Abstract
Fumarate, an electron acceptor in anaerobic respiration of Escherichia coli, has an additional function of assisting the flagellar motor to shift from counterclockwise to clockwise rotation, with a consequent modulation of the bacterial swimming behavior. Fumarate transmits its effect to the motor via the fumarate reductase complex (FrdABCD), shown to bind to FliG — one of the motor’s switch proteins. How binding of the FrdABCD respiratory enzyme to FliG enhances clockwise rotation and how fumarate is involved in this activity, have remained puzzling. Here we show that the FrdA subunit in the presence of fumarate is sufficient for binding to FliG and for clockwise enhancement. We further demonstrate by in vitro binding assays and super-resolution microscopy in vivo that the mechanism by which fumarate-occupied FrdA enhances clockwise rotation involves its preferential binding to the clockwise state of FliG (FliGcw). Continuum electrostatics combined with docking analysis and conformational sampling endorsed the experimental conclusions and suggested that the FrdA-FliGcw interaction is driven by the positive electrostatic potential generated by FrdA and the negatively charged areas of FliG. They further demonstrated that fumarate changes FrdA’s conformation to one that can bind to FliGcw. These findings also show that the reason for the failure of the succinate dehydrogenase flavoprotein SdhA (an almost-identical analog of FrdA shown to bind to FliG equally well) to enhance clockwise rotation is that it has no binding preference for FliGcw. We suggest that this mechanism is physiologically important as it can modulate the magnitude of ΔG0 between the clockwise and counterclockwise states of the motor to tune the motor to the growth conditions of the bacteria.
Keywords: flagellar motor, quinol-fumarate oxidoreductase, anaerobic complex II, respiratory complexes, FliG, protein electrostatics
Introduction
Bacteria such as Escherichia coli swim by rotating their flagella. Each flagellum rotates by a motor (reviewed in [1–4]), embedded in the cell membrane. The motor’s direction of rotation is the target of navigation control by the chemotaxis system because it dictates the swimming mode of the bacteria, with counterclockwise rotation pushing the bacterium forward and clockwise rotation causing cell reorientation [1–5]. The direction of rotation is determined by a ‘switch’, made of circular layers of three proteins, FliN, FliM and FliG.
An unexpected finding made over two decades ago was that fumarate, a dicarboxylate that serves as an electron acceptor in anaerobic respiration [6], acts as a switching and clockwise factor under aerobic conditions. Thus, in E. coli under aerobic conditions, fumarate enabled the flagella of cytoplasm-free cell envelopes to switch their direction of rotation [7,8] and enhanced switching and clockwise rotation in intact bacteria [9,10]. These effects were due, in part, to reduction of the standard free energy difference (ΔG0) between the clockwise and counterclockwise states of the switch [9], but they were neither due to the function of fumarate as an electron acceptor [8] nor to electron-transport-related energization [11]. The mechanism by which fumarate enhances clockwise rotation and switching has remained obscure. Years later the target of fumarate for these functions was identified. The membrane-bound enzyme fumarate reductase (also known as quinol-fumarate oxidoreductase and as anaerobic complex II) was found to interact with FliG at the switch [11]. This enzyme functions in the terminal step of the anaerobic electron transfer chain, at which electrons from the menaquinol cofactor are used to reduce fumarate [12]. Its expression level is 10–15-fold higher under anaerobic conditions than under aerobic conditions [13]. The presence of fumarate increases the expression level 1.5-fold further [13,14]. Fumarate reductase is a membrane-linked heterotetramer ([12] for a review), composed of FrdA — a cytoplasmic flavoprotein subunit, FrdB — a cytoplasmic iron-sulfur protein, and a transmembrane anchor domain that consists of two subunits — FrdC and FrdD (Figure 1A; the enzyme will be termed hereafter FrdABCD). FrdA is the subunit that binds fumarate [15]. Intriguingly, the almost identical aerobically expressed enzyme, succinate dehydrogenase (SdhCDAB), also binds to FliG but this binding has no effect on flagellar rotation [11]. Succinate dehydrogenase is a component of the tricarboxylic acid cycle, where it converts succinate to fumarate, and also a component of the aerobic electron transport chain, where it reduces quinone to quinol [12]. The enzyme is a structural and functional homolog of fumarate reductase, and both enzymes are considered to be evolutionarily related and part of the same enzyme family [12]. In fact, FrdABCD and SdhCDAB can functionally replace each other when overexpressed because they can catalyze the same reactions [76]. Due to the high degree of structural and functional similarity of the two enzyme complexes, they bind fumarate equally well [15]. It is unclear how one of them, FrdABCD, enhances clockwise rotation whereas the other, SdhCDAB, does not. This study looks for the cause of this difference and for the mechanism by which fumarate enhances switching and clockwise rotation.
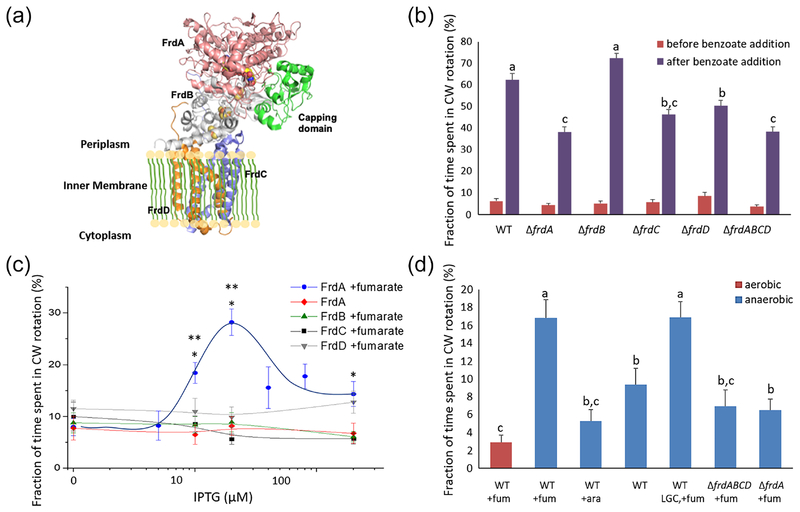
Figure 1.
FrdA alone can enhance clockwise rotation. A, Structure of E. coli FrdABCD. Based on PDB 3P4P. B, Clockwise rotation in mutants lacking individual FrdABCD subunits. See Table 2 for strains. The fraction of time spent in clockwise rotation was calculated in 1 min intervals immediately before and after the addition of benzoate (50 mM, pH 7.0). Columns marked with the same letter were statistically indifferent. Columns marked with dissimilar letters were significantly different (P<0.05) according to Kruskal Wallis statistical test (n = 113–190 cells). C, Effect of FrdA level on clockwise rotation. The expression of the subunits shown was done from the IPTG-induced plasmids listed in Table 2 in B275ΔfrdABCD background. Where indicated, fumarate (40 mM) was present in the medium during growth but not during the experiment. The curve was hand-drawn. *P<0.001 (for 10 and 20 μM IPTG) or <0.01 (for 200 μM IPTG) relative to absence of fumarate at the same IPTG concentration, and **P<0.05 or <0.001 (for 10 or 20 μM IPTG, respectively) relative to zero IPTG in the presence of fumarate according to Kruskal Wallis statistical test (n = 44–95 cells). D, Anaerobic growth in the presence of fumarate increases clockwise probability. The wild-type strain used was B275. See Table 1 for the other strains. Prior to tethering, the cells were washed and resuspended in fumarate-free motility buffer. Low-growth control stands for wild-type cells that grew to OD600 = 0.09, similar to the maximal growth OD of ΔfrdABCD cells under anaerobic conditions. Columns marked with the same letter were statistically indifferent. Columns marked with dissimilar letters were significantly different according to Kruskal Wallis test (P<0.001, except for the difference between aerobic WT+fum and ΔfrdA+fum, for which P<0.05; n = 70–170 cells). Abbreviations: ara, arabinose; fum, fumarate; LGC, low-growth control; WT, wild type.
Results
Intactness of the FrdABCD complex is not essential for clockwise enhancement
FrdABCD was shown to interact with FliG and be involved in multiple functions of the flagellar motor, including enhancement of clockwise rotation [11]. To determine whether the FrdABCD complex, as a whole, is required for this enhancement or whether one or more of its subunits are sufficient, we first examined how deletion of each subunit affects clockwise rotation. We focused on the repellent response of mutants lacking each of the four subunits of FrdABCD (one at a time) because the effect of FrdABCD deletion on clockwise rotation is best observed in response to a repellent [11]. To determine the direction of flagellar rotation we tethered individual cells via their flagellum to glass and recorded their direction of rotation around their immobilized flagella [16]. Consistent with Förster resonance energy transfer (FRET) studies suggesting that the binding site for FliG is on FrdA [17], we found that both frdA and frdABCD deletions similarly reduced the extent of clockwise rotation following stimulation with the repellent benzoate (Figure 1B). In contrast, a frdB deletion did not reduce the rotation in this direction and, perhaps, even increased it to a level slightly higher than that of the wild-type parent (though the increase was statistically insignificant). Since FrdB links FrdA to the membrane part of the complex, i.e., to the FrdCD units (Figure 1A) [12], the absence of FrdB means that FrdA is free in the cytoplasm. The ability of the ΔfrdB mutant to generate high clockwise levels therefore suggests that FrdA, which lacks enzymatic activity by itself [18], is nevertheless fully functional in generating clockwise rotation in its soluble, cytoplasmic form. This is consistent with the observation that the enhancement of clockwise rotation by fumarate, though done via FrdABCD, is independent of the electron-transfer activity of the latter [8,11]. The clockwise responses of the ΔfrdC and ΔfrdD mutants to benzoate were stronger than that of the ΔfrdA mutant (though the difference was statistically significant for the ΔfrdD mutant only). Yet, they were lower than the wild-type parent’s response, probably due to the futile interaction of FrdA with free FrdB in these mutants (FrdB cannot be linked to the membrane in the absence of either FrdC or FrdD [12]). Thus, it appears that an intact FrdABCD complex is not obligatory for proper clockwise rotation of the flagellar motor and that FrdA in its soluble form is sufficient for enhancing clockwise rotation.
Overproduced FrdA generates clockwise rotation
To validate the above conclusion, we examined the effect of overproduction of each Frd subunit (each tagged with 6×His at the N-terminus; for simplicity, the tag will not be mentioned hereafter) in the absence of the other subunits, thus ensuring that the measurement is of the subunit itself and not of the subunit in a complex. Aerobic expression of FrdA in a ΔfrdABCD mutant (strain B275ΔfrdABCD) to approximately the level measured in wild-type cells under anaerobic conditions (Supplementary Figure S1) generated a strong clockwise bias of the motor, provided that the cells had been grown in presence of fumarate (Figure 1C). This dependence on fumarate is consistent with the role of FrdABCD to transmit the effect of fumarate to the motor [11]. In contrast, expression of the other three subunits did not affect the level of clockwise rotation (Figure 1C). These results not only endorse the conclusion that FrdA is sufficient for clockwise rotation, they further suggest that when the concentration of soluble, fumarate-bound FrdA is sufficiently high as in cells under anaerobic conditions (conditions under which the FrdABCD complex is maximally expressed [13]), it can promote clockwise rotation even in the absence of any other stimulation.
Notably, expression of FrdA to higher levels decreased the fraction of time spent in clockwise rotation (Figure 1C). Two potential causes could account for this. One is reduced concentration of soluble FrdA due to FrdA aggregation at high concentrations. We put this possibility to the test by examining both the soluble and insoluble fractions of the lysed cells. Western blots of these fractions demonstrated that, even though the level of FrdA in the insoluble fraction increased with the IPTG concentration, its level in the soluble fraction did not decrease (Figure S1C), thus excluding reduced FrdA concentration due to aggregation as a cause of the observed decrease in clockwise rotation at high IPTG concentrations. The other potential cause is competition of fumarate-free FrdA with fumarate-bound FrdA for FliG. The fraction of fumarate-free FrdA molecules is expected to increase with the level of overexpression. Since fumarate is required for the effect of FrdA on clockwise rotation (Figure 1C) but, apparently, not for its binding to FliG (see below), it is probable that the fumarate-free FrdA molecules would compete with the fumarate-bound FrdA molecules for FliG but would not produce clockwise rotation. The consequence of this futile binding to FliG would be reduced clockwise rotation.
Anaerobic growth increases clockwise probability due to elevated FrdA
To determine whether or not the dependence of clockwise rotation on the FrdA level is physiologically relevant, we compared the rotation of tethered cells grown aerobically with the rotation of cells grown anaerobically, where the FrdABCD complex is expressed to higher levels [13]. Consistent with the results of FrdA expression described above, the clockwise level of anaerobically grown cells was significantly higher than that of aerobically grown cells (P<0.001; Figure 1D, two left columns). This effect was fumarate-dependent as absence of fumarate in the growth medium reduced clockwise rotation (P<0.001; Figure 1D, fourth column). As expected, the clockwise levels of the negative-control cells, anaerobically-grown ΔfrdA and ΔfrdABCD cells (Figure 1D, two right columns), were significantly lower than that of the anaerobically-grown wild-type cells (P<0.001), though they were somewhat higher than aerobically-grown cells, probably due to the anaerobic growth conditions. Since, under anaerobic conditions, ΔfrdABCD cells maximally grew to OD600 = 0.09, we performed a control with wild-type cells grown to the same low OD. The clockwise level of these control cells was similar to that of wild-type cells grown anaerobically to OD600 = 0.4 (Figure 1D, second and fifth columns). As another control, we studied wild-type cells that grew anaerobically in the presence of arabinose instead of fumarate. These cells grew similarly well in the presence of either one of these compounds (OD600 = 0.6 and 0.4 in the presence of arabinose and fumarate, respectively). Yet, the level of clockwise rotation was low when arabinose replaced fumarate (Figure 1D, third column). The switching frequency of the rotation followed the same trend as clockwise rotation, i.e., higher switching frequency when clockwise rotation was higher (Figure S2 versus Figure 1D), consistent with the function of fumarate as a clockwise and switching factor [7–10]. It should be noted that the rotation of anaerobically-grown cells was measured aerobically. We, therefore, performed a control to verify that FrdA, which can only be expressed to high levels under anaerobic conditions [13], had not degraded during the ~1 h aerobic tethering assay. We subjected lysates of cells exposed to air for 1.5 h after anaerobic growth to Western blots followed by mass spectrometry. Clearly, 1.5 h air exposure essentially did not affect the FrdA level (Figure S3). Thus, anaerobic growth conditions elevate the FrdA level with a resultant clockwise rotation, an effect that is maintained for at least 1.5 h after air exposure.
FrdA preferentially interacts with the clockwise conformation of FliG
Our observations that FrdA binding to FliG (in the presence of fumarate) is sufficient for clockwise enhancement raised two potential causes of this enhancement. One is that FrdA binding to the counterclockwise state of FliG (FliGccw) results in a shift of the latter to its clockwise state (FliGcw). The other is that FrdA preferentially binds to FliGcw with a consequent stabilization of this state. To examine whether any of these possibilities holds, we compared the binding of FrdA to FliGcw and FliGccw both in vitro and in vivo.
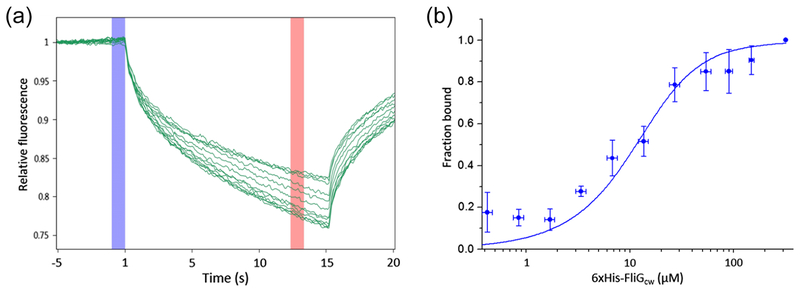
In vitro, we measured the binding of purified FrdA to purified wild-type FliG, which is essentially FliGccw [19], and to mutant FliG (containing a PAA deletion at positions 169–171) locked in the clockwise conformation [19], i.e., FliGcw. Employing MicroScale Thermophoresis (MST) [20], we found that FrdA bound to FliGcw with Kd ≈ 4 μM (Figure 2). Remarkably, this Kd is very similar to the Kd of CheY binding to FliM [21,22], suggesting that the interactions of FrdA and CheY with the motor are similarly strong. Unlike FliGcw, when we studied FliGwt at the same concentration range, we could neither obtain a consistent binding curve nor reach binding saturation. This could potentially be due to too weak FrdA binding to FliGccw, to the possibility that FliGccw, but not FliGcw, might oligomerize in vitro [23], or to the theoretical possibility that FliGwt, but not FliGcw, contains more than one conformation of FliG.
Figure 2.
Binding of FrdA to FliGcw monitored with MST. The FrdA concentration was kept constant at 15 μM and FliGcw was serially diluted 1:1 beginning with 138 μM. The natural fluorescence of the FAD cofactor was utilized for measuring the MST signal. A, MST traces of a representative experiment. Averaged values were taken before and after heating (blue and red areas). B, Binding curve obtained by averaging the values at each FliGcw concentration in 4 separate experiments. The curve is a fit (R2=0.955) according to the law of mass action equation [20], yielding Kd = 4±1 μM (±SEM, n = 4).
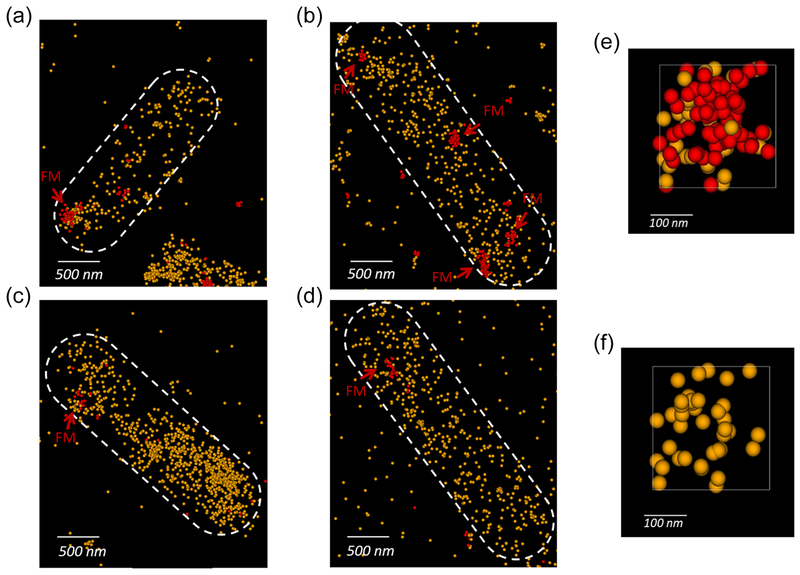
In vivo, we employed super-resolution microscopy, more specifically, photoactivated localization microscopy (PALM), which provides up to ~25 nm resolution [24]. We fluorescently labeled FrdA with the photoswitchable protein, Dendra2 [25], and expressed the frdABCD operon under anaerobic conditions from a low-copy number plasmid in the presence of fumarate. Additionally, we labeled FliM at the motor with Alexa 647 (detailed in Materials and Methods). To obtain FliGcw without a mutation in fliG, we expressed FliG in a motor that is almost constantly in the clockwise conformation due to mutation in fliM [fliM(R60C)] [26]. Representative examples are shown in Figure 3A–D. The number of FrdA-Dendra2 molecules in the cell seemed to vary largely between cell to cell (Table 1). A portion of the molecules were clustered, usually 1–3 clusters per bacterium (e.g., Figures 3A, C), as was reported for some other oxidative phosphorylation complexes in E. coli [27]. Such clusters were hardly detected in a negative control strain, in which Dendra2 was encoded in the same vector under the control of the same frdABCD promotor but without being fused to FrdA (Figure S4). Automated clustering analysis indicated an order of magnitude less clusters in the negative control (0.2 ± 0.4 clusters/cell, 269 ± 181 molecules/cell, mean ± SD of 22 control cells). Clearly, the density of FrdA-Dendra2 molecules near clockwise motors (i.e., within an area of 0.09 μm2 around the motor — see Materials and Methods), but not wild-type motors, was significantly higher than in other areas even though the total number of molecules was similar in both cell types (Table 1). In some of the cells we observed FrdA molecules arranged around the motor in the form of a ring (for example, Figure 3E, F). What can be concluded is that FrdA is indeed associated with the flagellar motor in vivo under anaerobic conditions, and the association is more intense with clockwise motors.
Figure 3.
Representative images of cells containing fluorescently labeled FrdA and FliM. A, B, Combined PALM/STORM imaging of cells containing wild-type motors (strain DFB190ΔfrdABCD containing the plasmids pH3-Dendra2-FrdABCD and pFliMWT-mPlum). Yellow dots represent individual Dendra2 (FrdA) molecules; red dots represent individual Alexa 647-labled wild-type FliM molecules; FM indicates the location of the flagellar motors. C, D, As in A, B but for cells containing motors with FliMcw molecules (strain DFB190ΔfrdABCD containing the plasmids pH3-Dendra2-FrdABCD and pFliMCW-mPlum). E, A ring structure of FrdA molecules formed around the motor. F, as in E, but with the motor fluorescence omitted to reveal the ring-like structure.
Table 1.
FrdA molecules are more crowded near clockwise motors.
| Wild-type motors | Clockwise motors | |
|---|---|---|
| FrdA-Dendra2 molecules / cella | 250 ± 86 | 230 ± 191 |
| Density near motors (molecules/μm2)b | 140 ± 98 | 250* ± 150 |
| Density in other areas (molecules/μm2)c | 120 ± 57 | 160 ± 75 |
Counting of molecules was done automatically.
The values shown are the mean number of FrdA-Dendra2 molecules ±SD in 28 wild-type and 32 clockwise cells (including cells in which Alexa 647-labled FliM molecules were not detected).
The values shown are the mean number of FrdA-Dendra2 molecules ±SD found within a 0.3×0.3 μm square around the Alexa 647 label for 24 and 15 wild-type and clockwise motors, respectively (in 19 and 10 cells, respectively).
The cells were those from the preceding row.
P<0.05 relative to density in other areas and relative to density near wild-type motors according to Student’s paired t-test and non-parametric Mann-Whitney test, respectively.
Molecular basis for the preferred binding to FliGcw
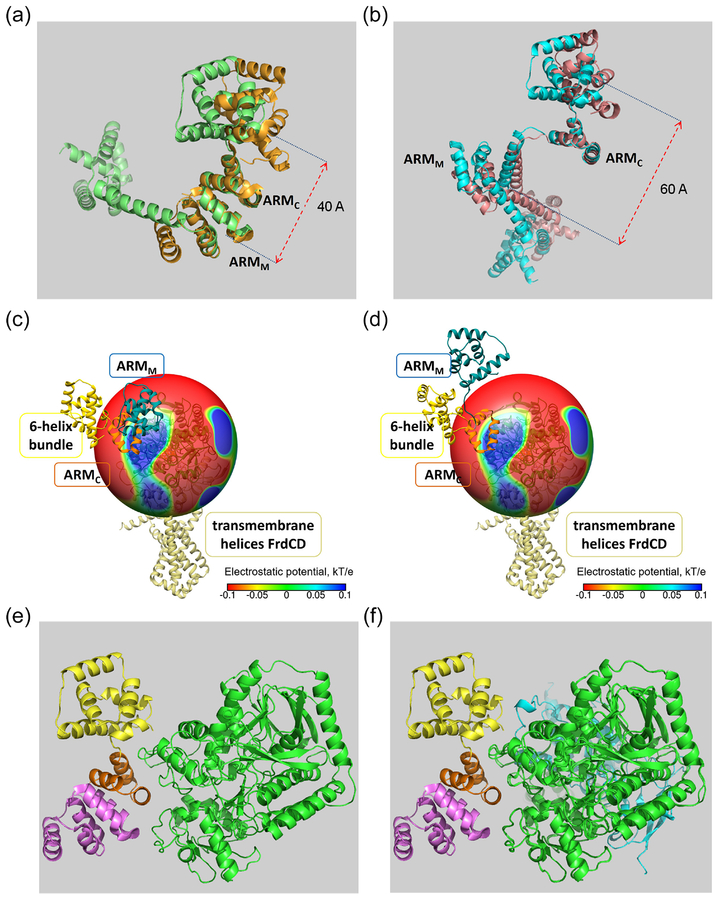
The results above suggested that FrdA is sufficient for clockwise enhancement and that it preferentially binds to the clockwise state of FliG. To obtain information for the structural causes of these results, we carried out molecular dynamics (MD) conformational sampling of the FliG protein combined with continuum electrostatics and docking analysis. Employing the structures of full-length FliG from Aquifex aeolicus (PDB code 3HJL) and of the middle-to-C-terminal domain of FliG from Thermotoga maritima (PDB codes 4FHR, 3AJC) as representatives of the counterclockwise (3HJL) and the clockwise (3AJC) states of E. coli FliG [28], we found multiple conformational transitions between the clockwise and counterclockwise states. These two structures are significantly different at the C-terminal domain, primarily due to the rotation of a six-helix bundle (helices C1–6) relative to the armadillo repeat motif (ARMC) in this domain [29] (Figure 4A, B). Conformational analysis of MD trajectories revealed that, of the many conformations obtained, only 4 conformation clusters appeared: FliGccw and FliGcw each in the compact (Figure 4A) and extended (Figure 4B) states (Supplementary Information for detailed analysis). The calculations further showed that FrdA preferentially interacts with compact FliGcw, that the interaction involves both ARMM and ARMC motifs, and that the binding of FrdA to FliGcw is driven by positive electrostatic potential generated by FrdA and the negatively charged areas of FliG (Figure 4C, D; Supplementary Information for details). These electrostatics calculations combined with docking analysis further suggested that the interaction between FliGcw and isolated FrdA (Figure 4E) is weaker than the interaction between FliGcw and FrdABCD (Figure 4F) due to the smaller average intermolecular contact surface area in the case of FrdA (estimated to be 990 versus 1200 Å2 for FrdA and FrdABCD, respectively). Our modeling indicates that the stronger binding of the whole complex to FliGcw is because FrdB also contributes to the electrostatic and hydrophobic interaction (Figure 4F; Supplementary Information for details). This stronger binding is well consistent with the measured Kd values for the binding of FrdA (Figure 2) and FrdABCD [11] to FliG (4 μM and 0.4 μM, respectively).
Figure 4.
Molecular basis for the preferred interactions as revealed by MD simulations combined with docking analysis. A, “Compact” conformation of FliGcw (green) and FliGccw (orange). B, “Extended” conformation of FliGcw (cyan) and FliGccw (salmon). Note the difference between compact (A) and the extended (B) conformations with respect to the distance between the center of the M domain and the center of the C-terminal domain. C, D, The compact conformation of FliGcw interacts with FrdA better than the extended conformation. The panels show the distribution of the electrostatic potential mapped on a sphere of a 40Å radius around a FrdA subunit interacting with FliG in its compact (C) and extended (D) conformations. FrdA values of the electrostatic potential on the color maps are given in kT/e units. It is evident that the extended conformation is less preferable for binding to FrdA (D). Higher binding affinity can be achieved in the compact conformation when both the ARMC and ARMM motifs of FliG enter the “blue area” around FrdA (C) at the initial step of the binding process, followed by the final stage (E, F). In these initial “encounter complexes” (C and D), minimal FliG-FrdA surface-to-surface distance (~15Å) is found between Lys121 and Lys281 of FrdA and Glu212 from ARMC helix_1 of FliG. Electrostatic interaction energies between FliGcw and FrdA are estimated to be in the range of −8 to −30 kJ/mol (Table S1), suggesting attraction and efficient steering at long distances. E, Proposed binding mode of a FrdA monomer to FliGcw. In this final step of the binding, helix_1 of the ARMC motif fits into the groove on the surface of FrdA surrounded by a “lysine cluster”. Green, FliG; yellow, C-terminal six-helix bundle; pink, the ARMM motif; orange, the ARMC motif. F, Proposed binding mode of FrdABCD to FliGcw, showing possible involvement of the loop Lys118-Pro137 (cyan) of FrdB in contacts with the C- terminal six-helix bundle of FliGcw.
A related question is what the function of fumarate is in FrdA-mediated clockwise enhancement. According to recent structural data [30], the presence of a dicarboxylate ligand of FrdA rotates the capping domain in relation to the flavin domain of FrdA. Our docking results suggest that residues 275–280 of the capping domain are exposed to the FliG binding area and interact with the ARMM motif. These data suggest that the fumarate-triggered rotation or shift of the capping domain increases the total contact surface area between FliGcw and FrdA with a consequent increased affinity between them.
Molecular basis for the effectiveness of FrdA versus SdhA in clockwise enhancement
One of the puzzles related to the interaction of FrdABCD with FliG is why the interaction of the structurally and functionally similar protein complex SdhCDAB, although binding with similar affinity to FliG, does not have any effect on clockwise rotation. To address this enigma, we compared between FrdABCD and SdhCDAB with respect to their predicted interaction with FliG. Our modeling showed that, unlike FrdA within FrdABCD, which binds specifically to FliGcw, SdhA binds equally well to both FliGcw and FliGccw (Supplementary Information and Figure S10). It thus seems that the mechanism by which FrdA enhances clockwise rotation is selective binding to FliGcw with a consequent stabilization of the motor’s clockwise state. In contrast, the similar binding of SdhA to both conformations of FliG stabilizes both directions of rotation, explaining the failure of SdhA binding to FliG to enhance clockwise rotation.
Discussion
Over two decades ago fumarate was found to be a switching and clockwise factor in E. coli [7–10], which acts by decreasing ΔG0 between the clockwise and counterclockwise states [9]. A decade later the enzyme FrdABCD, but not its aerobically-expressed homolog SdhCDAB, was identified as the protein that transmits the effect of fumarate to the motor [11]. These findings were intriguing because they neither offered clues to the mechanism by which fumarate and FrdABCD affect the direction of flagellar rotation nor they explained how both ‘twin’ enzymes similarly bind to the motor but, nevertheless, only one of them affects the rotation. The findings, made herein, appear to provide answers to these questions and are discussed below.
Function of FrdABCD at the motor
The observations, made in this study, of reduced repellent-stimulated clockwise enhancement in a ΔfrdA mutant similar to a ΔfrdABCD mutant, and elevated clockwise rotation in response to FrdA overproduction, but not to overproduction of other subunits (Figure 1), indicated that FrdA is the subunit responsible for the clockwise-enhancement effect of FrdABCD. The finding that clockwise enhancement was high in a ΔfrdB mutant, where FrdA is free in the cytoplasm, further suggested that FrdA, in its soluble form, is sufficient to enhance clockwise rotation. The results, however, do not distinguish between whether, under normal physiological conditions (i.e., intact FrdABCD), FrdA acts as a soluble molecule or within the FrdABCD complex. Hitherto, there is no information about whether or not FrdA exists in the cell under physiological conditions in a soluble form. (Obtaining such information is not trivial because preparation of a cell extract might release FrdA from the complex, making it difficult to distinguish how much free FrdA exists in vivo.) Nevertheless, the findings that the intact FrdABCD complex binds to FliG [11] suggest that, under physiological conditions, FrdA binds to, and acts on, the motor from within the intact FrdABCD complex. This conclusion is well in line with the ten-fold stronger binding of FrdABCD to FliG compared to that of FrdA, also revealed from our docking analysis combined with electrostatics calculations.
Mechanism of clockwise enhancement by FrdA and fumarate
An obvious question is how FrdA, which lacks any enzymatic activity [18], affects the rotation of the flagellar motor by merely binding to FliG and how fumarate affects FrdA. In principle, FrdA could affect the rotation by one of the following ways: (1) It could selectively bind to FliGcw and stabilize this conformation. (2) It could bind to FliGccw and stimulate its transition to the clockwise conformation. (3) It could bind to either conformation of FliG and affect somehow the entire ring organization with a resultant clockwise rotation. Our results, which demonstrated preferred FrdA binding to FliGcw both in vitro (Figure 2) and in vivo (Figure 3, Table 1), supported the first option. Furthermore, our MD conformational analysis indeed revealed that FrdA preferentially interacts with FliGcw in its compact conformation and that the interaction is electrostatic in nature (Figure 4). Importantly, our in-silico analysis revealed that, unlike the selective binding of FrdA to FliGcw, SdhA binds equally well to both FliGcw and FliGccw (Supplementary Information and Figure S10), explaining why SdhCDAB binding to FliG does not generate clockwise rotation [11]. Thus, the mechanism of clockwise enhancement by FrdA is probably preferred binding to FliGcw with a consequent stabilization of the motor’s clockwise state. As our docking results suggest, the function of fumarate is likely to cause a conformational change [30–33] that increases the total contact surface area between FrdA and FliGcw with a consequent increased affinity between them.
The proposed mechanism requires that, for fumarate being effective, at least one of the FliG subunits would be in its clockwise conformation. This is likely to be indeed the case at physiological temperatures, as evident from the occurrence of some clockwise rotation even in the absence of fumarate (Figure 1D). This clockwise rotation results from binding of the signaling molecule CheY to its target at the motor — the switch protein FliM ([2] for a review), followed by FliM interaction with FliG at a specific binding surface of the latter [34]. However, CheY per se is apparently neither involved in, nor necessary for, the functions of fumarate and FrdA. This is because, at temperatures of ~2°C, where some switching to clockwise rotation occurs even in the complete absence of CheY and of any other cytoplasmic chemotaxis protein, fumarate can still effectively enhance switching and clockwise rotation [9]. Consistently, at room temperature where clockwise rotation does not occur unless CheY is present, fumarate is effective only in the presence of CheY [9,10].
In vivo, FliM binds to to the middle domain of FliG (FliGM) in different bacterial species [35]. Analysis of available structural data [19,29,35,36] show that this binding is not affected by the arrangement of FliGM and FliGC, meaning that both the extended and compact states of FliG can bind to FliM similarly well. This binding has bearings on FrdA interaction with FliG because the movement of the latter is more restricted due to the FliG-FliMM packing and protein crowding within the switch complex, resulting in stronger interaction between FliG and FrdA. The lower amount of high-dielectric water and hydrophobic environment in the highly viscous switch complex should also enhance electrostatic interactions [37] in the FliG-FrdA binding.
Each flagellar motor contains ~25 FliG molecules arranged in a ring [38]. It is reasonable to assume that even in a counterclockwise motor, one or more of the FliG subunits would temporarily be in the clockwise conformation, depending on the level of active CheY in the cell, and that fumarate-occupied FrdA would bind to these FliGcw subunits and stabilize them. An expected outcome is that these stable FliGcw units in a counterclockwise ring would make the latter thermodynamically unfavorable and increase the probability of a ring shift to the clockwise state, as was shown earlier for genetically locked FliGcw [39]. As a result, ΔG0 between the clockwise and counterclockwise states would decrease in the presence of fumarate, as was indeed found experimentally [9].
Physiological significance
The results of this study demonstrated a remarkable case in which a subunit of a respiratory enzyme, which is mainly expressed under anaerobic conditions, directly interacts with a subunit of the flagellar motor when it is in a specific conformation, and affects its function. A legitimate question is what advantage this interaction confers on the bacterial cell. Since this interaction modulates the magnitude of ΔG0 between the clockwise and counterclockwise states of the motor [9] and depends on the level of FrdA (Figure 1C), it is reasonable that one of the functions of this interaction is to tune the motor according to the growth conditions of the bacteria. Anaerobic conditions elevate the expression level of FrdABCD [13] with a consequent elevation of clockwise rotation to moderate levels (Figure 1D). The outcome of such clockwise levels would be moderate tumbling frequencies of the cells, which are expected to better propagate in viscous media, such as agar, than cells with very low or very high tumbling frequencies [40]. This can be advantageous to E. coli in the mammalian digestive tract, where E. coli inhabits the polysaccharide-rich mucus layer lining the epithelium and where the environment is generally anaerobic [41,42] and respiration on fumarate via FrdABCD is essential [43,44]. Furthermore, in such energy-poor environments, swimming towards oxygen and energy sources is essential. This navigation, which is mediated by the chemotaxis receptor Aer [45,46], is expected to be more efficient with moderate tumbling frequencies, and the cells are expected to respond to the Aer-mediated signals with higher sensitivity. Consistent with this notion, an additional function of Aer is upregulation of the expression of anaerobic respiratory enzymes, including fumarate reductase [47].
Materials and Methods
Materials
Amino acids were purchased from Calbiochem (Darmstadt, Germany); ampicillin, kanamycin, chloramphenicol, tetracycline, fumarate (sodium salt), isopropyl β-D-1-thiogalactopyranoside (IPTG), L-arabinose, imidazole, benzoate (sodium salt), poly-L-lysine, resazurin (sodium salt), ethylenediaminetetraacetic (EDTA), and EDTA-free cOmplete™ Protease Inhibitor cocktail were purchased from Sigma-Aldrich (St. Louis, MO, USA); Phosphate-buffered saline (PBS) was purchased from Biological Industries USA, Inc. (Cromwell, CT,USA); Paraformaldehyde (PFA) was purchased from Thermo Scientific Inc. (Rockford, IL, USA); Vitamin B1 Hydrochloride from Fluka (Gillingham, England); BugBuster™ Protein Extraction Reagent (used to lyse cells for Western blots) was purchased from Novagen, Inc. (A Brand of EMD Biosciences, Inc., an Affiliate of Merck KGaA, Darmstadt, Germany); Bacto Casamino Acids and Bacto Agar was purchased from Difco (Franklin Lakes, NJ, USA). Bovine Trypsin was purchased from Promega (Madison, WI, USA). LC–MS/MS Solvents were purchased from Bio-Lab Chemicals (Jerusalem, Israel). All the compounds were of the highest purity grade available. Antibodies: anti-flagellin antibody (serotype H48) was a gift from the National Center for Enterobacteriaceae (Central Laboratories, Ministry of Health, Jerusalem, Israel). Anti-FrdABCD antibody was prepared in earlier studies in our lab [[11]; anti-mCherry monoclonal antibody was purchased from Abcam (Cambridge, MA, USA); and Alexa Fluor 647 anti-mouse antibody was purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA).
Bacterial strains
The strains and plasmids used in this study are listed in Table 2. For construction of B275ΔfrdABCD and DFB190ΔfrdABCD, a P1 phage lysate [48] prepared on RP437ΔfrdABCD was used to infect B275 and DFB190, respectively. Positive colonies were selected for tetracycline resistance and for their inability to grow on a fumarate and glycerol minimal medium under anaerobic conditions. For construction of B275ΔfrdA, B275ΔfrdB, B275ΔfrdC, and B275ΔfrdD, the P1 phage lysates, prepared on corresponding KEIO deletion strains (Table 2), were used to infect B275. All the insertions were verified by PCR amplification and sequencing.
Table 2.
Strains and plasmids used in this study
| Strain/plasmid | Relevant genotype/description | References |
|---|---|---|
| Strains | ||
| BL21(λDE3)pLysS | hsdS gal ompT rB− mB− [ΔC1857(Ts)indI Sam7 min5 lacUV5-T7 gene 1] | [70], Novagen |
| JW2865–3 | rrnB3 lacZ4787 hsdR514 (araBAD)567 (rhaBAD)568 rph-1ygfY (sdhE) | [71] |
| RP437 | his thr leu metE thi eda rpsL; wild type for chemotaxis | [72] |
| RP437ΔsdhΔfrd | ΔsdhΔfrd, RP437 parent | [11] |
| B275 | thr leu metE thi lac rpsL; wild type for chemotaxis | [73] |
| B275ΔfrdABCD | ΔfrdABCD, B275 parent | This study |
| B275ΔfrdA | ΔfrdA, B275 parent | This study |
| B275ΔfrdB | ΔfrdB, B275 parent | This study |
| B275ΔfrdC | ΔfrdC, B275 parent | This study |
| B275ΔfrdD | ΔfrdD, B275 parent | This study |
| DFB190 | ΔfliM, RP437 parent | [74] |
| DFB190ΔfrdABCD | ΔfliMΔfrdABCD, RP437 parent | This study |
| Plasmids | ||
| pCa24N | empty vector, CapR | [50] |
| pCa24N-FrdA | FrdA expression vector, N-terminal 6×His, pCa24N based, CapR | [50] |
| pCa24N-FrdB | FrdB expression vector, N-terminal 6×His, pCa24N based, CapR | [50] |
| pCa24N-FrdC | FrdC expression vector, N-terminal 6×His, pCa24N based, CapR | [50] |
| pCa24N-FrdD | FrdD expression vector, N-terminal 6×His, pCa24N based,CapR | [50] |
| pEWGI | FliG expression vector, N-terminal 10×His, AmpR | [11] |
| pCa24N-FliGcw | FliG(Δ169–171) expression vector, N-terminal 6×His, pEWG1 based, AmpR | This study |
| pH3 | FrdABCD, frdABCD operon with natural frd promoter, pBR322 derivative, low copy number plasmid AmpR | [75] |
| pH3-Dendra2-FrdABCD | Dendra2-FrdABCD expression vector (FrdA is linked to Dendra2) with natural frdABCD promoter, pH3 based, AmpR | This study |
| pH3-Dendra2 | Dendra2 expression vector with natural frdABCD promoter, pH3 based, AmpR | This study |
| pFliMWT-mPlum | FliM-mPlum expression vector with arabinose PBAD promoter | This study |
| pFliMCW-mPlum | FliM(R60C)-mPlum expression vector with arabinose PBAD promoter | This study |
Plasmids
The plasmid pH3-Dendra2-FrdABCD was constructed in two ligation steps. First, the plasmid pH3 (Table 2) was amplified by PCR using primers 5’-ACAGCGGTACCGTCGTGCAAACCTTTCAAGCCGATC-3’ and 5’-ATTCCTCCAGA-TTGTTTTTATCCCACAGCCACGTAC-3’. This added a KpnI site 5’ to frdA gene of the resulted linear DNA, which ends immediately downstream to the frdABCD promoter. Next, the sequence encoding for Dendra2 protein was amplified from pDendra2 Vector [Clontech Laboratories (Takara Bio Company), Mountain View, CA, USA], with the primer sequences 5’-GTGAACACCCCGGGAATTAACCTGATCAAG-3’ and 5’-ACAGCGGTACC-TGCAGGCGGCCGCCTCTTGTA-3’, while the start codon was replaced by GTG start codon as in frdA gene and its stop codon was removed. The amplified PCR fragment was digested with KpnI, leaving one end blunt, and ligated with KpnI predigested plasmid from the previous step, inserting dendra2 gene downstream the frd promoter and in-frame with the frdA gene. The functional status of Dendra2-FrdA fusion protein was verified by the ability of the DFB190ΔfrdABCD mutant, transformed with this plasmid, to grow under anaerobic conditions. The plasmid pH3-Dendra2 was constructed similarly but the stop codon was added at the 5’ of the dendra2 reverse primer.
The plasmid pCa24N-FliGCW was prepared by deleting residues 169–171 [19,49]. This was done by PCR amplification of the plasmid pCa24N-FliG [50] with the primers 5’-CTGGCGGAGCTGACCGAA GTACTGAATGG-3’ and 5’-TGCACGCCGCCAAAGGTGGCGATA-3’, which, upon ligation of the amplified sequence, resulted in a plasmid containing FliG without the sequence encoding for aforementioned residues.
To construct the plasmids pFliMWT-mPlum and pFliMCW-mPlum, the sequence encoding for mPlum fluorescent protein was amplified by PCR from pRSET-mPlum (Invitrogen™, Termo Fisher Scientific, Waltham, MA, USA) with the primers 5’-ACAGCAGATCTGGCATGGTGAGCAAGGGCGAGGAGGTC-3’ and 5’-ACGCAAGCTT-AGGCGCCGGTGGAGTGGC-3’, which add a BglII site upstream and stop codon and HindIII site downstream of the mPlum coding sequence. Next, the amplified PCR fragment was digested with BglII and HindIII and ligated with similarly digested plasmids pMWT (encoding Salmonella wild-type FliM) and pMCW1 [encoding Salmonella clockwise-locked FliM(R60C)] [51]. The functional status of FliG-mPlum fusion protein was verified by the ability of the DFB190 strain (Table 2), transformed with this plasmid, to form expanding rings on Tryptone broth semi-solid agar plates. Expression level of FliM-mPlum fusion protein in DFB190ΔfrdABCD was optimized by modulating the D-arabinose concentration in order to provide maximal restoration of motility during the anaerobic growth (2.5–3.5 μM arabinose). Transforming DFB190ΔfrdABCD with FliMCW-mPlum resulted in cells rotating clockwise 90% of the time.
All genetic constructs were confirmed by DNA sequencing. Fluorescence of the fusion proteins was verified by microscopic visualization. All FliM-mPlum fusions were found non-fluorescent due to the anaerobic conditions (the maturation process that makes mPlum fluorescent requires oxygen [52][53]).
Growth and media
All bacterial cultures for protein isolation and genetic manipulations were grown in Luria Bertani Broth [54] at 37°C. For determining the direction of flagellar rotation of aerobically-grown cells, overnight cultures were diluted 1:100 in fresh Tryptone broth and grown at 30°C to OD600 of ~0.5. For determining the direction of flagellar rotation of anaerobic cells and their control aerobic cells, overnight cultures were diluted 1:50 in fresh M9 salt medium supplemented with glycerol (20 mM), the required amino acids (1 mM each), and, where indicated, with sodium fumarate (40 mM) or L-arabinose (40 mM). For anaerobic conditions, the tube was filled to the end and tightly closed, then shaken at 30°C and 100 rpm. Conversion of colored resazurin (1μg/ml) to colorless resorufin in a parallelly-grown wild-type culture was used as an indication for reaching anaerobic conditions. For super-resolution experiments, the anaerobic bacterial cultures were similarly grown at 30°C but the M9 salt medium was supplemented with Casamino acids (0.5%), vitamin B1 (0.5 mg) and sodium fumarate (40 mM). For overexpression of Frd proteins, the overnight cultures were diluted 1:100 in fresh Tryptone broth supplemented with fumarate (40 mM) and grown aerobically at 30°C to OD600 of ~0.1, followed by further 3 h incubation with 0–1000 μM IPTG.
Protein isolation
For lysis, the bacterial cultures were suspended in buffer containing 50 mM Tris pH 8, 0.5 M NaCl, 5% glycerol, DNase, lysozyme, 1 mM PMSF, EDTA-free cOmplete™ Protease Inhibitor cocktail and lysed by sonication. All the proteins were concentrated by ultrafiltration (Amicon Ultra centrifugal filter units, Sigma-Aldrich, St. Louis, MO, USA).
His-tagged-FrdA
The pCA24N-FrdA vector was expressed in RP437ΔfrdΔsdhE (Table 2) by induction with 0.2 mM IPTG overnight at 15°C. His-FrdA was captured on a HisTrap_FF-crude_5ml column (GE Healthcare Life Sciences) equilibrated with 50 mM Tris (pH 8.0), 0.5 M NaCl, 20 mM imidazole and eluted with the same buffer containing 0.5 M imidazole. The pooled FrdA-containing fractions were loaded onto a HiLoad_16/60_Superdex column (GE Healthcare Life Sciences) equilibrated with PBS. The purification was carried out by the Dana and Yossie Hollander Center for Structural Proteomics (ISPC).
His-tagged-FliG
10×His-FliGwt and 6×His-FliGcw were expressed from the plasmids pEWG1 and pCa24N-FliGcw, respectively, after being transformed into strain BL21(lDE3)pLysS (Table 2). The expression was induced with IPTG (0.2–0.4 mM) at 15°C overnight. The proteins were purified successively by HisTrap-HP affinity chromatography (GE Healthcare Life Sciences), HiLoad 16/60 Superdex 75 prepgrade chromatography (GE Healthcare Life Sciences) both in PBS, and Tricorn Q 10/100GL chromatography (GE Healthcare Life Sciences) using 20 mM sodium phosphate pH 7.2 as a binding buffer. The proteins were eluted with a linear gradient to the same buffer containing 1 M NaCl. Some batches of 6×His-FliGcw were purified by HisTrap-HP affinity chromatography only.
Determination of the direction of flagellar rotation by tethering assay
The direction of flagellar rotation was determined by the tethering assay [16]. Bacterial cultures were transferred several times through a syringe needle to shear the flagella, washed twice in motility buffer (10 mM KPi pH 7.0, 0.1 mM EDTA and 0.1 mM L-methionine) and immobilized on a microscope coverslip using an anti-flagellin antibody. The coverslip was placed in a flow chamber and the rotation of the tethered cells was monitored with a phase-contrast microscope (Zeiss, Oberkochen, Germany) and recorded using uEye UI-2410-M-GL camera (IDS Imaging Development Systems GmbH; Obersulm, Germany) at 75 frames/s. The recordings were analyzed with a custom-made software [55].
MST
MST measurements were performed on a Monolith NT 115 instrument (NanoTemper Technologies GmbH, München, Germany). All measurements were performed at 90–60% LED power, 60% MST power with 30 s laser-on time and 5 s laser-off time. His-tagged-FrdA (~95% purity) was used at the concentration of 15 μM. The natural fluorescence of the flavin cofactor was utilized to follow the migration of the protein. His-tagged-FliGwt or His-tagged-FliGcw (~95% purity) was titrated in 1:1 dilutions beginning with 138 or 319 μM. The experiments were performed in PBS buffer with 0.05% Tween and measured in standard capillaries (NanoTemper Technologies). All binding reactions were incubated for 10 min at room temperature before they were loaded onto the capillaries. Curve fitting was carried out by Microcal™ Origin version 6 software (Northampton, MA, USA).
In-gel Digestion and Protein Identification by LC-ESI-MS/MS
Protein bands were excised from a Coomassie-stained SDS gel. They were subsequently reduced, alkylated and digested with bovine Trypsin, at a concentration of 12.5 ng/μl in 50 mM ammonium bicarbonate at 37°C, as described [56,57]. The peptide mixtures were extracted with 80% CH3CN and 1% CF3COOH, followed by evaporation of the organic solvent in a vacuum centrifuge. The resulting peptide mixtures were reconstituted in 80% formic acid and immediately diluted 1:10 with Milli-Q water before analysis. Liquid chromatography–tandem mass spectrometry (LC–MS/MS) was performed using a 15 cm reversed-phase fused-silica capillary column (inner diameter, 75 μm) made in-house and packed with 3 μm ReproSil-Pur C18AQ media (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The LC system, an UltiMate 3000 (Dionex, Sunnyvale, CA, USA), was used in conjunction with a LTQ Orbitrap XL (Thermo Fisher Scientific, Bremen, Germany) operated in the positive ion mode and equipped with a nanoelectrospray ion source. Peptides were separated with a two hour gradient from 5 to 65% acetonitrile (buffer A, 5% acetonitrile, 0.1% formic acid and 0.005% TFA; buffer B, 90% acetonitrile, 0.2% formic acid and 0.005%TFA). The voltage applied to the union to produce an electrospray was 1.2 kV. The mass spectrometer was operated in the data-dependent mode. Survey mass spectrometry scans were acquired in the Orbitrap (Thermo Fisher Scientific, Bremen, Germany) with the resolution set to a value of 60,000. The seven most intense ions per scan were fragmented and analyzed in the linear ion trap. Raw data files were searched with MASCOT (Matrix Science, London, UK) against the Uniprot E. coli database. Search parameters included a fixed modification of 57.02146 Da (carboxyamidomethylation) on Cys, and variable modifications 15.99491 Da (oxidation) on Met, and 0.984016 Da (deamidation) on Asn and Gln. The search parameters also included: maximum 2 missed cleavages, initial precursor ion mass tolerance 10 ppm, fragment ion mass tolerance 0.6 Da. Samples were further analyzed in Scaffold (Proteome software, Portland, Oregon, USA).
Super-resolution microscopy
The cells were first optimized with respect to growth conditions and fixation procedure by comparing the photoconversion efficiency of Dendra2 at each set of conditions before and after fixation, as recommended [58]. At the chosen conditions (see above for growth and below for fixation), the fluorescence intensity and the photoconversion efficiencies were similar before and after fixation. Immunostaining of FliM-mPlum fusion protein using anti-mCherry monoclonal antibody and Alexa Fluor 647-conjugated anti-mouse antibody was performed according to manufacturers’ instructions as follows. Immediately after air exposure, anaerobically-grown bacterial cultures were precipitated and resuspended in 4% (v/v) paraformaldehyde in PBS, followed by 5 min incubation with shaking for fixation and permeabilization. The cells were spun down (470×g, 10 min, room temperature) and then washed with PBS (2 centrifugations at 470×g for 3 min followed by centrifugation at 1300×g for 3 min). The cells were then blocked with 3% BSA in PBS for 30 min at room temperature with gentle rocking, followed by aspiration of the blocking buffer and 1 h incubation with the primary antibody anti-mCherry (5 μg/ml) in 1% BSA in PBS with gentle rocking. Following 3 washes in PBS (twice at 470×g and once at 1300×g, as above), the cells were incubated in the dark with gentle rocking for 45 min with the secondary antibody Alexa Fluor 647-conjugated anti-mouse antibody (1.4 μg/ml) in PBS, and washed with PBS 3 times as above. The cells were then immobilized for 20 min on pre-washed polylysine (0.2% w/v) coated coverslips, gently washed 3 times with 0.5 ml PBS, and covered with freshly prepared imaging buffer [59], which contains 10 mM NaCl, 10% (w/v) glucose, 5 mM cysteamine, and oxygen scavengers (7 μM glucose oxidase and 56 nM catalase) in 50 mM Tris (pH 8.0). Super-resolution images were recorded with Vutara SR 200 microscope (Bruker, Billerica, MA, USA). Images were recorded using a 60X, NA 1.2 water immersion objective (Olympus, Tokyo, Japan) and Evolve 512 EMCCD camera (Photometrics, Teledyne Technologies, Tucson, AZ, USA) with the gain set at 50. Combined PALM [24] and STORM [60] imaging was performed in two steps. First, STORM was recorded for 5000 frames while the photoblinking of Alexa Fluor 647 was induced with a 640 nm laser, set to 1.2 kW/cm2. Next, PALM was recorded. Photoswitching of Dendra2 was activated by a 405 nm laser, whose intensity was stepwise and slowly increased. The intensity of the exciting 561 nm laser was kept constant at 20 kW/cm2. All the recordings were carried out at 10 frames/s. Acquisition was carried out until all Dendra2 molecules were converted and detected (15,000–20,000 frames). Data were analyzed by Vutara SRX software (Bruker). To avoid over-counting in PALM records, single emission events detected in sequential frames and localized within the range of the resolution of a single-molecule were grouped as one detection event. In addition, to avoid over-counting because of photoblinking of single Dendra2 molecules, clusters of molecules whose time of appearance was too close to be individually detectable by the software were eliminated manually. Over-counting due to altered blinking behavior of Dendra2 in the imaging buffer was ruled out by preparing control samples for imaging in PBS [25] and finding that the number of molecules per bacterium in PBS (270 ± 20, mean ± SEM; n = 21 cells) was not different from the number of molecules detected in imaging buffer (cf. Table 1). Quantitative analysis of spatial distribution of Dendra2 molecules was performed using Image-Based Algorithm for two dimensions of Vutara SRX Software. Minimal cluster count was set to be 7 molecules. Minimal density threshold was separately set for each bacterium as 3 times the average density of Dendra2 in this cell. Clusters identified in this way were in good agreement with those detected visually. Motors were identified by groups of at least 12 single localizations forming two-dimension round or ring-like shapes. FrdA molecules were considered interacting with the motor if found within a 0.3×0.3μm square surrounding the center of the motor. This area was calculated on the basis of the diameter of FliM ring (~47 nm), the dimensions of the proteins involved (FrdA, 70 Å; Dendra2, 29.6 Å, according to the crystal structures available at the Protein Data Bank), and the resolution (35 nm).
Computational methods
MD (50–100 ns) [61–63] and detailed conformational analysis were performed for unbound E. coli FliG models. Homology models of E. coli FliG were built from the indicated structural data by using Modeller v8 [64] as published in ModBase [65]. In our computational modeling of possible interactions between FrdA(BCD) and FliG, we applied a rigid-body docking protocol by implementing Potential Smoothing and Search (PSS) algorithm, as described[66,67], to all conformational states of FliG identified in the MD study. For the FliG-FrdA complexes, we performed multiple short-time MD runs (10–20 ns) for refinement only. Docking was used in combination with Poisson-Boltzmann electrostatics methods ([68,69] and references cited therein). Multi-step binding process in the FliG-FrdA system was modeled as described [69].
Statistical analysis
All statistical analyses were carried out by Instat software (GraphPad Software, San Diego, CA).
Supplementary Material
Acknowledgments
We thank Dr. Hillary Voet, a statistician, for the statistical advices, Dr. Shira Albec from the Hollander Center for Structural Proteomics for protein purification, Dr. Irina Shin for carrying out the MST measurements, and Vladimir Kiss for assistance in fluorescence microscopy. Gary Cecchini was supported by the National Institutes of Health award GM61606. GC is also the recipient of a Senior Research Career Scientist award 1K6B004215 from the Department of Veterans Affairs.
Abbreviations used:
- ARM
armadillo repeat motif
- FliGccw
FliG in its counterclockwise state
- FliGcw
FliG in its clockwise state
- FrdABCD
fumarate reductase
- FRET
Förster resonance energy transfer
- MD
molecular dynamics
- MST
MicroScale Thermophoresis
- PALM
photoactivated localization microscopy
- SdhCDAB
succinate dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Berg HC, The rotary motor of bacterial flagella, Annu. Rev. Biochem 72 (2003) 19–54. [DOI] [PubMed] [Google Scholar]
- [2].Eisenbach M, Bacterial chemotaxis, in: Eisenbach M, Chemotaxis, Imperial College Press, London, 2004: pp. 53–215. [Google Scholar]
- [3].Terashima H, Kojima S, Homma M, Flagellar motility in bacteria structure and function of flagellar motor, Int Rev Cell Mol Biol 270 (2008) 39–85. [DOI] [PubMed] [Google Scholar]
- [4].Morimoto YV, Minamino T, Structure and function of the bi-directional bacterial flagellar motor, Biomolecules. 4 (2014) 217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Porter SL, Wadhams GH, Armitage JP, Signal processing in complex chemotaxis pathways, Nature Reviews. 9 (2011) 153–165. [DOI] [PubMed] [Google Scholar]
- [6].Janausch IG, Zientz E, Tran QH, Kroger A, Unden G, C-4-dicarboxylate carriers and sensors in bacteria, Biochim. Biophys. Acta 1553 (2002) 39–56. [DOI] [PubMed] [Google Scholar]
- [7].Barak R, Eisenbach M, Fumarate or a fumarate metabolite restores switching ability to rotating flagella of bacterial envelopes, J. Bacteriol 174 (1992) 643–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Barak R, Giebel I, Eisenbach M, The specificity of fumarate as a switching factor of the bacterial flagellar motor, Mol. Microbiol 19 (1996) 139–144. [DOI] [PubMed] [Google Scholar]
- [9].Prasad K, Caplan SR, Eisenbach M, Fumarate modulates bacterial flagellar rotation by lowering the free energy difference between the clockwise and counterclockwise states of the motor, J. Mol. Biol 280 (1998) 821–828. [DOI] [PubMed] [Google Scholar]
- [10].Montrone M, Eisenbach M, Oesterhelt D, Marwan W, Regulation of switching frequency and bias of the bacterial flagellar motor by CheY and fumarate, J. Bacteriol 180 (1998) 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cohen-Ben-Lulu GN, Francis NR, Shimoni E, Noy D, Davidov Y, Prasad K, et al. , The bacterial flagellar switch complex is getting more complex, Embo J 27 (2008) 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cecchini G, Schroder I, Gunsalus RP, Maklashina E, Succinate dehydrogenase and fumarate reductase from Escherichia coli, Biochim. Biophys. Acta 1553 (2002) 140–157. [DOI] [PubMed] [Google Scholar]
- [13].Jones HM, Gunsalus RP, Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate, J. Bacteriol 164 (1985) 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jones HM, Gunsalus RP, Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and thefnrgene product, J. Bacteriol 169 (1987) 3340–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sharma P, Maklashina E, Cecchini G, Iverson TM, Crystal structure of an assembly intermediate of respiratory Complex II, Nat Commun 9 (2018) 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Silverman M, Simon M, Flagellar rotation and the mechanism of bacterial motility, Nature. 249 (1974) 73–74. [DOI] [PubMed] [Google Scholar]
- [17].Li H, Sourjik V, Assembly and stability of flagellar motor in Escherichia coli, Mol. Microbiol 80 (2011) 886–899. [DOI] [PubMed] [Google Scholar]
- [18].Maklashina E, Rajagukguk S, Iverson TM, Cecchini G, The unassembled flavoprotein subunits of human and bacterial complex II have impaired catalytic activity and generate only minor amounts of ROS, J. Biol. Chem 293 (2018) 7754–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Minamino T, Imada K, Kinoshita M, Nakamura S, Morimoto YV, Namba K, Structural insight into the rotational switching mechanism of the bacterial flagellar motor, PLoS Biol 9 (2011) e1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S, Protein-binding assays in biological liquids using microscale thermophoresis, Nat Commun 1 (2010) 100. [DOI] [PubMed] [Google Scholar]
- [21].Sourjik V, Berg HC, Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer, Proc. Natl. Acad. Sci. U.S.a 99 (2002) 12669–12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sagi Y, Khan S, Eisenbach M, Binding of the chemotaxis response regulator CheY to the isolated, intact switch complex of the bacterial flagellar motor: Lack of cooperativity, J. Biol. Chem 278 (2003) 25867–25871. [DOI] [PubMed] [Google Scholar]
- [23].Kinoshita M, Furukawa Y, Uchiyama S, Imada K, Namba K, Minamino T, Insight into adaptive remodeling of the rotor ring complex of the bacterial flagellar motor, Biochem. Biophys. Res. Commun 496 (2018) 12–17. [DOI] [PubMed] [Google Scholar]
- [24].Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. , Imaging intracellular fluorescent proteins at nanometer resolution, Science. 313 (2006) 1642–1645. [DOI] [PubMed] [Google Scholar]
- [25].Lee S-H, Shin JY, Lee A, Bustamante C, Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM), Proc. Natl. Acad. Sci. U.S.a 109 (2012) 17436–17441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM, Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium, J. Bacteriol 174 (1992) 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Erhardt H, Dempwolff F, Pfreundschuh M, Riehle M, Schäfer C, Pohl T, et al. , Organization of the Escherichia coli aerobic enzyme complexes of oxidative phosphorylation in dynamic domains within the cytoplasmic membrane, Microbiology Open. 3 (2014) 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vartanian AS, Paz A, Fortgang EA, Abramson J, Dahlquist FW, Structure of flagellar motor proteins in complex allows for insights into motor structure and switching, J. Biol. Chem 287 (2012) 35779–35783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D, Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching, Nature. 466 (2010) 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Starbird CA, Tomasiak TM, Singh PK, Yankovskaya V, Maklashina E, Eisenbach M, et al. , New crystal forms of the integral membrane Escherichia coli quinol:fumarate reductase suggest that ligands control domain movement, J. Struct. Biol 202 (2018) 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC, Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site, J. Biol. Chem 277 (2002) 16124–16130. [DOI] [PubMed] [Google Scholar]
- [32].Hudson JM, Heffron K, Kotlyar V, Sher Y, Maklashina E, Cecchini G, et al. , Electron transfer and catalytic control by the iron-sulfur clusters in a respiratory enzyme, E. coli fumarate reductase, J. Am. Chem. Soc 127 (2005) 6977–6989. [DOI] [PubMed] [Google Scholar]
- [33].Starbird CA, Maklashina E, Sharma P, Qualls-Histed S, Cecchini G, Iverson TM, Structural and biochemical analyses reveal insights into covalent flavinylation of the Escherichia coli Complex II homolog quinol:fumarate reductase, J. Biol. Chem 292 (2017) 12921–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nishikino T, Hijikata A, Miyanoiri Y, Onoue Y, Kojima S, Shirai T, et al. , Rotational direction of flagellar motor from the conformation of FliG middle domain in marine Vibrio, Sci Rep 8 (2018) 17793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lam K-H, Lam WWL, Wong JY-K, Chan L-C, Kotaka M, Ling TK-W, et al. , Structural basis of FliG-FliM interaction in Helicobacter pylori, Mol. Microbiol 88 (2013) 798–812. [DOI] [PubMed] [Google Scholar]
- [36].Sircar R, Borbat PP, Lynch MJ, Bhatnagar J, Beyersdorf MS, Halkides CJ, et al. , Assembly States of FliM and FliG within the Flagellar Switch Complex, J. Mol. Biol 427 (2015) 867–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Glusker JP, The binding of ions to proteins, in: Allen G (Ed.), Protein, London, UK, 1999. [Google Scholar]
- [38].Kim EA, Panushka J, Meyer T, Carlisle R, Baker S, Ide N, et al. , Architecture of the Flagellar Switch Complex of Escherichia coli: Conformational Plasticity of FliG and Implications for Adaptive Remodeling, J. Mol. Biol 429 (2017) 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lele PP, Berg HC, Switching of bacterial flagellar motors is triggered by mutant FliG, Biophys. J 108 (2015) 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wolfe AJ, Berg HC, Migration of bacteria in semisolid agar, Proc. Natl. Acad. Sci. U.S.a 86 (1989) 6973–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chang D-E, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. , Carbon nutrition of Escherichia coli in the mouse intestine, Proc. Natl. Acad. Sci. U.S.a 101 (2004) 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI, Host-bacterial mutualism in the human intestine, Science. 307 (2005) 1915–1920. [DOI] [PubMed] [Google Scholar]
- [43].Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, et al. , Respiration of Escherichia coli in the mouse intestine, Infection and Immunity. 75 (2007) 4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, et al. , Anaerobic respiration of Escherichia coli in the mouse intestine, Infection and Immunity. 79 (2011) 4218–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bibikov SI, Biran R, Rudd KE, Parkinson JS, A signal transducer for aerotaxis in Escherichia coli, J. Bacteriol 179 (1997) 4075–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, et al. , The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior, Proc. Natl. Acad. Sci. U.S.a 94 (1997) 10541–10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Prüss BM, Campbell JW, Van Dyk TK, Zhu C, Kogan Y, Matsumura P, FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer, J. Bacteriol 185 (2003) 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Silhavy TJ, Berman ML, Enquist LW, Experiments with gene fusions, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1984. [Google Scholar]
- [49].Togashi F, Yamaguchi S, Kihara M, Aizawa SI, Macnab RM, An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB, J. Bacteriol 179 (1997) 2994–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, et al. , Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research, DNA Res 12 (2005) 291–299. [DOI] [PubMed] [Google Scholar]
- [51].Bren A, Eisenbach M, Changing the direction of flagellar rotation in bacteria by modulating the ratio between the rotational states of the switch protein FliM, J. Mol. Biol 312 (2001) 699–709. [DOI] [PubMed] [Google Scholar]
- [52].Moore MM, Oteng-Pabi SK, Pandelieva AT, Mayo SL, Chica RA, Recovery of red fluorescent protein chromophore maturation deficiency through rational design, PLoS ONE. 7 (2012) e52463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Piatkevich KD, Verkhusha VV, Guide to red fluorescent proteins and biosensors for flow cytometry, Methods Cell Biol 102 (2011) 431–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lennox ES, Transduction of linked genetic characters of the host by bacteriophage P1, Virology. 1 (1955) 190–206. [DOI] [PubMed] [Google Scholar]
- [55].Zarbiv G, Li H, Wolf A, Cecchini G, Caplan SR, Sourjik V, et al. , Energy complexes are apparently associated with the switch-motor complex of bacterial flagella, J. Mol. Biol 416 (2012) 192–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, et al. , Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry, Nature. 379 (1996) 466–469. [DOI] [PubMed] [Google Scholar]
- [57].Shevchenko A, Wilm M, Vorm O, Mann M, Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels, Anal. Chem 68 (1996) 850–858. [DOI] [PubMed] [Google Scholar]
- [58].Bach JN, Giacomelli G, Bramkamp M, Sample Preparation and Choice of Fluorophores for Single and Dual Color Photo-Activated Localization Microscopy (PALM) with Bacterial Cells, Methods Mol Biol 1563 (2017) 129–141. [DOI] [PubMed] [Google Scholar]
- [59].Chozinski TJ, Gagnon LA, Vaughan JC, Twinkle, twinkle little star: Photoswitchable fluorophores for super-resolution imaging, FEBS Lett 588 (2014) 3603–3612. [DOI] [PubMed] [Google Scholar]
- [60].Rust MJ, Bates M, Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nature Methods. 3 (2006) 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Berendsen H, van der Spoel D, van Drunen R, Gromacs - a message-passing parallel molecular-dynamics implementation, Comput Phys Commun 91 (1995) 43–56. [Google Scholar]
- [62].Lindahl E, Hess B, van der Spoel D, GROMACS 3.0: a package for molecular simulation and trajectory analysis, J Mol Model 7 (2001) 306–317. [Google Scholar]
- [63].van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC, GROMACS: fast, flexible, and free, J Comput Chem 26 (2005) 1701–1718. [DOI] [PubMed] [Google Scholar]
- [64].Eswar N, Eramian D, Webb B, Shen M-Y, Šali A, Protein structure modeling with MODELLER, Methods Mol Biol 426 (2008) 145–159. [DOI] [PubMed] [Google Scholar]
- [65].Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, et al. , ModBase, a database of annotated comparative protein structure models and associated resources, Nucleic Acids Res 42 (2014) D336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pappu RV, Hart RK, Ponder JW, Analysis and application of potential energy smoothing and search methods for global optimization, J Phys Chem B 102 (1998) 9725–9742. [Google Scholar]
- [67].Pappu RV, Marshall GR, Ponder JW, A potential smoothing algorithm accurately predicts transmembrane helix packing, Nature Structural Biology. 6 (1999) 50–55. [DOI] [PubMed] [Google Scholar]
- [68].Tworowski D, Safro M, The long-range electrostatic interactions control tRNA-aminoacyl-tRNA synthetase complex formation, Protein Sci 12 (2003) 1247–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tworowski D, Feldman AV, Safro MG, Electrostatic potential of aminoacyl-tRNA synthetase navigates tRNA on its pathway to the binding site, J. Mol. Biol 350 (2005) 866–882. [DOI] [PubMed] [Google Scholar]
- [70].Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW, Use of T7 RNA polymerase to direct expression of cloned genes, Methods Enzymol 185 (1990) 60–89. [DOI] [PubMed] [Google Scholar]
- [71].Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection, Mol Syst Biol 2 (2006) 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Parkinson JS, Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis, J. Bacteriol 135 (1978) 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hazelbauer GL, Adler J, Role of the galactose binding protein in chemotaxis of Escherichia coli toward galactose, Nature New Biol 230 (1971) 101–104. [DOI] [PubMed] [Google Scholar]
- [74].Tang H, Blair DF, Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators, J. Bacteriol 177 (1995) 3485–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Westenberg DJ, Gunsalus RP, Ackrell BA, Sices H, Cecchini G, Escherichia coli fumarate reductase frdC and frdD mutants. Identification of amino acid residues involved in catalytic activity with quinones, J. Biol. Chem 268 (1993) 815–822. [PubMed] [Google Scholar]
- [76].Maklashina E, Berthold DA, Cecchini G, Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth, J. Bacteriol 180 (1998) 5989–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.