Abstract
NTRK3-rearranged tumors other than infantile fibrosarcomas (IFSs) harboring the canonical ETV6-NTRK3 fusions are uncommon, and include mainly inflammatory myofibroblastic tumors and gastrointestinal stromal tumors. Herein, we describe an additional subset of seven tumors sharing NTRK3 gene rearrangements. The cohort included five females and two males (age range 1–67 years). Tumors were located in extremities, trunk, retroperitoneum, or intra-abdominal. In all tumors, fluorescence in situ hybridization (FISH) revealed rearrangements in NTRK3 accompanied by NTRK3 amplification in two cases. In three cases, RNA sequencing identified a fusion transcript composed of NTRK3 exon 14 fused to ETV6, TFG, and TPM4, respectively, retaining the NTRK3 kinase domain. All tumors were positive for pan-TRK by immunohistochemistry (IHC). Two cases showed low- to intermediate-grade histology composed of monomorphic spindle cells arranged in a patternless architecture, stromal bands, and perivascular rings of hyalinized collagen and coexpression of S100 and CD34. The remaining five cases were high-grade fascicular monomorphic spindle cell sarcomas, morphologically somewhat reminiscent of either malignant peripheral nerve sheath tumors (MPNSTs) or fibrosarcomas (FSs). Two high-grade NTRK3 sarcomas showed aggressive clinical behavior with development of lung metastases. Identification of high-grade NTRK3-rearranged sarcomas is clinically important, since the development of selective NTRK inhibitors has opened new avenues for targeted therapy. Although IHC for pan-TRK can be applied as a screening tool, molecular studies are recommended for a conclusive diagnosis of NTRK-rearranged neoplasms.
Keywords: fibrosarcoma, fusion, malignant peripheral nerve sheath tumor, NTRK3, sarcoma
1 |. INTRODUCTION
Neurotropic tropomyosin receptor kinase (NTRK) fusions were described recently in soft tissue tumors that are morphologically distinct from infantile fibrosarcoma (IFS).1,2 In particular, NTRK1-rearranged tumors are characterized by spindle cell morphology, presenting in patients of both sexes, with a wide range of age and anatomic locations, usually in extremities and trunk, and, more rarely, in intra-abdominal sites. These tumors show variable morphology and histologic grade, and frequently coexpress CD34 and S100 by immunohistochemistry (IHC), in addition to TRK-A and pan-TRK expression. Two histologic patterns have been described so far. First, in superficial locations, NTRK1-rearranged tumors infiltrate subcutaneous fat in a reticular fashion, a morphologic pattern reminiscent of lipofibromatosis (so-called lipofibromatosis-like neural tumor).3 The second group closely resembles malignant peripheral nerve sheath tumor (so-called MPNST-like tumors), spanning variable histologic grades. At one end of this morphologic spectrum are infiltrative MPNST-like tumors composed of haphazardly arranged monomorphic spindle cells with low mitotic activity and distinctive patterns of keloidal stromal and perivascular collagen deposition. At the other end of the spectrum, NTRK1-rearranged spindle cell sarcomas are composed of cellular fascicles of morphologically malignant spindle cells with a high mitotic rate (exceeding 10 mitotic figures [MFs]/10 high power fields [HPFs]) and these tumors are capable of developing (lung) metastases. In this study, we present a cohort of seven spindle cell sarcomas with NTRK3 gene rearrangements, which also display a wide range of histologic grade and clinical behavior.
2 |. MATERIALS AND METHODS
Seven NTRK3-rearranged soft tissue spindle cell tumors other than classic IFS were retrieved from the consultation files of two of the authors (C.R.A., C.D.F.). The case selection for molecular investigation was based on the initial reported observations that the morphologic spectrum of kinase fusion-positive sarcomas includes tumors resembling fibrosarcomas (FSs) or MPNSTs. A large collection of these various morphologies was available in our consultation files from prior molecular studies, which were previously screened and lacked abnormalities in NTRK1/2, BRAF, and RAF1 genes.1,3 Clinical data, including age, gender, anatomic site, and gross features of tumors were retrieved from pathology reports. All tumors had been surgically removed and hematoxylin and eosin-stained slides from resection specimens were reviewed by two of us (A.S., C.R.A.). Tumors were graded according to the FNCLCC grading system. IHC for CD34, S100, SOX10, pan-TRK (TRK-ABC), and H3K27me3 was performed according to standardized procedures on automated platforms. Staining was performed on a Leica-Bond-3 (Leica, Buffalo Grove, Illinois) or a Ventana Benchmark (Ventana Medical Systems, Tucson, Arizona) automated immunostaining platform using a heat-based antigen retrieval method and high pH buffer.
2.1 |. Targeted RNA sequencing
One case was analyzed by targeted RNA sequencing (Table 1). RNA was extracted from formalin-fixed paraffin embedded (FFPE) tissue using Amsbio’s ExpressArt FFPE Clear RNA Ready kit (Amsbio LLC, Cambridge, Massachusetts). The fragment length was assessed with an RNA 6000 chip on an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, California). RNAseq libraries were prepared using 20 to 100 ng total RNA with the TruSight RNA Fusion Panel (Illumina, San Diego, California). Targeted RNA sequencing was performed on an Illumina MiSeq platform. Reads were independently aligned with STAR (version 2.3) against the human reference genome (hg19) and analyzed by STAR-Fusion.
TABLE 1.
Clinicopathologic and molecular findings of tumors with NTRK3 fusions
| Patient | Location | cm | G | MC | NE | S100 | CD34 | Sox10 | H3 | TRK | Molecular | Clinical FU | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 67/F | Thigh | 4 | IG | 14 | − | pos | pos | neg | pos | pos | ETV6-NTRK3a | NA |
| 2 | 21/F | Paraspinal | 8 | IG | 11 | − | pos | pos | neg | pos | pos | TFG-NTRK3b | NED 5 mo |
| 3 | 1/M | Foot | 4 | HG | 13 | + | pos | pos | NA | NA | pos | EML4-NTRK3c | NA |
| 4 | 60/F | Popliteal | 8 | HG | 6 | + | pos | pos | neg | pos | neg | TPM4-NTRK3b | NED 36 mo |
| 5 | 11/M | Mesentery | 11 | HG | 28 | + | pos | pos | NA | NA | pos | NTRK3 b/aa | DOD 1 mo |
| 6 | 6/F | Retroperit | 11 | HG | 10 | + | neg | pos | neg | pos | pos | NTRK3 b/a +a | AWD 4 mo |
| 7 | 49/F | Abdomino-pelvis | 25 | HG | 28 | + | neg | pos | neg | pos | pos | NTRK3 b/a +a | NED 11 mo |
Abbreviations: cm, size in cm; G, grade; H3, H3K27me3; HG, high grade; IG, intermediate grade; MC, mitotic count/10 HPFs; NE, necrosis.
FISH; b/a, break-apart; +, amplification.
Archer FusionPlex assay.
Targeted RNA sequencing; FU, follow-up; NA, not available; NED, no evidence of disease; AWD, alive with disease; DOD, dead of disease; mo, months.
2.2 |. Anchored multiplex RNA sequencing (Archer Dx)
Two cases were studied by anchored multiplex RNA sequencing assay, the detailed procedure has been previously described.4 In short, unidirectional gene-specific primers were designed to target specific exons in 62 genes known to be involved in oncogenic fusions in solid tumors. In brief, RNA was extracted from FFPE specimens, followed by cDNA synthesis and library preparation. Anchored multiplex polymerase chain reaction amplicons were sequenced on Illumina Miseq, and the data were analyzed using the Archer software.
2.3 |. MSK-IMPACT assay
One case was investigated with the MSK-IMPACT assay.5 Briefly, MSK-IMPACT is a comprehensive molecular profiling assay that involves hybridization capture and deep sequencing of all exons and selected introns of up to 468 oncogenes and tumor-suppressor genes, allowing the detection of point mutations, small and large insertions or deletions, and rearrangements. In addition to capturing all coding regions of the genes, the assay also captures >1000 intergenic and intronic single-nucleotide polymorphisms (tiling probes), interspersed homogenously across the genome, aiding the accurate assessment of genome-wide copy number. In total, the probes target approximately 1.2 megabases of the human genome.
2.4 |. Fluorescence in situ hybridization
All cases were tested by FISH for NTRK3 gene abnormalities. Custom probes made by bacterial artificial chromosomes (BACs) clones flanking the genes of interest according to UCSC genome browser (http://genome.ucsc.edu) and obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, California; http://bacpac.chori.org).6 DNA from each BAC was isolated according to the manufacturer’s instructions. The BAC clones were labeled with fluorochromes (fluorescent-labeled dUTPs, Enzo Life Sciences, New York, New York) by nick translation and validated on normal metaphase chromosomes. The 4-μm-thick FFPE slides were deparaffinized, pretreated, and hybridized with denatured probes. After overnight incubation, the slides were washed, stained with 4’,6-diamidino-2-phenylindole, mounted with an antifade solution, and then examined on a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) controlled by the Isis 5 software (Metasystems).
3 |. RESULTS
The clinical presentation, gross and histologic features, IHC staining results, and molecular abnormalities of the seven NTRK3-rearranged spindle cell tumors are summarized in Table 1. The seven patients selected had a wide age range (1–67, median 21 years). Three tumors presented in children (range 1–11 years), one in a young adult (21 years) and three in older adults (range 49–67 years). Both genders were affected, including five women and two males. Anatomic sites were diverse with extremity, trunk, and retroperitoneal locations, whereas two high-grade sarcomas were located intra-abdominally, involving bowel wall. Tumor size ranged from 4 to 25 cm (mean 10 cm).
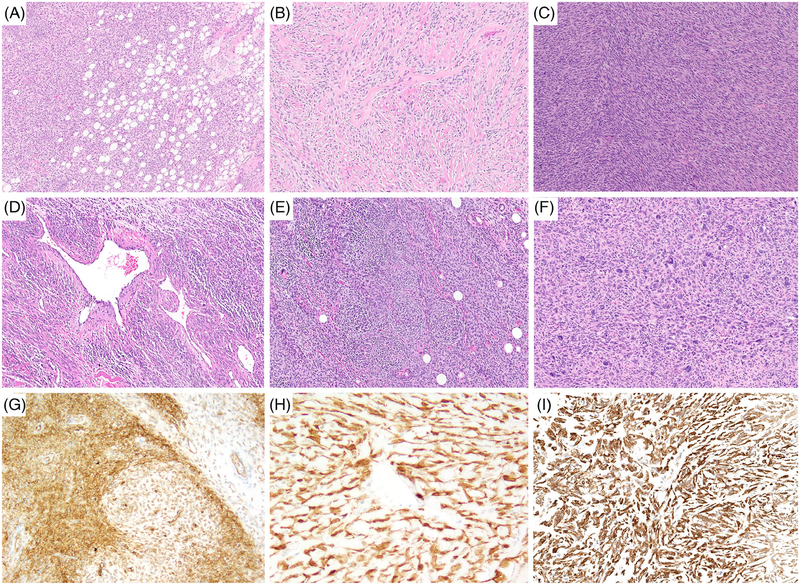
NTRK3-rearranged spindle cell tumors spanned a wide histologic spectrum. Two cases (cases 1 and 2) consisted of low- to intermediate-grade monomorphic spindle cell sarcomas with multifocal to diffuse expression of S100 protein. Both cases showed an infiltrative growth, with brisk mitotic activity in areas (>10 MF/10 HPFs) and lacked necrosis. Case 1 was superficial, while case 2 had a highly infiltrative pattern within skeletal muscle. The histology of both tumors displayed focally the presence of stromal hyalinization and perivascular collagen rings, reminiscent in part of the group of low-grade spindle cell neoplasms with NTRK1, RAF, and BRAF fusions recently described by our group1 (Figure 1A,B). Case 2 in addition showed a highly infiltrative growth within subcutaneous fat, closely resembling lipofibromatosis-like neural tumor3 (Figure 1A). In fact, this case showed a multitude of architectural patterns, including sweeping herring bone-like fascicles, hemangiopericytoma-like (HPC-like) staghorn vessels and perivascular myopericytoma-like whorling (Figure 1C–E). Moreover, hypocellular areas with perivascular and stromal depositions of hyalinized collagen alternated with hypercellular areas with monomorphic hyperchromatic spindle cells showing brisk mitotic activity. This case also showed focal myxoid change and an area with marked nuclear pleo-morphism (Figure 1F). By IHC, both tumors showed multifocal coexpression of CD34 and S100, in addition to pan-TRK (Figure 1G–I), whereas Sox10 was negative and H3K27me3 was retained.
FIGURE 1.
Morphologic spectrum of NTRK3 rearranged tumors with low- to intermediate-grade MPNST-like histological features. A-I, Intermediate-grade sarcoma with TGF-NTRK3 fusion presenting as a paraspinal tumor in a 21-year-old female (case 2); A, haphazardly arranged monomorphic spindle cell proliferation with reticular infiltration of fat morphologically resembling lipofibromatosis-like neural tumor; B, stromal and perivascular deposition of hyalinized collagen; C, cellular fascicular areas with monomorphic spindle cell sarcoma; D, hemangiopericytoma-like vasculature; E, concentric perivascular whorling as in myopericytoma; F, area with pleomorphic tumor; G-I, positive IHC staining for CD34, S100, and pan-TRK, respectively. IHC, immunohistochemistry; MPNST, malignant peripheral nerve sheath tumor [Color figure can be viewed at wileyonlinelibrary.com]
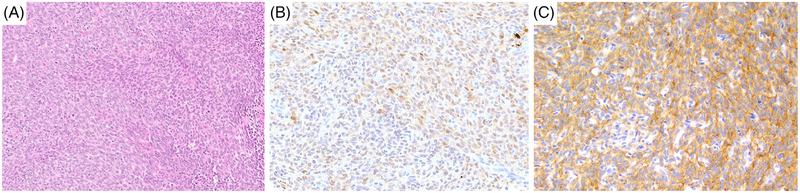
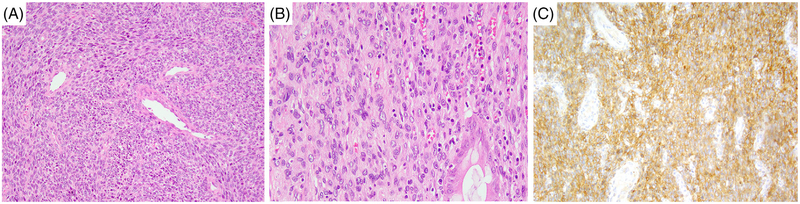
The remaining five cases consisted of high-grade spindle cell sarcomas showing increased mitotic activity and geographic areas of necrosis. Three tumors (cases 3–5) also showed focal S100 positivity. These tumors consisted of cellular fascicles of monomorphic hyper-chromatic spindle cells with a variable amount of palely eosinophilic cytoplasm (Figure 2A). Most cases showed relatively monomorphic cytomorphology, except for one case, which showed increased nuclear pleomorphism. The overall histologic features of this subset were somewhat reminiscent of high-grade MPNST showing multifocal coexpression of S100 (Figure 2B) and CD34. However, IHC for Sox10 was negative and nuclear H3K27me3 staining was retained. All three tumors were diffusely positive for pan-TRK, showing a cytoplasmic pattern (Figure 2C). The latter two cases (cases 6–7) showed a non-specific immunoprofile and had a fascicular growth with areas of necrosis (Figure 3A). The tumor cells had variably eosinophilic cytoplasm and case 7 showed patchy nuclear pleomorphism (Figure 3B). The two lesions showed CD34 and pan-TRK coexpression (Figure 3C), while S100 protein and SOX10 were negative. H3K27me3 expression was retained.
FIGURE 2.
Morphologic spectrum of NTRK3 rearranged sarcomas with a high-grade MPNST-like phenotype and S100 expression. A-C, High-grade MPNST-like sarcoma with EML4-NTRK3 fusion (case 3); A, cellular fascicular monomorphic spindle cell sarcoma; B, multifocal S100 expression; C, diffuse staining for pan-TRK. MPNST, malignant peripheral nerve sheath tumor [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 3.
Morphologic spectrum of NTRK3 rearranged high-grade sarcomas with a nonspecific immunophenotype. A-C, High-grade FS-like sarcoma with NTRK3 rearrangement and amplification presenting as a huge pelvic tumor in a 49-year-old female (case 7); A, cellular fascicular spindle cell sarcoma; B, tumor infiltrating colon; C, IHC expression of pan-TRK. FS, fibrosarcoma; IHC, immunohistochemistry [Color figure can be viewed at wileyonlinelibrary.com]
3.1 |. Molecular pathology
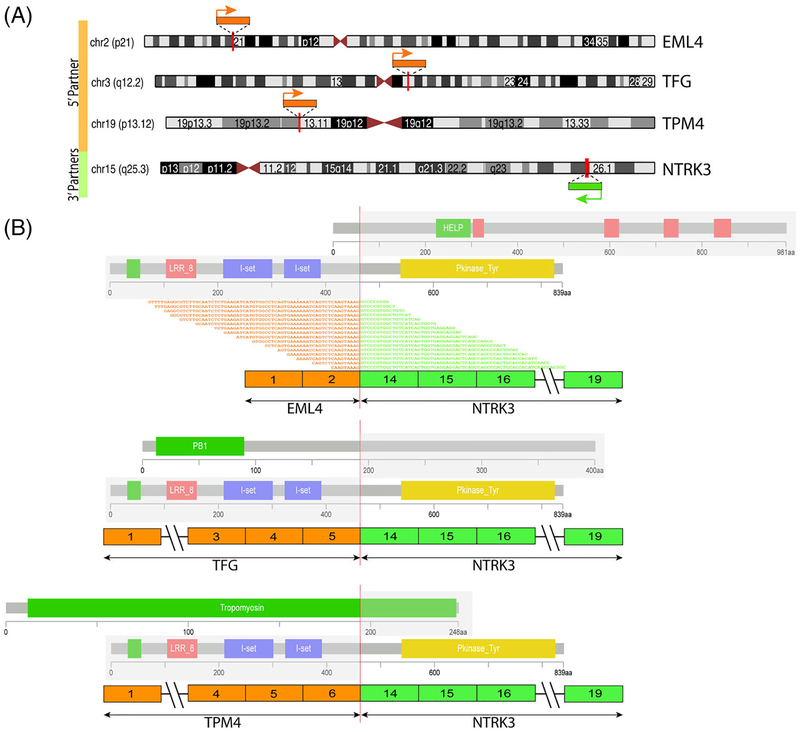
As shown in Table 1, four different NTRK3 fusion partners were identified, including ETV6-NTRK3 (detected by FISH), TFG-NTRK3 and TPM4-NTRK3 (by Archer Fusionplex RNA sequencing), and EML4-NTRK3 (by targeted RNA sequencing). In all three fusion transcripts detected by RNA sequencing, NTRK3 breakpoints were in exon 14 (fused with TFG exon 5, TPM4 exon 6, and EML4 exon 2), which retained the NTRK kinase domain in the projected fusion oncoprotein (Figure 4). By RNA sequencing, one case showed NTRK3 mRNA overexpression in contrast to other sarcoma types. All cases were confirmed by FISH to harbor NTRK3 gene rearrangements (Figure S1A); however, in two cases NTRK3 showed gene amplifications in addition to the break-apart signal (Figure S1B). By MSK-IMPACT, the intermediate-grade tumor with focal areas of anaplasia (case 2, TFG-NTRK3 fusion) contained two p53 mutations and CDKN2A/B deletions.
FIGURE 4.
Diagrammatic representation of three NTRK3 fusion-positive sarcomas studied with RNA sequencing. A, Chromosomal localization of NTRK3 on 15q25.3 and the three different gene partners: EML4, on 2p21; TFG, on 3q12.2; and TPM4, on 19p13.12; red vertical lines depict the genomic breakpoints. Arrows show the direction of transcription of each gene. B, Each of the three different NTRK3 fusion transcripts are depicted in relationship to their exonic composition. For the EML4-NTRK3 fusion detected by TruSight RNAseq, supporting fusion junction reads are also illustrated. In all cases, the fusion transcript included the 3’ of NTRK3 gene, starting from exon 14, thus retaining the kinase domain in the putative fusion oncoprotein. The protein domains of each of the genes involved are also schematically depicted. [Correction added on June 14, 2019 after first online publication: Figure 4 has been revised. The revised version is included here.] [Color figure can be viewed at wileyonlinelibrary.com]
3.2 |. Clinical follow-up
Follow-up information is summarized in Table 1. Given the limited number of cases with clinical follow-up and relatively short follow-up time, accurate prediction of prognosis of NTRK3-rearranged adult tumors is not feasible. Nevertheless, two tumors with high-grade morphology (cases 4 and 5) showed aggressive clinical behavior with metastatic spread to the lung. In case 4, lung metastases were excised and this patient has no evidence of disease after 3 years.
4 |. DISCUSSION
In recent years it has become evident that a small subset of spindle cell sarcomas (other than IFS with canonical ETV6-NTRK3 fusions) harbor NTRK fusions.1,6–9 Most of the reported tumors show NTRK1 gene fusions, with various partners, such as LMNA, TPM3, or TPR, whereas NTRK2 and NTRK3 rearrangements are relatively rare.1,10,11 NTRK1-positive spindle cell sarcomas span a wide clinical spectrum, with highly variable histology and tumor grade. Based on microscopic features, NTRK1-rearranged soft tissue sarcomas have been divided into low-grade tumors with haphazardly arranged monomorphic spindle cells and distinctive hyalinization and high-grade tumors with cellular fascicles of mitotically active hyperchromatic spindle cells, often with necrosis. Based on their additional immunoprofile, these tumors have been designated as FS-like (showing a nonspecific immunoprofile of vimentin or CD34 expression only)2 or MPNST-like (showing coexpression of S100 and CD34).1 In addition, a group of likely benign tumors with a highly infiltrative growth within fat, mimicking lipofibromatosis have also been decribed, harboring exclusively to date NTRK1 gene rearrangement.3
To date NTRK3 gene fusions have been described in only five sarcomas thought to represent fibrosarcomas (so-called FS-like sarcomas). Their clinicopathologic features, molecular genetics, and clinical follow-up being summarized in Table 2. Olson et al.10 found an EML4-NTRK3 fusion in a regional metastasis of an aggressive superficial spindle cell sarcoma occuring in the lower back of a 50-year-old female. Initially, this tumor was considered to represent fibrosarcomatous transformation in dermatofibrosarcoma protuberans (FS-DFSP). Yamazaki et al.11 detected a STRN3-NTRK3 fusion (in addition to a CDKN2A homozygous deletion) in a FS originating in the thigh of a 26-year-old man and a STRN-NTRK3 fusion in a low-grade FS originating in the radius. Moreover, two rare uterine FS-like sarcomas with EML4-NTRK312 and RBPMS-NTRK3 fusion have been reported.6 ETV6-NTRK3 fusions have also been described in rare examples of adult pulmonary inflammatory myofibroblastic tumors,13,14 as well as in rare gastrointestinal stromal tumors lacking genetic abnormalities in the KIT/PDGFRA/SDHx/BRAF pathways.15,16
TABLE 2.
Literature review of five NTRK3-rearranged adult fibrosarcomas
| Patient | Location | cm | Grade | MC | NE | S100 | CD34 | TRK | Molecular | Follow-up | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 26/M | Thigh | 11 | HG | 12 | + | neg | pos | pos | STRN3-NTRK3a | DOD 29 mo11 |
| 2 | 50/F | Back | 3 | HG | 15 | − | neg | pos | pos | EML4-NTRK3 | LN met 60 mo10 |
| 3 | 38/F | Radius | 9 | LG | 3 | − | neg | pos | pos | STRN-NTRK3 | AWD 113 mo11 |
| 4 | 26/F | Cervix | 12 | HG | 3 | + | pos | pos | pos | EML4-NTRK3a | AWD 52 mo12 |
| 5 | 47/F | Cervix | 14 | HG | 12 | + | pos | neg | pos | RBPMS-NTRK3 | DOD 79 mo6 |
Abbreviations: cm, size in cm; HG, high grade; MC, mitotic count/10 HPFs; NE, necrosis; TRK, pan-TRK IHC.
Additional homozygous CDKN2A deletion; mo; months; DOD, dead of disease; LN, lymph node; met, metastasis; AWD, alive with disease.
In this study, we investigate seven additional soft tissue spindle cell sarcomas with NTRK3 fusions, with morphology reminiscent of either MPNST or FS. Similar to NTRK1/2-rearranged tumors, those with NTRK3 fusions had a wide age distribution and occured in children and (young) adults, as well as in elderly patients. With respect to tumor location, in addition to originating in extremities or trunk, a significant number of NTRK3 rearranged sarcomas presented in abdominal locations. In contrast to NTRK1 fusion-positive tumors, none of the cases in this cohort showed benign or pure low-grade morphology. Although two cases showed focal areas of a lower grade component, the large majority of NTRK3-positive tumors in this series and those reported in the literature were high-grade sarcomas, and this translated into metastatic behavior (lung metastases) and poor clinical outcome (Tables 1 and 2).
In all three cases analyzed with RNA sequencing the breakpoint in NTRK3 was consistent in exon 14, resulting in a preserved tyrosine kinase domain. By RNA expression analysis, overexpression of NTRK3 was documented in one case. At the protein level, cytoplasmic pan-TRK was seen in all cases. However, pan-TRK immunoreactivity is not entirely specific,17 and therefore, additional molecular genetic testing is recommended for a conclusive diagnosis of NTRK-fused sarcomas. This has become particularly relevant, as these advanced stage tumors are potentially amenable to TRK inhibition, such as with Larotrectinib, a selective TRK inhibitor, which may reduce tumor volume as a preoperative strategy or improve survival in advanced stage disease.18,19
The mutational burden of sarcomas with NTRK fusions has not been studied in detail. However, one of our cases studied showed simultaneous p53 mutations and CDKN2A/B deletions. Interestingly, in two of five NTRK3-rearranged sarcomas described in the literature, CDKN2A deletions have been described11,12 (Table 2). Furthermore, two of the cases showed unbalanced NTRK3 gene rearrangements associated with NTRK3 amplification. The impact of these additional secondary genetic events of NTRK3-fusion-positive tumors on pathogenesis (tumor initiation and progression), histologic grade (mainly high grade), and aggressive outcome remains to be established.
Given the morphologic spectrum and nonspecific immunostaining for CD34 and S100, the differential diagnosis of NTRK3-rearranged sarcomas includes MPNST, FS, including FS-DFSP, solitary fibrous tumor, synovial sarcoma, among others.
Low-grade MPNST is often associated with neurofibromatosis type I (NF1) and usually, represents malignant transformation of a pre-existent neurofibroma. Low-grade MPNST contains numerous S100-positive and SOX10-positive Schwann cells, while losing most of the intermixed CD34-positive fibroblastic framework stroma, which is typically retained in the pre-existent neurofibroma component. In contrast, NTRK-rearranged tumors of low- to intermediate-grade often show coexpression of S100 and CD34, and consistently lack SOX10 reactivity. High-grade MPNST can be either sporadic, NF1-asociated or arise in association with prior radiation. Approximately 30 to 40% of high-grade MPNSTs show focal expression of the nerve sheath markers S100, GFAP, and SOX10. Moreover, diffuse loss of trimethylation of H3K27me3 is observed in the large majority of high-grade MPNSTs, which currently is the most sensitive and specific IHC marker.20,21 Notably, NTRK-rearranged sarcomas (including NTRK3-fused high-grade sarcomas) are not associated with NF1 or prior radiotherapy. Although these tumors may show reactivity for S100, they are always negative for SOX10 and retain H3K27me3 expression.
In superficial locations, CD34-positive NTRK-rearranged neoplasms showing a higher grade morphology may closely resemble FS ex-DFSP. However, the typical DFSP component usually has a repetitive storiform architecture and stains diffusely for CD34, whereas S100 is negative. CD34 may be lost in areas of fibrosarcomatous change. Importantly, the typical COL1A1-PDGFB fusion gene is retained in FS ex-DFSP.
The typical histologic features of solitary fibrous tumor are alternating hypocellular and hypercellular areas, HPC-like vessels, and fibromyxoid stroma containing haphazardly arranged primitive mesenchymal spindle cells. Some of these features overlap closely with those seen in NTRK-rearranged spindle cell neoplasm at the low-grade end of the spectrum. However, the molecular genetic hallmark of solitary fibrous tumor is NAB2-STAT6 gene fusion and the diffuse nuclear STAT6 staining can be used as a surrogate marker.22,23
By histology, typical features of monophasic synovial sarcoma are cellular fascicles of delicate monomorphic blue spindle cells with focal deposition of collagen and HPC-like vasculature. Tumors may focally express S100, but the majority are at least focally positive for epithelial membrane antigen and cytokeratins. An accurate diagnosis of monophasic synovial sarcoma is possible by molecular genetic techniques that detect the fusion genes SS18-SSX1/2.24
In conclusion, the seven NTRK3-rearranged soft tissue spindle cell sarcomas reported herein represent a morphologically diverse group of tumors with variable expression of CD34 and S100. Although NTRK3-rearranged lesions show histologic overlap with NTRK1 fusion-positive tumors, none of the cases were benign, being mostly at the high-grade end of the spectrum. To assist the practicing pathologist in a better pattern recognition, we have artificially divided these lesions in two groups, based on histologic features, grade and coexpression of S100 and CD34. First, tumors with low- to intermediate-grade histology were composed of monomophic spindle cells haphazardly arranged and associated with hyalinized collagen in stromal bands and perivascular rings, a hallmark feature that may help pathologists to recognize these heterogeneous NTRK-rearranged mesenchymal neoplasms. Moreover, by IHC, these NTRK-fused tumors typically show coexpression of S100 and CD34, in the absence of SOX10. At the other end of the spectrum were NTRK3-rearranged sarcomas with high-grade morphology, consisting of cellular fascicles with predominantly monomorphic hyperchromatic spindle cells (fascicular monomorphic blue spindle cell sarcomas). Some of the cases in this subset showed S100 positivity, while others had a nonspecific immunoprofile. This latter group resembled tumors that otherwise probably would have been classified either as high-grade MPNSTs or adult (or infantile) FSs. However, given the lack of specific diagnostic criteria for adult FS, we propose to classify these tumors simply as “NTRK3-rearranged spindle cell sarcomas”. Unlike classic high-grade MPNSTs, sarcomas with NTRK fusions are not associated with NF1 or a history of prior irradiation and, overall, IHC staining for SOX10 is negative, whereas H3K27me3 is retained. The clinical presentation of NTRK3-rearranged tumors (age and sex distribution and anatomic location) matches that of other NTRK-rearranged neoplasms and includes retroperitoneal, intra-abdominal, and uterine sites. Although selection bias cannot be ruled out, it appears that this group of NTRK3-rearranged tumors, compared to NTRK1-fused neoplasms, more often display high-grade morphology, which is often accompanied by aggressive clinical behavior and development of lung metastases. As highly effective targeted therapies with selective NTRK inhibitors are now available for clinical use, it is becoming important to corectly diagnose these high-grade NTRK-rearranged sarcomas. Although not entirely specific, IHC for pan-TRK remains very helpful in recognizing NTRK-rearranged sarcomas, which are otherwise misdiagnosed as MPNST, FS, or solitary fibrous tumor. Clearly, molecular studies are sometimes critical for a definitive diagnosis of NTRK-rearranged mesenchymal neoplasms.
Supplementary Material
Funding information
Kristin Ann Carr Foundation; Cycle for Survival; NIH Clinical Center, Grant/Award Numbers: P30 CA008748, P50 CA217694, P50 CA 140146–01
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Suurmeijer AJH, Dickson BC, Swanson D, et al. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Genes Chromosomes Cancer. 2018;57(12):611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis JL, Lockwood CM, Stohr B, et al. Expanding the spectrum of pediatric NTRK-rearranged mesenchymal tumors. Am J Surg Pathol. 2019;43(4):435–445. [DOI] [PubMed] [Google Scholar]
- 3.Agaram NP, Zhang L, Sung YS, et al. Recurrent NTRK1 gene fusions define a novel subset of locally aggressive lipofibromatosis-like neural tumors. Am J Surg Pathol. 2016;40(10):1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. [DOI] [PubMed] [Google Scholar]
- 5.Cheng DT, Mitchell TN, Zehir A, et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015; 17(3):251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang S, Cotzia P, Hyman DM, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol. 2018;42(6):791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis JL, Lockwood CM, Albert CM, Tsuchiya K, Hawkins DS, Rudzinski ER. Infantile NTRK-associated mesenchymal tumors. Pediatr Dev Pathol. 2018;21(1):68–78. [DOI] [PubMed] [Google Scholar]
- 8.Haller F, Knopf J, Ackermann A, et al. Paediatric and adult soft tissue sarcomas with NTRK1 gene fusions: a subset of spindle cell sarcomas unified by a prominent myopericytic/haemangiopericytic pattern. J Pathol. 2016;238(5):70–710. [DOI] [PubMed] [Google Scholar]
- 9.Pavlick D, Schrock AB, Malicki D, et al. Identification of NTRK fusions in pediatric mesenchymal tumors. Pediatr Blood Cancer. 2017;64(8): 1–5. [DOI] [PubMed] [Google Scholar]
- 10.Olson N, Rouhi O, Zhang L, et al. A novel case of an aggressive superficial spindle cell sarcoma in an adult resembling fibrosarcomatous dermatofibrosarcoma protuberans and harboring an EML4-NTRK3 fusion. J Cutan Pathol. 2018;45(12):933–939. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki F, Nakatani F, Asano N, et al. Novel NTRK3 fusions in fibrosarcomas of adults. Am J Surg Pathol. 2019;43(4):523–530. [DOI] [PubMed] [Google Scholar]
- 12.Croce S, Hostein I, Longacre TA, et al. Uterine and vaginal sarcomas resembling fibrosarcoma: a clinicopathological and molecular analysis of 13 cases showing common NTRK-rearrangements and the description of a COL1A1-PDGFB fusion novel to uterine neoplasms. Mod Pathol. 2019;32. [DOI] [PubMed] [Google Scholar]
- 13.Chang JC, Zhang L, Drilon AE, et al. Expanding the molecular characterization of thoracic inflammatory myofibroblastic tumors beyond ALK gene rearrangements. J Thorac Oncol. 2018;14(5):825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto H, Yoshida A, Taguchi K, et al. ALK, ROS1 and NTRK3 gene rearrangements in inflammatory myofibroblastic tumours. Histo-pathology. 2016;69(1):72–83. [DOI] [PubMed] [Google Scholar]
- 15.Brenca M, Rossi S, Polano M, et al. Transcriptome sequencing identifies ETV6-NTRK3 as a gene fusion involved in GIST. J Pathol. 2016; 238(4):543–549. [DOI] [PubMed] [Google Scholar]
- 16.Shi E, Chmielecki J, Tang CM, et al. FGFR1 and NTRK3 actionable alterations in “wild-type” gastrointestinal stromal tumors. J Transl Med. 2016;14(1):339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung YP, Fletcher CDM, Hornick JL. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology. 2018;73(4): 634–644. [DOI] [PubMed] [Google Scholar]
- 18.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DuBois SG, Laetsch TW, Federman N, et al. The use of neoadjuvant larotrectinib in the management of children with locally advanced TRK fusion sarcomas. Cancer. 2018;124(21):4241–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prieto-Granada CN, Wiesner T, Messina JL, Jungbluth AA, Chi P, Antonescu CR. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am J Surg Pathol. 2016;40(4):479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer IM, Fletcher CD, Hornick JL. Loss of H3K27 trimethylation distinguishes malignant peripheral nerve sheath tumors from histo-logic mimics. Mod Pathol. 2016;29(1):4–13. [DOI] [PubMed] [Google Scholar]
- 22.Robinson DR, Wu YM, Kalyana-Sundaram S, et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet. 2013;45(2):180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle LA, Vivero M, Fletcher CD, Mertens F, Hornick JL. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histo-logic mimics. Mod Pathol. 2014;27(3):390–395. [DOI] [PubMed] [Google Scholar]
- 24.Ten Heuvel SE, Hoekstra HJ, Suurmeijer AJ. Diagnostic accuracy of FISH and RT-PCR in 50 routinely processed synovial sarcomas. Appl Immunohistochem Mol Morphol. 2008;16(3):246–250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.