Abstract
Ribosome assembly is a complex process reliant on the coordination of trans-acting enzymes to produce functional ribosomal subunits and secure the translational capacity of cells. The endoribonuclease (RNase) Las1 and the poly-nucleotide kinase (PNK) Grc3 assemble into a multienzyme complex, herein designated RNase PNK, to orchestrate processing of precursor ribosomal RNA. RNase PNK belongs to the functionally-diverse HEPN nuclease superfamily, whose members rely on distinct cues for nuclease activation. To establish how RNase PNK coordinates its dual enzymatic activities, we solved a series of cryo-electron microscopy structures of Chaetomium thermophilum RNase PNK in multiple conformational states. The structures reveal that RNase PNK adopts a butterfly-like architecture harboring a composite HEPN nuclease active site flanked by discrete RNA kinase sites. We identify two molecular switches that coordinate nuclease and kinase function. Together our structures and corresponding functional studies establish a new mechanism of HEPN nuclease activation essential for ribosome production.
Introduction
Cells utilize multienzyme macromolecular complexes to drive fundamental biochemical reactions such as RNA processing, protein degradation, DNA remodeling, and the production of metabolites. Understanding how individual components of multienzyme complexes associate and communicate with one another is critical to establishing how these systems function as a whole1,2. One fundamental pathway that is dependent on multienzyme complexes is the biogenesis of ribosomes. Ribosome assembly is an intricate and energetically demanding pathway reliant on the exquisite coordination of numerous trans-acting enzymes that process, modify, and remodel pre-ribosome particles3-7. Precursor ribosomal RNA (pre-rRNA) is transcribed by RNA Pol I as a long poly-cistronic transcript known as the 35S in Saccharomyces cerevisiae in which the 18S, 5.8S, and 25S rRNAs are separated by two external (5′ ETS, 3′ ETS) and two internal (ITS1, ITS2) spacers. A series of endo- and exo-nucleolytic processing steps are required to remove the spacers and generate the mature rRNAs8,9. These transcribed spacers are critical for the spatial and temporal control of ribosome biogenesis and their processing is highly regulated to ensure that ribosomes are produced accurately and efficiently8.
Targeted removal of the ITS2, located between the 5.8S and 25S rRNAs, is one of the most complex pre-rRNA processing events. Processing of the ITS2 is initiated by the newly discovered endoribonuclease Las110,11. Las1 specifically cleaves at the C2 site within the ITS2, generating the precursors for the 5.8S and 25S rRNAs with 2′−3′ cyclic phosphate and 5′-hydroxyl ends, respectively (Figure 1a). Following cleavage, the RNA kinase Grc3 catalyzes the phosphorylation of the 5′-hydroxyl product11. The unique two-step process of C2 cleavage by Las1 and phosphorylation by Grc3 is responsible for triggering targeted decay of the ITS2 spacer. Phosphorylation by Grc3 is the signal for the 5′−3′ exonuclease Rat1 and its co-factor Rai1 to further process the 25S precursor11. The 3′ end of the 5.8S precursor is then subsequently degraded by the multienzyme nuclear exosome complex which has 3′−5′ exonuclease activity10,12.
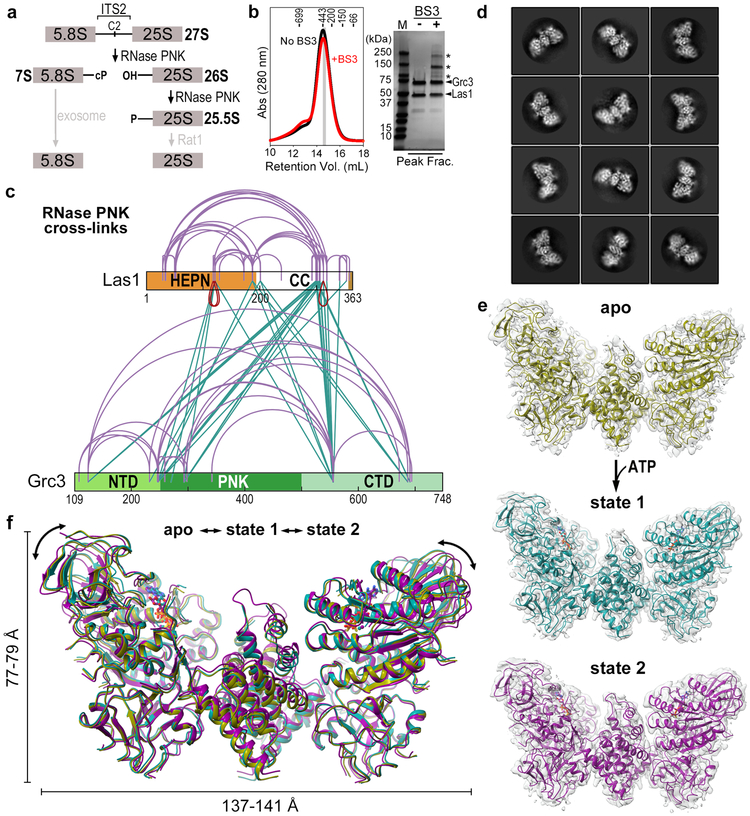
Fig. 1 ∣. RNase PNK Samples Multiple Conformational States.
a, Steps in the ITS2 pre-rRNA processing pathway that involve the RNase PNK components, Las1 and Grc3. Las1 cleaves at the C2 site leaving 2′-3′-cyclic phosphate (cP) and 5′-hydroxyl (OH) RNA ends followed by 5′-phosphorylation (P) by the Grc3 RNA kinase. Together this marks ITS2 for decay by the Rat1 exonuclease and the nuclear exosome. b, Size exclusion chromatogram of cross-linked (red) and non cross-linked (black) RNase PNK. Prior to gel filtration, BS3 (bis(sulfosuccinimidyl)suberate) cross-linker (50 μM) was incubated with 10 μM RNase PNK and incubated for 5 minutes at 22°C. Molecular weight (kDa) standards are indicated at the top. The gray bar corresponds to the peak fraction (Peak Frac.) shown on the SDS-PAGE gel which was used for cryo-EM and mass spectrometry analysis. Asterisks marks cross-linked RNase PNK species. Uncropped SDS-PAGE gel shown in Supplementary Data Set 1. c, BS3 cross-links identified within the RNase PNK complex. Purple arches represent intra- and inter-molecular Grc3-Grc3 and Las1-Las1 cross-links. Green lines represent inter-molecular Las1-Grc3 cross-links, while red arches represent inter-molecular Las1-Las1 cross-links assigned to the same lysine residue encoded within each Las1 protomer. d, Representative 2D classes of RNase PNK. e, Cryo-EM reconstruction (gray) and modeled RNase PNK determined in the absence of ligand (apo, yellow) and in the presence of ATP-γS (state 1, teal and state 2, purple). f, Superposition of apo (yellow), state 1 (teal) and state 2 (purple) RNase PNK ribbon diagrams. Double headed arrows mark the global displacement.
Las1 is an ancient nuclease conserved across eukaryotes and essential for cell viability13-15. Mutations within the LAS1L gene have been associated with several families of X-linked intellectual disability and a spinal motor neuron disease16,17. Thus, Las1 is part of the emerging theme that defects in RNA metabolism manifest as neurological dysfunction, highlighting the need to fully understand Las1 function and regulation18-20. Las1 was recently identified as an endoribonuclease based on a comprehensive classification of HEPN (higher eukaryote and prokaryote nucleotide binding) nuclease domains21. The amino acid sequences of α-helical HEPN nucleases are poorly conserved with the exception of a small RϕxxxH (where ϕ is typically H, D or N and x is any residue) motif important for nuclease activity21. Metal independent HEPN nucleases must dimerize to be active and are commonly associated with stress activated RNA processing pathways, including the unfolded protein response and CRISPR immune defense systems. As such, the activity of these nucleases is highly regulated by different cues such as ligand-induced dimerization or conformational changes that properly position the juxtaposed RϕxxxH motifs for catalysis21-24. Unlike stress activated HEPN nucleases, Las1 is vital for cell cycle progression13,25. Two years ago, we discovered that Las1 is reliant on its binding partner the Grc3 RNA kinase for nuclease activation26; yet the structural and molecular basis for this distinct regulation of the Las1 HEPN domain is completely unknown.
Las1 and Grc3 are dependent upon one another for stability, assembly, and enzyme activation suggesting a high degree of cooperativity between the two essential enzymes11,13,26,27. A low-resolution structure of Las1-Grc3 and accompanying biochemistry revealed that Las1 and Grc3 assemble into a dimer of Las1-Grc3 heterodimers that is a prerequisite for C2 cleavage and phosphorylation26. Moreover, we recently established that functional defects in either enzyme impairs nuclease and kinase function through an unknown mechanism of nuclease-kinase crosstalk26,27. Due to this functional interdependence, we christen the Las1 RiboNuclease Grc3 PolyNucleotide Kinase complex, RNase PNK. Given the indispensable role of RNase PNK in cell viability, ribosome assembly, and neurological function, high resolution structures are critical to define the molecular coordination between the nuclease and kinase active sites.
Here we report a series of cryo-EM derived maps of RNase PNK that allowed us to build de novo models of RNase PNK in multiple conformational states ranging from 2.9 to 3.2 Å resolution. The structures reveal the overall butterfly-like architecture of RNase PNK and the extensive interfaces between the nuclease and kinase components. Our findings reveal how the discrete nuclease and kinase active sites relay information to one another through two key molecular switches that coordinate their activity to drive ribosome production.
Results
Cryo-EM Reveals Multiple RNase PNK Conformational States
To obtain structural insights into the molecular architecture of RNase PNK, we determined cryo-EM structures of enzymatically active RNase PNK from Chaetomium thermophilum in three different states: apo, state 1, and state 2 (Table 1). Prior to vitrification, RNase PNK was cross-linked by the amine-reactive BS3 (bis(sulfosuccinimidyl)suberate) cross-linker (Figure 1b). Analysis of the cross-links from the apo sample by mass spectrometry revealed numerous inter- and intra-molecular cross-links between the Las1 nuclease and the Grc3 kinase (Figure 1c). We also detected three inter-molecular cross-links between the same lysine residue encoded within each Las1 protomer, two within the HEPN domain and another in the C-terminal half of the protein, indicating that these regions of Las1 must be near the dimerization interface (Figure 1c).
Table 1.
Cryo-EM data collection, refinement and validation statistics
| RNase PNK (EMD-20042, PDB 6OF4) |
ATP-γS bound RNase PNK State 1 (EMD-20041, PDB 6OF3) |
ATP-γS bound RNase PNK State 2 (EMD-20040, PDB 6OF2) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 75,000 | 75,000 | 75,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | ~40 | ~40 | ~40 |
| Defocus range (μm) | −1.0 to −2.5 | −1.0 to −2.5 | −1.0 to −2.5 |
| Pixel size (Å) | 1.08 | 1.08 | 1.08 |
| Symmetry imposed | C2 | C2 | C2 |
| Initial particle images (no.) | 761,763 | 703,078 | 703,078 |
| Final particle images (no.) | 102,753 | 74,582 | 70,561 |
| Map resolution (Å) | 3.2 | 3.0 | 2.9 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.5 to 9.0 | 2.24 to 12.1 | 2.24 to 12.1 |
| Refinement | |||
| Model composition | |||
| Nonhydrogen atoms | 11,000 | 11,098 | 11,064 |
| Protein residues | 1430 | 1434 | 1434 |
| Ligands | 0 | 2 | 2 |
| B factors (Å2) | |||
| Protein | 104.36 | 101.89 | 91.96 |
| Ligand | N/A | 111.87 | 104.67 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.010 | 0.008 | 0.006 |
| Bond angles (°) | 1.177 | 1.587 | 0.993 |
| Validation | |||
| MolProbity score | 2.65 | 1.89 | 2.23 |
| Clashscore | 11.15 | 3.95 | 8.01 |
| Poor rotamers (%) | 2.54 | 0.85 | 1.19 |
| Ramachandran plot | |||
| Favored (%) | 76.85 | 81.92 | 80.17 |
| Allowed (%) | 19.61 | 16.07 | 17.66 |
| Disallowed (%) | 3.55 | 2.01 | 2.17 |
We vitrified RNase PNK in the presence and absence of a non-hydrolysable ATP analogue and an RNA substrate encompassing part of the C. thermophilum ITS2 pre-rRNA (CT-ITS2-RNA, Table S1) and recorded two cryo-EM datasets. Initial 2D classification revealed striking butterfly shaped particles (Figure 1d), reminiscent of the approximate size and shape of the negative stain derived reconstruction of RNase PNK26. 3D classification was initially performed in the absence of symmetry and revealed numerous classes from each dataset (Figures S1, S2). Because we did not observe any asymmetric features, we imposed C2 symmetry for the final round of 3D refinement for the best classes from each dataset (Figures S1, S2). The dataset recorded in the absence of nucleotide resulted in one prominent state (apo state) and was solved to an overall resolution of 3.2 Å, sufficient for de novo model building (Figures 1e and S1, Table 1). The dataset recorded in the presence of ATP-γS and RNA revealed two prominent well-resolved RNase PNK reconstructions (state 1 and state 2) at resolutions of 2.9 Å and 3.0 Å, respectively. ATP-γS was bound in both kinase active sites, but no density for RNA was visible (Figures 1e and S2, Table 1, Videos S1 and S2). Overall the 3D architectures are similar, but there are noticeable differences (Figure 1f). State 2 closely resembles the apo state (root mean square deviation of 0.527 Å), while state 1 displays more prominent conformational changes along the periphery of RNase PNK (Figure 1f).
RNase PNK has a Butterfly-like Architecture
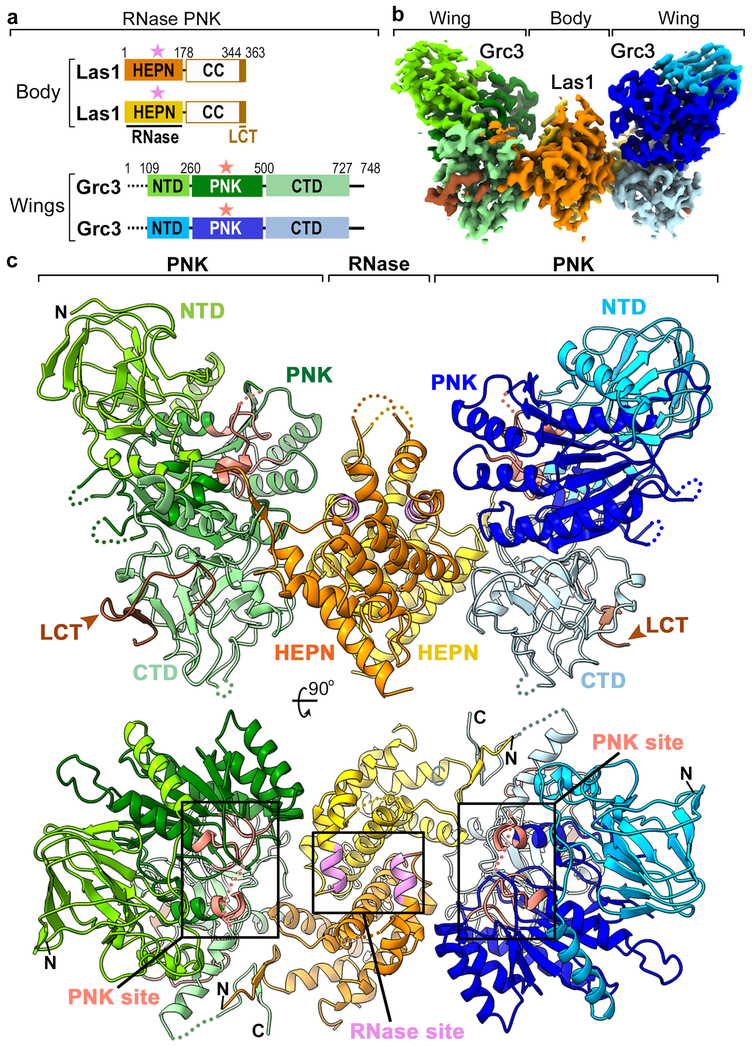
The structural architecture of RNase PNK in all three states resembles a butterfly in which the “body” is made up of a Las1 nuclease dimer flanked by two “wings” each consisting of a Grc3 kinase monomer (Figures 2a, 2b and 2c). Las1 is composed of a well conserved N-terminal α-helical HEPN nuclease domain (Figures 2a and S3a and Supplementary Note 1) followed by a poorly conserved C-terminal coiled coil (CC) domain. Aside from the last 20 residues of the Las1 C-terminal tail (LCT), the CC domain of Las1 is invisible in all three reconstructions suggesting that this domain is highly flexible. Correspondingly, we detected a number of inter-molecular cross-links between the CC domain and surface exposed residues encoded throughout the Grc3 protomer (Figure 1c). The lack of density describing the CC domain prevents LCT assignment to each Las1 protomer, as it is conceivable that the CC domain could span the distance between the LCT and either HEPN domain.
Fig. 2 ∣. Architecture of RNase PNK.
a, Domain architecture of C. thermophilum RNase PNK. Numbering above the diagram indicates the amino acid residue domain boundaries. Dashed lines indicate the proteolytic sensitive N-terminus of Grc3 that was removed prior to cryo-EM sample preparation. White boxes represent the CC domain of Las1 which was not visible in the structure. Colored stars mark the position of the catalytic centers. b, Apo cryo-EM reconstruction of RNase PNK with the individual domains colored as in 2a. c, Orthogonal views of the apo RNase PNK model shown as a cartoon and colored as in 2a. Las1 nuclease (RNase) and Grc3 kinase (PNK) active sites are highlighted in purple and red, respectively.
Grc3 is composed of three structured domains including a central PNK domain that is flanked by N- and C-terminal domains (NTD, CTD) (Figures 2a and S4 and Supplementary Note 2). In contrast to Las1, the two copies of the Grc3 kinase, found within the wings of the butterfly, do not contact each other. With the exception of a few loops and the end of the C-terminus, all three domains of Grc3 are well ordered in the structure (Figures 2c and S4). Grc3 shares a similar domain architecture with the ancestral polynucleotide kinase Clp128,29, however Grc3 is over 200 residues larger than Clp1 and the N- and C-terminal domains are structurally distinct (Figure S4). The NTD of Grc3 includes an antiparallel jellyroll fold with a small α-helical insertion, while the CTD of Grc3 has a novel fold made up of twisted antiparallel β-strands and three short α-helices (Figures 2c and S4). The PNK domain of Grc3 is comprised of a seven-stranded β-sheet surrounded by five α-helices and includes the elements crucial for phosphoryl transfer (Figures 2c and S5)27. All three domains of Grc3 come together along helix α7 from the PNK domain, through insertions of the NTD and CTD that are not found within Clp1 (Figure S4).
The interfaces within RNase PNK are extensive (Figure 2c). The CTD of Grc3 is responsible for the bulk of the interactions with Las1. Each CTD binds to both ends of Las1 through associations with the Las1 HEPN dimer and a single LCT (Figures 2c and S4). The LCT is characterized by several well conserved hydrophobic residues including W347, W355, and I360 and forms part of a β-sandwich with the Grc3 CTD (Figure S4b). These conserved hydrophobic residues all fit into shallow grooves formed by the Grc3 CTD. The LCT-Grc3 interface is reminiscent of Clp1 and Pcf11, subunits of a polyadenylation factor complex, however the Pcf11 binding surface lies within the PNK domain of Clp111,29. The isolated LCT can bind to Grc3 on its own and is critical for Grc3 stability in vitro, but this minimal Grc3-Las1 complex is monomeric and does not have kinase activity11,26. The PNK domain of Grc3 associates with a single Las1 HEPN domain while the NTD of Grc3 does not directly interact with Las1 (Figures 2c and S4). Altogether, the extensive interfaces within RNase PNK provide the structural framework for understanding the interdependence of Las1 and Grc3 for protein stability13.
RNase PNK has Distinct Nuclease and Kinase Active Sites
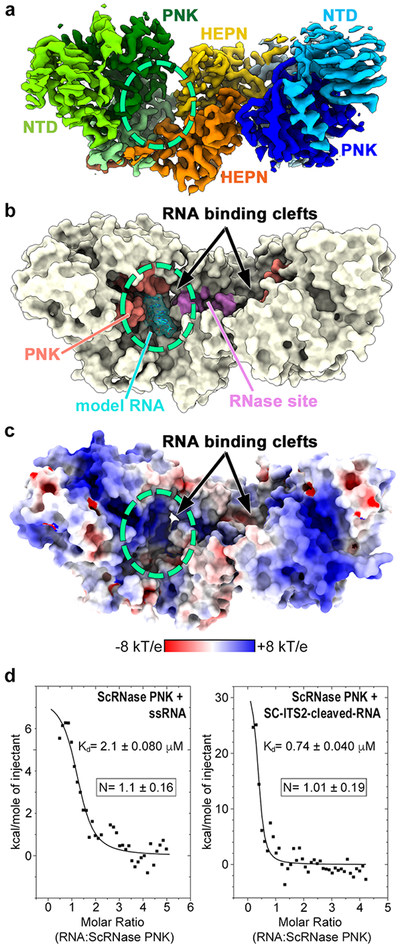
The Las1 HEPN dimer forms a composite nuclease active site within the head of the butterfly, while each wing contains a discrete Grc3 kinase active site (Figures 2c, 3a, and 3b). RNase PNK harbors symmetric RNA binding clefts at the nexus of the wings and body of the butterfly. The nuclease and kinase active sites lie along opposite faces of each cleft and are ~ 25 Å apart from each other. Each cleft is defined by a large electropositive surface wide enough to accommodate single or double-stranded RNA (Figures 3b and 3c). Superposition of states 1 and 2 revealed that conformational changes within RNase PNK alter the size of the cleft, with state 1 having a more open cleft (Video S3). Expansion and contraction of the cleft is likely important for RNA substrate binding, processing, and release. The structures suggest that RNase PNK has two distinct RNA binding clefts, however symmetric HEPN nucleases, such as Ire1 and RNase L, are known to engage a single RNA substrate asymmetrically23,30. To determine if RNase PNK can engage multiple RNA substrates, we carried out isothermal titration calorimetry (ITC) of S. cerevisiae RNase PNK with RNA substrates. We used two different RNA substrates including a small duplex that mimics the S. cerevisiae Las1 C2 cleavage product (SC-ITS2-cleaved-RNA, Table S1) and a single stranded RNA substrate (SS-RNA, Table S1) that we previously demonstrated is not cleaved by Las1, but can be phosphorylated by Grc3. ITC titrations with both RNA substrates revealed that only one RNA can be engaged by RNase PNK at a time (Figure 3d). Together this data establishes that in-line with other HEPN nucleases RNase PNK engages and processes RNA asymmetrically.
Fig. 3 ∣. RNase PNK Contains Dual RNA Binding Clefts.
a, Cryo-EM reconstruction of apo RNase PNK colored as in Figure 2a. The dashed circle marks the position of one of the symmetric RNA binding clefts. b, Surface representation of RNase PNK. The kinase active site is colored in red and the HEPN RNase site is colored in purple. Single stranded RNA (model RNA, cyan) was modeled into one of the RNA binding clefts using the coordinates from the RNA-bound crystal structure of Clp1 (PDB ID 4OHV). c, Electrostatic surface potential showing the RNA binding clefts (circled) formed at the interface of Las1 and Grc3. d, To determine whether both RNA binding clefts can be occupied by RNA simultaneously, we performed isothermal titration calorimetry (ITC) with S. cerevisiae RNase PNK and a minimal single-stranded RNA substrate (ssRNA) or a C2 cleaved RNA mimic of the S. cerevisieae ITS2 (SC-ITS2-cleaved-RNA). A representative ITC titration is presented along with the dissociation constant (Kd). The mean number of binding sites (N) and s.d. for ssRNA and SC-ITS2-cleaved-RNA were calculated from n=5 and n=3 replicates using independent protein preparations, respectively.
Las1 HEPN Domains Form a Composite Nuclease Active Site
Following asymmetric RNA binding, the HEPN nuclease domains of RNase PNK cleave the ITS2 at the C2 site. A defining characteristic of HEPN nucleases is that they are only active as dimers22-24,31-34. The Las1 HEPN core (α3-α7), lies along the two-fold axis of symmetry, forming a centralized active site at the juxtaposition of the RϕxxxH catalytic motifs found within helix α6 21 (Figures 4a, 4b and S3a). Despite sharing a similar secondary structure composition to other α-helical HEPN nucleases, the Las1 HEPN domains are structurally distinct due to different modes of dimerization leading to alternative tertiary and quaternary arrangements (Figure S6). The Las1 HEPN domains drive dimerization of RNase PNK, but cannot form a stable dimer on their own26. Las1 is dependent on Grc3 for stabilizing the HEPN-HEPN dimer interface, which in turn promotes nuclease activity26. Stabilization is partially mediated by an N-terminal extension (β1, α1-α2) of the Las1 HEPN domain important for Grc3 association (Figures 2c and S3). The constitutive dimerization of RNase PNK is in contrast to the two other eukaryotic HEPN nucleases Ire1 and RNase L in which ligand binding induces dimerization31.
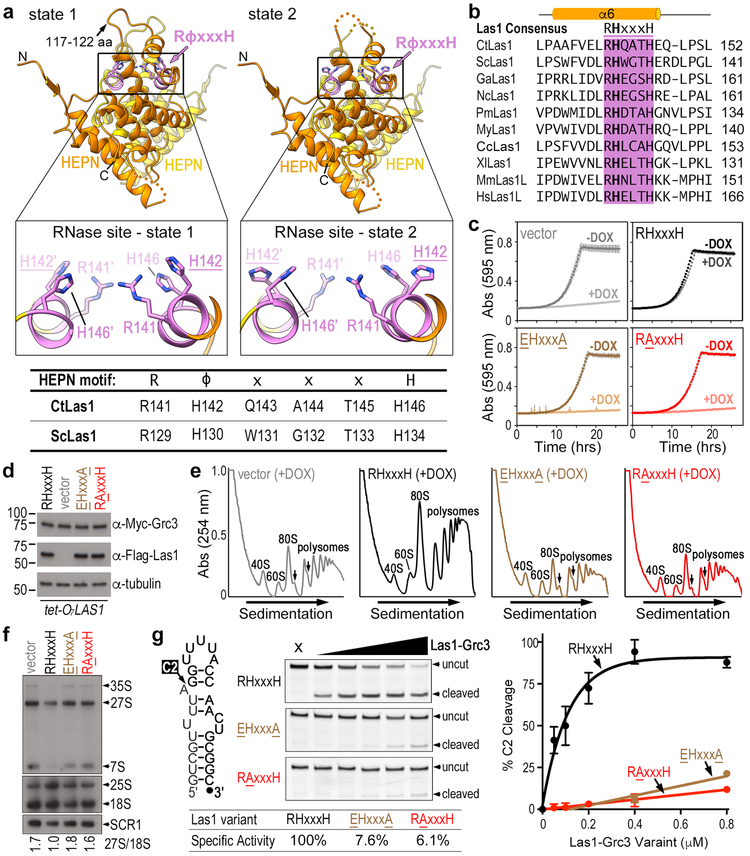
Fig. 4 ∣. Rearrangement of catalytic residue H142 within the Las1 HEPN nuclease site.
a, Ribbon diagram of the Las1 HEPN-HEPN dimer in state 1 and state 2. The conserved RϕxxxH motif responsible for Las1 RNA cleavage is purple and individual residues are shown as sticks. The boxes represent zoomed in views of the nuclease active site in state 1 and state 2. The table defines the equivalent RϕxxxH consensus residues in C. thermophilum Las1 (CtLas1) and S. cerevisiae Las1 (ScLas1). b, Amino acid sequence alignment of the HEPN RHxxxH motif from Las1 homologues. Chaetomium thermophilum (Ct), Saccharomyces cerevisiae (Sc), Gossypium arboretum (Ga), Noccaea caerulescens (Nc), Papilio machaon (Pm), Mizuhopecten yessoensis (My), Ceratitis capitata (Cc), Xenopus laevis (Xl), Mus Musculus (Mm), and Homo sapiens (Hs). c, Growth curves of S. cerevisiae tetO7-LAS1 strain transformed with plasmids harboring wild-type Las1 (RHxxxH), Las1 RAxxxH variant (ScLas1 H130A), Las1 EHxxxA variant (ScLas1 R129E, H134A) and empty YCplac vector (vector). Strains were grown in the presence (light) or absence (dark) of 40 μg/ml doxycycline at 30°C. Each curve was generated by plotting the mean and s.d. calculated from n=3 independent experiments. d, Endogenous expression of Las1 variants and Grc3 in transformed S. cerevisiae tetO7-LAS1 strain grown in YPD supplemented with 40 μg/ml doxycycline at 30°C. Cell lysate was analyzed by western blot using anti-Myc antibody (Grc3), anti-Flag (Las1) and anti-tubulin (loading control). Representative blots from n=2 independent experiments. e, Polysome profile of transformed tetO7-LAS1 strain grown in the presence of 40 μg/ml doxycycline at 30°C. Black arrows mark ribosome half-mers, which is a hallmark for defective ribosome maturation. Representative profiles from n=3 independent experiments. f, Northern blot analysis of transformed S. cerevisiae tetO7-LAS1 strains grown in YPD supplemented with 40 μg/ml doxycycline at 30°C. RNA loading was monitored using the 18S mature rRNA and SCR1. The integrative density of 27S pre-rRNA was normalized to 18S rRNA and reported below the blot. Representative blots from n=3 independent experiments. g, Cartoon of ITS2 mimicking RNA substrate (Sc-ITS2-RNA) used for in vitro C2 cleavage assays (left). Denaturing gels of C2 cleavage activity of ScRNase PNK variants (0-0.8 μM) incubated with Sc-ITS2 RNA (50 nM) (middle). C2 cleavage was quantified from n=3 technical replicates and the mean and s.d. were plotted (right). Specific activity normalized to wild-type ScRNase PNK C2 cleavage is reported. Source data for 4c, 4e, and 4g are available online. Uncropped images are shown in Supplementary Data Set 1.
Conformational Switching within the Nuclease Active Site
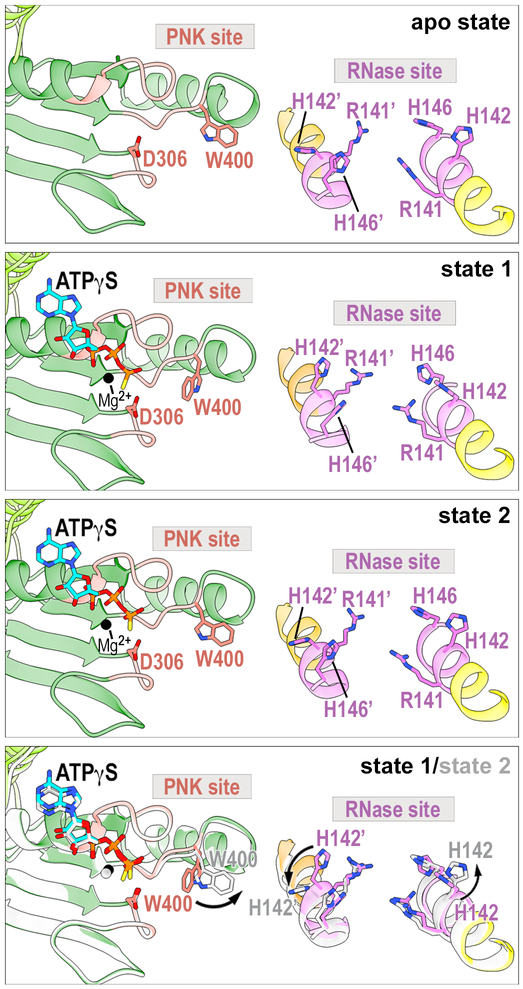
The HEPN dimer interface is largely unchanged between states, but we do observe hinge-like motions of the HEPN α-helical core (Video S3). The other noticeable difference between the states is the ordering of the loop (residues 117–122) between α4 and α5, which lies above the nuclease active site (Figures 4a and S3b). This loop is ordered in state 1, but disordered in state 2 and the apo state. The global hinge-like motion of the Las1 HEPN dimer interface is coupled with discrete rearrangements within the nuclease active site. The position of the catalytic Arginine (R141) and Histidine (H146) residues are largely unchanged in states 1 and 2, however we observe movement of the invariant Histidine (H142), adjacent to the catalytic Arginine (R141) (Figures 4a, 4b, and S3c). The residue adjacent to the catalytic Arginine within the RϕxxxH motif is most often an Asparagine, but some nucleases also harbor a Histidine or Aspartic Acid at this position21. The function of H142 from CtLas1 is unknown, but its conservation across Las1 homologues suggests an important role (Figure 4b). In state 1, H142 points towards the nuclease active site, while in state 2 H142 points towards the RNA binding cleft (Figure 4a inset). Conformational changes of H142 are coupled with the repositioning of another Las1 residue M124 that is found after the loop between α4 and α5 (Figure S3b). In state 2 M124 moves into the position of H142 from state 1 (Figure S3b). We modeled RNA into the nuclease active site using the coordinates from the RNase L RNA mimic engaged structure23. Based on our RNA modeling, state 1 appears primed for nuclease activity because H142 is positioned to coordinate the RNA substrate for cleavage (Figure S7). Our structures and RNA modeling suggest a critical role of H142 in acting as a switch for regulating nuclease activity.
Through a series of in vivo and in vitro assays, we determined that the HEPN Histidine Switch (CtLas1 H142) is critical for Las1 function. To determine the functional significance of the HEPN Histidine Switch, we created a tetO7-LAS1 S. cerevisiae strain in which the endogenous LAS1 gene is under the control of a doxycycline promoter. The tetO7-LAS1 strain was transformed with plasmids harboring wild-type Las1 (RHxxxH; ScLas1WT), a Las1 variant encoding a missense mutation to the HEPN Histidine Switch (RAxxxH; ScLas1H130A), a Las1 catalytic deficient variant (EHxxxA; ScLas1R129E/H134A)26, and empty YCplac vector. Strains were grown in the presence or absence of doxycycline at 30°C over a time course. Addition of doxycycline represses growth in the empty vector strain, but growth is restored in the strain harboring wild-type Las1 (Figure 4c). The strains harboring the HEPN Histidine Switch variant and the Las1 catalytic deficient variant do not grow in the presence of doxycycline indicating the significance of the Las1 RHxxxH motif for cell viability (Figure 4c). Western blot analysis confirmed that the Las1 variants do not hinder expression of either Las1 or Grc3 (Figure 4d). To determine the role of the HEPN Histidine Switch in ribosome assembly, we performed polysome profiling from the Las1 RAxxxH (ScLas1H130A) strain grown in the presence of doxycycline. The Histidine to Alanine mutation significantly impairs maturation of the large ribosomal subunit as we observe a decrease in the peak corresponding to the large ribosomal subunit and the formation of half-mers (Figure 4e). Nothern blot analysis further confirmed the importance of the Histidine Switch in pre-rRNA processing in vivo (Figure 4f). Moreover, the HEPN Histidine Switch variant is unable to cleave the ITS2 rRNA mimic in vitro (Figure 4g) confirming its importance for nuclease function. The in vivo and in vitro consequences of disrupting the HEPN Histidine Switch are equivalent to the double mutant of the catalytic Arginine and Histidine residues (Figure 4c-g), highlighting the significance of this residue for RNase PNK function. Our structural and functional studies establish that the Las1 nuclease is regulated by Grc3 stabilized HEPN dimerization, as well as, active site conformational rearrangements of an HEPN Histidine Switch. Intriguingly, CRISPR-Cas nucleases that are constitutively assembled into their respective HEPN dimers also rely on local conformational rearrangements of their RϕxxxH motif for nuclease activation22,32,35,36.
Conformational Switching within the Kinase Active Site
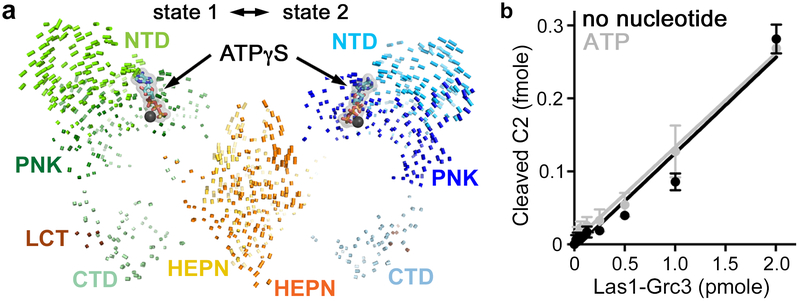
Following cleavage by the Las1 nuclease component, the RNA must move into one of the two Grc3 kinase active sites for phosphorylation. The Grc3 PNK active site contains four conserved structural motifs including the P-Loop, Walker B, Lid, and Clasp, that are important for catalysis27. ATP-γS binds into a pocket surrounded by the Grc3 NTD and the P-loop, Walker B, and Lid (Figures 5a and S5). Most of the interactions with the nucleotide are through the phosphate groups not the base, supporting earlier work which showed that Grc3 can utilize any nucleotide for phosphoryl transfer27. An overlay of the two ATP-γS bound states revealed that the most noticeable differences between the two states lie within the wings of the butterfly. The tips of the wings, predominately the NTD and PNK domains of Grc3, are able to adopt distinct conformations (Figure 5a). Morphing between the two states suggests a motion that is reminiscent of a butterfly fluttering its wings (Video S3). The interface between Las1 and the Grc3 CTD is largely unchanged likely because of its role in complex integrity (Figure 5a). To uncover whether the RNase PNK conformational landscape is driven by nucleotide binding, we measured C2 cleavage by RNase PNK in the presence and absence of ATP. There is no discernible difference in the rate of C2 cleavage in the presence or absence of ATP (Figure 5b), thus as we reported earlier, C2 cleavage is uncoupled from ATP binding26. Therefore nucleotide binding, while critical for the phosphorylation step, is not the trigger for the conformational changes between states 1 and 2.
Fig. 5 ∣. The Las1 RNase activity is uncoupled from ATP.
a, Vector map depicting the displacement of corresponding Cα atoms in ATP-γS bound RNase PNK states 1 and 2. Vector lengths correspond to domain motion. ATP-γS is shown as sticks. The metal, which is most likely magnesium27, is shown as a black sphere. b, RNA cleavage using an ITS2 RNA mimic substrate (SC-ITS2-RNA; 0.1 μM) with recombinant RNase PNK (0.8 μM) in the absence (no nucleotide) and presence (ATP, gray) of purified 2 mM ATP. The mean and s.d. were blotted from n=3 technical replicates. Source data for 5b is available online.
In addition to the movement of CtLas1 H142 from the nuclease active site, we also observed conformational changes within the kinase active site. Adjacent to the ATP binding pocket, the Grc3 kinase active site includes a Clasp motif ([T/S/L]xGW, where x is any residue) conserved in eukaryotic RNA kinases, Clp1 and Grc3, that is responsible for clamping the RNA into place27. RNA engaged structures of Clp1 revealed that the Tryptophan residue from the Clasp motif π-stacks with the incoming RNA base prior to phosphoryl transfer28. W400 from the CtGrc3 Clasp motif undergoes conformational changes and is found in alternative positions in states 1 and 2 (Figures 6 and S2f). To understand the functional significance of these conformational changes, we modeled RNA into the kinase active site using superimposed coordinates from Clp1 RNA engaged crystal structures (Figure S7)28. Based on our RNA modeling, state 2 appears primed for phosphorylation because W400 is correctly positioned to π-stack with the incoming base. In support of this model, we recently demonstrated that the S. cerevisiae equivalent to the Tryptophan Switch is essential for RNA phosphorylation27. Our cryo-EM reconstructions suggest an intricate interplay between the Tryptophan Switch from the kinase component and Histidine Switch from the nuclease component that allows for only one active site to be “on” at a time. State 1 is primed for nuclease activity while state 2 is primed for RNA phosphorylation (Figures 6 and 7).
Fig. 6 ∣. Conformational Changes within the RNase PNK Active Sites are Coordinated.
Local rearrangements within the nuclease and kinase active sites for the apo state, state 1 and state 2. Superposition of state 1 (color) and state 2 (gray) were generated by aligning Las1 α6 and highlights defined side chain rearrangements. In state 1, Las1 H142 is oriented towards the HEPN nuclease site (RNase site) while Grc3 W400 is arranged to π-stack with incoming RNA in state 2.
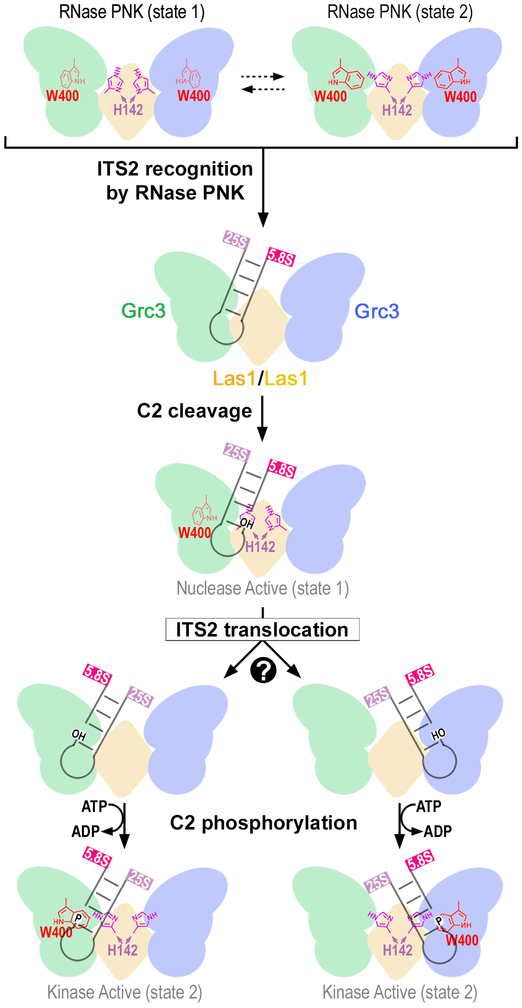
Fig. 7 ∣. Model of ITS2 processing by RNase PNK.
RNase PNK binds the ITS2 encoded pre-60S ribosome to initiate ITS2 processing. Global conformational changes to RNase PNK are coupled with local changes in the nuclease and kinase active sites driving ITS2 processing. RNase PNK adopts a spectrum of conformations including an extended state (Nuclease Active, state 1) where the catalytic Las1 H142 of RHxxxH points towards the nuclease active site for C2 cleavage of the ITS2 leaving 2′-3′ cyclic phosphate and 5′-hydroxyl (OH) RNA ends. Additional conformational changes reposition the 5′-hydroxyl RNA into one of the Grc3 RNA kinase active sites. Through a contracted conformation (Kinase Active, state 2), the catalytic W400 of the Grc3 kinase site adopts an alternative position to π-stack with an incoming RNA for phosphorylation (P). This coordinated process primes the 26S rRNA for processing by the Rat1-Rai1 exonuclease and the 7S rRNA for targeted RNA degradation by the nuclear exosome.
Discussion
ITS2 processing model
Together, our structural and functional analysis of the essential RNase PNK reveals its butterfly-shaped architecture and provides insight into the mechanism driving active site to active site crosstalk through local and global conformational changes. Our work supports the following model for targeted ITS2 pre-rRNA processing (Figure 7). After RNase PNK recruitment to pre-60S particles4,37-39, the ITS2 is asymmetrically engaged by RNase PNK and properly positioned relative to the Las1 HEPN catalytic residues for cleavage. The Las1 nuclease specifically cleaves the ITS2 at the C2 site generating the 7S pre-rRNA with a 2′−3′-cyclic phosphate and the 26S pre-rRNA with a 5′-hydroxyl. Following cleavage the 5′-hydroxyl cleavage product must move from the nuclease active site into one of the two distal kinase active sites. How the RNA transitions between active site remains unknown, but two possible scenarios include a rotation of the RNA within a single RNA-binding cleft or a larger-scale translocation to the opposite wing. Following RNA translocation, the 5′-hydroxyl is phosphorylated by the Grc3 kinase. The dual nuclease-kinase activities of RNase PNK prime the ITS2 for degradation by Rat1-Rai1 and the nuclear exosome10,12,40.
Alternative Mechanism of HEPN Nuclease Activation
Beyond providing insight into pre-rRNA processing this work establishes an alternative mode of HEPN nuclease activation and regulation. HEPN nucleases are found across all walks of life and participate in a variety of stress response pathways such as CRISPR RNA targeting pathways (Csm6, Cas13), the unfolded protein response (Ire1), and the type I interferon response (RNase L)21. For example recent work established that the CRISPR-associated HEPN nuclease Csm6 is allosterically activated by a cyclic oligoadenylate that is generated in response to phage infection41,42. An alternative mode of activation has been observed in Cas13 family members, which contain two HEPN domains within the same polypeptide. Cas13 HEPN nuclease domains are activated following pre-CRISPR RNA processing and activator RNA binding which triggers a conformational rearrangement that properly aligns the two RϕxxxH motifs for cleavage22,35,36,43. RNase PNK is the exception to the rule that HEPN nucleases are stress activated as it must be maintained in an active state to support its critical role in cell growth and proliferation13,25. Our work establishes the molecular basis for RNase PNK’s HEPN nuclease activation and regulation through its Grc3 kinase component.
The first level of nuclease regulation by the Grc3 kinase is that of structural support. The two copies of the Grc3 kinase within RNase PNK provide the structural framework to hold the Las1 HEPN nuclease domains in a dimeric arrangement, which is a requirement for HEPN nuclease activity. This structural support goes in both directions as the LCT of Las1 is critical for Grc3 stability. Kinase structural support is also observed in Ire1 and RNase L, however the complex architectures are widely different (Figure S6f). In Ire1 and RNase L, the protein kinase domains are part of the same polypeptide chain as the HEPN nuclease domain31. Ligand activation triggers dimerization of the ligand binding domains, kinase domains, and the HEPN domains resulting in extensive interfaces along the dimer axis23,24,30,34. Within RNase PNK, only the Las1 HEPN nuclease domains lie along the dimer axis. Rather than participating in dimerization with itself, Grc3 holds the HEPN domains in place through extensive interfaces with its CTD domain and, to a lesser extent, its PNK domain. This altered kinase-nuclease arrangement is likely in part because of the need to transfer the RNA substrate from the nuclease active site to the kinase active site.
The second level of nuclease regulation comes from the Grc3 kinase active site. A comprehensive characterization of Grc3 kinase mutants revealed that defects within the kinase active site hinder nuclease activation27. By capturing RNase PNK in multiple conformational states, we now understand the molecular basis for active site crosstalk. The addition of a non-hydrolysable ATP analogue allowed us to observe a state of RNase PNK in high resolution that we did not observe in the absence of nucleotide. While ATP binding is decoupled from C2 cleavage, nucleotide binding likely has a stabilizing effect on the Grc3 NTD. The NTD makes up part of ATP binding pocket and is more ordered in states 1 and 2 than in the apo reconstruction. The Grc3 kinase active site relays information to the nuclease active site resulting in the conformational changes that support nuclease activation. The conformational rearrangements in the Las1 nuclease domain are subtle, but similar in scale to the conformational changes observed in Cas13 nucleases following activation. Finally, we hypothesize that the final level of RNase PNK nuclease activation comes from the RNA substrate itself. In contrast to RNase L, Csm6, and Cas13 nucleases that cleave RNA non-specifically22,23,33, RNase PNK is a RNA sequence and structure specific nuclease26. Proper engagement of the ITS2 from a pre-60S particle likely triggers additional RNase PNK conformational changes that properly align the C2 site within the nuclease domain for cleavage.
Grc3 Polynucleotide Kinase Activation
The Las1 nuclease component of RNase PNK is in turn critical for Grc3 kinase activation. Las1 provides structural support of Grc3, through its conserved LCT. The LCT of Las1 however is not sufficient for activating the Grc3 kinase domain26. This activation instead comes from the Las1 HEPN nuclease domains, which provide additional structural support for Grc3. Our work identified W400 as a key residue from the kinase active site that changes conformation between states 1 and 2. W400 is part of a motif that is conserved across Grc3(Nol9) homologues and higher eukaryotic homologues of the related kinase Clp128. A series of crystal structures of Clp1 engaged with RNA revealed that this tryptophan is critical for base stacking with the ultimate base28. We postulate that this Tryptophan residue from Grc3 must move not only for nuclease activation, but also to accommodate the ITS2 pre-rRNA within the RNase PNK RNA-binding cleft. The Grc3 kinase component of RNase PNK lacks the specificity of the Las1 nuclease27. Similar to Clp1, Grc3 is capable of phosphorylating many RNA substrates in vitro. Therefore we suggest that RNase PNK is maintained in a kinase active state until engagement of the ITS2 pre-rRNA and/or some other unknown trigger for C2 cleavage.
In summary this work provides the first structural snapshots of an ancient multienzyme complex revealing how the individual components assemble together and coordinate with one another to drive an irreversible cascade of RNA decay. In-line with other multienzyme macromolecular machines, RNase PNK utilizes a series of intricate molecular cues to catalyze a fundamental two-step reaction. The interconnected nuclease and kinase components of RNase PNK support a communication network between the distant active sites that allows RNase PNK to coordinate its enzymatic activity through two key molecular switches. This sophisticated enzymatic coordination ensures both fidelity and directionality of ITS2 processing and is critical for the production of ribosomes.
Online Methods
Yeast Strains
The S. cerevisiae strain encoding a tetracycline-titratable promoter (tetO7) upstream of the endogenous LAS1 gene was generated from the parent strain CML476 (EUROSCARF) using the kanMX-tetO7 cassette as described previously44. A 3XMyc-tag was added upstream of endogenous GRC3 in the tetO7-LAS1 strain (yMP125). The yMP125 strain was transformed with plasmids encoding the N-terminal 3xFlag tagged Las1 wild type (pMP 580), Las1 H130A variant (pMP 655), Las1 R129E, H134A variant (pMP 581) or empty YCplac33 vector. Yeast strains are listed in the Table S2.
Cloning Las1 and Grc3 Variants
The expression plasmid for CtLas1-Grc3 (pMP 287) and ScLas1-Grc3 (pMP 001) were described previously26. The ScLas1-Grc3 H130A variant (pMP 458) and R129E, H134A variant (pMP 296) were generated using Q5 site-directed mutagenesis (NEB) using pMP 001. N-terminal Flag-tagged ScLas1 (pMP 580) was amplified along with 300 nt of flanking endogenous DNA sequence and inserted into YCplac3345 using KpnI and SacI. ScLas1 variants encoding the missense mutation H130A (pMP 655) and R129E, H134A (pMP 581) were amplified by overlap PCR. All constructs were verified by DNA sequencing (GeneWiz).
Expression and Purification of RNase PNK Variants
CtLas1-Grc3 was co-expressed in Rosetta 2 (DE3) pLacI (EMD Millipore) while ScLas1-Grc3 variants were expressed in LOBSTR (Kerafast) as previously described26. Las1-Grc3 constructs were purified as described earlier with minor modifications26. Briefly, Las1-Grc3 variants were loaded onto His60 Ni Superflow Resin (Clontech Laboratories Inc) using lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 5 mM MgCl2, 10% glycerol), washed with lysis buffer containing 30 mM imidazole and eluted using 200 mM imidazole. CtLas1-Grc3 and ScLas1-Grc3 variants were resolved by size exclusion chromatography using a HiLoad 16/600 Superdex 200pg column (GE Healthcare) equilibrated with 20 mM Hepes pH 7.7, 200 mM NaCl, 5 mM MgCl2, 5% glycerol and 20 mM Tris pH 8.0, 200 mM NaCl, 5 mM MgCl2, 5% glycerol, respectively.
Cryo-Electron Microscopy
Purified CtLas1-Grc3 complex (10 μM) was cross-linked with 50 μM bis(sulfosuccinimidyl)suberate (BS3; Sigma) in 20 mM Hepes pH 7.7, 200 mM NaCl, 5 mM MgCl2, 5% glycerol at room temperature for 5 minutes before quenching with 30 mM Tris pH 7.5 for 15 minutes at 4°C. Cross-linked reactions were resolved over a Superose 6 10/300 GL column (GE Healthcare) in 10 mM Tris pH 8.0, 200 mM NaCl, 5 mM MgCl2. CtLas1-Grc3 (0.15 mg/ml) was incubated in the absence (apo state) and presence (ATP-γS bound state) of 2 mM ATP-γS (Sigma) and 10 μM CT-ITS2-RNA (5′-UGUGUUGGGGdeoxyACCCGCGGCUGCUCG CGGGCCCUGAAAAGCA-3′) for 1 hour at 4°C. The CT-ITS2-RNA substrate represents part of the C. thermophilum ITS2 pre-rRNA sequence. Protein mixtures (3 μL) were applied to glow-discharged UltrAuFoil R1.2/1.3 300 mesh grids (Quantifoil) and vitrified after 5 sec blotting using the Automatic Plunge Freezer EM GP (Leica).
Data Acquisition
Images of CtLas1-Grc3 were collected using a Titan Krios electron microscope (FEI) operated at 300 keV and equipped with a Falcon 3EC direct electron detector. Movies were collected in counting mode at a nominal magnification of 75,000x corresponding to 1.08 Å/pixel. Overall, 1016 (apo state) and 833 (ATP-γS bound state) movies (30 frames/movie) were collected with defocus values ranging from −1.0 to −2.5 μm. Movies were recorded over 60 seconds resulting in a fluence of ~40 e-/A2 and flux of 0.8 e-/pixel/sec.
Image Processing
Recorded frames were aligned using MotionCor2 version 1.1.046 using a B-factor of 150 and was conducted with 5 × 5 tiles to correct for beam-induced motion. Dose-weighted aligned images were used to calculate the CTF parameters using CTFFIND447. Using RELION (version 3.0-beta)48, 761,763 particles from 1016 micrographs (apo state) and 703,078 particles from 833 micrographs (ATP-γS bound state) were selected using Laplacian-of-Gaussian. The particle stack was binned 4 × 4 (4.32 Å/pixel) and extracted with a pixel box size of 56. Particles from the apo state dataset were subjected to 2D classification and particles from the good classes were used to generate an initial model by stochastic gradient descent. Images were subjected to 3D classification resulting in a “clean” dataset of 393,892 particles (apo state) and 500,873 particles (ATP-γS bound state). 3D auto-refinement was performed followed by particle re-extraction (binned 2 × 2; 2.16 Å/pixel) with a pixel box size of 112. A second iteration of 3D classification (8 classes, tau=4) was performed prior to re-extracting particles (1.08 Å/pixel; pixel box size 224). An additional round of 3D refinement was performed followed by CTF refinement and particle polishing. Polished particles were imported into cisTEM (version 1.0.0 beta)49 and subjected to 40 iterations of 3D classification with and without imposing C2 symmetry. Thorough analysis of the classes generated from Las1-Grc3 pre-incubated with ATP-γS and RNA did not yield asymmetric classes nor unassigned density that could account for the presence of RNA. This is in-line with the weak binding affinity of RNase PNK for RNA (see Figure 3) and the low sample concentration used during cryo-EM grid preparations. Clear density was visible for the nucleotide in both kinase active sites prior to inforcing C2 symmetry. Since there were no asymmetric states observed, the most populated classes were subjected to 3D refinement with C2 symmetry and sharpened using LocalDeblur50. Local resolution was estimated using MonoRes51.
Atomic Modeling
RNase PNK cryo-EM reconstructions have an estimated resolution of 3.2 Å (apo state), 2.9 Å (ATP-γS bound state 1) and 3.0 Å (ATP-γS bound state 2). At this resolution we observe clear density for all the major features of the Las1 HEPN domain and LCT, as well as, Grc3 NTD, PNK and CTD domains. To build an initial model and ensure the correct handiness of the reconstruction of Grc3, we took the coordinates from the Caenorhabditis elegans Clp1 structure bound to Mg2+ and AMP-PNP (PDB ID 4OHV) and manually fit the subdomains using COOT52. The fit was then improved using rigid body refinement. To build an atomic model of Grc3, we generated a homology model for Grc3 PNK domain using Swiss Modeler 53. This model was then superimposed upon the initial model and the fit was improved with rigid body, phased reciprocal space refinement in CNS54 followed by iterative rounds of building in COOT and refinement in CNS. Due to the lack of homology between the N- and C-terminal domains of Grc3 and Clp1, the bulk of the N- and C-terminal domains were built de novo using COOT. Additionally, the Las1 HEPN domain and LCT were built de novo using COOT. Other minor adjustments were made to the model following iterative rounds of manual building and refinement in COOT and CNS. Molprobity55 was used to evaluate the model and the model statistics are listed in Table 1. Figures and videos were prepared with Pymol, Chimera and ChimeraX56-58.
Chemical Cross-linking Mass Spectrometry
CtLas1-Grc3 (10 μM) was cross-linked with 50 μM bis(sulfosuccinimidyl)suberate (BS3; Sigma) in 20 mM Hepes pH 7.7, 200 mM NaCl, 5 mM MgCl2, 5% glycerol at room temperature for 5 minutes before quenching with 30 mM Tris pH 7.5 for 15 minutes at 4°C. 10 μL of the cross-linked mixture was digest by the addition of 10 μL trypsin/LysC mix (0.1 μg/μL - Promega) overnight at 40°C. The digests were then stored at −80°C for subsequent MS analysis. Protein digests were analyzed by LC/MS on a Q Exactive Plus mass spectrometer (ThermoFisher Scientific) interfaced with an M-Class nanoAcquity UPLC system (Waters Corporation) equipped with a 75 μm x 150 mm BEH C18 column (1.8 μm particle, Waters Corporation) and a C18 trapping column (180 μm × 20 mm) with 5 μm particle size at a flow rate of 400 nL/min. The trapping column was in-line with the analytical column and upstream of a micro-tee union which was used for venting, trapping, and as a liquid junction. Trapping was performed using the initial solvent composition. 5 μL of digested sample was injected onto the column. Peptides were eluted by using a linear gradient from 99% solvent A (0.1% formic acid in water (v/v)) and 1% solvent B (0.1% formic acid in acetonitrile (v/v)) to 40% solvent B over 70 minutes. For the mass spectrometry, a top-ten data dependent acquisition method was employed with a dynamic exclusion time of 15 seconds and exclusion of singly charged species. The mass spectrometer was equipped with a nanoflex source with a stainless-steel needle and used in the positive ion mode. Instrument parameters were as follows: sheath gas, 0; auxiliary gas, 0; sweep gas, 0; spray voltage, 2.7 kV; capillary temperature, 275°C; S-lens, 60; scan range (m/z) of 375 to 1500; 1.6 m/z isolation window; resolution: 70,000 (MS), 17,500 (MS/MS); automated gain control (AGC), 3 × 10e6 ions (MS), 5 × 10e4 (MS/MS); and a maximum IT of 100 ms (MS), 50 ms (MS/MS). Mass calibration was performed before data acquisition using the Pierce LTQ Velos Positive Ion Calibration mixture (ThermoFisher Scientific). The LC/MS raw data were first converted to an MGF format using Mascot Distiller from Matrix Science and then analyzed using the Batch-Tag Web function of the ProteinProspector web-based software developed by the UCSF Mass Spectrometry Facility. The MGF file was searched against sequences for the recombinant CtGrc3 and CtLas1 by employing the User Protein Sequence field with other search parameters including: tryptic specificity and 3 missed cleavages; precursor charge range of 2, 3, 4, and 5; monoisotopic values; parent mass tolerance of 10 ppm and fragment mass tolerance of 50 ppm; oxidation of methionine as a variable modification; and in the Crosslinking field, the Link Search Type was defined as DSS. The putative cross-linked peptide output was triaged by limiting the mass error of putative cross-links to two standard deviations from the average error (about 4 ppm); requiring a Score Difference value >5 except for the cases of intermolecular cross-links of identical peptides; total expectation values below 1×10−4 with each peptide from the cross-link having expectation values below 0.1. Cross-link/mass spectrometry data was displayed using xiNET59.
Yeast Proliferation Assays and Western Blots
The S. cerevisiae strain encoding a tetracycline-titratable promoter (tetO7) upstream of the endogenous LAS1 gene was generated from the parent strain CML476 (EUROSCARF) using the kanMX-tetO7 cassette as described previously44. A 3XMyc-tag was added upstream of endogenous GRC3 in the tetO7-LAS1 strain (yMP125). The yMP125 strain was transformed with plasmids encoding the N-terminal 3xFlag tagged Las1 wild type (pMP 580), Las1 HEPN variants (pMP 581 and pMP 655) or empty YCplac33 vector. Transformed yeast strains (see Table S1) were pre-incubated in YPD supplemented with 40 μg/ml doxycycline for 24 hours at 22°C prior to performing proliferation assays. Addition of doxycycline represses the expression of endogenous Las1. Growth curves were generated by measuring the absorbance at 595 nm of 100 μL yeast cultures set to an OD600 of 0.05 and incubated at 30°C in YPD and YPD supplemented with doxycycline (40 μg/ml). Measurements were recorded every 15 minutes for 25 hours using an Infinite F200 Pro (Tecan) and i-control 1.11 software. The mean and standard deviation were calculated from three independent replicates. To confirm that the Las1 variants were expressed, Western blotting was performed as described previously27. Briefly, strains were grown to to mid-log phase, lysed with glass beads, and precipitated with trichloroacetic acid. Whole cell lysates were resolved by SDS-PAGE and proteins were detected using anti-Myc (Grc3; EMD Millipore), anti-Flag (Las1; Sigma), and anti-α-tubulin (Abcam).
Sucrose Gradient and Polysome Profiling
Sucrose gradient and polysome profiles were generated as described previously27 with minor modifications. Briefly, transformed tetO7-LAS1 strains (Table S1) were pre-incubated for 24 hours in YPD supplemented with 40 μg/mL doxycycline at 22°C. Cultures were grown at 30°C in the presence of doxycycline (40 μg/mL) to OD600 ~ 0.5 and incubated with 0.1 mg/mL cycloheximide for 5 minutes on ice. Cells were lysed in 20 mM Tris pH 7.4, 60 mM KCl, 10 mM MgCl2, 1 mM DTT, 1% (v/v) Triton X-100, 0.1 mg/mL cycloheximide, 0.2 mg/mL heparin using glass beads. Clarified lysates were resolved using a 7–47% sucrose gradient and fractionated by a gradient fractionator (Brandel).
Northern Blot Analysis
To monitor the accumulation of pre-rRNA intermediates, transformed yeast strains (see Table S1`) were pre-incubated in YPD supplemented with 20 μg/ml doxycycline for 12 hours at 22°C prior to growing the cells to mid-log phase in YPD supplemented with 40 μg/ml doxycycline at 30°C. RNA extraction and Northern blot analysis was performed as previously described26. Briefly, 3 μg of total RNA was resolved using a 1% formaldehyde agarose gel and transferred to a Hybond XL membrane (GE Healthcare) overnight. Digoxigenin conjugated probes that are previously described13 were used to detect pre-rRNA intermediates and mature rRNAs. RNA intermediates were quantified in ImageJ and RNA loading was normalized to the 18S rRNA. Digoxigenin conjugated SCR1 probe CGTGTCTAGCCGCGAGGAAGGATTTGTTCC/3Dig_N/ was included as an alternative loading control.
C2 RNA Cleavage Assays
C2 RNA cleavage was performed as described previously26,27 with minor changes. Full-length ScLas1-Grc3 variants (0–0.8 μM) were incubated with 1 mM EDTA and 50 nM fluorescein labeled C2 RNA mimic (5′-GUCGUUUUAGGUUUUACCAACUGCGGC/36-FAM/−3′) in the absence and presence of 2 mM nucleotide (ATP). Contaminating nucleotides were removed from commercial ATP (Sigma; A2383) using previously established nucleotide purification protocols60,61. Briefly, ATP (6 mg) was resuspended in 50 mM (NH4)HCO3 and bound to a MonoQ 4.6/100 PE (GE Healthcare) column equilibrated with 50 mM (NH4)HCO3. ATP was eluted using a 50–500 mM (NH4)HCO3 linear gradient over 45 mL. ATP fractions were lyophilized and resuspended to 10 mM using water. Unless specified otherwise, reactions were incubated for 1 hour at 37°C and quenched with urea-loading dye. Samples were resolved on 15% polyacrylamide (8 M urea) gels in 1x tris-borate-EDTA buffer and visualized using a Typhoon FLA 9500. Representative gels are shown from three independent replicates.
Isothermal Titration Calorimetry (ITC)
ITC was performed using a MicroCal iTC200 calorimeter (Malvern Panalytical). Prior to ITC analysis, SC-ITS2–3-RNA and SC-ITS2–5-RNA were annealed to create a duplex mimic of the Sc C2 cleavage product (SC-ITS2-cleaved-RNA). ScRNase PNK (20 μM) and the RNA substrates (SS-RNA 1 mM; SC-ITS2-cleaved-RNA 0.82 mM) were prepared in 20 mM Tris pH 8.0, 200 mM NaCl, 5 mM MgCl2, 5% glycerol. RNA was titrated into 0.36 mL of ScRNase PNK using 40 injections (0.5 μL/injection) with 120 second intervals at 25°C. Data acquisition and analysis were performed by the MicroCal Origin software package. Data analysis was performed by generating a binding isotherm and best fit using standard Levenberg-Marquardt methods to determine N (number of sites) and K (binding constant in M−1). The ITC thermograms were generated using the equations Θj = ([L]Kj)/(1+[L]Kj) and Lt = [L] + Pt Σ(njΘj) where Θ is the fraction of site j occupied by ligand, Lt is the total ligand concentration, [L] is the free ligand concentration, Pt is the total protein concentration, Kj is the binding constant of process j and nj is the total stoichiometric ratio of process j. Following data analysis, K (M−1) is then converted to Kd (µM). A representative replicate of 3–5 independent measurements is shown in Figure 3d.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-20042 (apo), EMD-20041 (ATP-γS bound state 1), and EMD-20040 (ATP-γS bound state 2). Atomic coordinates have been deposited in the Protein Data Bank under accession codes 6OF4 (apo), 6OF3 (ATP-γS bound state 1), and 6OF2 (ATP-γS bound state 2).The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014738. Source data for Figs. 4c, 4e, 4g, and 5b are available with the paper online. All other data will be made available upon reasonable request.
Supplementary Material
Acknowledgements
We thank Drs. Traci Hall, William Beard and Matthew Schellenberg, as well as, all the members of the Stanley Lab for their critical reading of this manuscript. We are grateful to all the members of the Molecular Microscopy Consortium for their help with Cryo-EM data collection and processing. We thank Dr. Leesa Deterding from the NIEHS Mass Spectrometry Research and Support Group for help with cross-linking mass spectrometry analysis. This work was supported by the US National Institute of Health Intramural Research Program; US National Institute of Environmental Health Sciences (NIEHS; ZIA ES103247 to R.E.S; ZIC ES103326 to M.J.B.) and the Canadian Institutes of Health Research (CIHR; 146626 to M.C.P).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Neuman N The Complex Macromolecular Complex. Trends Biochem Sci 41, 1–3 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Jones EY & Yeates TO Structure and function in complex macromolecular assemblies: some evolutionary themes. Curr Opin Struct Biol 22, 197–9 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Klinge S & Woolford JL Jr. Ribosome assembly coming into focus. Nat Rev Mol Cell Biol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler J & Hurt E Eukaryotic Ribosome Assembly. Annu Rev Biochem (2018). [DOI] [PubMed] [Google Scholar]
- 5.Woolford JL Jr. & Baserga SJ Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kressler D, Hurt E & Bassler J A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem Sci 42, 640–654 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Konikkat S & Woolford JL Jr. Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem J 474, 195–214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomecki R, Sikorski PJ & Zakrzewska-Placzek M Comparison of preribosomal RNA processing pathways in yeast, plant and human cells - focus on coordinated action of endo- and exoribonucleases. FEBS Lett (2017). [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Pevida A, Kressler D & de la Cruz J Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip Rev RNA 6, 191–209 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Fromm L et al. Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat Commun 8, 1787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasse L, Flemming D & Hurt E Coordinated Ribosomal ITS2 RNA Processing by the Las1 Complex Integrating Endonuclease, Polynucleotide Kinase, and Exonuclease Activities. Mol Cell 60, 808–15 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Schuller JM, Falk S, Fromm L, Hurt E & Conti E Structure of the nuclear exosomes captured on a maturing preribosome. Science 360, 219–222 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Castle CD et al. Las1 interacts with Grc3 polynucleotide kinase and is required for ribosome synthesis in Saccharomyces cerevisiae. Nucleic Acids Res 41, 1135–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castle CD, Cassimere EK & Denicourt C LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol Biol Cell 23, 716–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schillewaert S, Wacheul L, Lhomme F & Lafontaine DL The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol Cell Biol 32, 430–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu H et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 21, 133–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butterfield RJ et al. Congenital lethal motor neuron disease with a novel defect in ribosome biogenesis. Neurology 82, 1322–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapur M & Ackerman SL mRNA Translation Gone Awry: Translation Fidelity and Neurological Disease. Trends Genet 34, 218–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu EY, Cali CP & Lee EB RNA metabolism in neurodegenerative disease. Dis Model Mech 10, 509–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conlon EG & Manley JL RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev 31, 1509–1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anantharaman V, Makarova KS, Burroughs AM, Koonin EV & Aravind L Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct 8, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connell MR Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J Mol Biol 431, 66–87 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Han Y et al. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343, 1244–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KP et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132, 89–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castle CD, Cassimere EK, Lee J & Denicourt C Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol Cell Biol 30, 4404–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillon MC, Sobhany M, Borgnia MJ, Williams JG & Stanley RE Grc3 programs the essential endoribonuclease Las1 for specific RNA cleavage. Proc Natl Acad Sci U S A 114, E5530–E5538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillon MC, Sobhany M & Stanley RE Characterization of the molecular crosstalk within the essential Grc3/Las1 pre-rRNA processing complex. RNA 24, 721–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dikfidan A et al. RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase Clp1. Mol Cell 54, 975–86 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Noble CG, Beuth B & Taylor IA Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res 35, 87–99 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korennykh AV et al. Structural and functional basis for RNA cleavage by Ire1. BMC Biol 9, 47 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillon MC & Stanley RE Nuclease integrated kinase super assemblies (NiKs) and their role in RNA processing. Curr Genet (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L et al. Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell 168, 121–134 e12 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Niewoehner O & Jinek M Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H et al. Dimeric structure of pseudokinase RNase L bound to 2–5A reveals a basis for interferon-induced antiviral activity. Mol Cell 53, 221–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C et al. Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d. Cell 175, 212–223 e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knott GJ et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat Struct Mol Biol 24, 825–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kater L et al. Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes. Cell 171, 1599–1610 e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534, 133–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biedka S et al. Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J Cell Biol 217, 2503–2518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kummer E & Ban N Conformational Switching of the Nuclear Exosome during Ribosome Biogenesis. Biochemistry 57, 4765–4766 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Niewoehner O et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G & Siksnys V A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Liu L et al. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 170, 714–726 e10 (2017). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 44.Yen K, Gitsham P, Wishart J, Oliver SG & Zhang N An improved tetO promoter replacement system for regulating the expression of yeast genes. Yeast 20, 1255–62 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Gietz RD & Sugino A New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–34 (1988). [DOI] [PubMed] [Google Scholar]
- 46.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grant T, Rohou A & Grigorieff N cisTEM, user-friendly software for single-particle image processing. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramírez-Aportela E et al. Automatic local resolution-based sharpening of cryo-EM maps. bioRxiv, 433284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vilas JL et al. MonoRes: Automatic and Accurate Estimation of Local Resolution for Electron Microscopy Maps. Structure 26, 337–344 e4 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunger AT Version 1.2 of the Crystallography and NMR system. Nat Protoc 2, 2728–33 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Williams CJ et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrodinger L The PyMOL Molecular Graphics System Version 1.8. (2015). [Google Scholar]
- 58.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Combe CW, Fischer L & Rappsilber J xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics 14, 1137–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young C & Karbstein K Analysis of cofactor effects on RNA helicases. Methods Enzymol 511, 213–37 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Johnstone DB & Farr SB AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J 10, 3897–904 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-20042 (apo), EMD-20041 (ATP-γS bound state 1), and EMD-20040 (ATP-γS bound state 2). Atomic coordinates have been deposited in the Protein Data Bank under accession codes 6OF4 (apo), 6OF3 (ATP-γS bound state 1), and 6OF2 (ATP-γS bound state 2).The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014738. Source data for Figs. 4c, 4e, 4g, and 5b are available with the paper online. All other data will be made available upon reasonable request.