Abstract
Antigenic exposures at epithelial sites in infancy and early childhood are thought to influence the maturation of humoral immunity and modulate the risk of developing IgE-mediated allergic disease. How different kinds of environmental exposures influence B cell isotype switching to IgE, IgG, or IgA, and the somatic mutation maturation of these antibody pools, is not fully understood. We sequenced antibody repertoires in longitudinal blood samples in a birth cohort from infancy through the first three years of life and found that, whereas IgG and IgA show linear increases in mutational maturation with age, IgM and IgD mutation are more closely tied to pathogen exposure. Strikingly, IgE mutation frequencies are primarily increased in children with impaired skin barrier conditions such as eczema, suggesting that IgE affinity maturation could provide a mechanistic link between epithelial barrier failure and allergy development.
One Sentence Summary:
Distinct environmental exposures are associated with the maturation of antibody types in early childhood.
Introduction
B cell populations in humans harbor diverse B cell receptors (BCRs) that provide specific recognition and memory of antigens derived from pathogens and other sources. Upon interaction with the environment, the naïve B cell repertoire is altered by clonal expansion of antigen-specific B cells, and differentiation into specialized functional subsets such as memory B cells, plasmablasts and plasma cells. Antigen-stimulated B cells can switch their constant region isotype usage from IgM and IgD expressed in naïve cells to IgG, IgA or IgE isotypes that have distinct functional effector roles such as complement activation and interaction with specific constant region receptors expressed by other leukocytes. Antigen stimulation in an appropriate setting such as the germinal center of secondary lymphoid tissues also triggers somatic hypermutation (SHM) of the antibody genes, which is required for antibody affinity maturation. Studies of early immune system development in response to microbiome formation and infectious diseases have emphasized leukocyte subset changes and serological data (1–6). Changes in BCR repertoires driven by environmental exposures in early life are poorly understood.
Allergic conditions such as food and respiratory allergies are increasingly common in children and are mediated by allergen-specific IgE antibodies that bind to high-affinity receptors on mast cells and basophils, sensitizing these cells to degranulate upon allergen exposure (7–11). Isotype switching to IgE in humans can occur by “direct” switching from IgM-expressing B cells, or by “indirect” switching in B cells that have already switched to an IgG or IgA1 isotype prior to switching to IgE (12, 13). Early exposure to a broad diversity of antigens associated with pets or farm animals, or feeding with potentially allergenic foods, can be protective against allergy (14–16). Conversely, exposure to antigens through impaired skin barrier surfaces, such as that seen with eczema, may contribute to the development of allergic disease (17, 18). Allergen-specific IgE production is thought to be influenced by both host genetics and environmental exposures (7, 19), but the cellular mechanisms linking environmental stimuli to IgE development are unknown. The extent of SHM in IgE may be a critical determinant in the development of allergic disease, as more mutated antibody genes often correlate with greater antigen affinity (20). There is not yet consensus about the role of affinity maturation and antigen selection in IgE responses (21–26).
To analyze the molecular changes in BCR repertoires in young children developing under different environmental conditions, we carried out high-throughput sequencing (HTS) of Ig heavy chain (IgH) gene rearrangements of a sub-cohort of 51 children from the Stanford’s Outcomes Research in Kids (STORK) birth cohort (Table 1 and table S1) (27). IgH sequence features from yearly blood samples were correlated with clinical and epidemiological data. We propose that the generation of high-mutation IgE in infancy and early childhood provides a key mechanistic link between impaired skin barrier function and the development of pediatric allergic disease.
Table 1.
Demographic and clinical characteristics of the STORK study sample
| STORK study sample (n=51) | |
|---|---|
| Age in weeks at 1st time point, median (range) | 81 (49–126) |
| Sex | |
| Female | 24 |
| Male | 27 |
| Clinically diagnosed allergy/eczema | |
| Yes | 17 |
| No | 34 |
| Randomization arm of triclosan/triclocarban study | |
| Exposed | 27 |
| Unexposed | 22 |
| Not randomized | 2 |
| Household pets | |
| Yes | 16 |
| No | 35 |
Results
Antibody SHM increases with age, but IgE shows high variation between individuals in the first 3 years of life
The children in this cohort were followed from birth to three years of age with longitudinal yearly sampling of peripheral blood by venipuncture with subsequent isolation of peripheral blood mononuclear cells (PBMCs) (Fig. 1A). Ten children were sampled at all three years, 22 children were sampled at two yearly time points, and the remaining 19 children were sampled at one time point. PBMCs from 114 healthy human adults were used as controls. SHM of antibody V(D)J gene rearrangements is an irreversible genetic mark of antigen-driven proliferation of B cells. Fig. 1B shows the time-dependent linear accumulation of SHM in antibody genes of the STORK cohort subjects over the course of three years in antibodies of all subclasses except for IgE. A similar pattern of progressive SHM increase was observed within individuals from whom three longitudinal samples were obtained (Fig. 1C). Both switched (IgG and IgA) as well as IgM and IgD isotypes show this pattern over time, likely reflecting the development of class-switched as well as IgM+/IgD+, IgM+ or IgD+ memory B cells (28, 29). Mutated IgM and IgD B cells reach adult SHM frequencies by 2 years of age, whereas the IgG and IgA class-switched B cells progressively gain mutations but only reach 60–75% of adult SHM frequencies by age 3 years. In contrast, IgE SHM percentages are highly variable between children (Fig. 1B), although within 7 of the 10 children who were sampled at three timepoints, progressive SHM increase is noted (Fig. 1C). Clonality of B cells also increased with age and was significantly different between children and adults (p<10−15, two-sided Wilcoxon-Mann-Whitney test) (Fig. 1D).
Fig. 1. In years 1–3 of life, antibody SHM increases, but IgE shows high inter-individual variation.
(A) Data from 51 STORK children were collected with weekly phone surveys of parents, medical chart reviews every six months, household visits every four months, and venipuncture blood collection at approximately annual intervals. (B) Average SHM percentage for the IgH variable gene segment (IGHV) of expressed antibodies of the indicated isotype for each individual. Second and third samples from each subject are indicated by data point color. SHM ranges from 114 healthy adults are shown as box-whisker plots. The minimum, maximum, and median age of the healthy adults is 17, 87, and 52 years, respectively. Blue lines show linear regression for SHM frequencies and age, and the shaded region indicates the 95% confidence interval. (C) Longitudinal SHM data from the ten children who were sampled at three time points. Data for each individual are shown as dots of the same color, connected by lines. For both (B) and (C), SHM for each isotype in each subject are summarized as the median SHM of reads for each isotype within each clone, then taking the mean SHM frequency over all clones. (D) B cell clonality for 38 children, and 114 adult samples shown as a box-whisker. For the subset of children with sampling at two or more time points, second and third samples are indicated by color of the data point.
Children and adults show differing isotype frequencies but comparable selection for IGHV gene usage correlated with isotype genomic position
Analysis of age-dependent changes in the distribution of IgG and IgA subclasses (Fig. 2A) shows a significantly increased usage of the IgG1 and IgA1 subclasses in the STORK children (1–3 years) compared to the healthy adults (p<0.001). Overall, adult controls expressed more IgG2 and IgA2 than the children, and the usage of these subclasses seems to increase with age, with the exception of IgA2 usage in the oldest age group. IgG3 and IgG4 fractions are comparable across all ages and show the lowest usage of all subclasses.
Fig. 2. Children and adults differ in IgG and IgA subclass usage but show comparable IGHV gene selection correlated with isotype switching.
(A) The fraction of each IgG or IgA subclass used is shown. STORK specimen samples (n=89) are indicated as points in the age one to three year category, and the 114 healthy adults (age 17–87 years) are the same as those in Fig. 1. The sum of the subclass fractions is more than 1.0 because a clone can contain members expressing different subclasses. p values were determined by two-sided Wilcoxon-Mann-Whitney tests: *p < 0.05, **p < 0.01, and ***p < 0.001. IGHV gene usage is shown as the average for clones within each (B) child or (C) adult. Isotypes are plotted in their chromosomal ordering, from upstream isotypes IgM and IgD to the most downstream isotype, IgA2. IGHV ordering is based on the 20 most common IGHV genes in IgM in the STORK subjects. IgG4 and IgE are excluded due to low frequency. Dots represent outliers beyond 1.5-fold of the interquartile range. The plot axes were chosen to show the box-whiskers on a readable scale; rare outlier points with extreme values are not shown, but were included in all analyses.
To evaluate evidence of age-associated differences in IGHV gene usage, we determined the frequency of clones using each IGHV gene for each of the isotypes. A surprising finding was that a subset of IGHV genes was more commonly associated with isotypes that are located further downstream in the IGH locus, while another subset of IGHV genes showed the opposite pattern, with a progressive decrease in downstream isotypes. The same patterns were seen for both children and adults in all IGHV genes examined (Fig. 2, B and C). Because isotype switching is irreversible and unidirectional, these patterns suggest that there is progressive selection correlated with isotype switching that favors B cells expressing certain IGHV genes (IGHV3–23, IGHV3–21, IGHV3–7, IGHV3–48, IGHV3–30, IGHV3–11, IGHV3–74). In contrast, B cells expressing IGHV genes IGHV4–34, IGHV4–59, IGHV1–69, IGHV1–18, IGHV1–2, and IGHV1–46 appear to be consistently decreased in usage frequency in downstream isotypes in both children and adults. We note that all of the IGHV genes that are favored in this selection are of the IGHV3 group, whereas those that are progressively lost with downstream isotype switching are from the IGHV1 and IGHV4 groups.
B cells specific for vaccine antigens show isotype switching and SHM accumulation
To evaluate SHM and isotype data from clones specific for vaccine and pathogen-related antigens in the pediatric antibody repertoires, we generated antibody phage display libraries of single-chain variable fragment (scFv) antibodies from the first visit blood samples of two children who had received diphtheria-tetanus-pertussis (DTaP) vaccination. Phage were panned against tetanus toxoid (TT) antigen for two rounds of enrichment, then isolated as individual phage clones, sequenced, and tested by ELISA for binding to TT (Fig. 3; fig. S1, A and B). Members of the TT-specific antibody clones were detected in the total IgH repertoire data from each subject. TT-binding clones included examples expressing only a single isotype category (IgM, IgA subclasses, or IgG subclasses) as well as a clone containing IgM, IgG and IgA class-switched members. Clonal lineage trees from clones that contained multiple isotypes are shown in fig. S2. Notably, even the IgM-containing clones showed members that had acquired SHM, although the isotype-switched clone members generally had higher SHM frequencies than the IgM. These SHM frequencies were within the ranges observed for each of these isotypes for the total IgH repertoires from the children (Fig. 1B).
Fig. 3. SHM frequencies in B cell clones with members that bind TT in antibody phage display.
scFv antibody phage display libraries were generated from IgH, Igκ, and Igλ transcripts from subjects 2072 and 2074. Members of the TT-specific clonal lineages were identified from phage clones in the total IgH repertoire sequence data sets generated for each antibody isotype from the PBMCs of the children, and the isotype and SHM percentages in the IGHV gene of all clone members were plotted. Each dot represents a read from a TT-binding clone member, with each clone plotted in a different color.
IgM and IgD SHM frequencies are correlated with higher rates of upper respiratory infection
We evaluated the IgH repertoires of the children based on two categories of infectious disease: 1) illnesses with fever, diarrhea, and/or vomiting (FDV), and 2) upper respiratory infection (URI) defined by the presence of cold, cough, and/or ear-pulling. SHM frequencies were determined from the final blood sample obtained from the child. For each category of illness, we calculated the proportion of surveyed days during which the child showed signs of illness up to the final blood sample. We observed that having a higher proportion of URI days but not FDV days correlated with more SHM in IgM and IgD subclasses (p=0.04 and p=0.002, respectively, with IgM adjusted R-value=0.06 and IgD adjusted R-value=0.16; Fig. 4, A and B). In contrast, IgG and IgA SHM showed no relationship with infectious disease days. For both healthy children (sick for fewer than 10% of days surveyed) and those with higher frequencies of illness (sick for 10% or more of days surveyed), the SHM in IgG showed evidence of antigen-driven selection, as gauged by an excess of non-synonymous mutations in complementarity-determining region (CDR) sequences beyond the expected amount for random mutations in an antibody at the observed SHM frequency (fig. S3).
Fig. 4. IgM and IgD SHM are correlated with days of URI.
Each point represents a STORK subject (n=51) and shows the average SHM percentage for the IGHV gene segment of expressed antibodies of the indicated isotype as a function of the proportion of (A) FDV and (B) URI sick days. SHM data was from each subject’s final blood draw. SHM for each isotype in each subject were summarized as the median SHM of reads expressed as the indicated isotype within each clone, then taking the mean SHM percentage over all clones. Linear regression lines were plotted with 95% confidence intervals.
Microbicide usage and household cleanliness affect switched isotype SHM frequencies
STORK households were evaluated by investigators using a standardized scoring of cleanliness that was evaluated as the average of 4 main axes, scored from 0 (cleaner) to 3 (least clean): i) floors and carpets; ii) walls, visible furniture, and windowsills; iii) bathroom and toilet; and iv) kitchen. Decreased household cleanliness was associated with lower IgA2 SHM (p=0.029) but showed no notable association with cleanliness otherwise. We note that the overall variability in cleanliness among households was low (Fig. 5A).
Fig. 5. Household cleanliness and TC usage show differing associations with IGH SHM.

(A) Data show results for 48 households. Household cleanliness was assessed on 4 axes with scores given from 0 (clean) to 3 (very dirty). Linear regression lines are shown with 95% confidence intervals in gray. (B) Forty-nine households were randomized to TC usage (n=27) or avoidance (n=22). (C) Children randomized to TC and non-TC groups, and the effect on FDV and URI sickday proportions. SHM for each isotype in each subject were summarized as the median SHM of reads expressed as the indicated isotype within each clone, then taking the mean SHM frequency over all clones. Wilcoxon-Mann-Whitney was used to test for significance.
Some prior reports have described an association between the use of commercial triclosan- and triclocarban- TC-containing microbicides and the development of allergic disease in children (30, 31) and in a mouse model of asthma (32). Households in the STORK intervention had been randomized to receive either TC-containing (n=27) or non-TC containing (n=22) household and personal cleaning products, as previously described (two households chose not to participate in the randomization portion of the study) (27). Children randomized to the TC arm had higher SHM percentages in all IgG subclasses as well as in IgD (Fig. 5B). In addition, IgE SHM was significantly higher in the non-TC group (p=0.024) and remained significant even after removal of data from allergic children from the analyses (p=0.046). Interestingly, children randomized to the TC-using group experienced fewer days with FDV compared to children in the TC-avoiding group. Randomization to TC usage did not affect the rate of URI (Fig. 5C).
IgE SHM is significantly elevated in subjects with eczema or allergy.
We next evaluated SHM frequencies in each isotype for the STORK children with eczema or allergic diseases (asthma, hives, food allergy, and/or eczema diagnosed by a physician and recorded in the medical chart) compared to non-allergic children (Fig 6). IgG and IgA SHM did not differ by eczema status (Fig. 6A) or other allergies (Fig. 6B). However, IgE SHM was elevated in subjects with eczema and/or other allergies (p=0.030 and p=0.017, respectively). We found that SHM in IgE showed evidence of antigen-driven selection, assessed as an excess of non-synonymous mutations in IgE CDR sequences compared to a random mutation model without selection (fig. S4, A and B). In addition, IgD SHM was significantly lower in children with eczema and/or other allergies (p=0.027). We considered whether previously described allergy-protective environmental factors might contribute to the differences in IgE SHM frequencies. Of the subjects studied here (n=51), 16 children came from households with one or more pets (i.e. cats, dogs, other small mammals, birds, reptiles, and/or fish). We did not observe any significant difference in IgE SHM percentages between children with pets in the home compared to children with no pets (fig. S5, p=0.213).
Fig. 6. STORK subjects with eczema and other allergies show increased IgE SHM.
IgH repertoires from children with known allergy (asthma, hives, food allergy, and/or eczema; table S2) were analyzed. In (A), subjects with and without eczema were compared (n=14 and n=37, respectively). In (B), subjects with any allergy (n=17) were grouped as having allergy symptoms and compared to subjects with no allergic symptoms (n=34). SHM for each isotype in each subject were summarized as the median SHM of reads expressed as the indicated isotype within each clone, then taking the mean SHM percentage over all clones. p values were determined using Wilcoxon-Mann-Whitney test.
Isotypes expressed by other members of IgE+ B cell clones differ between healthy and allergic subjects
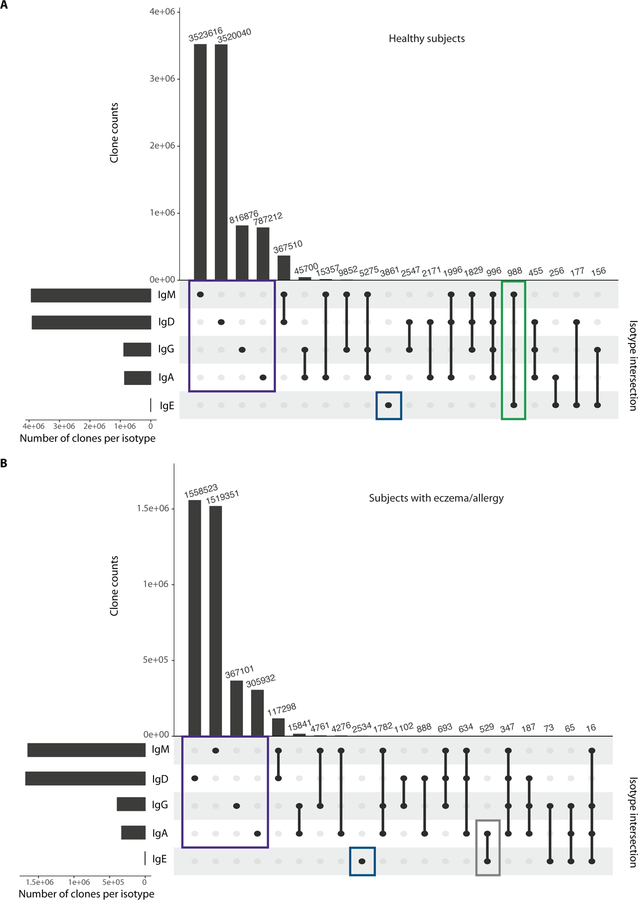
HTS of IgH repertoires can detect multiple B cells belonging to a clone, potentially revealing intra-clonal isotype switching. We hypothesized that there might be differences in the isotypes expressed by clones containing IgE members in healthy subjects compared to eczematous or allergic children, related to the mechanism of IgE development. IgM was the most common isotype found within IgE+ clonal lineages in healthy children, whereas IgA was the most common isotype within IgE+ clonal lineages of those with eczema or allergic diagnoses (Fig. 7, A and B). We note, however, that there were also clones containing IgE and IgA in healthy children. Some representative lineage trees of clones containing IgE members are shown in fig. S6. These data, together with the SHM frequencies measured in IgE, are consistent with healthy children more often having IgE+ clone members derived from direct isotype switching, while eczematous or allergic individuals more often generate IgE+ clone members by indirect switching, potentially from IgA1-expressing clone members.
Fig. 7. B cell clones from healthy subjects most commonly co-express IgE and IgM, while allergic subjects co-express IgE and IgA.
UpSet plot showing the size of set intersections for clones expressing IgM, IgD, IgA, IgG, and IgE. Bars indicate the number of clones that express the given isotype(s). The vertical lines that connect the points represent set intersections. Analyses were performed on (A) 34 healthy subjects and (B) 17 subjects with eczema or allergies. Clones expressing only a single isotype are shown: IgM, IgD, IgA, or IgG (purple boxes), and IgE-only clones (dark blue boxes). The green box shows clones in healthy children that contain IgE+ and IgM+ members, whereas the grey box shows B cell clones from children with eczema or allergies that contain both IgE+ and IgA+ members. The 20 most frequent intersections are shown for both healthy and allergic children.
Discussion
Understanding the effects of environmental exposures on childhood immune system development has been constrained by the difficulty in obtaining longitudinal blood samples from birth cohorts, and the small blood volumes that can be collected. Clinical findings such as the protection from peanut allergy provided by feeding peanut-containing food to infants in the first year of life in the LEAP trial (15, 16) and the importance of the skin barrier in preventing IgE-mediated allergic sensitization (17, 18) have spurred efforts to better understand the factors influencing adaptive immunity in infants and children. We used HTS of IgH gene rearrangements to obtain detailed longitudinal data from thousands of distinct B cell clones per child, with the caveat that our data are derived entirely from peripheral blood B cells, and thus cannot address immune development in other tissue sites. From the limiting volume of blood available in each sample, we prioritized capturing full IgH repertoire data from the children rather than sorting and isolating individual subsets of B cells defined phenotypically, or sorting rare antigen-specific B cells.
STORK children showed a consistent increase in SHM during the first three years of life for all isotypes except IgE, which showed large variation between individuals. By three years of age, IgG and IgA SHM reached approximately 60–75% of adult SHM frequencies, highlighting the rapid shaping of the antigen-experienced B cell repertoire during early childhood, and the limited changes during adulthood and old age. These SHM frequencies were correlated with the clonality of B cells measured in the children compared to adults, consistent with less overall accumulated antigen-driven clonal expansion in the children. Infectious diseases causing URI symptoms correlated with IgM and IgD SHM percentages, suggesting that these exposures drive SHM of antigen-experienced IgM+ and/or IgD+ B cells. The lack of correlation between IgG SHM and URI symptoms or IgA SHM and GI symptoms was somewhat unexpected since these antibody subclasses are thought to play a major role in protective immunity against viruses and pathogenic bacteria. However, many other antigenic exposures occur during the first three years of life, so it is possible that pathogenic organisms contribute only a portion of the increases in IgG and IgA SHM. Notably, the B cell clones specific for TT that we identified by antibody phage display included not only isotype-switched examples, but also clones expressing only IgM that showed a wide range of SHM frequencies, highlighting an important role for IgM in the early childhood responses to this vaccine antigen.
Other effects of selection in shaping the antibody repertoire were seen in a striking correlation between the IGHV gene usage frequency and antibody isotype subclass, when the subclasses were examined in their chromosomal ordering. V genes of the VH1 and VH4 families showed a consistent and progressive decrease in usage fraction in B cells that have switched to progressively further downstream isotypes, while VH3 family genes showed the opposite pattern. These data suggest that class switching to downstream isotypes is coupled to progressively stronger selection for or against particular VH genes. This pattern was particularly clear in the infant and young child repertoires but was also readily detectable and consistent in adult specimens. Whether these changes in usage frequency are related to propensity for autoreactivity, antibody stability, or other phenotypic features of the B cells expressing these V genes, will require further targeted study.
Some antibody isotypes are preferentially used in responses to particular pathogens; notably, anti-carbohydrate antibodies such as those targeting bacterial capsular antigens are often IgG2 in humans. STORK children showed increased usage of the IgG1 subclass and decreased IgG2 compared to adults, consistent with prior clinical observations of impaired responses against polysaccharide-only vaccines in infants and young children (33, 34). The decreased fraction of IgG2+ B cells did not vary markedly between age one and three, suggesting that IgG2+ B cell frequencies increase toward adult proportions later in childhood. Prior histologic reports describe decreased marginal zone B cells, thought to produce anti-carbohydrate antibodies, in the lymph nodes and spleen of infants (33, 35). However, while splenic marginal B cell populations increase to adult SHM frequencies by two years of life (33), we find that circulating IgG2+ B cells have not yet increased, indicating this circulating B cell pool is slower to develop.
Surprisingly, STORK children from households randomized to receive TC-containing bactericidal products had higher antibody SHM in all IgG subclasses compared to children from non-TC households. We expected less IgG SHM in the TC group, under the simple hypothesis that the killing of environmental or pathogenic bacteria would decrease antigenic exposures. TC effects on microbiota are inconsistent in the literature: some studies report TC perturbations of microbial communities (36), others found no effect on microbial species richness or overall diversity (37). In the full STORK cohort, children in TC households had fewer medical visits where antibiotics were prescribed for possible or definite bacterial infection, but there was no change in infectious disease incidence (38). In our sub-study, we observed that the children from TC households had lower FDV rates (Fig. 3) but unchanged rates of URI (typically associated with viral pathogens). Increased IgG SHM in the TC group may be related to changes in commensal bacteria, giving rise to new antigenic exposures. Interestingly, children in the TC group had significantly lower IgE SHM than non-TC children (p=0.024), even when children with diagnosed allergies were removed from the analysis (n=9 in the TC group and n=7 in the non-TC group). Effects of TC-containing products on rates of allergy are unresolved in the literature (30, 31, 39).
Impaired skin barriers in infants or children with eczema or mutations in filaggrin genes are implicated in allergic sensitization to food allergens (17, 18, 40), but the cellular mechanisms for this are unclear. We find that children with eczema and other allergic conditions have significantly elevated IgE SHM frequencies compared to healthy children (eczema-only: p=0.03; eczema/allergy: p=0.017). Allergen-binding IgE antibodies in allergic individuals have elevated frequencies of SHM (22, 25, 41). IgE+ B cells can be formed by isotype switching directly from IgM+ precursors, or indirectly from B cells that express IgG or IgA (12, 13). Indirect switching should produce IgE antibodies with higher SHM, as IgG and IgA1 antibodies typically have higher SHM frequencies than IgM. We find that the IgE antibodies observed in STORK children show evidence of affinity maturation. Children with eczema or allergic symptoms show greater frequencies of clones containing IgE+ members and IgA+ members, and also have elevated frequencies of SHM in IgE, whereas non-allergic children showed higher frequencies of IgM+ B cells in the same clones with IgE+ members. These findings could indicate that the impaired barrier function of eczematous skin may increase the rate of B cell indirect isotype switching to IgE from precursors including IgA+ B cells, yielding higher SHM in IgE+ B cells. Easier access of IgE+ B cells to environmental allergens through the impaired barrier of the eczematous skin could, as others have suggested, eventually contribute to allergic sensitization and the development of other allergic disease symptoms (42–44). Oral ingestion of peanut beginning in the first year of life has been shown to protect against peanut allergy (15, 16). Future studies should address whether oral ingestion alters the repertoires of allergen-specific IgE antibodies compared to those seen in children whose only exposure is via a disrupted skin barrier. An analysis of the IgE+ B cell clones from such a study would help to clarify which of these isotype-switching and SHM phenotypes is most associated with development of symptomatic allergy in childhood.
In summary, the detailed epidemiological and clinical data gathered from the STORK birth cohort provided an opportunity to test associations between infectious disease and other environmental exposures on antibody repertoire development. IgE+ B cells show elevated SHM in children with eczema, suggesting a key mechanistic link between impaired skin barrier function and IgE SHM that could contribute to allergic sensitization and disease. Replication of these results in independent and larger infant cohorts will be necessary to confirm and extend our findings and possibly reveal additional associations between childhood B cell development, environmental exposures, and protective or pathogenic humoral immune responses.
Materials and Methods
Study design
The STORK prospective observational birth cohort study was designed to assess the effect of infectious diseases on immune system development and growth, with a nested randomized intervention of TC-containing wash products to determine whether TCs decreased infection during the babies’ first year (38). Yearly venipuncture blood samples from healthy children of approximately one through three years of age were evaluated from a subset of the full STORK cohort. B cell receptor repertoires amplified from PBMCs were analyzed with high-throughput sequencing, with one library generated for each isotype category (IgM, IgD, IgG, IgA, and IgE) per sample. Healthy adult blood was used as a control. Recruitment of STORK subjects, documentation of informed consent, collection of blood specimens, and experimental measurements were carried out with Institutional Review Board approval (IRB-17756) from both Stanford University and the Santa Clara Valley Medical Center (SCVMC). Data included assessment of household cleanliness every four months, illness every few weeks, and non-blinded randomized intervention of triclosan and triclocarban-containing (TC) cleaning products. Primary data are reported in data file S1.
STORK sub-cohort details
The multiethnic cohort of mothers and their babies was followed from the second trimester of pregnancy through the babies’ third birthday (27). Healthy women aged 18–42 years with single-fetus pregnancies who wished to participate were enrolled. The total number of enrolled babies was 136. Relative to the overall population of the United States, there is an over-selection of Hispanic families, and families of lower socioeconomic status in the overall cohort as well as in the sub-cohort presented here (table S1). The only additional criterion affecting the inclusion of infants in this sub-study was the willingness of families to participate in the blood-based immunological study and consent to blood collection from the infant or child. Blood was collected by venipuncture in two EDTA tubes (total of 6–7 mL per baby). About 1–2 mL of blood from each child was allocated to the present study and processed (plasma and PBMC isolation) within 24 hours of blood draw. Participants in this study were representative of the larger cohort (table S1). We included 51 children ranging in age from 49 weeks (first visit) to 160 weeks (third visit), with up to three longitudinal time points per child (blood samples: n=93) (table S2). Blood was collected at one time point for 19 babies, two time points for 22 children, and all three time points for ten children.
Households were visited every 4 months. At the initial household visit, the mothers were given a detailed household questionnaire including demographic information, information on household structure (including inhabitants, rooms, pets, vermin), and indicators of socioeconomic status. Pets from subjects studied here were cats, dogs, other small mammals, birds, reptiles, and/or fish. The households with only fish as pets (n=2) were excluded from the pet-containing group (n=16). In addition, the households are assessed for cleanliness and given a score, which is calculated based on four axes, each scored from 0 for clean, to 3 for dirty. The 4 axes measure cleanliness of: i) floors and carpets; ii) walls, visible furniture, and windowsills; iii) bathroom and toilet; and iv) kitchen. Forty-eight households were evaluated for cleanliness on all 4 axes while 3 households were evaluated on 3 axes. We calculated the final score by adding the scores from the evaluation and dividing with the number of evaluated axes. Weekly automated telephone or email surveys gathered during the child’s first years of life were used to assess for signs of infectious disease in the children and recorded total days per week of vomiting, fever, diarrhea, and cold/cough/ear-pulling, the latter an indication of upper respiratory infection (URI). If the children were reported as healthy, parents were asked about milk intake and amount of sleep, to ensure that equal amounts of time were spent answering questions regardless of the child’s health status. Only reported physician diagnosed instances of eczema, hives, asthma, and food allergy were used for atopic disease classification in this study.
For the non-blinded randomized intervention of triclosan and triclocarban-containing (TC) household and personal cleaning products, 60 households were randomized to receive TC-containing and 72 households received non-TC containing products. In the sub-cohort of STORK children presented here, there were 27 TC households and 22 non-TC households.
Statistical analysis
Given the rarity of IgE+ B cells in blood, we combined the sequence data from all available time points for each child for the household cleanliness and TC usage analyses (Fig. 5), and the eczema and allergy analyses (Fig. 6 and fig. S2A, S3). The distribution of IgE SHM frequencies, which showed no systematic increase with age, supported the approach of combining data across all available time points for SHM analysis. For the UpSet plot, we combined clone data from all children with no allergic disease and clone data from all children with eczema or allergic disease. Graphics were produced with R (45) using the ggplot2 package (46) and UpSetR (47). Statistical analyses were also performed using R, and p values were calculated by one-sided Wilcoxon-Mann-Whitney test, or where indicated, two-sided (using the R function wilcox.test). Box-whisker plots show median (horizontal line), interquartile range (box), and 1.5 times the interquartile range (whiskers). Linear regression lines were plotted with 95% confidence intervals calculated using the ggplot2 function stat_smooth.
Supplementary Material
Acknowledgements:
We thank the families of the STORK cohort for their participation and commitment to this research. We would also like to thank Thomas D. Haggerty, the late Shu-Fang Yang, and Ankur Mathur for excellent technical assistance, and Lars Peter Nielsen from Statens Serum Institut, Denmark, for gifting reagents.
Funding:
The study was supported by Stanford Center for Clinical and Translational Education and Research (SCCTER) NIH grant 1UL1 RR025744 from the National Center for Research Resources, NIH grant U19 AI090019, grants from the Ellison Medical Foundation to M.M.D. and S.D.B., the Child Health Research Institute and the Stanford NIH-NCATS-CTSA (grant no. UL1 TR001085) to S.C.A.N., NIH grant 1R01 AI125567 to S.D.B., a gift from Robert C. and Mary Ellen Waggoner, Ulla og Mogens Folmer Andersens Fond (S.C.A.N.), a Big Data for Human Health Seed Grant from the Li Ka Shing Foundation (S.D.B., S.A.J., and R.T.), and an endowment to S.D.B. from the Crown Family Foundation.
Footnotes
List of Supplementary Materials
Fig. S1. Tetanus toxoid-binding scFvs confirmed by ELISA.
Fig. S2. Clonal lineages of tetanus toxoid-binding B cells.
Fig. S3. Proportion of IgG+ clone members that show evidence of antigen selection.
Fig. S4. Proportion of IgE+ clone members that show evidence of antigen selection.
Fig. S5. Household pet exposure and antibody SHM percentages.
Fig. S6. Clonal lineage trees with IgE-containing members.
Table S1: Demographic and household characteristics in this sub-cohort (n=51) and the full cohort (n=136).
Table S2: Clinical and household information for STORK children in this study.
Table S3: Primer sequences for heavy and light chain V(D)J amplicon generation for phage display.
Data file S1. Primary data
Competing interests: The authors declare that they have no competing interests. S.D.B. has consulted as an expert witness for Regeneron and Sanofi on topics unrelated to the present study.
Data and materials availability:
All data associated with this study are present in the paper or Supplementary Materials. The IgH repertoire data for this study have been deposited to SRA as BioProjects with the following accession numbers: STORK data PRJNA503602, and healthy adult control data PRJNA491287.
References and Notes:
- 1.von Mutius E, Vercelli D, Farm living: effects on childhood asthma and allergy. Nat. Rev. Immunol 10, 861–868 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Lundell AC, Bjornsson V, Ljung A, Ceder M, Johansen S, Lindhagen G, Tornhage CJ, Adlerberth I, Wold AE, Rudin A, Infant B cell memory differentiation and early gut bacterial colonization. J. Immunol 188, 4315–4322 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Rudin A, Lundell AC, Infant B cell memory and gut bacterial colonization. Gut Microbes 3, 474–475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjogren YM, Tomicic S, Lundberg A, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E, Jenmalm MC, Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy 39, 1842–1851 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Holbreich M, Genuneit J, Weber J, Braun-Fahrlander C, Waser M, von Mutius E, Amish children living in northern Indiana have a very low prevalence of allergic sensitization. J. Allergy Clin. Immunol 129, 1671–1673 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J, Gustafsson A, Bernhardsson AK, Zhang C, Bohlin K, Brodin P, Stereotypic Immune System Development in Newborn Children. Cell 174, 1277–1292 e1214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L, The biology of IGE and the basis of allergic disease. Annu. Rev. Immunol 21, 579–628 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Branum AM, Lukacs SL, Food allergy among children in the United States. Pediatrics 124, 1549–1555 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Jackson KD, Howie LD, Akinbami LJ, NCHS Data Brief, Number 121, May 2013. NCHS data brief, 1–8 (2013). [PubMed] [Google Scholar]
- 10.Bock SA, Sampson HA, Food allergy in infancy. Pediatr. Clin. North Am 41, 1047–1067 (1994). [DOI] [PubMed] [Google Scholar]
- 11.de Silva IL, Mehr SS, Tey D, Tang ML, Paediatric anaphylaxis: a 5 year retrospective review. Allergy 63, 1071–1076 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Gould HJ, Beavil RL, Vercelli D, lgE isotype determination: ε-germline gene transcription, DNA recombination and B-cell differentiation. Br. Med. Bull, (2000). [DOI] [PubMed] [Google Scholar]
- 13.Gould HJ, Takhar P, Harries HE, Durham SR, Corrigan CJ, Germinal-centre reactions in allergic inflammation. Trends Immunol 27, 446–452 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Gern JE, Reardon CL, Hoffjan S, Nicolae D, Li Z, Roberg KA, Neaville WA, Carlson-Dakes K, Adler K, Hamilton R, Anderson E, Gilbertson-White S, Tisler C, Dasilva D, Anklam K, Mikus LD, Rosenthal LA, Ober C, Gangnon R, Lemanske RF Jr., Effects of dog ownership and genotype on immune development and atopy in infancy. J. Allergy Clin. Immunol 113, 307–314 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, Brough HA, Phippard D, Basting M, Feeney M, Turcanu V, Sever ML, Gomez Lorenzo M, Plaut M, Lack G, Team LS, Randomized trial of peanut consumption in infants at risk for peanut allergy. N. Engl. J. Med 372, 803–813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Toit G, Sayre PH, Roberts G, Sever ML, Lawson K, Bahnson HT, Brough HA, Santos AF, Harris KM, Radulovic S, Basting M, Turcanu V, Plaut M, Lack G, Immune Tolerance Network L-OST, Effect of Avoidance on Peanut Allergy after Early Peanut Consumption. N. Engl. J. Med 374, 1435–1443 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Lack G, Fox D, Northstone K, Golding J, Avon P Longitudinal Study of, T. Children Study, Factors associated with the development of peanut allergy in childhood. N. Engl. J. Med 348, 977–985 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Brough HA, Liu AH, Sicherer S, Makinson K, Douiri A, Brown SJ, Stephens AC, Irwin McLean WH, Turcanu V, Wood RA, Jones SM, Burks W, Dawson P, Stablein D, Sampson H, Lack G, Atopic dermatitis increases the effect of exposure to peanut antigen in dust on peanut sensitization and likely peanut allergy. J. Allergy Clin. Immunol 135, 164–170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strachan DP, Hay fever, hygiene, and household size. BMJ 299, 1259–1260 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson KJL, Wang Y, Collins AM, Human immunoglobulin classes and subclasses show variability in VDJ gene mutation levels. Immunol. Cell Biol 92, 729–733 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Davies JM, O’Hehir RE, Immunogenetic characteristics of immunoglobulin E in allergic disease. Clin. Exp. Allergy 38, 566–578 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Janezic A, Chapman CJ, Snow RE, Hourihane JO, Warner JO, Stevenson FK, Immunogenetic analysis of the heavy chain variable regions of IgE from patients allergic to peanuts. J. Allergy Clin. Immunol 101, 391–396 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Lim A, Luderschmidt S, Weidinger A, Schnopp C, Ring J, Hein R, Ollert M, Mempel M, The IgE repertoire in PBMCs of atopic patients is characterized by individual rearrangements without variable region of the heavy immunoglobulin chain bias. J. Allergy Clin. Immunol 120, 696–706 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hoh RA, Joshi SA, Liu Y, Wang C, Roskin KM, Lee JY, Pham T, Looney TJ, Jackson KJL, Dixit VP, King J, Lyu SC, Jenks J, Hamilton RG, Nadeau KC, Boyd SD, Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J. Allergy Clin. Immunol 137, 157–167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahlke I, Nott DJ, Ruhno J, Sewell WA, Collins AM, Antigen selection in the IgE response of allergic and nonallergic individuals. J. Allergy Clin. Immunol 117, 1477–1483 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Jackson KJL, Davies J, Chen Z, Gaeta BA, Rimmer J, Sewell WA, Collins AM, IgE-associated IGHV genes from venom and peanut allergic individuals lack mutational evidence of antigen selection. PLoS One 9, e89730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ley C, Sanchez Mde L, Mathur A, Yang S, Sundaram V, Parsonnet J, Stanford’s Outcomes Research in Kids (STORK): a prospective study of healthy pregnant women and their babies in Northern California. BMJ Open 6, e010810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein U, Rajewsky K, Kuppers R, Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med 188, 1679–1689 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duchamp M, Sterlin D, Diabate A, Uring-Lambert B, Guerin-El Khourouj V, Le Mauff B, Monnier D, Malcus C, Labalette M, Picard C, B-cell subpopulations in children: National reference values. Immun Inflamm Dis 2, 131–140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savage JH, Matsui EC, Wood RA, Keet CA, Urinary levels of triclosan and parabens are associated with aeroallergen and food sensitization. J. Allergy Clin. Immunol 130, 453–460 e457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertelsen RJ, Longnecker MP, Lovik M, Calafat AM, Carlsen KH, London SJ, Lodrup Carlsen KC, Triclosan exposure and allergic sensitization in Norwegian children. Allergy 68, 84–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson SE, Franko J, Kashon ML, Anderson KL, Hubbs AF, Lukomska E, Meade BJ, Exposure to triclosan augments the allergic response to ovalbumin in a mouse model of asthma. Toxicol. Sci 132, 96–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timens W, Boes A, Rozeboom-Uiterwijk T, Poppema S, Immaturity of the human splenic marginal zone in infancy. Possible contribution to the deficient infant immune response. J. Immunol 143, 3200–3206 (1989). [PubMed] [Google Scholar]
- 34.Rijkers GT, Sanders EA, Breukels MA, Zegers BJ, Infant B cell responses to polysaccharide determinants. Vaccine 16, 1396–1400 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Luscieti P, Hubschmid T, Cottier H, Hess MW, Sobin LH, Human lymph node morphology as a function of age and site. J. Clin. Pathol 33, 454–461 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Looft T, Allen HK, Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 3, 463–467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole AC, Pischel L, Ley C, Suh G, Goodrich JK, Haggerty TD, Ley RE, Parsonnet J, Crossover Control Study of the Effect of Personal Care Products Containing Triclosan on the Microbiome. mSphere 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ley C, Sundaram V, Sanchez ML, Desai M, Parsonnet J, Triclosan and triclocarban exposure, infectious disease symptoms and antibiotic prescription in infants-A community-based randomized intervention. PLoS One 13, e0199298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy SB, Antibacterial household products: cause for concern. Emerg. Infect. Dis 7, 512–515 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brough HA, Simpson A, Makinson K, Hankinson J, Brown S, Douiri A, Belgrave DC, Penagos M, Stephens AC, McLean WH, Turcanu V, Nicolaou N, Custovic A, Lack G, Peanut allergy: effect of environmental peanut exposure in children with filaggrin loss-of-function mutations. J. Allergy Clin. Immunol 134, 867–875 e861 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coker HA, Durham SR, Gould HJ, Local somatic hypermutation and class switch recombination in the nasal mucosa of allergic rhinitis patients. J. Immunol 171, 5602–5610 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Marenholz I, Nickel R, Ruschendorf F, Schulz F, Esparza-Gordillo J, Kerscher T, Gruber C, Lau S, Worm M, Keil T, Kurek M, Zaluga E, Wahn U, Lee YA, Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J. Allergy Clin. Immunol 118, 866–871 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Spergel JM, Paller AS, Atopic dermatitis and the atopic march. J. Allergy Clin. Immunol 112, S118–127 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Novak N, Bieber T, Leung DY, Immune mechanisms leading to atopic dermatitis. J. Allergy Clin. Immunol 112, S128–139 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Team RC, A language and environment for statistical computing. R Foundation for Statistical Computing (Vienna, Austria, 2015). [Google Scholar]
- 46.Wickham H, ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; New York, 2016). [Google Scholar]
- 47.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H, UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 20, 1983–1992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA, Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia 17, 2257–2317 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Roskin KM, Simchoni N, Liu Y, Lee JY, Seo K, Hoh RA, Pham T, Park JH, Furman D, Dekker CL, Davis MM, James JA, Nadeau KC, Cunningham-Rundles C, Boyd SD, IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci. Transl. Med 7, 302ra135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Looney TJ, Lee JY, Roskin KM, Hoh RA, King J, Glanville J, Liu Y, Pham TD, Dekker CL, Davis MM, Boyd SD, Human B-cell isotype switching origins of IgE. J. Allergy Clin. Immunol 137, 579–586 e577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magoc T, Salzberg SL, FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye J, Ma N, Madden TL, Ostell JM, IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 41, W34–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson EH, Measurement of Diversity. Nature 163, 688–688 (1949). [Google Scholar]
- 54.Collins AM, Ikutani M, Puiu D, Buck GA, Nadkarni A, Gaeta B, Partitioning of rearranged Ig genes by mutation analysis demonstrates D-D fusion and V gene replacement in the expressed human repertoire. J. Immunol 172, 340–348 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Rondot S, Koch J, Breitling F, Dubel S, A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol 19, 75–78 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G, By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol 222, 581–597 (1991). [DOI] [PubMed] [Google Scholar]
- 57.Mole CM, Bene MC, Montagne PM, Seilles E, Faure GC, Light chains of immunoglobulins in human secretions. Clin. Chim. Acta 224, 191–197 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Rader C, Steinberger P, Barbas CF, Barbas C. F. r., Burton DR, Scott JK, Silverman GJ, Eds. (Cold Spring Harbor Laboratory Press, 2001), pp. 10.12–10.15. [Google Scholar]
- 59.Bradbury ARM, Marks JD, in Phage Display, Clackson T, Lowman HB, Eds. (Oxford University Press, 2009), pp. 277–279. [Google Scholar]
- 60.Li W, Godzik A, Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.