Abstract
The biology of primary lytic Kaposi’s sarcoma-associated herpesvirus (KSHV) infection is still not well understood, which is largely attributed to the lack of cell lines permissive to robust lytic KSHV infection in vitro. Our study demonstrates that primary human dermal lymphatic microvascular endothelial cells (HDLMEC) support lytic KSHV replication following de novo infection, resulting in robust KSHV production, indicating that HDLMECs are suitable for studying the regulation of primary lytic KSHV infection. Importantly, by utilizing lytically infected HDLMECs, we show for the first time that the KSHV latent genes LANA and viral cyclin are required for lytic replication during de novo lytic infection, a function of these latent genes that has not yet been recognized. Since Kaposi’s sarcoma is considered to be originated from infected lymphatic endothelial cells, HDLMECs represent a valuable in vitro cell culture model for investigating lytic KSHV infection, which has been understudied in KSHV pathogenesis.
Keywords: KSHV, lytic infection, lymphatic endothelial cells, latent genes, herpesvirus
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a human gammaherpesvirus, which can cause a number of cancers that are derived from either latently infected lymphatic endothelial cells such as Kaposi’s sarcoma (KS) or latently infected B cells such as primary effusion lymphoma (Cesarman, 2011; Mesri et al., 2010). While latent infection is essential for the development of KSHV-associated tumors, the lytic phase is also crucial to many aspects of the pathogenesis of KSHV infection (Cavallin et al., 2014; Giffin and Damania, 2014). The lytic cycle, which is controlled by the viral replication and transcription activator protein called RTA, can be initiated either following primary infection or by reactivation from latently infected cells (Duus et al., 2004; Gao et al., 2003; Nakamura et al., 2003; Sun et al., 1998). The viral proteins expressed during the lytic phase have a wide range of functions besides driving virus production. They can regulate cell survival, cell cycle, and viral immune evasion, while some of the lytic proteins possess oncogenic activity whose expression is often detected in a subset of cancer cells in KSHV-associated tumors, suggesting that these lytic proteins may also contribute to the transformation and/or persistence of infected cells (Cavallin et al., 2014; Giffin and Damania, 2014). Further evidence that the lytic cycle of KSHV plays a role in viral oncogenesis came from the discovery that anti-herpesviral drugs that block lytic replication, but not latent replication, resulted in a reduction of KS lesion size in patients and an impaired progression to KS for those at risk (Martin et al., 1999; Mocroft et al., 1996). Finally, it is also appreciated that the continual lytic replication of KSHV may produce virus to re-seed a latently infected cell population, which can promote the growth of KSHV-associated cancers (Grundhoff and Ganem, 2004). Therefore, studying KSHV genes in the regulation of the lytic cycle is crucial to fully understand KSHV pathogenesis.
Since the default pathway of KSHV infection in vitro is the establishment of viral latency, the function of viral genes in the lytic phase is mostly assessed during lytic reactivation, which is triggered by either RTA overexpression or chemical inducers in latently infected cells (Bechtel et al., 2003; Nakamura et al., 2003; Purushothaman et al., 2015). While these lytic reactivation cell culture models provided vast information about the role of viral factors in KSHV lytic cycle, they have limitations. For example, if viral latent genes may have critical roles during both the latent and lytic phases, they would be difficult to study in the lytic cycle because their role in latency could also affect lytic reactivation. Thus, it is imperative that we study the contribution of viral genes directly in lytic infection. Although oral epithelial cells and human umbilical vein endothelial cells (HUVEC) have been shown to support lytic replication to some degree following de novo infection, neither oral epithelial cells nor HUVECs proved to be permissive to robust KSHV lytic infection (Duus et al., 2004; Gao et al., 2003). Thus, we still lack a permissive cell culture model, which could be used to study primary lytic KSHV infection similarly to alpha- and beta-herpesviruses.
Recently, it has been reported that primary lymphatic endothelial cells (LEC) stably infected by KSHV for 2 weeks displayed not only latent, but also widespread lytic gene expression and a very low amount of virus production (Chang and Ganem, 2013). Since KSHV is known to undergo a temporary and limited lytic phase during de novo infection, even in cells in which KSHV ultimately establishes tight latency (Krishnan et al., 2004), we hypothesized that primary LECs may support a potent lytic viral replication program following de novo infection. This hypothesis prompted us to test whether or not primary LECs can support a more robust lytic KSHV replication and virus production following de novo infection, as opposed to stably infected LECs. In this report, we show that de novo KSHV infection of human dermal lymphatic microvascular endothelial cells (HDLMEC) results in robust lytic replication and virus production by 4 days postinfection. The quantity of infectious virions, which was produced from de novo infected HDLMECs, was at least 400 times greater than the amount of virus produced by stably infected LECs as reported previously (Chang and Ganem, 2013). Furthermore, we present evidence for the first time that the KSHV latent genes LANA and viral cyclin, in addition to their roles in regulating latency, are essential for KSHV replication during lytic infection.
Materials and Methods
Primary cells and cell lines
Primary human adult, dermal lymphatic microvascular endothelial cells (HDLMEC) were purchased from Lonza (CC-2810) and cultured in microvascular endothelial cell growth medium, which contained 5% FBS and a combination of growth factors (CC-3202). HDLMECs were used between passages 6 and 9 for the experiments. The 293T cells and the iSLKBAC16 cell lines, which were used for the production of wild type and the latent gene knockout BAC16 mutants, were maintained in DMEM containing 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. The TRExBCBL1-3xFLAG-RTA cell line was cultured in RPMI containing 10% Tet-approved FBS (Clontech), 100 U/ml penicillin, and 100 µg/ml streptomycin (Papp et al., 2018).
Virus Production and infection of HDLMECs
The construction of latent gene knockout BAC16 viruses and the establishment of their corresponding iSLK cell lines have been published previously (Toth et al., 2016). KSHV production has been described in detail in our previous study (Toth et al., 2017). To generate wild type and latent gene knockout KSHV, iSLKBAC16 cell lines were reactivated using 1 µg/ml doxycycline and 1 mM sodium butyrate. To produce KSHV from the TRExBCBL1-3xFLAG-RTA cell line, cells were treated with 1 µg/ml doxycycline. Four days after initiating lytic reactivation, the supernatants were collected and concentrated by ultracentrifugation. For infection, 1.5×105 HDLMECs were plated per well on six-well tissue culture plates. 24 hours later, the cells were infected by spinoculation (2000 rpm, 45 min, 30°C) at different multiplicities of infection (MOI). 2 hours later, the media was removed, the cells were washed with PBS, and fresh medium was added to the cells.
Microscopy and immunofluorescence analysis
At 48 hours postinfection (hpi), infected HDLMECs were fixed with 4% paraformaldehyde, permeabilized by 0.5% Triton X-100, washed twice with washing buffer (PBS with 0.2% Tween-20), and then incubated in blocking buffer (5% FBS, 0.2% fish skin gelatin, and 0.2% Tween-20 in PBS) for 30 min. Rabbit anti-ORF6 antibody and rabbit IgG (negative control) were diluted in blocking buffer and used to stain the cells for 2 hours at room temperature. Subsequently, the cells were washed three times with washing buffer and then incubated with anti-rabbit Alexa Fluor 568 antibody (Invitrogen) for 45 min at room temperature. Cells were washed with washing buffer three times and then stained with DAPI to visualize the nuclei. For GFP imaging and the immunofluorescence analyses, a Revolve fluorescence microscope (Echo Laboratories) was used.
Antibodies and immunoblot assay
Antibodies utilized in this study: mouse anti-Tubulin (GTU-88, Sigma), mouse anti-ORF45 (sc-53883, Santa Cruz), mouse anti-K8 (sc-57889, Santa Cruz), mouse anti-K8.1 (sc-65446, Santa Cruz), mouse anti-ORF26 (2F6B8) (NBP1-47357, Novus Biologicals). Rabbit anti-ORF6 was generously provided by Dr. Gary S. Hayward (Johns Hopkins University). HDLMECs were lysed in 2x Laemmli buffer (Bio-Rad) containing 2-mercaptoethanol, and protein concentration was measured by using a Pierce 660nm Protein Assay Kit. Proteins separated on a 10% acrylamide gel were transferred onto a PVDF membrane, which was then blocked with 5% non-fat dry milk in PBS with 0.1% Tween-20 (PBST), and incubated overnight with primary antibody at 4°C. The next day, the PVDF membrane was washed three times with PBST, incubated with HRP-linked secondary antibody for 1–2 hours at room temperature, washed three times in PBST, and then developed using SuperSignal West Femto (Thermo Scientific), and imaged on a Bio-Rad ChemiDoc Imager.
Quantification of viral RNA and DNA
Total RNA was extracted by Trizol (Sigma) from infected cells. cDNA was prepared from 500 ng of DNase I-treated RNA using the iScript cDNA Synthesis Kit (Bio-Rad). Viral mRNA expression was quantified by qPCR in a CFX96 real time PCR machine using SYBR Green supermix (Bio-Rad). Viral gene expression was normalized to the level of 18S using the 2−∆Ct method. To prepare DNA, KSHV-infected cells were lysed in RIPA buffer and sonicated in a Bioruptor Pico (Diagenode) for 30 sec. Total DNA was isolated using the phenol-chloroform extraction method. 10 ng of total DNA was measured in qPCR and the fold change in the viral DNA load was calculated by the 2−∆∆Ct method. Primer sequences have been published previously (Toth et al., 2016). An unpaired student’s two-tailed T test was used to determine statistical significance.
Flow cytometry to determine virus titer
To calculate the virus titer, KSHV-infected HDLMEC-derived supernatant was used to infect 293T cells. One day prior to infection, 2.5×105 293T cells were plated per well on six-well tissue culture plates. 293T cells were infected with 2 ml of media containing 0.4–1 ml of HDLMEC-derived supernatant using spinoculation. 24 hours after infection, the cells were harvested, fixed in 4% paraformaldehyde, and resuspended in FACS buffer (1% FBS and 4 mM EDTA in PBS). The percentage of GFP+ cells was quantified on a BD Accuri C6 flow cytometer (BD Biosciences), which was used to calculate the yield of KSHV as infectious units (IU)/ml.
Results and Discussion
Characterization of lytic KSHV infection in human dermal lymphatic microvascular endothelial cells
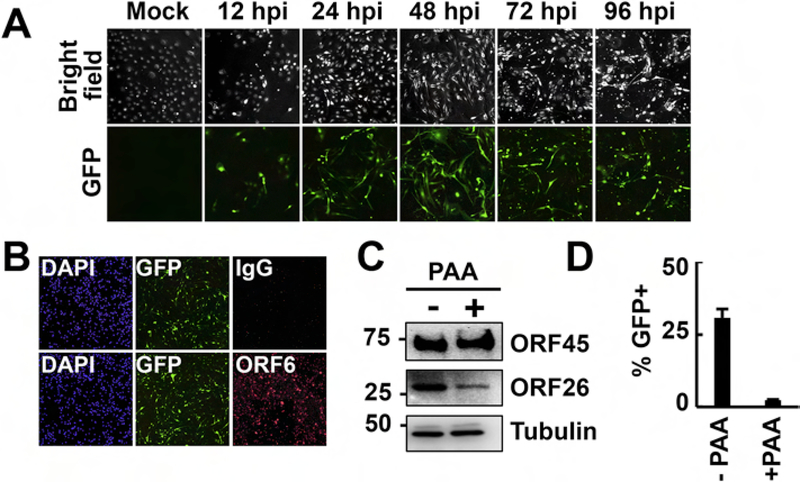
We infected HDLMECs with the KSHV clone BAC16, which constitutively expresses a GFP as a marker for the identification of infected cells (Brulois et al., 2012). While spindle cell conversion, a phenotype induced by KSHV infection of endothelial cells, could be observed by 24 hours postinfection (hpi), we also noticed an increasing number of cells that were rounded up by 72–96 hpi, which was reminiscent of a cytopathic effect due to advanced lytic infection (Fig. 1A). In fact, immunofluorescence analysis of BAC16-infected HDLMECs revealed that the early viral protein ORF6 was expressed in the majority of infected cells at 48 hpi, indicating lytic infection (Fig. 1B). To show whether or not HDLMECs indeed support lytic viral replication following de novo KSHV infection, we measured the virus production in infected HDLMECs at 72 hpi (Fig. 1C and D). Since the lytic replication and virus production of KSHV depends on viral DNA polymerase-mediated viral DNA synthesis, the virus production was tested both in the absence and the presence of 100 µM of viral DNA polymerase inhibitor phosphonoacetic acid (PAA). We found that while the PAA treatment did not affect the production of the early viral protein ORF45, it did greatly reduce the expression of late protein ORF26 (Fig. 1C) and virus production (Fig. 1D), confirming the lytic infection of HDLMECs by KSHV.
Figure 1. Evidences for lytic de novo KSHV infection of HDLMECs.
(A) Time course imaging of GFP+ cells in HDLMECs infected with KSHV BAC16 (MOI=1). GFP+ cells indicate infected cells. (B) Immunofluorescence detection of the early KSHV protein ORF6 in infected HDLMECs at 48 hpi. (C) Immunoblot analysis of viral protein expression in HDLMECs, which were infected with BAC16 for 72 hours in the absence or presence of 100 µM PAA. (D) Measurement of infectious virion production in samples described in panel C. 293T cells were infected with supernatants harvested from infected HDLMECs and the number of GFP+ 293T cells was determined by flow cytometry at 24 hpi, based upon which the virus titer was calculated. Virus titer in the -PAA sample was 168,850 IU/ml.
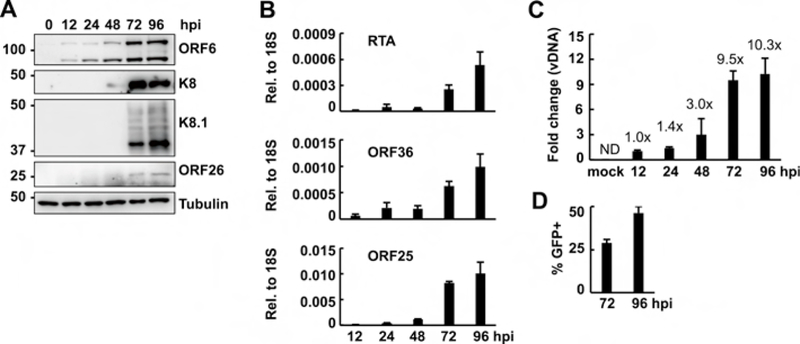
To analyze the progress of lytic KSHV infection in HDLMECs, we performed a time course KSHV infection and analyzed lytic viral gene expression, viral DNA replication, and virus production. In Figure 2A, immunoblot analyses show that early (ORF6, K8) and late (K8.1, ORF26) viral proteins were expressed in infected HDLMECs in a temporally ordered manner, which is characteristic for lytic herpesviral infection. The gradual induction of KSHV genes in the course of HDLMEC infection was also confirmed by RT-qPCR measurement of viral mRNAs (Fig. 2B). In line with the increasing viral gene expression over time, we also detected increasing viral DNA copy number (Fig. 2C) and infectious virion production (Fig. 2D).
Figure 2. Characterization of lytic KSHV infection of HDLMECs.
(A) Time course immunoblot analysis of the expression of KSHV proteins in HDLMECs infected with BAC16 (MOI=1). (B) RT-qPCR measurement of viral mRNA expression in HDLMECs in the course of KSHV infection. (C) Viral DNA load was determined by KSHV ORF11-specific qPCR. The fold change of viral DNA level was calculated relative to the input viral DNA level at 12 hpi. (D) Infectious virion production from BAC16-infected HDLMECs was determined at 72 and 96 hpi. The virus titer was 158,950 IU/ml at 72 hpi.
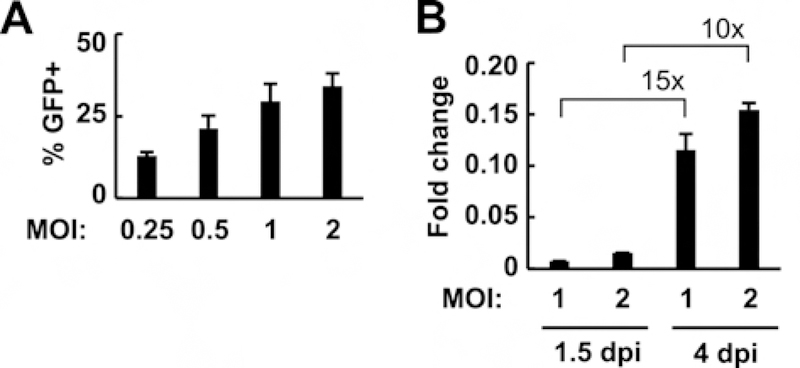
Importantly, when HDLMECs were infected with an increasing amount of KSHV BAC16, the cells responded with increased virus production, supporting the notion that HDLMECs are permissive to lytic KSHV infection (Fig. 3A). We calculated that we could get more than 400,000 IU/ml virus titer at 96 hpi by spin infection of 150,000 HDLMECs with KSHV at an MOI of 1. In comparison, stably infected LECs were reported to produce ~1000 IU/ml (Chang and Ganem, 2013). We note, however, that in line with the previous study by Chang and Ganem, we could also obtain a stably infected HDLMEC cell culture after 10 days postinfection (data not shown), which is likely derived from a fraction of cells that successfully survived infection, and could proliferate to form a latently infected cell population (Chang and Ganem, 2013). To verify that the lytic de novo infection of HDLMECs is not specific for the recombinant KSHV clone BAC16, HDLMECs were also infected with wild-type (WT) KSHV derived from a KSHV+ primary effusion lymphoma cell line (BCBL1) (Fig. 3B). We found that BCBL1-derived KSHV infection of HDLMECs also led to virus production. We note that primary effusion lymphoma cell line-derived KSHV has been used before for infection of primary dermal microvascular endothelial cells (HDMEC), but robust lytic replication was not observed following de novo KSHV infection (Ciufo et al., 2001). Importantly, HDMECs isolated from donors are usually composed of a mixed population of blood vascular endothelial cells (BEC) and lymphatic endothelial cells (LEC), and it has been shown that KSHV infection of BECs results in latency, while LEC infection is more prone to show widespread induction of lytic genes (Chang and Ganem, 2013). Thus, the different ratio of BECs and LECs in a HDMEC culture can affect the outcome of de novo KSHV infection, which can be one of the reasons that different investigators may observe varying degrees of lytic gene expression and virus production in KSHV-infected HDMECs. In our experiments we used LEC-enriched HDLMECs for de novo KSHV infection. In addition, it is also known that lymphatic endothelial cells have a unique gene expression program driven by a set of specific transcription factors such as Prox1, Sox18, and COUP-TFII (Watabe, 2012). Thus, it is also conceivable that depending on how LECs were isolated, and the cell culture conditions, these may affect the expression of the LEC transcription factors and their target genes, which can also influence the susceptibility of LECs to lytic KSHV infection. Further studies will be required to test these possibilities. Altogether, our data support the notion that HDLMECs are permissive to lytic KSHV infection, supporting the completion of the full viral lytic cycle following de novo infection.
Figure 3. Lytic infection of HDLMECs with increasing titer of KSHV.
(A) HDLMECs were infected with BAC16 at increasing multiplicity of infection. The supernatants were collected at 72 hpi and used for infection of 293T cells to determine the virus titer. The titer was 70,950 IU/ml by infecting at an MOI of 0.25. (B) HDLMECs were infected with an increasing amount of BCBL1-derived KSHV. Supernatants of infected HDLMECs were collected at 1.5 dpi and 4 dpi and used for infection of 293T cells, in which the viral DNA load was measured by qPCR at 24 hpi.
Analysis of the requisite of latent genes for KSHV lytic infection
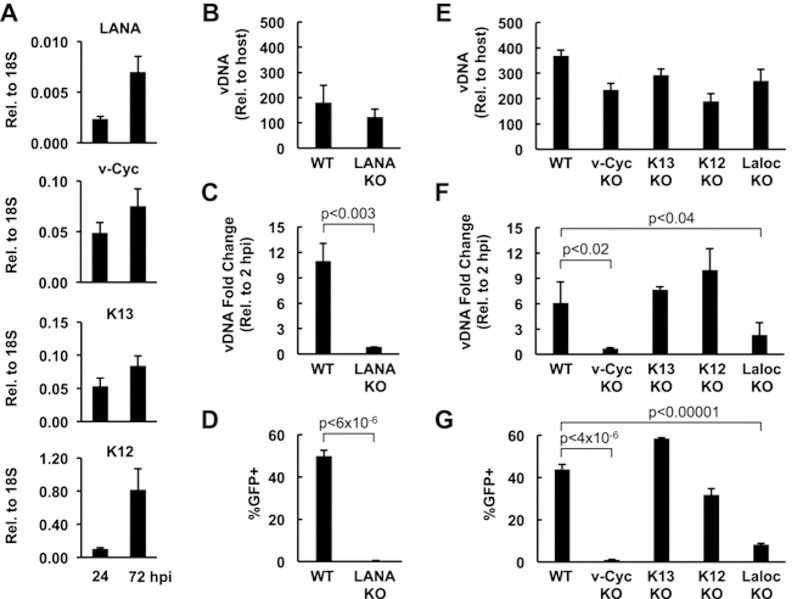
To further validate that HDLMECs can be used to test the function of KSHV genes in lytic KSHV infection, we tested the replicative fitness of latent gene-knockout BAC16 mutants in comparison to WT BAC16. The latent genes are a set of four protein-coding ORFs and 12 viral miRNAs expressed from the latency locus of the KSHV genome, which have been implicated in the suppression of the lytic cycle and the regulation of the establishment and maintenance of viral latency (Plaisance-Bonstaff et al., 2014; Toth et al., 2016; Ye et al., 2011). However, it is also known that the latent genes of KSHV, which are constitutively expressed in infected cells, can regulate cellular pathways that intersect both latency and lytic reactivation. It is possible that the latent viral factors, whose expression increases in HDLMECs during de novo KSHV infection, can also affect KSHV replication following primary lytic infection, a possibility that has not yet been investigated before (Fig. 4A). To test this idea, we produced WT, latent gene knockout (KO), and latency locus deletion (Laloc KO) BAC16 viruses using iSLK KSHV-producing cell lines as published previously (Toth et al., 2016). HDLMECs were infected with the same titer of WT and BAC16 mutants, and then we measured the viral DNA copy number at 2 hpi (input viral DNA load) and at 96 hpi (replicated viral DNA) as well as virus production (Fig. 4B–G). We found that while the input viral DNA levels of BAC16 mutants were comparable with that of WT BAC16 at 2 hpi (Fig. 4A and D), viral DNA replication (Fig. 4B and E) and virus production (Fig. 4C and F) were significantly diminished in the latency-associated nuclear antigen (LANA) KO and viral Cyclin (v-Cyc) KO BAC16-infected cells and also reduced in the case of the Laloc KO BAC16 infection. Importantly, these experiments demonstrate for the first time that LANA and v-Cyc (homolog to cellular cyclin D) are essential for KSHV DNA replication and virus production during primary lytic infection. In contrast, studies on LANA during latent infection showed a repressive role for LANA where it can block the lytic cycle by various mechanisms such as recruiting polycomb proteins to the incoming viral DNA during de novo latent infection, inhibiting KSHV lytic gene expression, or directly suppressing oriLyt-dependent viral DNA replication during latency (Li et al., 2008; Rossetto et al., 2009; Toth et al., 2016; Verma et al., 2006; Ye et al., 2008). Interestingly, the homolog of KSHV LANA in MHV68 was reported to facilitate lytic viral replication and virus production, which was partly attributed to the inhibition of cellular stress by MHV68 LANA during infection (Forrest et al., 2007). KSHV LANA was also shown to promote the proliferation and the survival of infected cells, which can contribute to its positive effect on lytic viral infection (Wei et al., 2016). We note, however, that we did not see increased cell death in LANA KO BAC16-infected cells compared to WT BAC16-infected cells, which would explain the lack of virus production in LANA KO KSHV-infected cells (data not shown). Importantly, KSHV LANA interacts with not only transcription repressors, but also activators, which can be involved in LANA-mediated lytic viral infection (Hu et al., 2014; Wong et al., 2004). Similarly, KSHV v-Cyc has been shown to control latently infected cells, while MHV68 v-Cyc was found to be critical for lytic reactivation of MHV68 in mice (Sarek et al., 2010; van Dyk et al., 2000). Since KSHV v-Cyc can interact with and boost the activity of several cyclin-dependent kinases involved in transcriptional regulation, it will be important to test in the future if CDK/v-Cyc interactions play any role in facilitating lytic KSHV infection through viral gene regulation (Chang and Li, 2008; Li et al., 1997). Interestingly, we found that while the replication and virus production of Laloc KO BAC16 was significantly less relative to WT BAC16, they were higher compared to LANA KO or v-Cyc KO, suggesting that the latent genes may have both positive and negative roles in lytic KSHV infection. We note that both LANA KO and v-Cyc KO KSHV can be produced from latently infected iSLK cells upon lytic reactivation (Toth et al., 2016). However, virus production from iSLKBAC16 cell lines requires the overexpression of RTA along with the addition of a histone deacetylase inhibitor to the cells, which creates a robust condition for strong lytic reactivation. In contrast, HDLMEC infection reflects a natural infection, which is less efficient compared to the iSLK system. Additional experiments will be required to determine the role of LANA and v-Cyc in natural lytic KSHV infection, which can be done by using lytically infected HDLMECs.
Figure 4. Analysis of the replication fitness of latent gene knockout KSHV mutants in lytic viral infection.
(A) RT-qPCR measurement of the expression of latent viral genes in WT KSHV-infected HDLMECs (MOI=1) at 24 and 72 hpi. (B) Input viral DNA level was determined by KSHV ORF11-specific qPCR relative to host DNA level in WT and LANA KO BAC16-infected HDLMECs at 2 hpi. HDLMECs were infected with KSHV at an MOI of 1. (C) Viral DNA load produced by viral replication was calculated at 96 hpi relative to the viral DNA level at 2 hpi. (D) Measurement of virus production. Supernatants of infected HDLMECs were collected at 96 hpi and 600 ul from each was used for infection of 293T cells, and the resulting number of GFP+ infected cells was determined by flow cytometry at 24 hpi. Virus titer of WT KSHV was 458,333 IU/ml. (E) qPCR measurement of viral DNA input in WT and BAC16 mutant infected HDLMECs at 2 hpi. The cells were infected at an MOI of 2. (F) The viral DNA replication was calculated as a fold change at 96 hpi relative to the viral DNA level at 2 hpi. (G) Virus production was measured as described in panel C. Virus titer of WT KSHV was 402,416 IU/ml.
In summary, we found that in contrast to stably infected LECs, de novo infection of HDLMECs resulted in robust lytic KSHV replication and virus production by 4 dpi. We also demonstrated that lytic infection of HDLMECs can be used to reveal novel functions of viral genes in the lytic cycle of KSHV such as the previously unrecognized role of latent genes in promoting lytic KSHV infection. Since lymphatic endothelial cells are relevant to KS pathophysiology, HDLMECs prove to be a valuable cell culture model for investigating lytic KSHV infection in KSHV pathogenesis.
Highlights.
Lymphatic microvascular endothelial cells support primary lytic KSHV infection.
Primary lytic infection results in robust KSHV replication and virus production.
Latent genes LANA and viral cyclin are required for primary lytic KSHV infection.
Acknowledgements
We thank Dr. Jae U. Jung (University of Southern California) for providing the iSLK cell line. We are grateful to Dr. Gary S. Hayward (Johns Hopkins University) for providing the ORF6 antibody. We also thank Dr. Bernadett Papp (University of Florida) and members of the Toth lab for their valuable comments and discussions. This study was supported by the American Cancer Society Research Scholar Grant (RSG-18-221-01-MPC), NIH grant R01AI132554, UF Health Cancer Center A1 Accelerator Grant, and UF Research Opportunity Seed Fund. GG was supported by NIH Training Grant T90DE021990.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There is no conflict of interest.
References
- Bechtel JT, Liang Y, Hvidding J, Ganem D, 2003. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. Journal of virology 77, 6474–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU, 2012. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. Journal of virology 86, 9708–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallin LE, Goldschmidt-Clermont P, Mesri EA, 2014. Molecular and cellular mechanisms of KSHV oncogenesis of Kaposi’s sarcoma associated with HIV/AIDS. PLoS pathogens 10, e1004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett 305, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Ganem D, 2013. A unique herpesviral transcriptional program in KSHV-infected lymphatic endothelial cells leads to mTORC1 activation and rapamycin sensitivity. Cell host & microbe 13, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PC, Li M, 2008. Kaposi’s sarcoma-associated herpesvirus K-cyclin interacts with Cdk9 and stimulates Cdk9-mediated phosphorylation of p53 tumor suppressor. Journal of virology 82, 278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciufo DM, Cannon JS, Poole LJ, Wu FY, Murray P, Ambinder RF, Hayward GS, 2001. Spindle cell conversion by Kaposi’s sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. Journal of virology 75, 5614–5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duus KM, Lentchitsky V, Wagenaar T, Grose C, Webster-Cyriaque J, 2004. Wild-type Kaposi’s sarcoma-associated herpesvirus isolated from the oropharynx of immune-competent individuals has tropism for cultured oral epithelial cells. Journal of virology 78, 4074–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest JC, Paden CR, Allen RD 3rd, Collins J, Speck SH, 2007. ORF73-null murine gammaherpesvirus 68 reveals roles for mLANA and p53 in virus replication. Journal of virology 81, 11957–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SJ, Deng JH, Zhou FC, 2003. Productive lytic replication of a recombinant Kaposi’s sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. Journal of virology 77, 9738–9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin L, Damania B, 2014. KSHV: pathways to tumorigenesis and persistent infection. Advances in virus research 88, 111–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Ganem D, 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J Clin Invest 113, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Yang Y, Turner PC, Jain V, McIntyre LM, Renne R, 2014. LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex. PLoS pathogens 10, e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B, 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi’s sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. Journal of virology 78, 3601–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lee H, Yoon DW, Albrecht JC, Fleckenstein B, Neipel F, Jung JU, 1997. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. Journal of virology 71, 1984–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou F, Ye F, Gao SJ, 2008. Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DF, Kuppermann BD, Wolitz RA, Palestine AG, Li H, Robinson CA, 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med 340, 1063–1070. [DOI] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C, 2010. Kaposi’s sarcoma and its associated herpesvirus. Nat Rev Cancer 10, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocroft A, Youle M, Gazzard B, Morcinek J, Halai R, Phillips AN, 1996. Anti-herpesvirus treatment and risk of Kaposi’s sarcoma in HIV infection. Royal Free/Chelsea and Westminster Hospitals Collaborative Group. Aids 10, 1101–1105. [PubMed] [Google Scholar]
- Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU, 2003. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. Journal of virology 77, 4205–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Motlagh N, Smindak RJ, Jang SJ, Sharma A, Alonso JD, Toth Z, 2018. Genome-wide identification of direct RTA targets reveals key host factors for KSHV lytic reactivation. Journal of virology [DOI] [PMC free article] [PubMed]
- Plaisance-Bonstaff K, Choi HS, Beals T, Krueger BJ, Boss IW, Gay LA, Haecker I, Hu J, Renne R, 2014. KSHV miRNAs decrease expression of lytic genes in latently infected PEL and endothelial cells by targeting host transcription factors. Viruses 6, 4005–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman P, Uppal T, Verma SC, 2015. Molecular biology of KSHV lytic reactivation. Viruses 7, 116–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto C, Yamboliev I, Pari GS, 2009. Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 K-bZIP modulates latency-associated nuclear protein-mediated suppression of lytic origin-dependent DNA synthesis. Journal of virology 83, 8492–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarek G, Jarviluoma A, Moore HM, Tojkander S, Vartia S, Biberfeld P, Laiho M, Ojala PM, 2010. Nucleophosmin phosphorylation by v-cyclin-CDK6 controls KSHV latency. PLoS pathogens 6, e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G, 1998. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proceedings of the National Academy of Sciences of the United States of America 95, 10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Papp B, Brulois K, Choi YJ, Gao SJ, Jung JU, 2016. LANA-Mediated Recruitment of Host Polycomb Repressive Complexes onto the KSHV Genome during De Novo Infection. PLoS pathogens 12, e1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Smindak RJ, Papp B, 2017. Inhibition of the lytic cycle of Kaposi’s sarcoma-associated herpesvirus by cohesin factors following de novo infection. Virology 512, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyk LF, Virgin H.W.t., Speck SH, 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. Journal of virology 74, 7451–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SC, Lan K, Choudhuri T, Robertson ES, 2006. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen modulates K1 expression through its cis-acting elements within the terminal repeats. Journal of virology 80, 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T, 2012. Roles of transcriptional network during the formation of lymphatic vessels. J Biochem 152, 213–220. [DOI] [PubMed] [Google Scholar]
- Wei F, Gan J, Wang C, Zhu C, Cai Q, 2016. Cell Cycle Regulatory Functions of the KSHV Oncoprotein LANA. Front Microbiol 7, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LY, Matchett GA, Wilson AC, 2004. Transcriptional activation by the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. Journal of virology 78, 10074–10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Lei X, Gao SJ, 2011. Mechanisms of Kaposi’s Sarcoma-Associated Herpesvirus Latency and Reactivation. Adv Virol 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Zhou FC, Xie JP, Kang T, Greene W, Kuhne K, Lei XF, Li QH, Gao SJ, 2008. Kaposi’s sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-kappaB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. Journal of virology 82, 4235–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]