Abstract
Lewy bodies, hallmarks of Parkinson’s disease, contains C-terminally truncated (ΔC) α-synuclein (α-syn). Here, we report fibril structures of three N-terminally acetylated (Ac) α-syn constructs, Ac1–140, Acl–122, and Ac1–103, solved by cryoelectron microscopy. Both ΔC-α-syn variants exhibited faster aggregation kinetics and Ac1–103 fibrils efficiently seeded the full length protein, highlighting their importance in pathogenesis. Interestingly, fibril helical twists increased upon the removal of C-terminal residues and can be propagated through cross-seeding. Compared to that of Ac1–140, increased electron densities were seen in the N-terminus of Ac1–103, whereas the C-terminus of Ac1–122 appeared more structured. In accord, the respective termini of ΔC-α-syn exhibited increased protease resistance. Despite similar amyloid core residues, distinctive features were seen for both Ac1–122 and Ac1–103. Particularly, Ac1–103 has the tightest packed core with an additional turn, likely attributable to conformational changes in the N-terminal region. These molecular differences offer insights into the effect of C-terminal truncations on α-syn fibril polymorphism.
Keywords: CryoEM, TEM, thioflavin T, amyloid, Raman spectroscopy
A hallmark of Parkinson’s disease (PD) is the presence of Lewy bodies (LBs), intracytoplasmic inclusions defined by the presence of aggregated, β-sheet rich α-synuclein (α-syn) fibrils [1, 2]. Normally, α-syn is 140-residues in length; however, posttranslational C-terminal truncations of α-syn are also found in LBs [3]. Impairment and/or overburden of the cellular degradation machinery are likely responsible [4]. The removal of the C-terminus accelerates α-syn fibril formation both in vitro [5–9] and in vivo [10, 11], implicating a role for C-terminally truncated (ΔC) species in PD pathogenesis. While the C-terminus of α-syn is peripheral to the amyloid core [12–18], this region is involved in the process of amyloid formation [19, 20]. Here, to elucidate the effect of C-terminal truncations on fibril structures, differences between N-terminally acetylated (Ac) full length (Ac1–140) and two LB-derived ΔC-α-syn species [21, 22], Ac1–122 and Ac1–103 (Fig. 1A), were investigated by cryoelectron microscopy (cryoEM), the prevailing technique to solve amyloid structures [15–17, 23–31], coupled to measurements of aggregation kinetics, Raman spectroscopic characterization, and limited-proteolysis experiments.
Fig. 1.
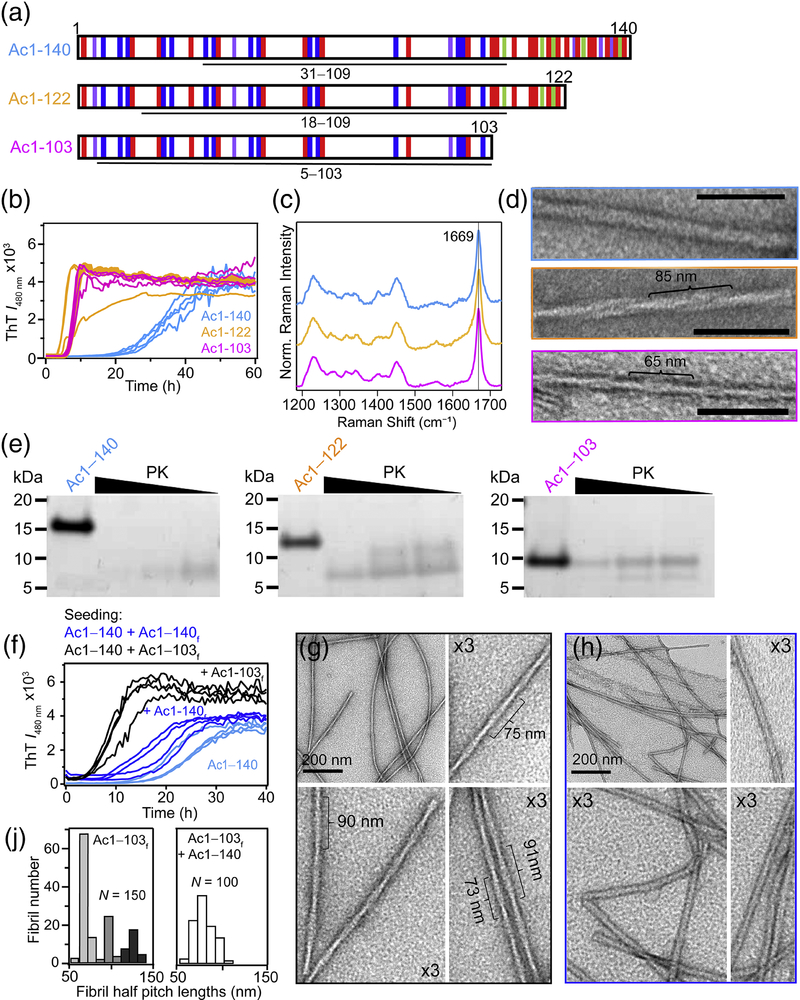
Structural characterization and aggregation of C-terminal α-syn truncations. (A) Schematic representation of the primary amino acid sequence of Ac1–140, Ac1–122 and Ac1–103 showing basic (blue), acidic (red), aromatic (purple) and proline (green) residues. Underline regions correspond to PK-resistant cores determined by LC-MS (Supplemental Table S1). (B) Aggregation kinetics monitored by ThT ([α-syn] = 50 μM and [ThT] = 10 μM in 10 mM NaPi, 140 mM NaCl, pH 7.4, 37 °C, λex = 440 nm and λobs = 480 nm). (C) Raman spectra showing the characteristic amide-I band (1669 cm−1) for β-sheet. (D) Representative TEM images. Estimated helical half pitch (defined as a 180° helical turn) is ~85 and ~65 nm for Ac1–122 and Ac1–103. Ac1–140 could not be determined. Scale bar is 100 nm. (E) Limited protease digestions of α-syn fibrils (40 μM) with decreasing PK (4, 0.8, and 0.4 ng for 20 h at 37 °C) visualized by SDS-PAGE. (F) Seeding reactions of Ac1–140 (50 μM) in the absence and in the presence of 5% Ac1–140 or Ac1–103 fibrils monitored by ThT (same conditions as panel B). TEM images of Acl–140 seeded with (G) Ac1–103 or (H) Ac1–140 fibrils. (J) Histograms of measured half pitch lengths from Ac1–103 fibrils (left) (N = 150) and Ac1–140 seeded with Ac1–103 fibrils (right, N = 100). The subscript f denotes fibrils.
The aggregation propensity of α-syn was first assessed by thioflavin-T (ThT), an extrinsic fluorophore which increases in intensity upon amyloid binding [32]. Aggregation kinetics of Ac1–122 and Ac1–103 had significant reductions in lag and growth phases compared to Ac1–140 (Fig. 1B), consistent with prior work on various ΔC-fragments [5–9]. Raman spectra for all three α-syn fibrils (Fig. 1C) exhibited indistinguishable amide-I bands (C=O stretch) characteristic for β-sheet conformation (1669 cm−1), indicating little secondary structural differences [33, 34].
At the ultrastructural level, fibril differences were revealed by negative stain transmission electron microscopy (TEM, Fig. 1D). While all three fibrils contained two protofilaments (~5 nm each), increased helical twists (decreased pitch length) were seen for Ac1–122 and Ac1–103, suggesting that the C-terminus imposes a steric hinderance and restricts the degree of twisting. Another possibility is that the twist is established during nucleation, at the initiation step of fibril formation. Limited proteolysis experiments were then performed using proteinase-K (PK), a broad spectrum protease widely used to evaluate amyloid structures (Fig. 1E) [35] and endoproteinase Glu-C, a protease selective for Asp and Glu residues (Fig. S1). SDS-PAGE analysis clearly showed that Ac1–103 is the most stable, as intact Ac1–103 (~10 kDa band) persisted under all conditions examined. Liquid-chromatography mass spectrometry analysis (Supplemental Tables S1 and S2) indicated that there is greater resistance to cutting at residues A17/A18, E28/A29, and A30/G31 (where / indicates the cleavage site) for both Ac1–122 and Ac1–103, suggesting that the N-terminus is more ordered in ΔC-α-syn. These results demonstrate that the removal of C-terminal residues influences both α-syn aggregation kinetics and fibril structure.
Next, we asked whether a more twisted Ac1–140 fibril polymorph can be made by seeding with Ac1–103. Cross- and self-seeding reactions were compared by adding either pre-formed Ac1–103 (Supplemental Fig. S2) or Ac1–140 fibrils to Ac1–140 (Fig. 1F). Ac1–140 aggregation was stimulated by both seeds with cross-seeding by Ac1–103 appearing superior. Negative stain TEM confirmed that a greater twist was indeed propagated in the cross-seeded Ac1–140 fibrils (Fig. 1G) compared to the self-seeded fibrils (Fig. 1H). Upon cross-seeding, Ac1–140 has an increased helical twist (half pitch ~77 nm), albeit not as twisted as the Ac1–103 seeds themselves (Fig. 1J). Since the Ac1–103 fibrils can efficiently seed Ac1–140 and generate greater twisted fibrils, the presence of C-terminal residues does not preclude fibril twisting and thus, additional factors are modulating the observed periodicity. Collectively, these results reinforce the pathological relevance of C-terminal truncations in promoting amyloid formation and templating distinctive fibril polymorphs.
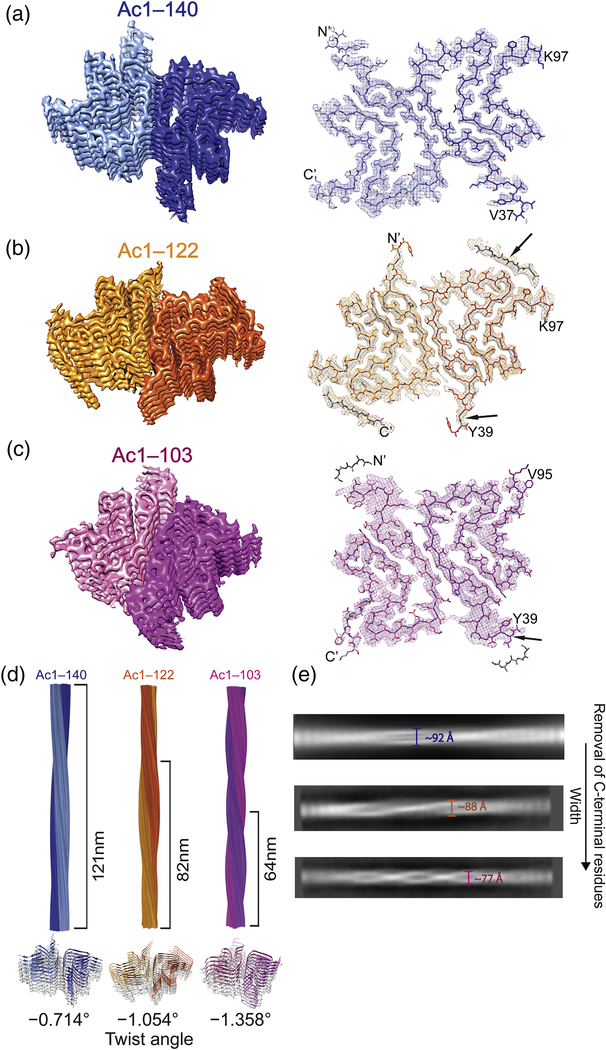
To elucidate their differences, Ac-α-syn fibril structures were determined by cryoEM at near-atomic resolution (Supplemental Table S3). The density maps show that all three structures are composed of two protofilaments intertwining along an approximate 21 screw axis into a left-handed helix (Fig. 2 and Supplemental Fig. S3). Each of the two protofilaments stack along the axis with a helical rise of 4.8 Å, determined by the layer line from the power spectrum of the micrographs (Supplemental Fig. S4). Fibril structures were determined to an overall resolution of 2.8 Å for Ac1–140 (Fig. 2A), 3.0 Å for Ac1–122 (Fig. 2B), and 3.4 Å for Ac1–103 (Fig. 2C). At this resolution, most of the side chains are well resolved, allowing for unambiguous molecular modeling (see Supplemental Information). Similar number of residues were determined for all three structures.
Fig. 2.
CryoEM 3D reconstructions and atomic models of the α-syn fibrils. (A) Ac1–140 (PDB code 6OSJ), (B) Ac1–122 (PDB code 6OSL), and (C) Ac1–103 (PDB code 6OSM). In each panel, left: a surface-rendered view of the 3D reconstruction with two protofilaments colored in different shades of blue (Ac1–140), gold (Ac1–122), or magenta (Ac1–103); right: atomic models. Arrows indicate differences at N- and C-terminal regions. (D) 3D models of α-syn fibrils. Values of half pitch length are as indicated (top). Tilted views of each fibril depicted with distinct helical twist angles (bottom). (E) 2D averages. Fibril diameter (defined as width) decreases upon the removal of C-terminal residues.
Interestingly, there are lower-resolution densities that cannot be reliably modeled at both N- and C-terminal regions (Figs. 2B, 2C, Supplemental Figs. S3, and S5), likely attributable to conformational flexibility, consistent with previous studies [15, 17]. The extra densities seen for the C-terminal and N-terminal region in Ac1–122 and Ac1–103, respectively, are supported by the limited-proteolysis data. Specifically, at the C-terminal end, more resistance was observed for Ac1–122 at Q109/E110, whereas restricted access to N-terminal cleavage sites (A17/A18, E28/A29, and A30/G31) were seen for Ac1–103 (Supplemental Tables S1 and S2).
An increase of the helical twist from −0.714° to −1.054° to −1.358° (Fig. 2D) is seen with decreasing α-syn length, similar to our negative stain TEM data. Accordingly, the width of the fibrils measured through 2D class averages also decreases approximately from 92 to 88 to 77 Å (Fig. 2E), consistent with previous observations where helical pitch tends to increase with fibril width [36].
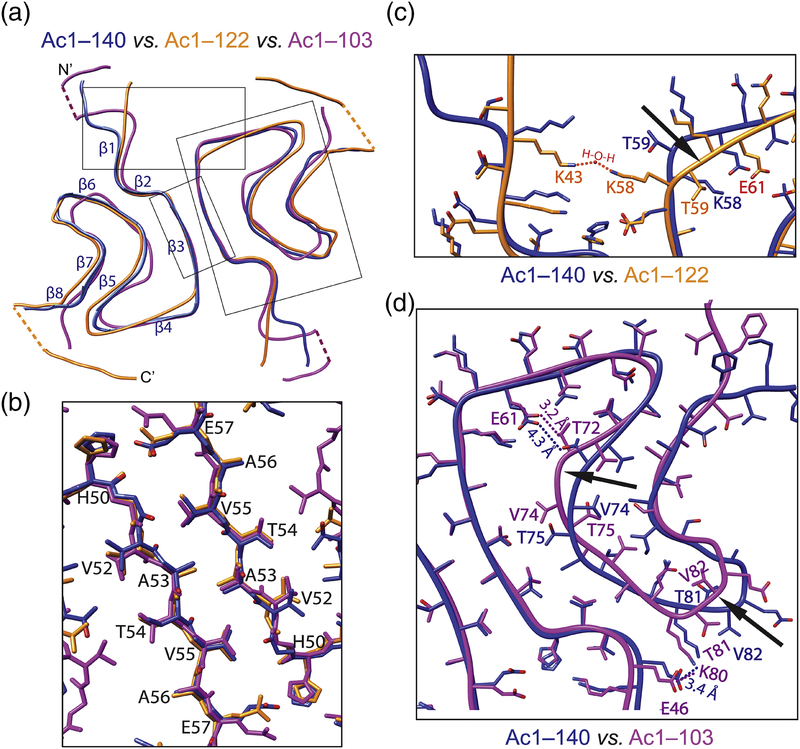
In the fibrillar state, Ac-α-syn folds into a β-strand-rich architecture with a Greek-key-like structure (Fig. 3A). The hydrophobic residues H50–E57 form a stable steric-zipper interface between two protofilaments (Figs. 3B and S6), which is further stabilized by the intermolecular interaction involving E57 from one and H50 together with K45 of another protofilament. Comparison of our Ac1–140 with other published full length cryoEM structures including acetylated (PDB 6A6B [16]) and non-acetylated α-syn (PDB 6CU7 [15]) as well as the solid-state NMR structure (2N0A [12]) is shown in Supplemental Fig. S7. The two Ac1–140 (ours and PDB 6A6B [16]) structures are nearly identical (RMSD = 0.75 Å). The acetylated and non-acetylated cryoEM structures are also similar with one noted exception of a stabilizing, internal salt bridge between K58 and E61 for the acetylated form, which leads to a different arch region between β3 and β4. In the non-acetylated structure [15], K58 faces outward, unavailable for interaction. While our helical twist is −0.714° (or 179.643° with 21 screw symmetry) reminiscent of the other acetylated structure (179.63°) [16], the non-acetylated fibril [15] is more twisted (179.53°). Unsurprisingly, the solid-state NMR structure [12] of a single α-syn protofilament is the most conformationally different as it lacks the interfacial hydrophobic interactions.
Fig. 3.
Comparison of atomic structures of α-syn fibrils. (A) Backbone overlays with individual β-strands as indicated. Overall RMSD values are 2.58 and 4.07 Å comparing Ac1–140 (blue) with Ac1–122 (gold) and Ac1–103 (magenta). Boxed regions are expanded in panels B-D. Dashed lines indicate limited EM densities. (B) Interface between the two protofilaments are composed of residues H50–E57. (C) Sidechain orientations of K58 and T59 are flipped in Ac1–122 vs. Ac1–140, causing differences between β3 and β4 (indicated by the arrow). Red dash lines denote potential interactions between putative H2O and both K43 and K58 in Ac1–122. (D) Residues V74 and T75, T81 and V82 are facing opposite directions in Ac1–103 vs. Ac1–140, creating differences in regions between β5–β6 and β6–β7, respectively (indicated by arrows). Hydrogen bond between E61–T72 and salt bridge between E46–K80 in Ac1–103 are also indicated.
Comparing Ac1–140, Ac1–122, and Ac1–103, there are little perturbations at the fibril interface (Fig. 3B); however, the removal of C-terminal residues impacted both N- and C-termini (Fig. 3A). In Ac1–122, the orientations of sidechains of K58 and T59 are flipped (Fig. 3C), leading to a notable change between β3 and β4 region. An internal salt bridge between K58 and E61 which exists in Ac1–140 and Ac1–103 are thus broken (Fig. 3C and Supplemental Fig. S8), which may destabilize the Ac1–122 fibril structure. However, we hypothesize the presence of compensatory interactions between the two protofilaments, facilitated by a water molecule, forming hydrogen bonds with K43 and K58 from the protomers (Fig. 3C).
Clear differences in the region spanning residues E46–V95 are also revealed between Ac1–140 and Ac1–103 (Fig. 3D). In particular, residues V74, T75, T81, T82 are clearly oriented in opposite directions, creating conformational changes of the backbone in regions between β5–β6 and β6–β7. A compaction is seen with an additional turn in β5, bringing the turn region between β6 and β7 inward to the central core (Fig. 3D, indicated by arrows). We surmise that upon the removal of C-terminal residues, sharper turns are stabilized, which impact the formation of the arch regions, giving rise to conformation changes in both N- and C-terminal regions (Figs. 2B and C, indicated by arrows). Moreover, the interactions associated with E61⋯T72 showed hydrogen bonding between E61 and T72 for Ac1–103. Together with the salt bridge between E46 and K80, these interactions would correlate to a more hydrophobic environment, implicating that the N-terminus of Ac1–103 is in close proximity to this region. This hypothesis is supported by the observation of limited EM density proximal to β6 (Supplemental Fig. S9), suggestive of a folding back of the N-terminal residues, which would then explain why the N-terminus of Ac1–103 fibrils are protected from PK-digestion.
Taken together, the loss of C-terminal residues strongly correlates to accelerated aggregation and increased helical twist of α-syn fibrils. Within the commonly observed Greek-key like topology, molecular differences and interaction changes generate a more compact core for the twisted Ac1–103 fibril structure. Interestingly, Ac1–103 fibrils are almost as twisted as the recently reported “twister” structure for non-acetylated 1–140 [15], which has a completely different core and interfacial residues. This work highlights a more complex picture of α-syn fibril polymorphism from the residue to the ultrastructural level, where the helical twist is not dictated by the core structure. Rather, we suggest that fibril twist is modulated by both N- and C-terminal regions, involving polypeptide conformational changes and steric hinderance, respectively. Finally, since Ac1–103 efficiently seeds Ac1–140, this twisted fibril polymorph poses a greater threat in promoting α-syn amyloid formation and explains their presence in LBs in PD.
Supplementary Material
Highlights.
Effects of C-terminal truncations on α-syn fibril formation were studied
Fibril helical twists increased upon the removal of C-terminal residues
Fibril structures of full length and ΔC-α-syn were solved by cryoEM
Conformational differences were revealed at N- and C-termini
Structural insights were gained on α-syn fibril polymorphism
Acknowledgment
Supported by the Intramural Research Program at the NIH, NHLBI. TEM, LC-MS, and cryoEM were performed on instruments maintained by the NHLBI EM Core, NHLBI Biochemistry Core, and the NIH Multi-institute CryoEM Facility (MICEF), respectively. CryoEM data processing was performed using the NIH High-Performance Computing (HPC) Biowulf cluster (http://hpc.nih.gov).
Abbreviations
- PD
Parkinson’s disease
- LBs
Lewy bodies
- α-syn
α-synuclein
- ΔC
C-terminally truncated
- Ac
N-terminally acetylated
- cryoEM
cryoelectron microscopy
- ThT
Thioflavin T
- TEM
transmission electron microscopy
- PK
proteinase-K
Footnotes
Appendix A. Supplementary information
Supplementary information to this article can be found online at https://doi.org/10.1016/j.jmb.2019.07.001.
Accession numbers: The cryoEM density maps for all three α-syn fibrils (Ac1-140, Ac1-122, and Ac1-103) were deposited in the Electron Microscopy Data Bank with the accession number EMD-20183, EMD-20185 and EMD-20186 respectively. The associated atomic models were deposited in the RCSB Protein Data Bank with the entry code 6OSJ, 6OSL and 6OSM respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. U. S. A 95 (1998) 6469–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lucking CB, Brice A. α-synuclein and Parkinson’s disease. Cell. Mol. Life Sci 57 (2000) 1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Schmid AW, Fauvet B, Moniatte M, Lashuel HA. α-synuclein post-translational modifications as potential biomarkers for Parkinson disease and other synucleinopathies. Mol. Cell. Proteomics 12 (2013) 3543–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Xilouri M, Brekk OR, Stefanis L. α-synuclein and protein degradation systems: a reciprocal relationship. Mol. Neurobiol 47 (2013) 537–551. [DOI] [PubMed] [Google Scholar]
- [5].Murray IVJ, Giasson BI, Quinn SM, Koppaka V, Axelsen PH, Ischiropoulos H, et al. Role of α-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry. 42 (2003) 8530–8540. [DOI] [PubMed] [Google Scholar]
- [6].van der Wateren IM, Knowles TPJ, Buell AK, Dobson CM, Galvagnion C. C-terminal truncation of α-synuclein promotes amyloid fibril amplification at physiological pH. Chem. Sci 9 (2018) 5506–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Crowther RA, Jakes R, Spillantini MG, Goedert M. Synthetic filaments assembled from C-terminally truncated α-synuclein. FEBS Lett. 436 (1998) 309–312. [DOI] [PubMed] [Google Scholar]
- [8].Levitan K, Chereau D, Cohen SIA, Knowles TPJ, Dobson CM, Fink AL, et al. Conserved C-terminal charge exerts a profound influence on the aggregation rate of α-synuclein. J. Mol. Biol 411 (2011) 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hoyer W, Cherny D, Subramaniam V, Jovin TM. Impact of the acidic C-terminal region comprising amino acids 109–140 on α-synuclein aggregation in vitro. Biochemistry. 43 (2004) 16233–16242. [DOI] [PubMed] [Google Scholar]
- [10].Ulusoy A, Febbraro F, Jensen PH, Kirik D, Romero-Ramos M. Co-expression of C-terminal truncated α-synuclein enhances full-length α-synuclein-induced pathology. Eur. J. Neurosci 32 (2010) 409–422. [DOI] [PubMed] [Google Scholar]
- [11].Liu CW, Giasson BI, Lewis KA, Lee VM, DeMartino GN, Thomas PJ. A precipitating role for truncated α-synuclein and the proteasome in α-synuclein aggregation - Implications for pathogenesis of Parkinson disease. J. Biol. Chem 280 (2005) 22670–22678. [DOI] [PubMed] [Google Scholar]
- [12].Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol 23 (2016) 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, Verel R, et al. The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. U. S. A 105 (2008) 8637–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen M, Margittai M, Chen J, Langen R. Investigation of α-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem 282 (2007) 24970–24979. [DOI] [PubMed] [Google Scholar]
- [15].Li BS, Ge P, Murray KA, Sheth P, Zhang M, Nair G, et al. Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun 9 (2018) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li YW, Zhao CY, Luo F, Liu ZY, Gui XR, Luo ZP, et al. Amyloid fibril structure of α-synuclein determined by cryoelectron microscopy. Cell Res. 28 (2018) 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guerrero-Ferreira R, Taylor NMI, Mona D, Ringler P, Lauer ME, Riek R, et al. Cryo-EM structure of α-synuclein fibrils. eLife. 7 (2018) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McGlinchey RP, Dominah GA, Lee JC. Taking a bite out of amyloid: mechanistic insights into a-synuclein degradation by cathepsin L. Biochemistry. 56 (2017) 3881–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yap TL, Pfefferkorn CM, Lee JC. Residue-specific fluorescent probes of α-synuclein: detection of early events at the N- and C-termini during fibril assembly. Biochemistry. 50 (2011) 1963–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Flynn JD, Jiang Z, Lee JC. Segmental C-13-labeling and Raman microspectroscopy of α-synuclein amyloid formation. Angew. Chem.-Int. Edit. 57 (2018) 17069–17072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang ZT, Kang SS, Liu X, Ahn EH, Zhang ZH, He L, et al. Asparagine endopeptidase cleaves α-synuclein and mediates pathologic activities in Parkinson’s disease. Nat. Struct. Mol. Biol 24 (2017) 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kellie JF, Higgs RE, Ryder JW, Major A, Beach TG, Adler CH, et al. Quantitative measurement of intact α-synuclein proteoforms from post-mortem control and Parkinson’s disease brain tissue by intact protein mass spectrometry. Sci Rep. 4 (2014) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Iadanza MG, Silvers R, Boardman J, Smith HI, Karamanos TK, Debelouchina GT, et al. The structure of a β(2)-microglobulin fibril suggests a molecular basis for its amyloid polymorphism. Nat. Commun 9 (2018) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol 19 (2018) 755–773. [DOI] [PubMed] [Google Scholar]
- [25].Gremer L, Scholzel D, Schenk C, Reinartz E, Labahn J, Ravelli RBG, et al. Fibril structure of amyloid-beta(1–42) by cryo-electron microscopy. Science. 358 (2017) 116–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 547 (2017) 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Radamaker L, Lin YH, Annamalai K, Huhn S, Hegenbart U, Schonland SO, et al. Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis. Nat. Commun 10 (2019) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Swuec P, Lavatelli F, Tasaki M, Paissonil C, Rognoni P, Maritanl M, et al. Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Nat. Commun 10 (2019) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liberta F, Loerch S, Rennegarbege M, Schierhorn A, Westermark P, Westermark GT, et al. Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids. Nat. Commun 10 (2019) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Falcon B, Zhang WJ, Murzin AG, Murshudov G, Garringer HJ, Vidal R, et al. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature. 561 (2018) 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. 568 (2019) 420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].LeVine H Quantification of β-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 309 (1999) 274–284. [DOI] [PubMed] [Google Scholar]
- [33].Flynn JD, McGlinchey RP, Walker RL, Lee JC. Structural features of α-synuclein amyloid fibrils revealed by Raman spectroscopy. J. Biol. Chem 293 (2018) 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Flynn JD, Lee JC. Raman fingerprints of amyloid structures. Chem. Commun 54 (2018) 6983–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bousset L, Pieri L, Ruiz-Arlandis G, Gath J, Jensen PH, Habenstein B, et al. Structural and functional characterization of two α-synuclein strains. Nat. Commun 4 (2013) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Close W, Neumann M, Schmidt A, Hora M, Annamalai K, Schmidt M, et al. Physical basis of amyloid fibril polymorphism. Nat. Commun 9 (2018) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.