Abstract
Photoreceptor phosphodiesterase (PDE6) is the central effector enzyme in the visual excitation pathway in rod and cone photoreceptors. Its tight regulation is essential for the speed, sensitivity, recovery and adaptation of visual signaling. The rod PDE6 holoenzyme (Pαβγ2) is composed of a catalytic heterodimer (Pαβ) that binds two inhibitory γ subunits. Each of the two catalytic subunits (Pα and Pβ) contains a catalytic domain responsible for cGMP hydrolysis and two tandem GAF domains, one of which binds cGMP noncatalytically. Unlike related GAF-containing PDEs where cGMP binding allosterically activates catalysis, the physiological significance of cGMP binding to the GAF domains of PDE6 is unknown. To elucidate the structural determinants of PDE6 allosteric regulators, we biochemically characterized PDE6 complexes in various allosteric states (Pαβ, Pαβ-cGMP, Pαβγ2 and Pαβγ2-cGMP) with a quantitative cross-linking/mass spectrometry approach. We employed a normalization strategy to dissect the cross-linking reactivity of individual residues in order to assess the spatial cross-linking propensity of detected pairs. In addition to identifying cross-linked pairs that undergo conformational changes upon ligand binding, we observed an asymmetric binding of the inhibitory γ-subunit and the noncatalytic cGMP to the GAFa domains of rod PDE6, as well as a stable open conformation of Pαβ catalytic dimer in different allosteric states. These results advance our understanding of the exquisite regulatory control of the lifetime of rod PDE6 activation/deactivation during visual signaling, as well as providing a structural basis for interpreting how mutations in rod PDE6 subunits can lead to retinal diseases.
Keywords: phosphodiesterase 6 (PDE6), chemical cross-linking, quantitative mass spectrometry, visual transduction, allosteric regulation
Introduction
The conversion of light stimuli into neuro-electrical signals in photoreceptor cells of the vertebrate retina is mediated by a central effector enzyme, cyclic nucleotide phosphodiesterase-6 (PDE6). The photoreceptor-specific PDE6 is activated by the heterotrimeric G-protein, transducin, upon photoactivation of the G-protein coupled receptor, rhodopsin. Light-induced, PDE6-catalyzed cGMP hydrolysis leads a rapid decrease of cellular cGMP levels on the millisecond time scale, causing the closure of cGMP-gated ion channels, outer segment membrane hyperpolarization, and signal transmission to retinal neurons [1–4].
The rod photoreceptor is remarkably sensitive to light, and its ability to detect single photons enables scotopic (nighttime) vision. The cellular anatomy of rods, from the densely packed disk membrane structures in rod outer segment (ROS) to the high concentrations of membrane-confined phototransduction components, ensures both efficient absorption of photons and elaborate orchestration of transduction, amplification, and attenuation of visual signals [2].
As the central effector enzyme of rod and cone visual transduction, PDE6 possesses some salient characteristics that distinguish it from other members of the phosphodiesterase super-family. For instance, the isoform of PDE6 in the rod photoreceptor consists of a catalytic dimer of two homologous catalytic subunits, α and β (Pαβ), whereas cone PDE6 and the other ten PDE families are homodimeric. In addition, the PDE6 holoenzyme contains two tightly bound inhibitory γ-subunits (Pγ), which are displaced by activated transducin during visual excitation [1].
Structural models for several members of the phosphodiesterase super-family have been reported [5–7], including the crystal structure of the nearly full-length PDE2 catalytic dimer, which contains N-terminal tandem GAF domains (GAFa and GAFb) that allosterically bind cGMP and alter enzyme activity in the C-terminal catalytic domain [8]. In addition, crystal structures of the catalytic and tandem GAFab domains of PDE5 (the most closely related PDE family to PDE6) [9, 10], the GAFa domain of cone PDE6 [11], as well as a PDE5/6 chimeric catalytic domain [12] have been determined. The molecular architecture of the PDE6 holoenzyme has been determined by integrating high-density chemical cross-linking data with individual domain structures and negative-stain electron microscopy (EM) data [7]. The resulting model is in good agreement with more recently reported cryo-EM maps [13, 14], and the Gulati et al. study has provided additional structural evidence supporting a highly extended conformation of the inhibitory Pγ subunit [14], in marked contrast to the disordered structure of free Pγ in solution [15].
The regulatory Pγ subunit inhibits cGMP hydrolysis by direct binding of its last ten amino acid residues at the entrance to the active site [12, 16–18]. The affinity of Pγ for Pαβ catalytic dimer is allosterically modulated in a reciprocal manner by noncatalytic cGMP binding in the GAFa domain [19]. Pγ interacts with both GAF and catalytic domains of Pαβ and links the regulatory and catalytic domains of PDE6 in two ways: (1) Pγ binding to the catalytic dimer enhances the binding affinity of cGMP to the GAF domain [20]; (2) cGMP occupancy of the GAF domain enhances Pγ affinity to Pαβ [21, 22]. Direct allosteric communication between the GAF domains and catalytic domains of PDE6 has also been observed independent of Pγ binding [23]. Despite the abundance of biochemical data through functional assays, little is known about the molecular basis for this allosteric regulation.
Here, we utilize chemical cross-linking and quantitative mass spectrometric analysis (qCX-MS) to structurally characterize the allosteric regulation of PDE6 holoenzyme by the Pγ subunit and by cGMP binding to the regulatory GAF domains. This chemical cross-linking approach can identify amino acid residues in spatial proximity but not necessarily close in primary sequence, providing intermediate resolution information on relative domain positions and orientations [24–26]. In addition, quantitative information from mass spectrometric analysis can reveal conformational changes or shifts in conformational equilibrium that are challenging to detect with other approaches to structural determination [27]. To understand the structural determinants involved in allosteric regulation of PDE6, we carried out qCX-MS analysis of PDE6 in four different allosteric states and quantified the cross-linking propensity of 22 cross-linked pairs. Our results suggest that even though the binding of cGMP or Pγ does not cause major alterations in the molecular architecture of PDE6, substantial conformational changes take place, some of which are distant from ligand binding sites. In addition, our results reveal differences in the conformational changes of the Pα and Pβ subunits upon cGMP or Pγ binding, providing insights into the functional significance of the rod PDE6 catalytic heterodimer.
Results
Quantitative analysis of cross-linking of PDE6 in different liganded states
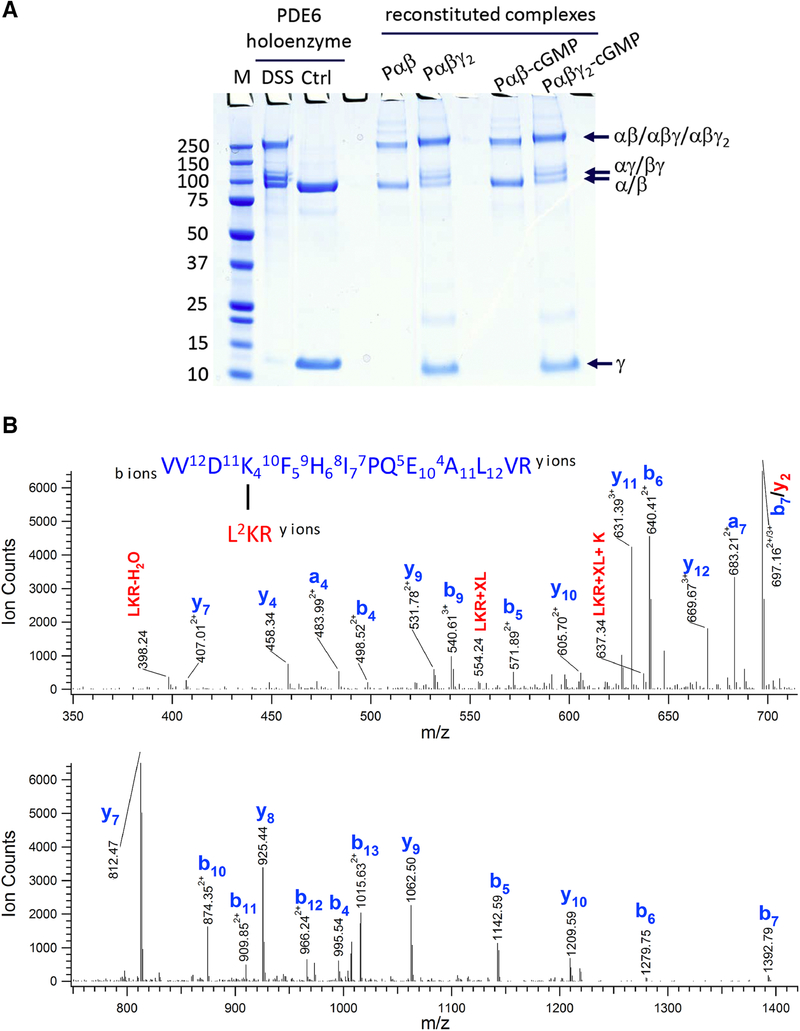
Due to the lack of an effective heterologous expression system for full-length rod Pα and Pβ subunits, we resorted to purification of native rod PDE6 from bovine rod outer segments. To investigate the conformation of PDE6 complexes in various allosteric states, we proteolytically digested the tightly bound Pγ subunits of the PDE6 holoenzyme [28] and re-purified the Pαβ catalytic dimer before reconstituting the following complexes with cGMP and/or Pγ: Pαβγ2, Pαβ with bound cGMP (Pαβ-cGMP) and Pαβγ2 with bound cGMP (Pαβγ2-cGMP). The catalytic activity of Pαβ catalytic dimer and various reconstituted PDE6 complexes were evaluated to ensure their functional and structural integrity [29]. When treated with chemical cross-linkers, the native PDE6 holoenzyme and the reconstituted PDE6 complexes yielded similar gel shift patterns, migrating at ~110 kDa (αγ, βγ) and 200–250 kDa (αβ, αβγ and αβγ2) in SDS-PAGE gels (Fig. 1A).
Figure 1.
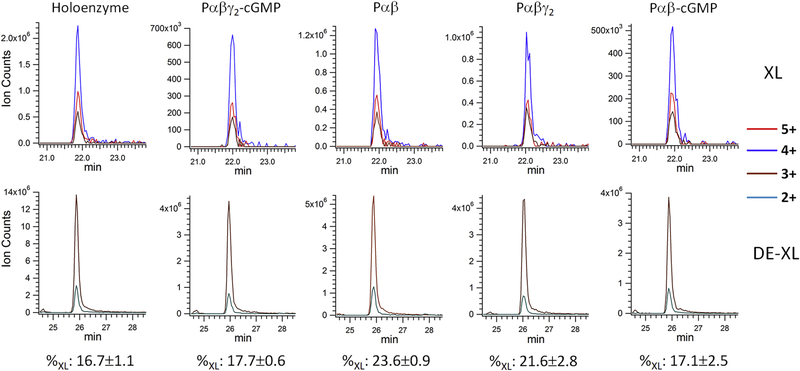
Chemical cross-linking of the PDE6 complexes in various allosteric states. A) Purified PDE6 holoenzyme or the Pαβ catalytic dimer reconstituted with Pγ or/and cGMP were cross-linked with DSS in 200-fold molar excess. A control sample (Ctrl) was treated identically except that the DSS cross-linker was omitted. Gel bands of uncross-linked subunits (γ and α/β) and cross-linked subunits (αγ or βγ at ~110 kDa; αβ or αβγ or αβγ2 at ~220 kDa) are indicated. B) Tandem MS spectrum of a cross-linked peptide from the 220 kDa band, with fragments from peptide V531-R544 and L580-R582. C) Extracted ion chromatograms (XIC) of the Lys534-Lys581 cross-linked peptides (XL) in charge state +5, +4 and +3 from various PDE6 complexes were overlaid (top panel). XIC of Lys534 dead-end cross-link (DE-XL) in charge state +3 and +2 from various PDE6 complexes were overlaid (bottom panel). MS signals of XL and DE-XL were used to calculate the cross-linking percentage (%XL) of Lys534-Lys581 in various PDE6 complexes.
We chose the amine-reactive, bifunctional cross-linker disuccinimidyl suberate (DSS), which yielded informative cross-linked pairs in our previous cross-linking/mass spectrometric analysis of PDE6 holoenzyme [7]. DSS mediates the formation of covalent bonds between lysine residues that are in close proximity, which can be identified by tryptic digestion and mass spectrometric analysis [26]. Furthermore, quantitative analysis of the cross-linked products can potentially elucidate conformational changes or shifts in conformational equilibrium [27]. After the initial cross-linking reaction, the second NHS-ester functionality of DSS can also react with water in a competing reaction to produce dead-end cross-links (DE-XL). Although the amount of cross-linked products relies on the concentration of cross-linkers, proteins and protein 3D structure, the relative amount of cross-linked products and DE-XLs is predominantly determined by the accessibility of a cross-linkable amino acid for the second NHS-ester functionality. It is reasonable to assume that the percentage of a productive cross-link after the initial cross-linking reaction mainly reflects the distance of cross-linked residues as well as a spatial orientation suited for the cross-linking reaction.
As one example, we identified an intra-subunit cross-link between Lys534 and Lys581 of Pα subunit (designated Lys534Pα-Lys581Pα) in the catalytic domain of the Pα subunit. The tandem MS spectrum of the peptide contained a nearly complete ladder of sequence ions for Lys534 (Fig. 1B, colored in blue) and four fragment ions for the short Lys581 peptide moiety (Fig. 1B, colored in red). Together with an accurate mass measurement (<5 ppm) of the peptide, we conclusively identified the cross-link between Pα Lys534 and Lys581. To calculate the cross-linking percentage of these two Lys residues, we used the sum of MS ion intensity of the cross-linked peptide or DE-XLs in all charge states (Fig. 1C). The DE-XL for Lys581 peptide moiety was not detected, because its molecular weight (three amino acids) was lower than the mass window for our LC-MS detection.
Since the Lys534Pα-Lys581Pα cross-link was observed in multiple gel bands (~90 kDa, ~110 kDa and 200–250 kDa) of cross-linked PDE6 complexes in various allosteric states, we calculated the average and standard deviation of the cross-linking percentage (Fig. 1C). As expected, the Lys534Pα-Lys581Pα cross-linking percentage of reconstituted Pαβγ2-cGMP complex was in excellent agreement with that of PDE6 holoenzyme, suggesting that the reconstitution of PDE6 holoenzyme (from purified Pαβ mixed with Pγ) maintained the structural integrity of native PDE6 holoenzyme purified from rod photoreceptor cells. Furthermore, the small standard deviations in the cross-linking percentage suggested small technical variations in our MS sample preparation and analysis.
We treated various liganded states of PDE6 with 200-fold (Table 1) or 70-fold (Supplemental Table 1) molar excess of DSS cross-linker relative to PDE6 holoenzyme. Data from 70-fold molar excess of DSS cross-linking (70×) samples generated significantly fewer cross-linked pairs and weaker signals. Only cross-links involving highly reactive Lys residues were detected in the 70× sample, consistent with a previous study under one-hit cross-linking conditions [30]. For those instances where quantitative analysis of cross-linked peptides from the 70× sample was feasible, similar cross-linking percentages were observed to the PDE6 holoenzyme with a 200-fold molar excess of DSS. For example, although the ion intensity of the Lys534Pα-Lys581Pα cross-linked peptide from 70× sample (Fig. S1; 2.0×105) was substantially lower than that from 200× sample (Fig. 1C; 2.0×106), similar cross-linking percentages were obtained from both samples (70×: 15.8 ± 0.0; 200×: 16.7 ± 1.1). Although the 70× data set was more limited than the 200× sample, these results indicate that our cross-linking percentage values are independent of cross-linker concentrations over this range of DSS concentrations.
Table 1.
Cross-linking propensity of PDE6 intra-subunit cross-links.
| Lys 1 | Lys 2 | Holoenzyme (%XL) | Pαβγ2-cGMP (%XL) | Pαβ (%XL) | Pαβγ2 (%XL) | Pαβ-cGMP (%XL) |
|---|---|---|---|---|---|---|
| Pα | ||||||
| 76 | 84 | 27.7±4.3 | 19.1±2.3 | 8.2±2.0* | 23.2±1.4 | 21.6±9.9 |
| 534 | 581 | 16.7±1.1 | 17.7±0.6 | 23.6±0.9** | 21.6±2.8 | 17.1±2.5 |
| 677 | 683 | 40.4±3.1 | 44.7±0.7 | 51.9±4.1* | 36.4±0.8** | 52.8±5.4** |
| 765 | 819 | 1.3±0.3 | 3.7±0.6 | 3.5±0.2 | 1.5±0.2** | 5.1±0.8** |
| 765 | 827 | 7.7±0.1 | 15.7±1.1 | 15.0±0.1 | 6.5±0.5** | 24.6±1.8** |
| 819 | 827 | 62.6±1.2 | 60.3±2.6 | 59.8±0.5 | 66.7±5.7 | 53.6±0.7 |
| Pβ | ||||||
| 490 | 532 | 7.1±2.4 | 8.0±0.9 | 4.8±1.2 | 6.2±2.6 | 8.1±1.9 |
| 675 | 681 | 30.3±4.1 | 30.0±2.1 | 46.4±0.7** | 21.8±1.9* | 50.2±8.0** |
| 763 | 826 | 2.6±0.1 | 2.6±0.4 | 2.4±0.6 | 1.8±0.2 | 2.9±0.1 |
| 763 | 827 | 1.2±0.4 | 0.7±0.1 | 0.8±0.1 | 0.5±0.1 | 0.9±0.1 |
| 763 | 817 | 14.0±1.5 | 36.6±1.0 | 32.9±3.1 | 19.9±0.4** | 47.9±1.0** |
| 763 | 832 | 1.9±0.3 | 0.2±0.0 | 0.3±0.0 | 0.1±0.1 | 0.3±0.0 |
| 817 | 827 | 10.5±0.3 | 6.5±1.3 | 6.9±2.0 | 7.4±1.0 | 4.9±0.8 |
| 817 | 826 | 28.8±3.6 | 29.3±1.1 | 31.3±1.1* | 37.2±0.1** | 23.2±0.4** |
| 817 | 832 | 9.6±0.4 | 3.2±0.8 | 2.3±0.0 | 4.1±0.6 | 4.8±0.8* |
| 826 | 832 | 0.6±0.1 | 3.2±0.3 | 3.3±0.1 | 3.1±0.2 | 2.9±0.3 |
Statistical significance was evaluated by ANOVA analysis followed by Tukey’s test. Conformational states that significantly deviated from the reconstituted PDE6 holoenzyme (Pαβγ2-cGMP) are annotated by asterisks (** for 0.05 α value; * for 0.1 α value).
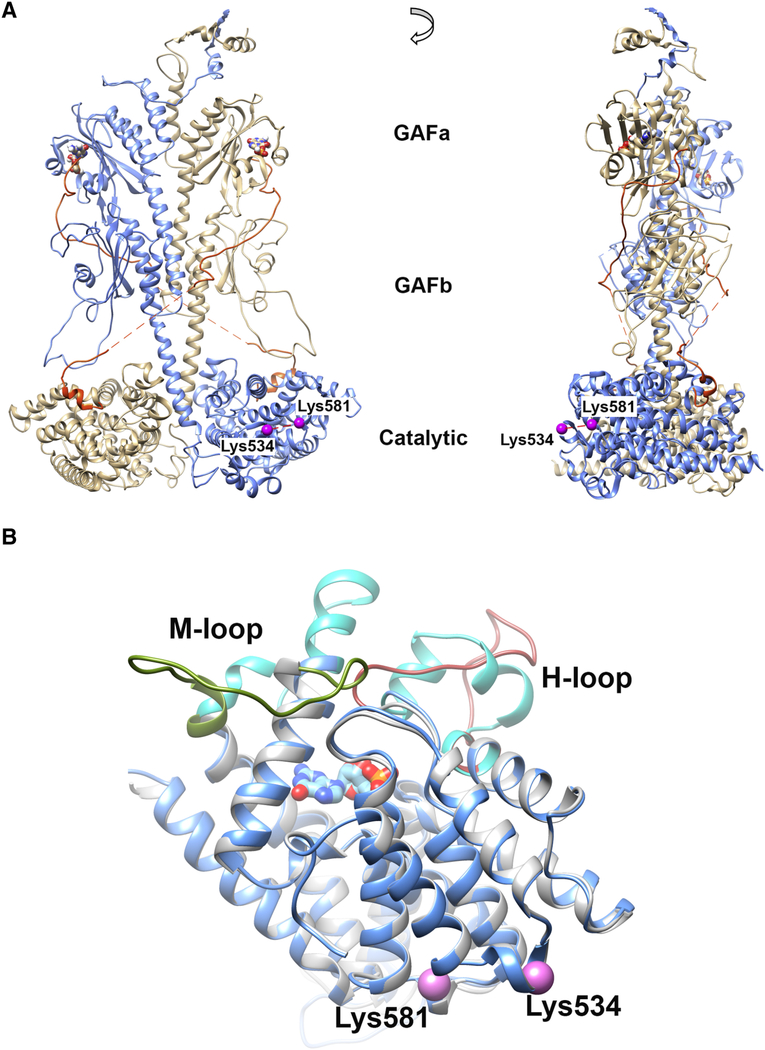
The amino acid residues Lys534Pα and Lys581Pα localize in the catalytic domain of the Pα subunit, on the opposite side of the opening to the cGMP catalytic site (Fig. 2A). A significant increase in Lys534Pα-Lys581Pα cross-linking propensity was detected in Pαβ catalytic dimer, and the Pαβγ2 complex compared with the two reconstituted PDE6 structures with cGMP bound to the GAFa domains (Fig. 1C). To examine the consequence of cGMP binding on the structure of the Pα catalytic domain, we compared structural models of the Pα catalytic domain in the apo (PDB ID: 2H40 as template; [9]) or cGMP-bound state (PDB ID: 1T9S, as template; [31]). Superimposition of representative models in apo and cGMP-bound states suggest that the major structural differences reside in the vicinity of the catalytic site, where the H-loop (Fig. 2B, colored in brown) and M-loop (Fig. 2B, colored in green) in the apo structure (Fig. 2B, colored in gray) need to swing away from the catalytic site to allow the binding of nucleotide in the 5’-GMP-bound structure (Fig. 2B, colored in blue and cyan for H- and M-loop). In contrast, secondary structures distal from the catalytic site retain similar conformations with or without cGMP bound, including the small helices around Lys534 and Lys581 (Fig. 2B, violet spheres). Therefore, the decrease in Lys534Pα-Lys581Pα cross-linking propensity upon cGMP binding is most likely due to a conformational change induced by the binding of cGMP to noncatalytic binding sites in the GAFa domain. Due to the much lower binding affinity of cGMP to the active site (cGMP hydrolysis by activated PDE6 occurs at the diffusion-controlled limit; [32]), it is unlikely that the observed conformational change results from the binding of cGMP or its reaction product (5’-GMP) in the catalytic pocket.
Figure 2.
Location of Lys534 and Lys581 in 3D models. A) Lys534 and Lys581 (colored in violet) locate to the catalytic domain of Pα (blue structure) on the opposite side of the catalytic site, as illustrated on the PDE6 holoenzyme model built from a cryo-EM study (PDB ID: 6MZB) [14]. The cGMP molecules (bound to GAFa domain) are represented by ball-and-stick heteroatoms. B) Superimposition of comparative models for Pα catalytic domain with (blue; template PDB ID: 1T9S) or without (gray; template PDB ID: 2H40) cGMP binding. The M-loop and H-loop are colored in olive green and brown in apo structure; whereas the M-loop and H-loop are colored in cyan in 5’-GMP-bound structure. The cross-linked Pα residues Lys534 and Lys581 are indicated as violet spheres.
Structural differences of PDE6 complexes in different liganded states
Overall, we were able to identify sufficient quantities of 22 cross-linked peptides present in all four liganded states (see Table 1 for intra-subunit cross-links and Table 2 for inter-subunit cross-links). All 22 cross-linked pairs reported here were also identified in a previous CX-MS study on PDE6 holoenzyme that led to a medium-resolution structural model [7], in good agreement with two recently reported cryo-EM structures [13, 14]. We mapped our cross-linked pairs onto the cryo-EM structure of Gulati et al. study (PDB ID: 6MZB) [14] to gain structural insights on allosteric regulation of PDE6 enzyme by cGMP and Pγ subunit.
Table 2.
Cross-linking propensity of PDE6 inter-subunit cross-links.
| Lys 1 | Lys 2 | Holoenzyme (%XL) | Pαβγ2-cGMP (%XL) | Pαβ (%XL) | Pαβγ2 (%XL) | Pαβ-cGMP (%XL) |
|---|---|---|---|---|---|---|
| Pα | Pβ | |||||
| 447 | 440 | 24.2 | 26.9 | 33.4 | 16.5 | 34.8 |
| 447 | 618 | 32.7 | 38.8 | 27.4 | 31.2 | 35.2 |
| Pα | Pγ | |||||
| 76 | N-terma | --- | 51.9 | --- | 33.9 | --- |
| 76 | 7a | 30.4 | --- | --- | --- | --- |
| Pβ | Pγ | |||||
| 78 | N-terma | --- | 62.3 | --- | 62.9 | --- |
| 78 | 7a | 16.4 | --- | --- | --- | --- |
In the endogenous PDE6 holoenzyme, the N-terminus of Pγ subunit was acetylated. In the reconstituted PDE6 complexes, the N-terminus of Pγ subunit was not acetylated. Since the pKa of the N-terminal amide group is 2 unit smaller than the pKa of Lys residues, the N-terminus of Pγ subunit became the preferred cross-linked Pγ amino acid residue from reconstituted samples.
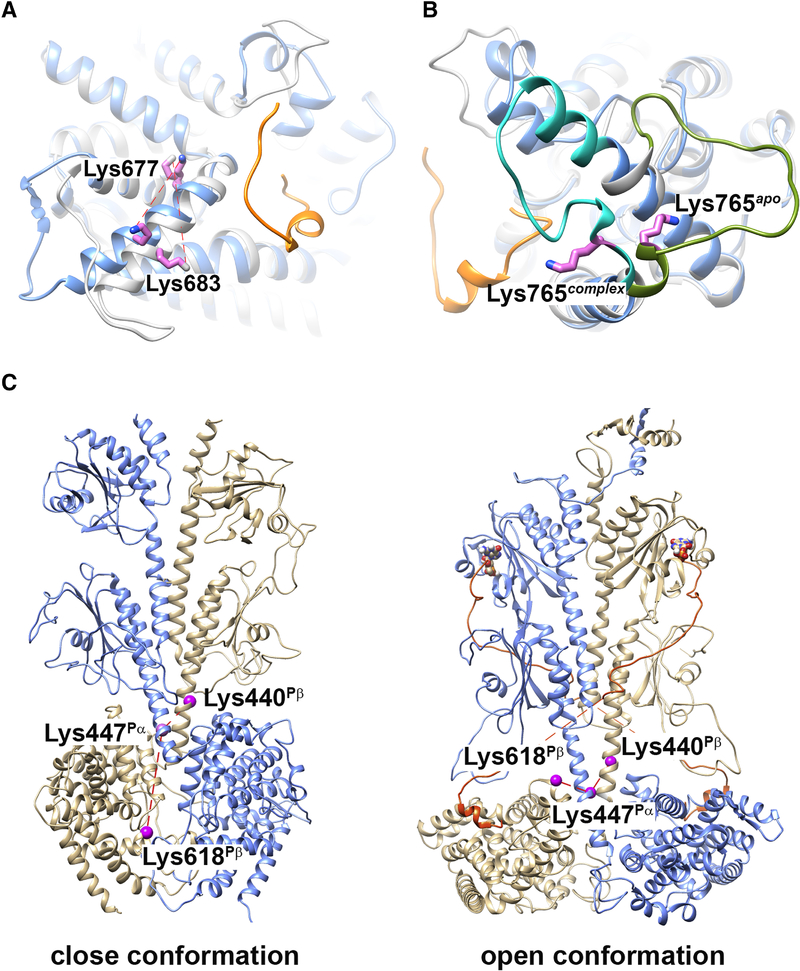
The Lys677Pα-Lys683Pα cross-linked pair in the primary sequence is the homolog of the Lys675Pβ-Lys681Pβ cross-linked pair in the Pαβ subunit, both of which show a decrease in cross-linking efficiency when Pγ is reconstituted with Pαβ. Although specific cross-linking percentage values were different for the two subunits, they yielded similar trends in the cross-linking propensity for various allosteric states (Table 1). Homologous residues Lys677Pα/Lys675Pβ and Lys683Pα/Lys681Pβ localize to the helix-α12 of the catalytic domain (Fig. 3A), in close proximity to the binding site for the C-terminal region of the Pγ subunit that has been shown to block catalysis by restricting access to substrate [33]. Direct binding of Pγ subunit to the catalytic domain likely constrains the side-chain packing of helix-α12 in Pγ-bound state (Fig. 3A, colored in blue) than apo state (Fig. 3A, colored in gray), decreasing the cross-linking efficiency of Lys683Pα/Lys681Pβ and Lys677Pα/Lys675Pβ pairs.
Figure 3.
Structural elements identified by cross-linking propensity of PDE6 complexes in various allosteric states. (A and B) Superimposition of comparative models for Pα catalytic domain with (blue; PDB ID: 6MZB) or without (gray; template PDB ID: 2H40) a fragment of Pγ bound. The C-terminal segment of Pγ in the PDE6 holoenzyme structure is colored in orange. Cross-linked Pα residues Lys683-Lys677 (A) and Lys765 (B) are indicated as violet sticks. C) Comparative models for Pαβ catalytic dimer without (left; template PDB ID: 3IBJ) or with (right; PDB ID: 6MZB) ligand and Pγ binding. Pα subunit is colored in blue and Pβ subunit in tan. The C-terminal segment of Pγ is colored in orange. Cross-linked Pα residues Lys447Pα, Lys440Pβ and Lys618Pβ are indicated as violet spheres.
Another set of homologous cross-links were identified between Lys 765 with Lys 817 in the Pα-subunit and Lys 763 and Lys 815 in the Pβ-subunit. When compared with the unliganded Pαβ state, both subunits showed an increase in cross-linking percent upon cGMP binding and a decrease upon Pγ binding. Lys765Pα and Lys763Pβ are located within the M-loop, which participates in regulating the catalytic activity of PDE6 through its interaction with the C-terminal residues of Pγ [12]. Lys819Pα and Lys817Pβ reside in the α16 terminal helix of the catalytic domain. Comparison of apo (PDB ID: 2H40 as template; [9]) and Pγ-bound (PDB ID: 6MZB; [14]) structures shows that the movement of the M-loop (Fig. 3B, colored in olive green for apo state, colored in cyan for Pγ-bound state) upon Pγ binding accounts for the decrease in cross-linking efficiency of Lys765Pα/Lys819Pα and Lys763Pβ/Lys817Pβ pairs. Binding of cGMP to the distant GAFa domain enhance cross-linking efficiency, which could be explained by the well-established ability of noncatalytic cGMP binding to affect the affinity with which Pγ binds to Pαβ [20, 23].
Although only two inter-subunit Pαβ cross-linked pairs generated quantifiable data for all four conformational states, their cross-linking propensity was fairly consistent through various allosteric states. The Lys447Pα-Lys440Pβ pair localize to the long helices at the C-terminal end of the GAFb domains, a region that contributes to the dimerization interface between the Pα and Pβ subunit (Fig. 3C). The Lys447Pα-Lys618Pβ pair cross-links the long GAFb helix of Pα with the flexible H-loop of the Pβ catalytic domain (Fig. 3C). The results in Table 2 do not show evidence of significant changes in cross-linking propensity in any of the four allosteric states we studied. In contradiction to the open conformation of the PDE6 holoenzyme model observed in cGMP/Pγ-bound state (Fig. 3C, right structure), the model built from the crystal structure of apo PDE2A (PDE ID: 3IBJ) placed Lys618Pβ at the dimer interface excluded from solvent (Fig. 3C, left structure). Our qCX-MS data preclude the possibility of an apo PDE2A-like conformation adapted by the Pαβ catalytic dimer, even in the absence of cGMP and Pγ subunit. Rather, a comparable cross-linking propensity of Lys447Pα-Lys440Pβ and Lys447Pα-Lys618Pβ cross-linked pairs in Pαβ suggested a similar Pα and Pβ dimerization interface near the catalytic domain in Pαβ catalytic dimer.
In this qCX-MS study, the cross-links between Pγ and Pαβ are identified for all four states in the N-terminal region of Pγ with the GAFa domains of Pα and Pβ (Table 2). A substantial increase (> 50% increase) in cross-linking propensity between Lys76Pα (helix α2) and the N-terminus of Pγ was observed when cGMP occupied its Pα GAFa binding site. This observation highlights a role of noncatalytic cGMP in regulating the interaction of Pγ and Pα GAFa domain. However, the corresponding Lys78Pβ-N-termPγ cross-linked pair was highly cross-linked regardless of the occupancy of cGMP in the Pβ GAFa domain. This asymmetric binding of Pγ to the GAFa domain of the Pα and Pβ subunits was consistent with our previous results with PDE6 holoenzyme [7]; however, the structural basis for this asymmetric behavior is unclear.
Discussion
In this study, we used an innovative quantitative CX-MS analysis to evaluate conformational changes in PDE6 that are critical to the allosteric regulation of this central effector of the visual signaling pathway in retinal photoreceptor cells. By using dead-end cross-links that formed during cross-linking reactions, we were able to determine the relative amount of cross-linked peptides in four distinct allosteric states: Pαβ, Pαβ-cGMP, Pαβγ2, and Pαβγ2-cGMP. This approach has the following advantages over direct quantitation of cross-linked products: 1) it reduces biochemical variations caused by minor differences in protein and cross-linker concentrations, as well as technical variations in MS sample handling; 2) it minimizes the effects of residue reactivities towards cross-linking propensity, thus reflects the distance and local structures of cross-linked residues; 3) it measures protein behavior in solution, thus can reveal differences in conformational dynamics especially in less structured regions that are subject to functional regulation. The relative small errors in our measurements of cross-linking propensity (Table 1 and Supplemental Table 1) suggest the reproducibility and robustness of our protein preparations and cross-linking experiments, as well as minor technical variations in MS sample handling and analysis. The similarity in the quantitative results of cross-linked pairs obtained from native PDE6 holoenzyme and reconstituted PDE6 holoenzyme (i.e. Pαβγ2-cGMP) also support the biochemical and functional relevance of the reconstituted samples used in this study.
The several cross-links that were sensitive to cGMP or Pγ binding represent residues localized primarily in less structured regions of the catalytic domain, such as Lys534Pα, Lys581Pα, Lys765Pα and Lys763Pβ (both locate in the M-loop near the opening to the active site) [7]. By identifying ligand-dependent conformational changes in the PDE6 catalytic dimer by qCX-MS, this work also has revealed several structural elements implicated in allosteric communication induced by cGMP and/or Pγ binding to the PDE6 catalytic dimer. Furthermore, two inter-subunit cross-links between Pα and Pβ subunits, namely Lys447Pα-Lys440Pβ (in the long α-helix at the end of the GAFb domain) and Lys447Pα-Lys618Pβ (between the GAFb long α-helix and the catalytic domain) revealed similar cross-linking propensity of these pairs in all four liganded states (Table 2). These results as well as other reports [14, 34] support a model in which the overall molecular architecture of PDE6 does not undergo major changes (in contrast to PDE2; [8]) and that less structured elements of the catalytic dimer are responsible for the allosteric communication between the regulatory and catalytic domains [14].
Moreover, our qCX-MS analyses reveal dissimilarities in the ability of the Pα and Pβ subunits to react with cross-linking reagents. For example, the homologous pairs Lys765Pα– Lys819Pα and Lys763Pβ–Lys817Pβ exhibit markedly different propensities to form cross-links with the β-subunit (ranging from 20–48% for the four liganded states) compared with the α-subunit (1–5%). Interestingly, of the intramolecular sites undergoing statistically significant changes in percent cross-linking, twice as many were observed within the α-subunit compared with the β-subunit (Table 1, Supplemental Table 2). Whereas the catalytic domain of the β-subunit may provide greater accessibility to chemical cross-linkers, the α-subunit may play a dominant role in ligand-dependent allosteric communication. Our observed structural and conformational asymmetry of the two catalytic subunits may underlie the reported the biphasic activation mechanism of transducin during the first steps in the visual signaling pathway [34].
Each of the two GAFa domains of rod PDE6 catalytic dimer contains a noncatalytic cGMP binding site as well as sites of interaction with the Pγ subunit. The interactions of Pγ and noncatalytic cGMP with the GAFa domain act in a synergistic manner, with Pγ enhancing cGMP binding affinity and vice versa [35]. In addition, the noncatalytic cGMP binding sites of Pα and Pβ exhibit different affinities for cGMP [32], but the identity of the lower and higher affinity sites to specific catalytic subunits is unknown. In this study, we observed a significant increase in Lys76Pα-N-termPγ cross-linking in the presence of cGMP, an effect not observed in the homologous cross-linking pair on the β-subunit, Lys78Pβ-N-termPγ (Table 2). Asymmetric interactions of the γ-subunit with the GAFa domains of Pα and Pβ was also reported in our previous CX-MS study where a subunit-specific interaction between Pγ and the ‘lid’ region of noncatalytic cGMP binding site on Pα was observed for the PDE6 holoenzyme [7]. Taken together, we speculate that the GAFa domain of the rod PDE6 Pα subunit may be the locus for the higher affinity binding site for both Pγ and cGMP that underlies the synergistic action when both ligands bind to the GAFa domain. Such high affinity, synergistic interaction provides a strong anchor for Pγ to rapidly activate and deactivate rod PDE6 catalysis during visual excitation and recovery, as well as potentially modulating the activated lifetime of PDE6 during light adaptation (reviewed in [1]). This asymmetry in Pγ and cGMP binding to the PDE6 catalytic subunits may also underlie the asymmetric binding of activated transducin to the PDE6 holoenzyme [34].
Our identification of structural elements undergoing ligand-dependent changes in PDE6 conformation provides insights into the molecular etiology of retinal degenerative diseases and visual disorders that result from point mutations in the catalytic and inhibitory subunits of PDE6. For example, several of the conformationally-sensitive sites found in this study (e.g., α-subunit residues Arg102, Lys581, Lys677, Lys683; and β-subunit residues Lys618 and Lys681) are located within 10 A of residues identified as missense mutations of the PDE6 catalytic subunits associated with retinal diseases (e.g., Ser573Pro [36]; Val685Met [37]). Another report identified PDE6 somatic mutations associated with cancer [38] that map to conformationally dynamic regions of the PDE6 catalytic subunits revealed by this study. Whereas some known diseasecausing PDE6 mutations can be easily ascribed sites involved in the catalytic mechanism of cyclic nucleotide hydrolysis (e.g., Arg560Cys [39]), identification of novel, conformationally dynamic regions of the PDE6 catalytic subunits associated with disease-causing mutations offers the potential of developing allosteric inhibitors of PDE6 to treat retinal degenerative diseases, in analogy to the use of allosteric inhibitors of human PDE4 to treat polycystic kidney disease [40].
Material and Methods
Materials
Bovine retinas were purchased from W. L. Lawson, Inc. Trypsin was purchased from Promega, and disuccinimidyl suberate (DSS) was purchased from Pierce.
Protein preparation
PDE6 holoenzyme and Pαβ catalytic dimer were prepared as described previously [29]. Recombinant Pγ was expressed in E. coil BL21/DE3 cells, purified through a SP-Sepharose column and then by reverse-phase chromatography [7, 17]. For the reconstitution of the Pαβγ2 complex, purified Pαβ (0.2 nM −1 nM) was incubated with stoichiometric amounts of purified Pγ at room temperature for 20 min. For preparation of of the Pαβ-cGMP complex, purified Pαβ (0.2 nM −1 nM) was pre-incubated with 100 μM vardenafil (a PDE5/6-specifc inhibitor) at room temperature for 20 minutes before 1 mM cGMP and 10 mM EDTA were added. The reconstitution of Pαβγ2-cGMP complex was similar to that of Pαβ-cGMP complex, except that purified Pαβ was pre-incubated with both 100 μM vardenafil and purified Pγ at room temperature for 20 minutes before cGMP and EDTA were added. The catalytic activity of purified Pαβ and reconstituted PDE6 complexes was measured by the phosphate release assay [41]. The inhibition potency (IC50) was calculated from curve fitting the results to a 3-parameter logistic equation.
Chemical cross-linking, in-gel digestion, and mass spectrometric analysis
Chemical cross-linking reactions were carried out following the manufacturer’s instruction. PDE6 complexes were buffer-exchanged into HEPES buffer (20 mM HEPES, 100 mM NaCl, 5mM MgCl2, pH 8.0) and cross-linked with 200-fold (or 70-fold) molar excess of DSS cross-linker at room temperature for 1 h. After the cross-linking reaction was quenched with 1 μL 0.8M ammonium hydroxide, proteins were concentrated by rotary evaporation under vacuum, separated by SDS-PAGE, and visualized with Coomassie Brilliant Blue G-250.
Cross-linked products were in-gel digested and analyzed by LC-MS and LC-MS-MS as described previously [42]. Briefly, 1μl aliquot of the digestion mixture was injected into a Dionex Ultimate 3000 RSLCnano UHPLC system (Dionex Corporation, Sunnyvale, CA), and separated by a 75 μm × 25 cm PepMap RSLC column (100 Å, 2 μm) at a flow rate of ~450 nl/min. The eluant was connected directly to a nanoelectrospray ionization source of an LTQ Orbitrap XL mass spectrometer (Thermo Scientific, Waltham, MA). LC-MS data were acquired in an information-dependent acquisition mode, cycling between a MS scan (m/z 315–2,000) acquired in the Orbitrap, followed by low-energy CID analysis on the 3 most intense multiply charged precursors acquired in the linear ion trap.
Cross-linked peptide identification
Cross-linked peptides were identified using an integrated module in Protein Prospector, based on a bioinformatic strategy described previously [43, 44]. The score of a cross-linked peptide was based on number and types of fragment ions identified, as well as the sequence and charge state of the cross-linked peptide. Only results where the score difference is greater than 0 (i.e. the cross-linked peptide match was better than a single peptide match alone) are considered. The expectation values are calculated based on matches to single peptides, so should be treated as another score, rather than a statistical measure of reliability. Cross-linked peptides identified from various samples were combined together, and the cross-linked percentage for each peptide pair was calculated using the following formula:
where the peak height (PH) was the sum of apex peak height of a peptide in all charge states, and dead-end XLs were cross-linker modified where only one NHS-ester function of DSS was cross-linked to a Lys residue and the other NHS-ester function was hydrolyzed by water.
Structural visualization and prediction
Comparative modeling with Modeller [45] was carried out at the interface of UCSF Chimera structural visualization platform [46]. The templates used to build comparative models included the apo state of the PDE5 catalytic domain (PDB ID: 2H40) or 5’-GMP-bound state (PDB ID: 1T9S). The 3.4 Å cryo-EM structure of the native rod PDE6 holoenzyme (PDB ID: 6MZB) [14] was also used for structural analysis.
Supplementary Material
Highlights.
PDE6 is regulated by the binding of inhibitory Pγ and noncatalytic cGMP.
cGMP binding in GAFa domains change the conformation of catalytic domains.
Pγ and cGMP display asymmetric binding to the GAFa domains of rod PDE6.
The Pαβ catalytic dimer adapts an open conformation in different allosteric states.
Acknowledgements
We thank Robert Chalkley, Peter Baker and Al Burlingame for access to a developmental version of the Protein Prospector software. This work was supported by NSF CLF 1307367, NIH HD-093783 (to FC), NIH EY-05798 (to RHC) and the UNH Collaborative Research Excellence (CoRE) fund (to FC and RHC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cote RH. Photoreceptor phosphodiesterase (PDE6): a G-protein-activated PDE regulating visual excitation in rod and cone photoreceptor cells In: Beavo JA, Francis SH, Houslay MD, editors. Cyclic Nucleotide Phosphodiesterases in Health and Disease. Boca Raton, FL: CRC Press; 2006. p. 165–93. [Google Scholar]
- [2].Wensel TG. Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 2008;48:2052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Arshavsky VY, Burns ME. Photoreceptor signaling: supporting vision across a wide range of light intensities. J Biol Chem. 2012;287:1620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Palczewski K, Orban T. From atomic structures to neuronal functions of g protein-coupled receptors. Annu Rev Neurosci. 2013;36:139–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ke H, Wang H. Crystal structures of phosphodiesterases and implications on substrate specificity and inhibitor selectivity. Curr Top Med Chem. 2007;7:391–403. [DOI] [PubMed] [Google Scholar]
- [6].Francis SH, Blount MA, Corbin JD. Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol Rev. 2011;91:651–90. [DOI] [PubMed] [Google Scholar]
- [7].Zeng-Elmore X, Gao XZ, Pellarin R, Schneidman-Duhovny D, Zhang XJ, Kozacka KA, et al. Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. J Mol Biol. 2014;426:3713–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS. Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc Natl Acad Sci U S A. 2009;106:18225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang H, Liu Y, Huai Q, Cai J, Zoraghi R, Francis SH, et al. Multiple conformations of phosphodiesterase-5: implications for enzyme function and drug development. J Biol Chem. 2006;281:21469–79. [DOI] [PubMed] [Google Scholar]
- [10].Wang H, Robinson H, Ke H. Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. J Biol Chem. 2010;285:38149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Martinez SE, Heikaus CC, Klevit RE, Beavo JA. The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. J Biol Chem. 2008;283:25913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barren B, Gakhar L, Muradov H, Boyd KK, Ramaswamy S, Artemyev NO. Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit. Embo J. 2009;28:3613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Z, He F, Constantine R, Baker ML, Baehr W, Schmid MF, et al. Domain organization and conformational plasticity of the G protein effector, PDE6. J Biol Chem. 2015;290:17131–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gulati S, Palczewski K, Engel A, Stahlberg H, Kovacik L. Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases. Sci Adv. 2019;5:eaav4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Song J, Guo LW, Muradov H, Artemyev NO, Ruoho AE, Markley JL. Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc Natl Acad Sci U S A. 2008;105:1505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Granovsky AE, Natochin M, Artemyev NO. The gamma subunit of rod Cgmp-phosphodiesterase blocks the enzyme catalytic site. J Biol Chem. 1997;272:11686–9. [DOI] [PubMed] [Google Scholar]
- [17].Zhang XJ, Gao XZ, Yao W, Cote RH. Functional mapping of interacting regions of the photoreceptor phosphodiesterase (PDE6) gamma-subunit with PDE6 catalytic dimer, transducin, and regulator of G-protein signaling9–1 (RGS9–1). J Biol Chem. 2012;287:26312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang Z, Artemyev NO. Determinants for phosphodiesterase 6 inhibition by its gamma-subunit. Biochemistry. 2010;49:3862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yamazaki A, Bartucci F, Ting A, Bitensky MW. Reciprocal effects of an inhibitory factor on catalytic activity and noncatalytic cGMP binding sites of rod phosphodiesterase. Proc Natl Acad Sci U S A. 1982;79:3702–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cote RH, Bownds MD, Arshavsky VY. cGMP binding sites on photoreceptor phosphodiesterase: role in feedback regulation of visual transduction. Proc Natl Acad Sci U S A. 1994;91:4845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Arshavsky VY, Dumke CL, Bownds MD. Noncatalytic cGMP-binding sites of amphibian rod cGMP phosphodiesterase control interaction with its inhibitory gamma-subunits. A putative regulatory mechanism of the rod photoresponse. J Biol Chem. 1992;267:24501–7. [PubMed] [Google Scholar]
- [22].Mou H, Cote RH. The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory g subunit. JBC. 2001;276:27527–34. [DOI] [PubMed] [Google Scholar]
- [23].Zhang XJ, Cahill KB, Elfenbein A, Arshavsky VY, Cote RH. Direct allosteric regulation between the GAF domain and catalytic domain of photoreceptor phosphodiesterase PDE6. J Biol Chem. 2008;283:29699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rappsilber J The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Singh P, Panchaud A, Goodlett DR. Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal Chem. 2010;82:2636–42. [DOI] [PubMed] [Google Scholar]
- [26].Chu F, Thornton DT, Nguyen HT. Chemical cross-linking in the structural analysis of protein assemblies. Methods. 2018;144:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Street TO, Zeng X, Pellarin R, Bonomi M, Sali A, Kelly MJ, et al. Elucidating the mechanism of substrate recognition by the bacterial Hsp90 molecular chaperone. J Mol Biol. 2014;426:2393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Catty P, Deterre P. Activation and solubilization of the retinal cGMP-specific phosphodiesterase by limited proteolysis--Role of the C- terminal domain of the β-subunit. Eur J Biochem. 1991;199:263–9. [DOI] [PubMed] [Google Scholar]
- [29].Pentia DC, Hosier S, Collupy RA, Valeriani BA, Cote RH. Purification of PDE6 isozymes from mammalian retina. Methods Mol Biol. 2005;307:125–40. [DOI] [PubMed] [Google Scholar]
- [30].Guo X, Bandyopadhyay P, Schilling B, Young MM, Fujii N, Aynechi T, et al. Partial acetylation of lysine residues improves intraprotein cross-linking. Anal Chem. 2008;80:951–60. [DOI] [PubMed] [Google Scholar]
- [31].Zhang KY, Card GL, Suzuki Y, Artis DR, Fong D, Gillette S, et al. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol Cell. 2004;15:279–86. [DOI] [PubMed] [Google Scholar]
- [32].Mou H, Grazio HJ, Cook TA, Beavo JA, Cote RH. cGMP binding to noncatalytic sites on mammalian rod photoreceptor phosphodiesterase is regulated by binding of its g and d subunits. JBC. 1999;274:18813–20. [DOI] [PubMed] [Google Scholar]
- [33].Granovsky AE, Artemyev NO. A conformational switch in the inhibitory gamma-subunit of PDE6 upon enzyme activation by transducin. Biochemistry. 2001;40:13209–15. [DOI] [PubMed] [Google Scholar]
- [34].Qureshi BM, Behrmann E, Schoneberg J, Loerke J, Burger J, Mielke T, et al. It takes two transducins to activate the cGMP-phosphodiesterase 6 in retinal rods. Open Biol. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cote RH, Bownds MD, Arshavsky VY. cGMP binding sites on photoreceptor phosphodiesterase: Role in feedback regulation of visual transduction. Proc Natl Acad Sci U S A. 1994;91:4845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dryja TP, Rucinski DE, Chen SH, Berson EL. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1999;40:1859–65. [PubMed] [Google Scholar]
- [37].Corton M, Blanco MJ, Torres M, Sanchez-Salorio M, Carracedo A, Brion M. Identification of a novel mutation in the human PDE6A gene in autosomal recessive retinitis pigmentosa: homology with the nmf28/nmf28 mice model. Clin Genet. 2010;78:495–8. [DOI] [PubMed] [Google Scholar]
- [38].Maryam A, Vedithi SC, Khalid RR, Alsulami AF, Torres PHM, Siddiqi AR, et al. The Molecular Organization of Human cGMP Specific Phosphodiesterase 6 (PDE6): Structural Implications of Somatic Mutations in Cancer and Retinitis Pigmentosa. Comput Struct Biotechnol J. 2019;17:378–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang T, Reingruber J, Woodruff ML, Majumder A, Camarena A, Artemyev NO, et al. The PDE6 mutation in the rd10 retinal degeneration mouse model causes protein mislocalization and instability and promotes cell death through increased ion influx. J Biol Chem. 2018;293:15332–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Omar F, Findlay JE, Carfray G, Allcock RW, Jiang Z, Moore C, et al. Small-molecule allosteric activators of PDE4 long form cyclic AMP phosphodiesterases. Proc Natl Acad Sci U S A. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cote RH. Kinetics and regulation of cGMP binding to noncatalytic binding sites on photoreceptor phosphodiestrase. Methods Enzymol. 2000;315:646–72. [DOI] [PubMed] [Google Scholar]
- [42].Chu F, Shan SO, Moustakas DT, Alber F, Egea PF, Stroud RM, et al. Unraveling the interface of signal recognition particle and its receptor by using chemical cross-linking and tandem mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:16454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chu F, Baker PR, Burlingame AL, Chalkley RJ. Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol Cell Proteomics. 2010;9:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Trnka MJ, Baker PR, Robinson PJ, Burlingame AL, Chalkley RJ. Matching Cross-linked Peptide Spectra: Only as Good as the Worse Identification. Mol Cell Proteomics. 2014;13:420–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. [DOI] [PubMed] [Google Scholar]
- [46].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.