white-red gradient (right panels). Top five-ranked rotamers of each substituted residue are modeled using the Dunbrack backbone-dependent rotamer library [Dunbrack 2002] and interaction was analyzed by Find Clashes/Contacts tool in a molecular modeling software, USCF Chimera software (Pettersen et al. 2004). Hydrogen bonds formed with Thr73 (C), Arg147 (D), Ser244 (E), and Arg274 (H) are shown by dashed lines (left panels). Estimated hydrogen bonds formed with Thr244 in Ser244Thr (G), Gln274 in Arg274Gln (H) are shown by dashed lines (right panels).
Fig. 7. Simulation analyses of the docking of PAPS and pregnenolone into the active site of SULT2B1b allozymes.
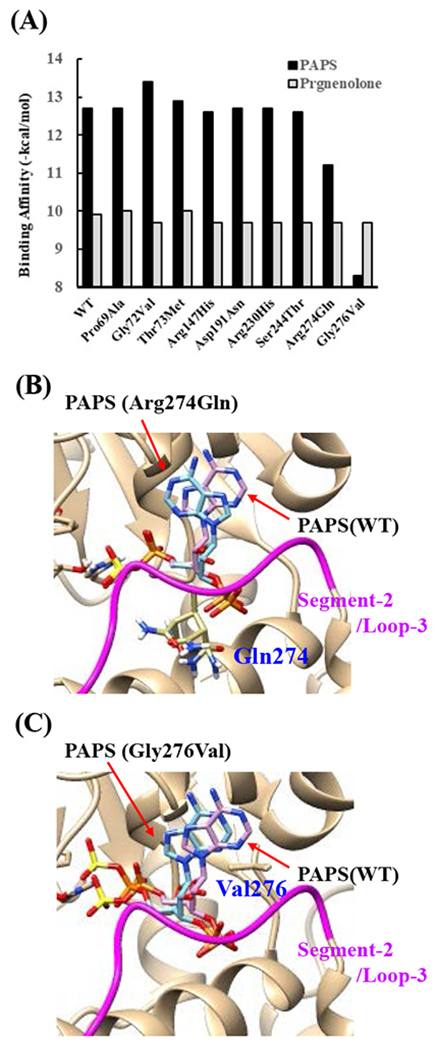
(A) Minimum binding energy of PAPS (black scale) and pregnenolone (grey scale) in the active site of SULT2B1 allozymes. Conformation of mutated amino acid residues of allozymes simulated using the Dunbrack backbone-dependent rotamer library was used for docking analyses as the template protein structures. (B) Stereo view of the PAPS docked into the active site of Arg274Gln. Superpositions of PAPS docked in Arg274Gln and wild-type are shown in blue and purple backbone, respectively. (C) Stereo view of the PAPS docked into the active site of Gly276Val. Superpositions of PAPS docked in Gly276Val and wild-type are shown in blue and purple backbone, respectively.
Fig. 8. Snapshots of molecular dynamics simulations of SULT2B1b cSNPs.
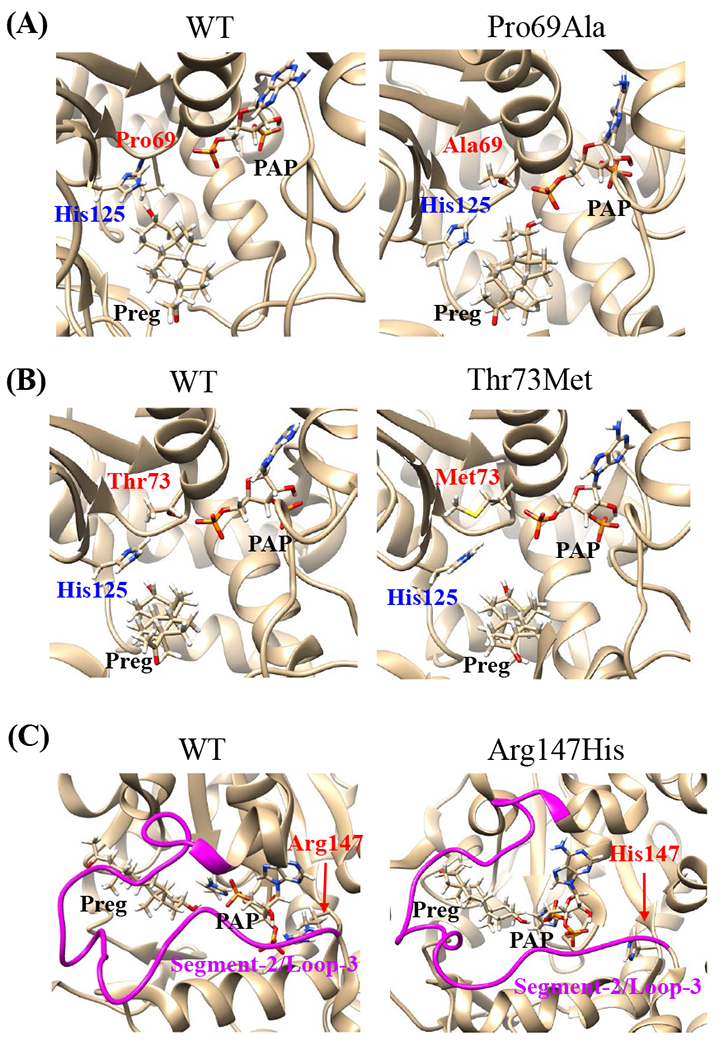
Snapshots of most clustered conformation of allozymes, Pro69Ala (A), Thr73Met (B), Arg147His (C), are shown in comparison with that of the wild-type. (A,B) Active site of SULT2B1b structures are shown with His125 (catalytic residue), Preg (pregnenolone), and PAP, as well as the original and substituted residues. (C) Conformations of segment-2/loop-3 structures (magenta color) are shown with Preg (pregnenolone) and PAP, as well as the original and substituted residues.
4. Discussion
In humans, DHEA and its sulfate ester, DHEA-S, are known to be the most abundant steroids in circulation (Falany and Rohn-Glowacki, 2013). DHEA is synthesized and secreted mainly from the adrenal glands and, to a lower extent, in brain, gonads, and skin (Davis et al., 2011). DHEA has been proposed to be effective in reducing cardiovascular risk, alleviating insulin resistance, stimulating endothelial proliferation, and improving memory and cognitive function (Labrie, 2010; Traish et al., 2011). More importantly, DHEA is a major precursor for the biosynthesis of sex steroid hormones (Schiffer et al., 2018). In peripheral tissues, the adrenally secreted DHEA is taken up and converted to estrogens and androgens via a process known as intracrinology (Schiffer et al., 2018). SULT2B1b has been shown to be capable of sulfating specifically 3β-hydroxysteroids including cholesterol, DHEA, and pregnenolone, and is highly expressed in steroid hormone-responsive tissues such as prostate, breast, placenta, and endometrium (Falany et al., 2006). Genetic polymorphisms of SULT2B1 have been correlated with the progression and proliferation of several different types of cancer including prostate cancer, esophageal squamous cell carcinoma, hepatocellular carcinoma, gastric cancer, and colorectal cancer (Hyland et al., 2013; Levesque et al., 2014; Mostaghel, 2013; Yang et al., 2013; Hu et al., 2015; Vickman et al., 2016; Chen et al., 2016). SULT2B1b mRNA has been detected in a number of cancerous human tissues and cell lines including prostate adenocarcinoma cells, LNCaP prostate adenocarcinoma cells, as well as T47D and MCF-7 breast cancer cell lines (He and Falany, 2007). While the exact role of SULT2B1b in different type of cancers has not been fully elucidated, studies have suggested that its involvement in the sulfation of hydroxysteroids may limit their availability for the sex steroids biosynthesis and their capacity to bind to corresponding androgen receptors (He and Falany, 2007; Seo et al., 2013). Interestingly, the down-regulation of SULT2B1b in prostate cancer has been proposed to be the reason behind prostate cancer progression due to the lack of the protective effect of SULT2B1b in decreasing steroid hormone precursors like DHEA (He and Falany, 2007; Seo et al., 2013).
The current study was designed to investigate the functional relevance of SULT2B1 cSNPs on the sulfating activity of the resulting SULT2B1b allozymes toward two major steroids precursors, DHEA and pregnenolone. Ten SULT2B1b allozymes, previously prepared via site-directed mutagenesis and bacterial expression using the pGEX gene fusion system (Alherz et al., 2018), were analyzed. (It is noted that while several of the SULT2B1 cSNPs studied exhibit very low allele frequencies (cf. Table 1), one of them that causes an Arg274Gln substitution (SNP ID: rs762765702) has been linked to autosomal-recessive congenital ichthyosis, a genetic skin disorder (Heinz et al., 2017). The functional relevance of other SULT2B1 cSNPs studied still awaits clarification from epidemiological studies.) As shown in the Results section, three of the ten allozymes (SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val) showed no detectable activity with both DHEA and pregnenolone. The other seven allozymes (SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, SULT2B1b-Ser244Thr, SULT2B1b-Arg274Gln, and SULT2B1b-Pro345Leu) exhibited significant decrease in their sulfating activity toward DHEA and pregnenolone. It should be pointed out that the sulfating activity of SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, and SULT2B1b-Pro345Leu in this study were noticeably lower than the sulfating activity of the same allozymes previously reported using DHEA as a substrate (Ji et al., 2007). It is possible that this discrepancy could have been due to the use of different enzyme preparation (purified recombinant enzymes vs. enzymes expressed in COS-1 cells (Ji et al., 2007)). Subsequent kinetic analysis showed that amino acid variations in the seven allozymes caused significant decrease in the catalytic activity and efficiency toward DHEA, whereas with pregnenolone, only four allozymes (SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, SULT2B1b-Ser244Thr, SULT2B1b-Arg274Gln, and SULT2B1b-Pro345Leu) showed significant decrease in the Kcat/Km. These results indicate clearly that SULT2B1b cSNPs indeed have great effect on the enzyme function. It is interesting to note that the amino acid substitution in SULT2B1b-Pro345Leu occurs in a proline- and serine-rich carboxyl terminal region of the SULT2B1b molecule, which has been shown to be subjected to phosphorylation on serine residue(s) (He and Falany, 2006). Whether the phosphorylation status of the enzyme may affect the kinetic properties of SULT2B1b-Pro345Leu, as well as other SULT2B1b allozymes, will be an intriguing issue for future investigation.
The crystal structure of human SULT2B1b determined previously has revealed a number of structural elements that are critical to the functioning of the enzyme (Lee et al., 2003). In relation to the interaction with the co-substrate, PAPS, the elements include a 3’-phosphate-binding region, a 5’-phosphosulphate-binding (PSB) loop, and a PAP adenine-binding region (Lee et al., 2003). The PSB loop is composed of the conserved amino acid sequence 67TYPKSGT73, of which the amino acid residues Lys70, Ser71, Gly72, and Thr73, as well as Thr74, are involved in binding the 5’-phosphate of PAPS. The amino acid residues Arg274, Lys275, Gly276, Arg147, and Ser155 are involved in binding the 3’-phosphate of PAPS. Moreover, Ser244, Tyr210, Trp75, and Phe246 are involved in the interaction with the adenine group of the PAPS molecule (Lee et al., 2003). It is noted that seven (SULT2B1b-Pro69Ala, SULT2B1b-Gly72Val, SULT2B1b-Thr73Met, SULT2B1b-Arg147His, SULT2B1b-Ser244Thr, SULT2B1b-Arg274Gln, and SULT2B1b-Gly276Val) of the ten SULT2B1b allozymes examined in this study contain amino acid variations in the PAP/PAPS-binding pocket (Fig. 5) (Lee et al., 2003). Of these seven SULT2B1b allozymes, the amino acid changes in three (SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val) were found to exert the most drastic effects, leading to the complete loss of sulfating activity toward DHEA or pregnenolone. For SULT2B 1b-Gly72Val and SULT2B1b-Gly276Val, Gly72 and Gly276 in the wild-type enzyme have been proposed to form hydrogen-bonding with the O4P and O2P phosphate oxygens of the co-substrate, PAPS, respectively (Fig. 6). Replacement of these glycine residues, which have more conformational flexibility, with valine residues, which carry a bulkier side chain, may place more conformational restriction and interfere with the interaction, and thus the binding, with PAPS. Furthermore, the replacement of glycine with valine may affect the interaction of Gly276 and Phe246, leading to the alteration of the PAPS orientation and less effective catalysis. In the case of SULT2B1b-Arg147His, the substitution of the arginine residue with histidine may also affect the hydrogen-bonding to the oxygen atom of the 3’phosphate of the PAPS and thus the loss of the sulfating activity (Lee et al., 2003; Betts and Russell, 2003). These three substitutions, therefore, may directly affect the interaction with PAPS rather than with substrate. In contrast to the three SULT2B1b allozymes mentioned above, the amino acid changes in SULT2B1 b-Pro69Ala, SULT2B1 b-Thr73Met, SULT2B 1b-Ser244Thr, and SULT2B1b-Arg274Gln caused only differential decreases in the sulfating activity. Among them, SULT2B1b-Ser244Thr resulted with the smallest reductions (24% and 40%, respectively) in the catalytic efficiency toward DHEA and pregnenolone, compared with the wild-type enzyme. As shown in Fig. 6 (E), the substitution of serine with threonine may enable hydrogen bonding with the a14-helix, possibly affecting the loop-3 structure. These relatively small decreases in sulfating activity could have been due to the replacement of the serine residue with a highly similar amino acid residue, threonine, which also carries a hydroxyl group in the side chain (Betts and Russell, 2003). On the other hand, the amino acid changes in SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, and SULT2B1b-Arg274Gln caused more dramatic differences in the sulfating activity. For SULT2B1b-Pro69Ala, the substitution of proline, which has no direct interaction with PAPS in the wild-type enzyme, with alanine may affect the interaction between Pro69 and His125 and Phe186 that interact with the substrate, leading to less effective catalysis and substrate recognition. Conformational change of the substrate binding pocket in SULT2B1b-Pro69Ala may be the reason behind the dramatic decrease (with a more than 79% and 90%, respectively) in the catalytic efficiency toward both pregnenolone and DHEA and compared to the wild-type. In the case of SULT2B1b-Thr73Met, the decrease in the sulfating activity was due to the replacement of the threonine residue that carries a polar side chain with methionine that carries a hydrophobic side chain, which might have disrupted the hydrogen bonding with the O4P oxygen atom of the 5’-phosphate of the PAPS (Lee et al., 2003; Betts and Russell, 2003). Moreover, the substitution of the threonine residue with methionine may affect the hydrogen bond formation between Thr73 and Thr67, Trp103, and His125, leading to less effective catalysis and substrate recognition. For SULT2B1b-Arg274Gln, the substitution of arginine with glutamine may alter the interaction between O3P and O1P oxygen atoms of the PAPS molecule with the positively charged nitrogen atom of arginine, as well as the interaction of the Arginine274 and both Leu217 and His158, causing a dramatic decrease in the sulfating activity toward DHEA or pregnenolone. Interestingly, SULT2B1b-Arg274Gln has recently been linked to autosomal-recessive congenital ichthyosis, a genetic skin disorder (Heinz et al., 2017). On the other hand, amino acid variations in SULT2B1b-Asp191Asn and SULT2B1b-Arg230His allozymes seem not to affect the interaction of the amino acid residues at these locations and the surrounding residues, which could be the reason for the smaller effect of the amino acid variation in theses allozymes compared to the wild-type SULT2B1b. Therefore, it appears that most of the allozymes that showed the decrease in sulfating activity may have altered interactions with PAPS due to substitutions of the corresponding residues. Docking simulation analyses of the docking of PAPS into the active site of SULT2B1b allozymes suggested the low binding affinity of Arg274Gln and Gly276Val with PAPS (Fig. 7). Substitution of those residues may affect the superposition of PAPS in the structure as predicted by docking simulation analysis (Fig 7B).The superpositions of PAPS docked into Arg274Gln and Gly276Val allozymes were also shifted in comparison with the superposition of PAPS docked into the wild-type enzyme. Other allozymes showed similar binding affinity with PAPS and superposition of PAPS docked into the active site of respective allozymes (Fig. 7A). Among allozymes tested in this study, the substitution of Pro63 and Thr73 with Ala and Met, respectively, may affect the interaction with substrate. It should be noted, however, that rotamer analysis of the substituted residues was performed by fixing the backbone structure. Potential effects of the substitutions on the overall structure and the interaction with substrate can not be ruled out. Molecular dynamic analysis was performed to further investigate the effect of the substitutions in the overall structure of SULT2B1b (Fig. 8). For Pro69Ala allozyme, superposition of pregnenolone differed from that of the wild-type enzyme (Fig. 8A). For Thr73Met allozyme, the location of His125 was slightly shifted toward substrate side (Fig. 8B). These two observations may be (one of) the reasons for the dramatic decrease in their catalytic activity. Interestingly, segment-2 conformation of Arg147His allozyme was highly differed from that of the wild-type, which may affect the stable binding of PAPS (Fig. 8C). It has been reported that PAPS binding shifts the conformation of the SULT molecule from a “closed state” to an “open state”, leading to alteration in the substrate-binding profiles (Tibbs et al., 2015). In the case of SULT2A1, PAPS binding has been reported to restrict the docking of raloxifene to the active site, while having no effect on the docking of DHEA (Cook et al., 2012). Although the structure of SULT2B1b without PAP has not been resolved, PAPS binding, like with other SULTs, may change the conformation of SULT2B1b, leading to alteration in the binding of DHEA or pregnenolone. The structure of SULT2B1b in an “open state”, if available, may provide the detailed effects of the substitution on the catalytic activity of SULT2B1b with DHEA, pregnenolone, or other substrates. It should be noted that SULT2B1b exhibits the extended N-terminus and C-terminus, whose structures remain to be resolved (Fuda et al., 2002, Lee et al., 2002). Previous studies also showed that N-terminal sequence in SULTs may play an important role in the substrate entry (Fuda et al., 2002, Lee et al., 2002). Further structure analysis may eventually resolve the conformation of N- and C-terminal sequences in SULT2B1b.
Fig. 5. Ribbon diagram of the structure of human SULT2B1b-pregnenolone-PAP complex showing the locations of amino acid residues involved in the SULT2B1 cSNPs.
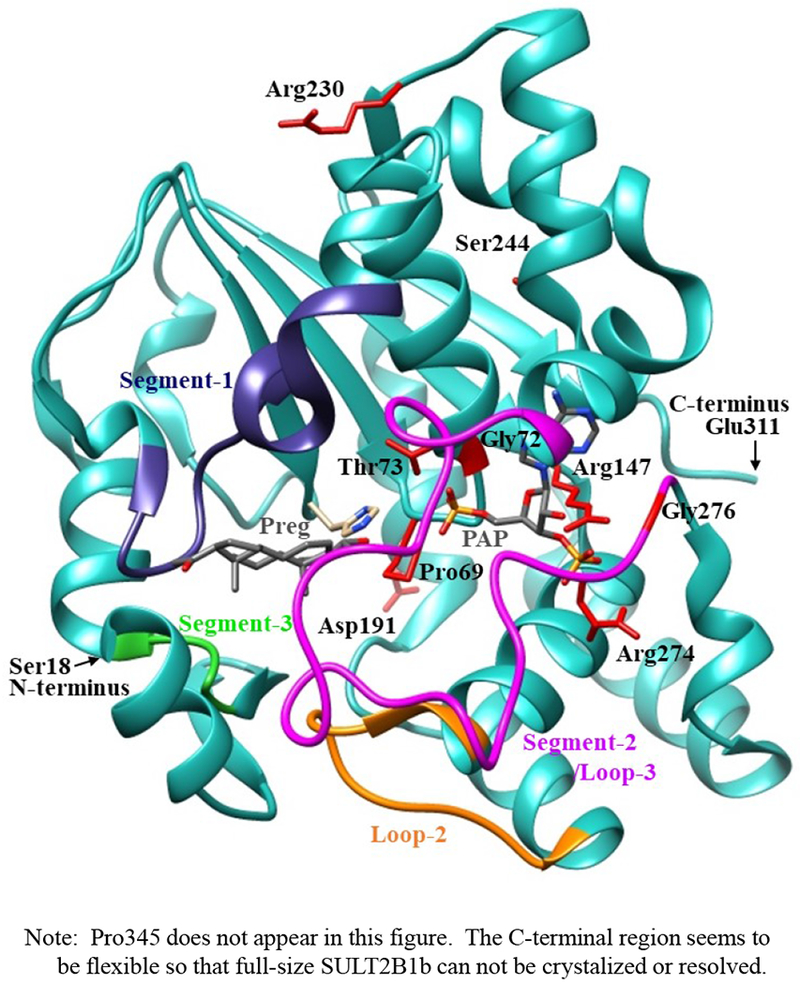
The structure of SULT2B1b complexed with pregnenolone and PAP (Protein Data Bank code: 1Q20), containing the polypeptides from Ser18 (N-terminus) to Glu311 (C-terminus), was shown by ribbon diagram. Pregnenolone (Preg) and PAP molecules in the structure are shown by bond structures. Segement-1, Segment-2 (Loops 3), Segment-3, and loop-2 refer to Arg93-Gly109 (dark blue), Asn252-Ala271 (magenta color), Val41-Ser45 (Green), and Ile-Thr171 (orange color) previously reported to form a gate for substrate entry [Cook et al. 2012, Tibbs et al. 2015]. Side chains of the amino acid residues involved in the SULT2B1b cSNPs, Pro69, Gly72, Thr73, Arg147, Asp191, Arg230, Ser244, Arg274, and Gly276 are indicated by bond structures (red color).
To conclude, the current study demonstrated clearly the differential sulfating activity of human SULT2B1b allozymes toward DHEA and pregnenolone. Kinetic analysis revealed further the differences in substrate affinity and catalytic activity between different SULT2B1b allozymes. These results showed unambiguously the effect of human SULT2B1 genetic polymorphisms on the enzymatic characteristics of the resulting SULT2B1b allozymes. Further studies are warranted in order to clarify the impact of SULT2B1 genetic polymorphisms on steroid metabolism, as well as the risk for SULT2B1b-associated disorders such as colorectal cancer and autosomal-recessive congenital ichthyosis.
Highlights.
Ten SULT2B1b allozymes, corresponding to SULT2B1b single nucleotide polymorphisms selected based on the locations and potential importance of resulting amino acid substitutions, were bacterially expressed and affinity-purified.
Pregnenolone- and dehydroepiandrosterone- sulfating activities of SULT2B1b allozymes, in comparison with the wild-type enzyme, were analyzed.
Kinetic constants of SULT2B1b allozymes, as well as the wild-type enzyme, in mediating the sulfation of Pregnenolone or dehydroepiandrosterone were determined.
Acknowledgements:
This work was supported in part by a grant from National Institutes of Health (Grant # R03HD071146).
Abbreviations:
- DHEA
dehydroepiandrosterone
- PAPS
3’-phosphoadenosine 5’-phosphosulfate
- SULT
cytosolic sulfotransferase
- TLC
thin-layer chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests: The authors declare no conflict of interests.
References
- Alherz FA, Abunnaja MS, El Daibani AA, Bairam AF, Rasool MI, et al. , 2018. On the role of genetic polymorphisms in the sulfation of cholesterol by human cytosolic sulphotransferase SULT2B1b. J. Biochem 164, 215–221. http://doi:10.1093/jb/mvy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MJ, Russell RB, 2003. Amino acid properties and consequences of substitutions, in: Barnes WR and Gray IC (Eds.), Bioinformatics for Geneticists. John Wiley & Sons, Ltd., Chichester, England, pp. 289–316. [Google Scholar]
- Chen W, Zhou, Ye, Zhan B, 2016. Overexpression of SULT2B1b Promotes Angiogenesis in Human Gastric Cancer. Cell. Physiol. Biochem 38, 1040–1054. http:// [DOI] [PubMed] [Google Scholar]
- Cook I, Wang T, Falany CN, Leyh TS, 2012. A nucleotide-gated molecular pore selects sulfotransferase substrates. Biochemistry. 51, 5674–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Almo SC, Kim J, Falany CN, et al. , 2012. The gate that governs sulfotransferase selectivity. Biochemistry. 52, 415–424. http://doi:10.1159/000443055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SR, Panjari M, Stanczyk FZ, 2011. Clinical review: DHEA replacement for postmenopausal women. J. Clin. Endocrinol. Metab 96, 1642–1653. http://doi:10.1210/jc.2010-2888 [DOI] [PubMed] [Google Scholar]
- Dunbrack RL Jr., Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol 12, 431–440. [DOI] [PubMed] [Google Scholar]
- Falany CN, 1997. Enzymology of human cytosolic sulfotransferases. FASEB J. 11, 206–216. http://doi:10.1016/s0959-440x(02)00344-5 [DOI] [PubMed] [Google Scholar]
- Falany CN, He D, Dumas N, Frost AR, Falany JL, 2006. Human cytosolic sulfotransferase 2B1: isoform expression, tissue specificity and subcellular localization. J. Steroid Biochem. Mol. Biol 102, 214–221. http://doi:10.1016/j.jsbmb.2006.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany CN, Rohn-Glowacki KJ, 2013. SULT2B1: unique properties and characteristics of a hydroxysteroid sulfotransferase family. Drug Metab. Rev 45, 388–400. http://doi:10.3109/03602532.2013.835609 [DOI] [PubMed] [Google Scholar]
- Falany CN, Roth J, 1993. Properties of human cytosolic sulfotransferases involved in drug metabolism, in: Jeffery EH (Ed.), Human Drug Metabolism: From Molecular Biology to Man. CRC Press Inc., Florida, pp. 101–115. [Google Scholar]
- Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM, 2004. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J. 4, 54–65. http://doi:10.1038/sj.tpj.6500223 [DOI] [PubMed] [Google Scholar]
- Fuda H, Lee YC, Shimizu C, Javitt NB & Strott CA (2002) Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. J Biol Chem 277, 36161–36166. [DOI] [PubMed] [Google Scholar]
- Geese WJ, Raftogianis RB, 2001. Biochemical characterization and tissue distribution of human SULT2B1. Biochem. Biophys. Res. Commun 288, 280–289. http://doi:10.1006/bbrc.2001.5746 [DOI] [PubMed] [Google Scholar]
- Glatt H, Boeing H, Engelke CE, Ma L, Kuhlow A, et al. , 2001. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat. Res 482, 27–40. http://doi:10.1016/s0027-5107(01)00207-x [DOI] [PubMed] [Google Scholar]
- He D, Falany CN, 2006. Characterization of proline-serine-rich carboxyl terminus in human sulfotransferase 2B1b: immunogenicity, subcellular localization, kinetic properties, and phosphorylation. Drug Metab. Dispos 34, 1749–1755. http://doi:10.1124/dmd.106.011114 [DOI] [PubMed] [Google Scholar]
- He D, Falany CN, 2007. Inhibition of SULT2B1b expression alters effects of 3beta-hydroxysteroids on cell proliferation and steroid hormone receptor expression in human LNCaP prostate cancer cells. Prostate. 6, 1318–1329. http://doi:10.1002/pros.20615 [DOI] [PubMed] [Google Scholar]
- Heinz L, Kim GJ, Marrakchi S, Christiansen J, Turki H, et al. , 2017. Mutations in SULT2B1 cause autosomal-recessive congenital ichthyosis in humans. Am. J. Hum. Genet 100, 926–939. http://doi:10.1016/j.ajhg.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, et al. , 1998. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 53, 284–295. http://doi:10.1006/geno.1998.5518 [DOI] [PubMed] [Google Scholar]
- Hu L, Yang GZ, Zhang Y, Feng D, Zhai YX, et al. , 2015. Overexpression of SULT2B1b is an independent prognostic indicator and promotes cell growth and invasion in colorectal carcinoma. Lab Invest. 95, 1005–1018. http://doi:10.1038/labinvest.2015.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui Y, Liu MC, 2015. Sulfation of ritodrine by the human cytosolic sulfotransferases (SULTs): effects of SULT1A3 genetic polymorphism. Eur. J. Pharmacol 761, 125–129. http://doi:10.1016/j.ejphar.2015.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland PL, Freedman ND, Hu N, Tang ZZ, Wang L, et al. , 2013. Genetic variants in sex hormone metabolic pathway genes and risk of esophageal squamous cell carcinoma. Carcinogenesis. 34, 1062–1068. http://doi:10.1093/carcin/bgt030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Moon I, Zlatkovic J, Salavaggione O, Thomae B, et al. , 2007. Human hydroxysteroid sulfotransferase SULT2B1 pharmacogenomics: gene sequence variation and functional genomics. J. Pharmacol. Exp. Ther 322, 529–540. http://doi:10.1124/jpet.107.122895 [DOI] [PubMed] [Google Scholar]
- Labrie F, 2010. DHEA, important source of sex steroids in men and even more in women. Prog. Brain Res 182, 97–148. http://doi:10.1016/S0079-6123(10)82004-7 [DOI] [PubMed] [Google Scholar]
- Lee KA, Fuda H, Lee YC, Negishi M, Strott CA, et al. , Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone and 3’-phosphoadenosine 5’-phosphate. Rationale for specificity differences between prototypical SULT2A1 and the SULT2B1 isoforms. J. Biol. Chem 278, 44593–44599. http://doi:10.1074/jbc.M308312200 [DOI] [PubMed] [Google Scholar]
- Levesque E, Laverdiere I, Audet-Walsh E, Caron P, Rouleau M, et al. , 2014Steroidogenic germline polymorphism predictors of prostate cancer progression in the estradiol pathway. Clin. Cancer Res 20, 2971–2983. http://doi:10.1158/1078-0432.CCR-13-2567 [DOI] [PubMed] [Google Scholar]
- Mostaghel EA, 2013. Steroid hormone synthetic pathways in prostate cancer. Transl. Androl. Urol 2, 212–227. http://doi:10.3978/j.issn.2223-4683.2013.09.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI), National Library of Medicine. Database of Single Nucleotide Polymorphisms (dbSNP). http://www.ncbi.nlm.nih.gov/SNP/ (accessed 4 April 2019)
- Neunzig J, Bernhardt R, 2014. Dehydroepiandrosterone sulfate (DHEAS) stimulates the first step in the biosynthesis of steroid hormones. PloS one. 9, e89727 http://doi:10.1371/journal.pone.0089727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordam C, Dhir V, McNelis JC, Schlereth F, Hanley NA et al. , 2009. Inactivating PAPSS2 mutations in a patient with premature pubarche. N. Engl. J Med 360, 2310–2318. http://doi:10.1056/NEJMoa0810489. [DOI] [PubMed] [Google Scholar]
- Otterness DM, Weinshilboum RM, 1994. Human dehydroepiandrosterone sulfotransferase: molecular cloning of cDNA and genomic DNA. Chem. Biol. Interact 92, 145–159. http://doi:10.1016/0009-2797(94)90060-4 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. , 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. http://doi:10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Schiffer L, Arlt W, Storbeck KH, 2018. Intracrine androgen biosynthesis, metabolism and action revisited. Mol. Cell. Endocrinol 465, 4–26. http://doi:10.1016/j.mce.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YK, Mirkheshti N, Song CS Kim S, Dodds S, et al. , 2013. SULT2B1b sulfotransferase: induction by vitamin D receptor and reduced expression in prostate cancer. Mol. Endocrinol 27, 925–939. http://doi:10.1210/me.2012-1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomae BA, Eckloff BW, Freimuth RR, Wieben ED, Weinshilboum RM, 2002. Human sulfotransferase SULT2A1 pharmacogenetics: genotype-to-phenotype studies. Pharmacogenomics J. 2, 48–56. http://doi:10.1038/sj.tpj.6500089 [DOI] [PubMed] [Google Scholar]
- Tibbs ZE, Rohn-Glowacki KJ, Crittenden F, Guidry AL, Falany CN, 2015. Structural plasticity in the human cytosolic sulfotransferase dimer and its role in substrate selectivity and catalysis. Drug Metab. Pharmacokinet 30, 3–20. http://doi:10.1016/j.dmpk.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Traish AM, Kang HP, Saad F, Guay AT, 2011. Dehydroepiandrosterone (DHEA)-A precursor steroid or an active hormone in human physiology. J. Sex. Med 8, 2960–2982. http://doi:10.1111/j.1743-6109.2011.02523.x [DOI] [PubMed] [Google Scholar]
- Trott O Olson AJ., 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee M, Mayo W, Le Moal M, 2001. Role of pregnenolone, dehydroepiandrosterone and their sulfate esters on learning and memory in cognitive aging. Brain Res. Rev 37 (2001) 301–312. http://doi:10.1016/s0165-0173(01)00135-7 [DOI] [PubMed] [Google Scholar]
- Vickman RE, Crist SA, Kerian K, Eberlin L, Cooks RG, et al. , 2016. Cholesterol sulfonation enzyme, SULT2B1b, modulates AR and cell growth properties in prostate cancer. Mol. Cancer Res 14, 776–786. http://doi:10.1158/1541-7786.mcr-16-0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness D, 1994. Sulfotransferase enzymes, in: Kauffman FC (Ed.), Handbook of Experimental Pharmacology, Vol. 112 Springer-Verlag, Berlin, Heidelberg, pp. 45–78. [Google Scholar]
- Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, et al. , 1998. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5’-phosphosulfate kinase enzyme, Biosci. Biotechnol. Biochem 62, 1037–1040. http://doi:10.1271/bbb.62.1037 [DOI] [PubMed] [Google Scholar]
- Yang X, Xu Y, Guo F, Ning Y, Zhi X, Yin L, et al. , 2013. Hydroxysteroid sulfotransferase SULT2B1b promotes hepatocellular carcinoma cells proliferation in vitro and in vivo. PloS one 8, e60853 http://doi:10.1371/journal.pone.0060853. [DOI] [PMC free article] [PubMed] [Google Scholar]


