Abstract
Mutations of the Ras proteins HRAS, KRAS4A, KRAS4B, and NRAS are associated with a high percentage of all human cancers. The proteins are composed of highly homologous N-terminal catalytic or globular domains, plus C-terminal hypervariable regions (HVRs). Post-translational modifications of all RAS HVRs helps target RAS proteins to cellular membrane locations where they perform their signaling functions. For the predominant KRAS4 isoform, KRAS4B, post-translational farnesylation and carboxymethylation, along with a patch of HVR basic residues help foster membrane binding. Recent investigations implicate membrane-bound RAS dimers, oligomers and nanoclusters as landing pads for effector proteins that relay RAS signals. The details of these RAS signaling platforms have not been elucidated completely, in part due to the difficulties in preparing modified proteins. We have employed properly farnesylated and carboxymethylated KRAS4B in lipid monolayer incubations to examine how the proteins assemble on membranes. Our results reveal novel insights as to how KRAS4B may organize on membranes.
Keywords: RAS, oligomerization, nanocluster, membrane, monolayer
INTRODUCTION
Mutations of the KRAS, HRAS and NRAS genes are associated with a high percentage of all human cancers (1–2). The proteins encoded by these genes are regulated by their binding to either GTP or GDP (1–4). GTP-bound RAS (RAS-GTP) correlates with the active form of the protein, and activation can be stimulated by GEFs that exchange GTP for GDP on the GDP-bound form of RAS (RAS-GDP), or by resistance to the actions of GAPs, that stimulate the conversion of RAS-GTP to RAS-GDP (1–4). RAS mutations at codons 12, 13 and 61 reduce GTPase activity, compromise GAP-assisted activity, and stabilize RAS-GTP (Figure 1; 1–7). In turn, the switch I (residues in the region of 30–40) and switch II (residues 60–76) elements of RAS-GTP proteins are able to bind to effector proteins such as PI3K and RAF (1–2,5–7) that respectively promote the AKT/mTOR and MAPK signaling pathways (Figure 1; 1–2, 5–7).
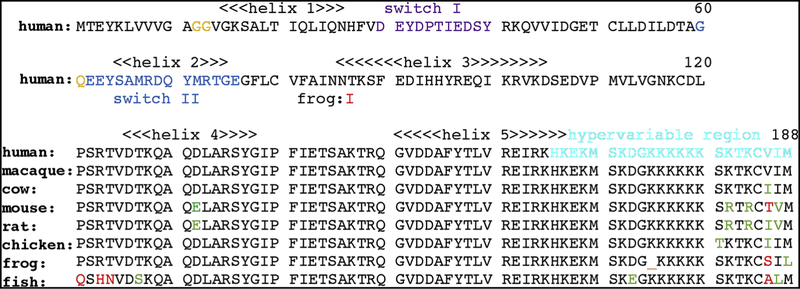
Figure 1. Alignment of KRAS4B sequences.
Shown is the coded proteins sequence of human KRAS4B (NCBI sequence ID: NP_004976.2) with helices 1–5 as indicated, switches I and II respectively colored in purple and blue, residues associated with activating mutations in gold, and the hypervariable region (HVR) in cyan. Note that the KRAS4B HVR is post-translationally processed by farnesylation at cysteine 185, proteolytic removal of residues 186–188, and carboxymethylation of the farnesylated C-terminal cysteine. Note also that residues 1–120 of human KRAS4B are identical to macaque (Macaca mulatta; NP_00124844.1), cow (Bos taurus; NP_001103471.1), mouse (Mus musculus; NP_067259.4), rat (Rattus norvegicus; NP_113703.1), chicken (Gallus gallus; NP_001243091.1), frog (Xenopus laevis; XP_01811022.1) and zebra fish (Danio rerio; NP_001003744.1) residues 1–120 except for the indicated frog residue 87. Sequence variations among the shown vertebrate KRAS4B proteins through residues 121–188 are indicated in green for conservative changes and red for non-conservative replacements.
Nucleotide, GEF, GAP and effector binding occur on the highly conserved catalytic or G domains of RAS proteins (residues 1–165), but these interactions occur at the PM due to the membrane affinity of the C-terminal HVRs (residues 166–188; 8–12). Whereas all four RAS isoforms, HRAS, NRAS, KRAS4A, and KRAS4B are C-terminally farnesylated and carboxymethylated, their HVR sequences are not conserved. Three of the variants are singly (NRAS, KRAS4A) or doubly (HRAS) palmitoylated, while the KRAS4B HVR is not palmitoylated, but carries a lysine rich stretch which is postulated to bind to acidic phospholipids at the inner leaflet of cell PMs (Figure 1; 1–2, 11, 70).
The signaling cascades that involve PM-bound RAS may be governed by interactions of single RAS monomers, but increasing evidence suggests the potential importance of RAS oligomers (6–7, 14–29). In particular, a number of investigations using a variety of techniques have implicated RAS dimers as the active signaling form of the protein (14–21). This correlates well with the observation that RAF effectors require dimerization to promote downstream signaling (14, 30–31). Researchers also have found that membrane-bound RAS proteins assemble oligomeric nanoclusters that appear to serve as platforms for RAS signaling, although the specific nature of these nanoclusters remains to be determined (21–29). While structures of monomeric RAS on lipid nanodiscs recently have been reported (32–34), determination of how RAS proteins associate with each other on membranes has been hampered by difficulties in the purification and analysis of KRAS proteins with properly modified HVRs (35–36). We have circumvented these problems by purifying the dominant KRAS variant, KRAS4B, from insect cells in its properly farnesylated and carboxymethylated form (KRAS4B-FMe; 35–36). KRAS4B-FMe proteins loaded with either GDP (KRAS-GDP) or the GTP analogue GppNHp (KRAS-GppNHp) were assembled onto lipid monolayers (37–44) containing PS and examined by transmission EM. Our results yield novel insights as to how KRAS may organize in signaling nanoclusters on membranes.
RESULTS
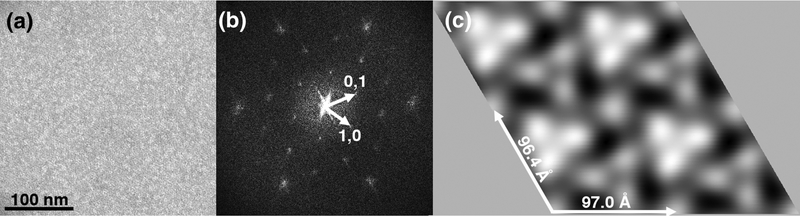
To examine how KRAS proteins organize on membranes, we adapted a lipid monolayer assembly system (37–44). Farnesylated and carboxymethylated KRAS4B proteins loaded with either GDP (KRAS-GDP) or a GTP analogue (KRAS-GppNHp) were incubated beneath lipid monolayers containing 60% (w/w) PC, 20% cholesterol, and 20% DOPS, lifted onto EM grids, stained, and examined by transmission EM. In testing a variety of incubation parameters, we found conditions that yielded organized patches of membrane-bound KRAS-GppNHp (Figure 2), but so far, we have not determined conditions that have given organized KRAS-GDP arrays. Successful KRAS-GppNHp incubations to date have required the inclusion of 5% PEG, which is incompatible with low contrast cryo-EM imaging (45–46), so we opted to examine the organization of membrane-bound KRAS-GppNHp by negative stain EM (37–44).
Figure 2. Organization of KRAS-GppNHp proteins on membranes.
KRAS-GppNHp proteins were assembled onto membranes composed of 60% PC, 20% cholesterol and 20% PS, lifted onto an EM grid, stained and imaged (A). In panel B, the Fourier transform of the image is depicted as a power spectrum, showing a lattice of reflections, with the 1,0 and 0,1 reflections as indicated. In panel C, the reflections in the Fourier transform were masked and back-transformed with no symmetry constraints (p1), yielding a unit cell with real space dimensions of a = 97.0Å, b = 96.4Å, and γ = 119.3°. In this rendering, KRAS-GppNHp proteins are viewed perpendicular to the membrane, and appear as bright white and fainter white trimer units surrounding trigonally symmetric membrane regions.
Surprisingly, power spectra of Fourier transforms from membrane-bound KRASGppNHp repeatedly showed hexagonal patterns (Figure 2B). In the representative example shown, simple masking of reflections and back-transformation without any symmetry constraints (p1) yielded a Fourier filtered (37–44) image with an obvious trigonal appearance (Figure 2C). As shown, the unit cell here was a = 97.0Å, b = 96.4Å, γ = 119.3°, and the arrays contained two distinct types of protein trimer units (depicted in white, perpendicular to the membrane). To validate this unexpected finding, Fourier transforms from multiple untilted images of membrane-bound KRAS-GppNHp were subjected to space group analysis, in which phases of symmetry related reflections are compared for all 2D crystal space groups (37–44, 47). The output of such an analysis is given as phase residuals for phase matching for each space group, where a phase residual of 0° is a perfect match, and a residual of 90° reflects a random match (47). As illustrated in Table 1, the trigonal (p3) space group clearly resulted in the best match, and every individual KRAS-GppNHp 2D array examined gave p3 as the optimal space group.
Table 1. Space Group Fitting.
Diffraction patterns obtained as calculated Fourier transforms from seven KRAS-GppNHp untilted 2D crystals of proteins assembled onto PC-cholesterol-PS membranes were indexed, boxed, and unbent as described in the Methods section. Statistics for space group fitting were obtained using the 2dx ALLSPACE program, which compares the phases of diffraction pattern reflections for internal consistency with 2D space groups, and outputs phase residuals for each space group fit. Using this algorithm, a perfect fit gives a phase residual of 0°, while a random fit yields a 90° phase residual. Note that because internal phase residual comparisons are not relevant with the primitive (p1) space group, the phase residual for p1 is on the basis of signal-to-noise ratios of observed amplitudes. Each untilted 2D crystal examined gave p3 as the optimal space group and, as shown, the averaged phase residual for the p3 space group is significantly less than all other space groups.
| group | residual | group | residual | group | |
|---|---|---|---|---|---|
| residual | |||||
| p1 | 17.2± 1.3 | c12a | 54.5± 9.6 | p422 | |
| 53.9± 6.1 | |||||
| p2 | 55.8± 6.7 | p222 | 56.4± 8.4 | p4212 | |
| 63.2± 3.9 | |||||
| p12b | 51.5± 10.4 | p2221b | 60.6± 5.3 | p3 | |
| 7.8± 4.0 | |||||
| p12a | 54.5± 9.6 | p2221a | 68.0± 3.1 | p312 | |
| 38.5± 2.6 | |||||
| p121b | 58.8± 4.2 | p22121 | 63.3± 3.9 | p321 | |
| 37.1± 7.0 | |||||
| p121a | 53.7± 7.3 | c222 | 56.4± 8.4 | p6 | |
| 37.7± 7.1 | |||||
| c12b | 51.5± 10.4 | p4 | 46.3± 5.9 | p622 | |
| 50.0± 6.0 | 6.0 | ||||
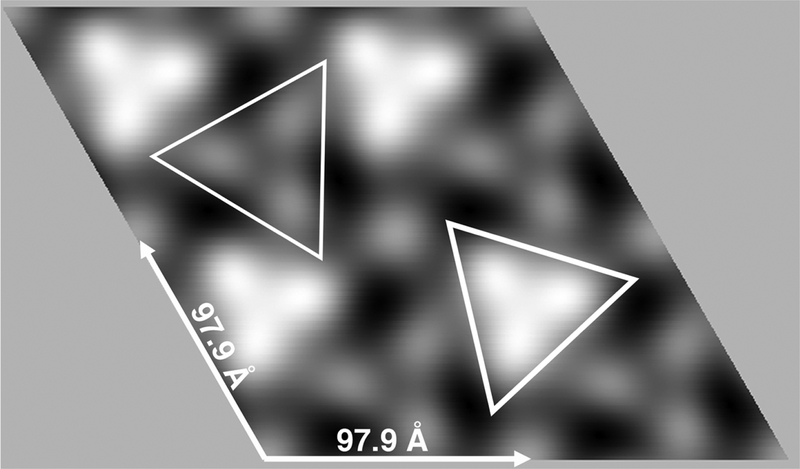
Based on the space group analysis above, we performed a merge of untilted datasets, assuming p3 symmetry as described in the Methods section. The averaged p3 unit cell was a = b = 97.9 + 5.0 Å, γ = 120.0 + 4.8°, and the merge to 20Å resolution gave a phase residual (47) of 8.1°. Our merged 2D projection image (Figure 3) was consistent with our initial results (Figure 1). As viewed perpendicular to the membrane with protein regions rendered in white, two types of trimer units again were apparent, one of higher density (indicated by the thick lined triangle) and another of lower density (indicated by the thin lined triangle). Based on these data, we speculated that the higher density units might represent globular or catalytic domain trimers, while the lower density units might be HVR trimers.
Figure 3. 2D reconstruction of KRAS-GppNHp proteins assembled on membranes.
2D crystals of KRAS-GppNHp proteins assembled on PS-containing membranes were prepared, processed and imaged described in the Methods section. From a set of seven crystals, Fourier transform amplitude and phase data were merged assuming p3 symmetry. The averaged unit cell was a = b = 97.9 ± 5.0 Å, γ = 120.0 + 4.8°. The merge was performed to a resolution of 20Å using reflections of IQ≤6, and the phase residual for the merge was 8.1°, where 0° indicates perfect matching, and 90° indicates random matching. Protein areas appear white, and are viewed perpendicular to the membrane. Bright and faint trimer units respectively are outlined with thick- and thin-lined triangles.
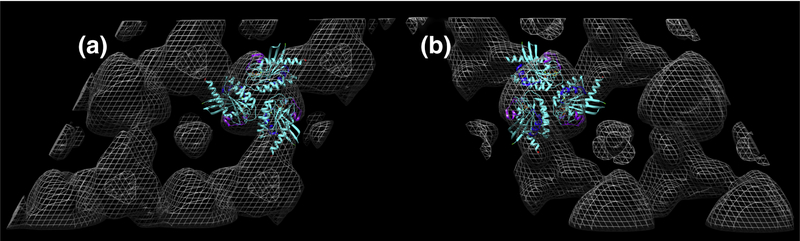
We undertook a 3D reconstruction of the membrane-bound KRAS-GppNHp arrays using standard methodologies (see Methods; 39–43, 47). Briefly, data from 55 images taken from 0–59.2° tilt were merged assuming p3 symmetry to 20Å resolution, yielding a phase residual of 8.2°. To verify membrane orientation, we performed a 3D merge in parallel of the myristoylated HIV-1 MA protein, for which the membrane orientation is known (43–44). Final 3D reconstruction volumes are depicted in Figure 4. At high stringency (2σ) putative KRASGppNHp globular (G) and HVR (H) densities viewed perpendicularly from the membrane side (Figure 4B) appeared as disconnected trimer units. However, on reduction of the contour level to 1σ, connections between G and H units became apparent (Figure 4A). These results support the interpretation that membrane-bound KRAS-GppNHp globular units trimerize with different partners than their HVRs. Figure 4, panel C shows the KRAS-GppNHp surfaces depicted in mesh, viewed from the membrane side, slightly tilted, while panel D gives the view parallel to the membrane, with the membrane side up. As rendered here, the putative HVR units appear ~1.5 nm closer to the plane of the membrane than the globular units.
Figure 4. 3D reconstruction of membrane-bound KRAS-GppNHp.
Crystalline areas of KRAS-GppNHp proteins assembled on membranes were imaged at defocus values of −0.6 to −1.4 microns, and tilt angles up to 59.2°. Data from 55 images were merged assuming p3 symmetry, and back-transformed to yield a 3D density map at 20Å resolution with a phase residual of 8.2°, where 0° indicates perfect matching, and 90° indicates random matching. Membrane orientation was determined by performing a merge on a known dataset in parallel. Panels A and B are viewed perpendicularly from the membrane side at contours of 1 and 2 σ, respectively. Panels C and D are contoured at 1 σ and viewed from the membrane side, slightly tilted (C), and from the side, with the membrane side up (D). Based on membrane proximity and density values, one putative globular (G) domain trimer and one hypervariable (H) domain trimer in each panel are indicated. Note that this arrangement shows putative HVRs trimerizing with different partners than putative globular domain trimers.
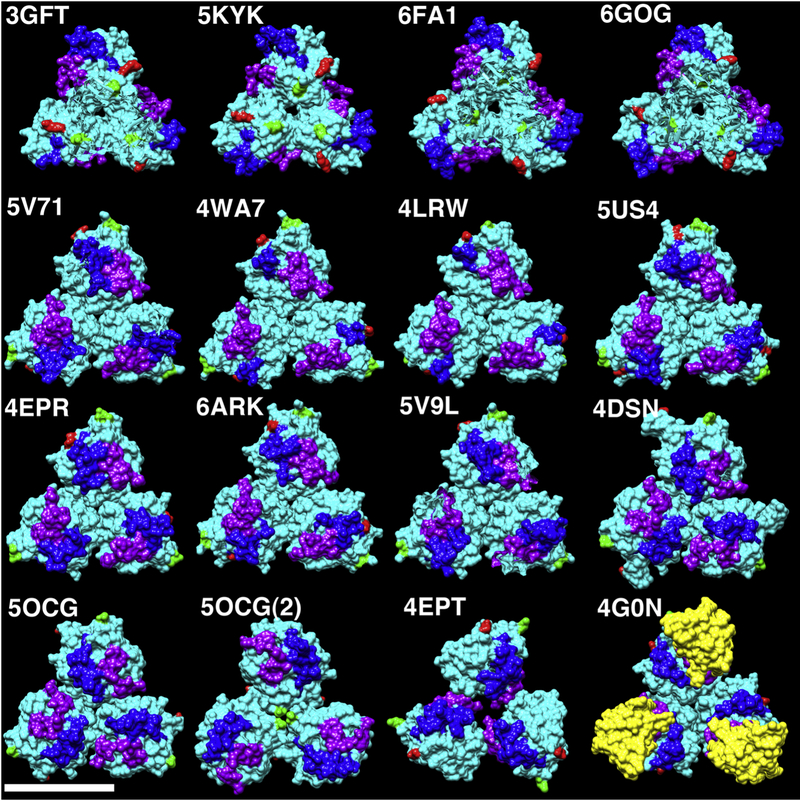
Given the unexpected observation of membrane-bound KRAS-GppNHp trimers, we examined 249 KRAS, HRAS, and NRAS protein crystal structures from the RCSB database (48) for the occurrence of trimers (see Methods section). Of the 112 KRAS structures examined, 46 from seven different space groups showed symmetric KRAS trimers of four different types (5, 49–60) that we refer to as trimers A-D. Examples are shown in Figure 5, where the top row represents A type trimers, the second and third rows plus 5OCG (54) represent B type trimers, C type trimers are illustrated by 5OCG(2)--a second type of trimer in the 5OCG unit cell, and 4EPT (50) is the only representative of D type trimers. Symmetric trimers also were observed in one of the two NRAS crystal structures, and in 55 of 135 HRAS structures (see Methods). Notably all HRAS trimers, whether liganded or not, are of type B, and 51 of the 55 are of space group H 3 2. An example of one HRAS-GppNHp trimer bound to the RBD (61) of RAF is illustrated in the bottom left panel of Figure 5 (4G0N), with the RBD colored in yellow.
Figure 5. Comparison of RAS protein trimers.
Globular RAS protein trimers extracted from the unit cells of the indicated PDB files are depicted as solid cyan surfaces. N-terminal residues are colored green, C-terminal residues are colored red, switch I residues are colored purple, and switch II residues are colored blue. The bottom right structure (4G0N) derives from the crystal structure of wild type HRAS-GppNHp bound to the RBD of Raf (colored in yellow). All others derive from crystal structures of KRAS4B reported in multiple publications representing six different 3D crystal space groups as described in the Methods section. The size bar (bottom left) indicates 50Å. Note that 5OCG and 5OCG(2) are two different trimer forms corresponding respectively to 5OCG unit cell chains 4–6 and 1–3. Due to similarities in trimer packing, we refer to the top row as representative of trimer type A, the second and third rows (plus 5OCG) as trimer type B, 5OCG(2) as C, and 4EPT as D.
At the resolution of our membrane-bound KRAS-GppNHp reconstruction, it is impossible to determine what type of KRAS trimer might correspond to our observed densities. Nevertheless, to get a rough estimate as to how Xray crystal trimers might be accomodated by the membrane-bound KRAS-GppNHp volumes, we hand-fitted the B type trimer from chains 7–9 of the unit cell of G12D KRAS-GppNHp (PDB 6GOF; 60) to our putative globular domain KRas-GppNHp densities such that switch I and II residues point away from the membrane (Figure 6). Even given the coarseness of this approach, and without accounting for the shrinkage of processed negatively stained structures relative to fully hydrated structures (62–66), we believe it is evident that the trimer units we observe correspond to KRAS trimers. These results are discussed below.
Figure 6. Fitting of KRAS4B to the 3D density map of membrane-bound KRAS-GppNHp.
Chains 7–9 of the unit cell of G12D KRAS-GppNHp (PDB 6GOF) are depicted as cyan ribbons, with N-terminal residues in green, C-terminal residues in red, switch I residues in purple, and switch II residues in blue. They were hand-fitted to putative globular KRAS-GppNHp volumes such that switch I and II residues face away from the membrane. Panel A is viewed perpendicularly from the membrane side, while panel B is viewed from the opposite side. Note that we have not corrected for shrinkage of negatively stained samples.
DISCUSSION
In our analysis of KRAS proteins assembled on membranes composed of PC, cholesterol and PS, we were unable to find conditions where KRAS-GDP assembled organized arrays, but found that KRAS-GppNHp assembled 2D crystalline areas that were compatible with image analysis (Figures 2–4). While our inability to obtain organized membrane-bound KRAS-GDP structures may have implications as to the nature of how KRAS-GDP binds membranes, further efforts will be needed to test this possibility, particularly given that KRAS-GDP trimers have been observed in Xray crystal structures (see Methods section). For KRAS-GppNHp, the obvious unexpected finding was the observation of trimer units in which putative KRAS globular domains associate with different partners than the putative HVRs.
To date, high resolution structures for membrane-bound RAS oligomers are not available, and some results suggest that RAS remains monomeric on membranes (71). Nevertheless, accumulated evidence supports the notion that RAS proteins organize as signaling nanoclusters on membranes (14–29). Notably, RAS immunogold EM labeling results suggest that nanoclusters are approximately 10 nm in diameter, and modeling efforts have implicated RAS residues K101 and E107 in clustering (21, 23, 29). Imaging, biochemical and genetic investigations suggest that RAS dimers may be the basic units of signaling nanoclusters (6–7, 21–29), although higher order oligomers have been modeled (29). A number of different dimer interfaces have been proposed (7, 18–21, 29), with recent evidence supporting the importance of dimers involving RAS helices 3 and 4 (α3-α4; 29, 34) or helices 4 and 5 (α4-α5; 15, 18–20). Nevertheless, ample evidence also supports arguments for the dynamic remodeling of RAS complexes through different conformations and orientations (21, 27, 29).
Given the available data, we envision several alternatives for our observation of membrane-bound KRAS-GppNHp trimers. A trivial explanation is that our results do not reflect a biological reality, because we employed purified proteins in an artificial system. One argument against this is that our incubations occurred in aqueous solution, beneath a conventional membrane of PC, chlolesterol and PS; and that similar approaches with other proteins have yielded biologically relevant structural data (37–47, 67). A second point worth note is that of the 249 RAS structures we examined from the RCSB databank, 102 from seven different space groups showed symmetric trimers, while non-symmetric RAS trimers also were observed.
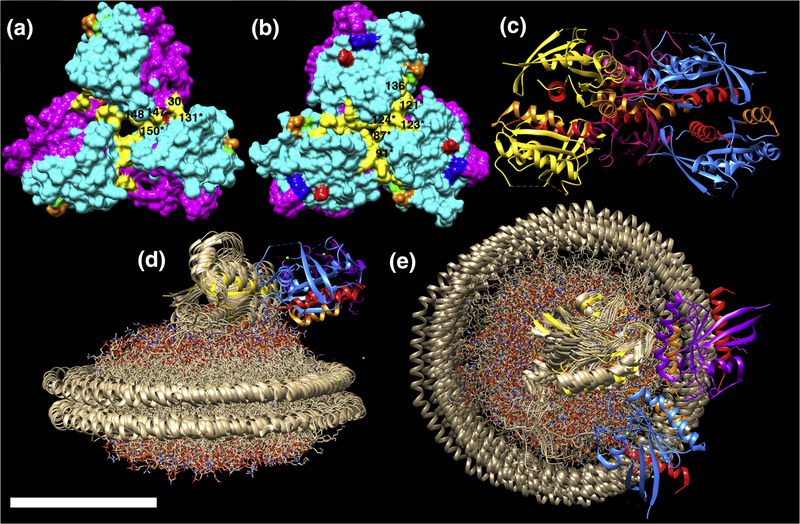
Assuming that membrane-bound KRAS trimers are biologically relevant, a simple model is that they correspond to active signaling platforms. Except for the type D trimers where switch I regions form trimer interfaces (see Figure 5, 4EPT), all other symmetric RAS trimers can be oriented so as to present switch I and II residues as landing pads for effector binding and oligomerization (Figure 5). In this orientation, occupation of two of the three trimer effector binding sites by RAF RBDs may be envisioned to result in RAF dimerization and activation (1–4, 14, 30–31). The predominant KRAS trimer A and B forms both have available effector binding site regions, but differ in their interface regions. As shown in Figure 7, A type trimer interface residues (in yellow) include Q25, N26, H27, D30, Q131, K147, T148, and Q150, while B type trimer interface residues include T87, K88, E91, P121, S122, R123, T124, AND S136. Both of these sets of interface residues are largely conserved among vertebrate KRAS4B homologues, except for a residue 87 T to I substitution in the frog homologue, and a residue 124 T to N substition in the zebra fish form (Figure 1). Also, for both trimer A and B forms, residues K101 (green) and E107 (orange), predicted to facilitate enhanced signaling via nanocluster formation (23, 29), are accessible for possible interactions with other trimers. In contrast, residues D154 (red) and R161 (blue) in the α4-α5 region localize to the interfaces of type A trimers (Figure 7A), but are accessible in type B trimers (Figure 7B). Indeed, a number of KRAS crystal structures, such as that for KRAS(G12A)-GppNHp (Figure 7C; 5) show members of type B trimers connected to additional KRAS proteins via α4-α5 dimer interfaces.
Figure 7. Analysis of trimer interfaces.
The structures of KRAS trimers type A (A, 6GOG) and B (B, 5OCG chains 4–6) are depicted with effector lobes (residues 1–86) in magenta, allosteric lobes (residues 87–186) in cyan, and with previously predicted oligomer interface residues indicated as follows: K101, green; E107, orange; D154, red; R161, blue. Trimer interface residues are indicated in yellow. For A, these correspond to Q25, N26, H27, D30, Q131, K147, T148, Q150. For B, they are T87, K88, E91, P121, S122, R123, T124, and S136. Labels of visible interface residues correspond to residue numbers without asterisks when the residue derives from the top monomer, and with asterisks when the residue derives from the bottom right monomer. Panel C shows a ribbon diagram of KRAS(G12A)-GppNHp (PDB 5VPY) chains 7A,B (purple), 8A,B (yellow), 9A,B (blue), with B chains on top. As shown, upper and lower trimer units dimerize via α4 (orange) plus α5 (red) interfaces. Panels D-E show only chains 7B, 8B, and 9B from PDB 5VPY fitted to the structure of the lipid nanodisc-bound KRAS monomer in the exposed form (PDB 2MSC; in bronze), viewed parallel (D) and perpendicular (E) to the membrane. Chains are colored as in panel C, but with α4 helices in orange, and α5 helices in red. Note that for panels D and E, switch regions are oriented away from the lipid nanodisc. The size bar indicates 50Å for all panels.
Remarkably, the same α4-α5 interface that is modeled to foster RAS dimerization (15, 18–20) also has been reported to mediate one of two types of RAS binding to membranes (28, 32–33). Specifically, FRET and MD studies, as well as NMR analyses of RAS bound to nanodiscs indicate two separate modes of RAS binding to membranes (28, 32–33). In this “balance” model (28), the “occluded” form, has membrane interface residues that include β1, β2, β3, α2, and the C-terminal portion of α3; but the α4-α5 interface is available for dimerization (28, 32–33). In contrast, the “exposed,” or “signaling-competent” form places α4-α5 against the membrane, but leaves switch I and II residues exposed (28, 32–33; Figures 7D–E). Of particular interest is that B form trimers readily match the exposed form of membrane-bound RAS. Indeed, the same KRAS(G12A)GppNHp trimer from Figure 7C appears to form an acceptable fit to the KRAS bound to lipid nanodiscs in the exposed form (32; Figures 7D–E). Notably, such an orientation also appears compatible with KRAS trimer binding to membrane-associated RAF CRDs in models based on solution data of RAS-CRD associations (69).
What might be the role of KRAS trimerization in the exposed state? We speculate that trimer assembly could function to lock KRAS monomers into the exposed, signaling-competent conformation. In contrast, if it is assumed that α4-α5 dimers act as signaling platforms (15, 18–20), B form trimerization might squelch signaling. In either case, trimer based nanoclustering also might be involved. In this regard, it is important to note that effector proteins and constituents of membrane microdomains are likely to be major regulators of these processes (31, 72–76). At this point, we believe a number of extensions to our studies are worth pursuing. We are actively investigating alternative lipid monolayer incubation parameters with RAS protein variants and effectors to find other crystal forms, and conditions compatible with cryo-EM analysis. It also would seem worthwhile to investigate mutations at residues predicted to be at trimer interfaces for their effects on oligomerization and RAS signaling.
METHODS
Protein Preparation.
Recombinant farnesylated and carboxymethylated KRAS4B (KRAS-FMe) was expressed in Tni-FNL insect cells as outlined by Gillette et al. (35). Cells lysis, and protein purification followed the protocol of Agamasu et al. (36). As purified, KRAS-FMe is bound with GDP (KRAS-GDP). Exchange into GppNHp to generate KRAS-GppNHp was performed as described Agamasu et al. (36), and the efficiency of exchange was evaluated by HPLC (35). KRAS4B-FMe-GppNHp (KRAS-GppNHp) and KRAS4B-FMe-GDP (KRAS-GDP) proteins were stored at −80° in 20 mM Hepes pH 7.3, 300 mM NaCl, 1 mM TCEP, 5 mM MgCl2.
Membrane incubations and electron microscopy.
For membrane monolayer incubations (37–44), 8 ul drops of 64 uM KRAS-GDP or KRAS-GppNHp protein in buffer (25 mM sodium acetate pH 6.0, 375 mM NaCl, 5 mM Hepes, 1.25 mM MgCl2, 1.25 mM BME, 0.25 mM TCEP, 5% PEG6000) were deposited onto glass slides and overlayed with 1.1 ul of lipid mix composed of 150 ug/ml egg PC (Avanti), 50 ug/ml DOPS (Avanti) and 50 ug/ml cholesterol (Sigma) in 1:1 chlorofom:hexane. Incubations were performed for 18–72 h at 4°C in sealed 15 cm diameter plastic dishes, humidified with blotter paper wetted with 12.5 mM sodium acetate pH 6.0, 150 mM NaCl. After incubations, samples were lifted onto lacey EM grids (Ted Pella) rinsed 30 s on drops of distilled water, wicked, stained 45–60 s with 1.33% (wt/vol) uranyl acetate (Sigma), wicked and dried.
Samples were imaged under low dose conditions at 120 keV on the OHSU Multiscale Microscopy Core FEI Tecnai transmission electron microscope (TEM), which is equipped with an Eagle 4 megapixel CCD multiscan camera, and has a spherical aberration (Cs) of 6.3 mm. Images were taken at tilt angles from 0–59.2°, at 3.19Å/pixel, and at defocus values between −600 to −1400 nm, in which the first zero of the CTF is beyond 20Å resolution. Images were collected as TIFF files and analyzed as described below.
Image analysis.
Image analysis steps followed previously described procedures (37–47), employing the 2dx image processing package (47). Note that while not ordered areas of KRAS-GDP proteins werer observed in our trials, numerous crystalline patches of KRAS-GppNHp proteins were observed on multiple grids. Ordered areas of KRAS-GppNHp proteins in 55 images were boxed, Fourier transformed, indexed, and unbent. For generation of Fourier filtered, unsymmetrized images of untilted samples (Figure 2), APH files were back-transformed to a resolution of 20Å. Space group analysis to 20Å was calculated in 3° phase origin search steps for seven zero tilt images using the 2dx version of ALLSPACE (47), which compares the phases of Fourier transform reflections for internal consistency with 2D space groups (or by amplitude comparisons for the p1 space group), and outputs a phase residual, where a perfect fit gives a residual of 0° and a random fit gives a residual of 90°. Each untilted 2D crystal examined was assigned p3 as the optimal space group, and average space group fits are presented in Table 1.
Trigonal (p3) unit cell parameters were calculated from the positions of 1,0 and 0,1 reflections in the power spectra of zero tilt images, and yielded an averaged real space unit cell of a = b = 97.9 + 5.0 Å, γ = 120.0 + 4.8°. 2D merging of untilted data to 20Å resolution was performed using reflections of IQ≤6 (47) and phase origin search steps starting at 3° and ending at 0.5°. The final phase residual for the 2D merge was 8.1°, and the averaged 2D unit cell was depicted from 2dx output with protein areas indicated in white. For generation of the KRAS-GppNHp 3D map, the 55 tilted plus untilted image files were merged as above, assuming p3 symmetry, a z window of 0.0002 Å-1, and a z thickness of 250Å. Phase origin search steps initially were 3.0°, and then refined to 0.5°. For the 3D reconstruction, a phase residual of 8.2° was obtained, and merged lattice line data were binned using 2dx, and exported into CCP4 MAP image format for visualization using the Chimera software package (68), which was used for the generation of Figures 3–6. To verify membrane orientation, we performed a parallel merging on a dataset collected for the myristoylated HIV-1 matrix (MA) protein, for which the membrane orientation is known (43–44). The final 3D volumes were depicted as solid or mesh surfaces in 2 × 2 unit cells unit cells at the indicated threshold (sigma, σ) values.
For comparison with Xray crystal structure data, the unit cells for all available KRAS4B structures in the RCSB protein data bank (48), whether liganded or not, were examined for the occurence of symmetric trimer types A-D, in which interface regions are defined roughly as follows: A: α4-β6-α5 residues with neighbor α1, switch 1, loop 118–122, and loop 145–151 residues; B: central α3-α4 residues with neighbor loop 118–125 and N-terminal α3-α4 residues; C: α5 N-terminal residues with neighbor α5 C-terminal residues, and β1 and β3 N-terminal residues, and β2 C-terminal residues with their neighbors; D: switch 1 residues with neighbor loop 116–125 and N-terminal α3 residues. Thus, the following PDB files (in unit cell form) were examined, and are superscripted a, b, c, or d, according to the trimer type observed: 3GFTa, 4DSNb,c, 4DSOb,c, 4DSTb,c, 4DSUb,c, 4EPRb, 4EPTd, 4EPV, 4EPW, 4EPX, 4EPY, 4L8Gb, 4LDJ, 4LPKb, 4LRWb, 4LUC, 4LV6, 4LYF, 4LYH, 4LYJ, 4M1O, 4M1S, 4M1T, 4M1W, 4M1Y 4M21, 4M22, 4NMM, 4OBE, 4PZY, 4PZZ, 4Q01, 4Q02, 4Q03, 4QL3, 4TQ9, 4TQA, 4WA7b, 5F2E, 5KYKa, 5MLAb, 5MLB, 5O2S, 5O2T, 5OCGb,c, 5OCOa, 5OCTa, 5TAR, 5TB5, 5UFE, 5UFQ, 5UK9, 5UQWb, 5US4b, 5USJ, 5V6S, 5V6V, 5V71b, 5V9Lb, 5V9Ob, 5V9U, 5VBM, 5VBZ, 5VP7b, 5VPIb, 5VPYb, 5VPZb, 5VQ0b, 5VQ1b, 5VQ2b, 5VQ6b, 5VQ8b, 5W22b, 5WHA, 5WHB, 5WHD, 5WHE, 5WLB, 5WPL, 5WPM, 5XCO, 5YXZ, 5YY1, 6ARKb, 6ASA, 6ASE, 6B0V, 6B0Y, 6BP1, 6EPL, 6EPM, 6EPN, 6EPO, 6EPP, 6F76a, 6FA1a, 6FA2a, 6FA3a, 6FA4a, 6GODb,c, 6GOEb,c, 6GOFb,c, 6GOGa, 6GOMa, 6GQTa, 6GQWa, 6GQXa, 6GQYa, 6H46, 6H47, 6N2J, 6N2K. Note that the 46 PDB files showing KRAS4B trimers (out of 112 PDBs) represent the seven space groups C 1 2 1, H 3, H 3 2, P 21 21 21, P 2 3, P 3, P 63. Note additionally that all (15 of 15) of the KRAS4B A type trimers were bound to GTP or GTP analogues, as were 13 of 30 B trimers, and 7 of 8 C trimers, while the single D type trimer was GDP-bound. We also observed one of two NRAS PDB files (5UHV, 3CON) showing B type trimers (3CON; P 63), as well as 55 out of 135 HRAS PDB files showing B type trimers, as follows: 1IAQ, 1K8Rb, 1NVU, 1NVV, 1NVW, 1NVX, 1P2S, 1P2T, 1P2U, 1P2V, 1XCM, 1XD2, 1XJ0b, 1ZVQb, 1ZW6b, 2C5L, 2CE2, 2CL0b, 2CL6, 2CL7, 2CLC, 2CLD, 2EVW, 2QUZb, 2RGAb, 2RGBb, 2RGCb, 2RGD , 2RGEb, 2RGG, 2UZI, 2VH5, 2X1Vb, 3DDC, 3I3Sb, 3K8Yb, 3K9L, 3K9N, 3KKMb, 3KKN, 3KUDb, 3L8Yb, 3L8Zb, 3LBHb, 3LBIb, 3LBNb, 3LO5b, 3OIUb, 3OIVb, 3OIWb, 3RRYb, 3RRZb, 3RS0b, 3RS2b, 3RS3b, 3RS4b, 3RS5b, 3RS7b, 3RSLb, 3RSOb, 3TGP, 3V4Fb, 4DLRb, 4DLSb, 4DLTb, 4DLUb, 4DLVb, 4DLWb, 4DLXb, 4DLYb, 4DLZb, 4DOT, 4DPZ, 4EFL, 4EFM, 4EFN, 4FA0, 4G0Nb, 4G3Xb, 4K81, 4L9Sb, 4L9Wb, 4NYI, 4NYJ, 4NYM, 4Q21, 4RSG, 4URU, 4URV, 4URW, 4URX, 4URY, 4URZ, 4US0, 4US1, 4US2, 4XVQ, 4XVR, 5B2Zb, 5B30b, 5E95, 5VBEb, 5VBZ, 5WDOb, 5WDPb, 5WDQb, 5WFO, 5WFP, 5WFQ, 5WFR, 5WPL, 5X9S, 6AMB, 6AXG, 6BVI, 6BVJ, 6BVK, 6BVL, 6BVM, 6CUO, 6CUP, 6CUR, 6D55, 6D56, 6D59, 6D5E, 6D5G, 6D5H, 6D5J, 6D5L, 6D5M, 6D5V, 6D5W, 6E4F, 6Q21. Of the HRAS crystals showing trimers, 51 are space group H 3 2, two are P 3 2 1, and one each are P 63 and I 2 2 2. Note that α4-α5 interface dimers (15, 18–20) were also observed for all of the HRAS unit or supercells that showed trimers, with the exception of PDB 2X1V. In contrast, only thirteen of the 46 KRAS crystals that show trimers also show α4-α5 interface dimers (4EPR, 5US4, 5VP7, 5VPI, 5VPY, 5VPZ, 5VQ0, 5VQ1, 5VQ2, 5VQ6, 5VQ8, 5W22, 6ARK).
For presentation, fifteen KRAS4B trimer units reported in multiple separate publications (5, 49–60) and representing six space groups were extracted from the unit cells in PDB files, and depicted as solid cyan surfaces with N-terminal residues colored green, C-terminal residues colored red, switch I residues (30–40) colored purple, and switch II residues (60–76) colored blue. We also represented the trimer unit from wild type HRAS-GppNHp bound to the RBD of RAF (61) using the same color scheme, but with the RBD colored yellow. For fitting of the trimer unit from G12D KRAS-GppNHp (chains 7–9 of PDB 6GOF; space group P 3 2 1; 60), the proteins were represented as ribbons using the same color scheme as described above. Trimers were hand-fitted to the putative KRAS-GppNHp globular volume derived from our 3D reconstruction such that switch I and II residues face away from the membrane. Note that we have not corrected for the shrinkage of negatively stained samples during preparation and imaging (62–66), relative to fully hydrated samples.
HIGHLIGHTS.
The manner by which KRAS4B proteins organize on membranes is not known.
We examined KRAS4B assembled on membranes containing phosphatidylserine.
Unexpectedly, KRAS4B proteins organized as trimers on membranes.
Our results reveal novel insights as to how KRAS4B may cluster on membranes.
ACKNOWLEDGMENTS
These investigations were supported by the OHSU Knight Cancer Institute NCI Cancer Center Support Grant P30CA069533 and by the OHSU Center for Spatial Systems Biomedicine. EB, AOS, RLB and AA also received support from NIH grant R01 GM060170. This project was funded in part with federal funds from the National Cancer Institute, NIH Contract HHSN261200800001E. We thank Vanessa Wall, Angela Carter, Kelly Snead, and Peter Frank from the NCI-RAS Initiative for production of farnesylated and methylated KRas4b protein used in this work. We are grateful for helpful advice and assistance from James Chen, David Farrens, Claudia Lopez, Xiaolin Nan, Jonathan Pruneda, Ujwal Shinde, Philip Stork and Robin Woolman. The content of this publication does not necessarily reflect the view or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the US government.
ABBREVIATIONS
- 2D
two dimensional
- 3D
3 dimensional
- APH
amplitudes and phases
- BME
β-mercaptoethanol
- CCD
charge coupled device
- CRD
cysteine-rich domain
- Cs
spherical aberration
- CTF
contrast transfer function
- DOPS
dioleoyl phosphatidylserine
- EM
electron microscopy
- FRET
fluorescence resonance energy transfer
- G
globular
- GAP
GppNHp, guanosine-5′-([β,γ]-imido)-triphosphate, GTPase-activating protein
- GDP
guanosine diphosphate
- GEF
guanine exchange factor
- GTP
guanosine triphosphate
- HIV-1
human immunodeficiency virus type 1
- HPLC
high pressure liquid chromatography
- HVR
hypervariable region
- keV
kiloelectron volt
- MA
matrix
- MD
molecular dynamics
- NCBI
National Center for Biotechnology Information
- NMR
nuclear magnetic resonance spectrometry
- PC
phosphatidyl choline
- PDB
protein data bank
- PEG
polyethyleneglycol
- PI3K
phosphoinositide 3-kinase
- PM
plasma membrane
- PS
phosphatidylserine
- RBD
RAS binding domain
- TCEP
tris(2-carboyethyl)phosphine
- TEM
transmission electron microscopy
- TIFF
tagged image format file
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
REFERENCES
- 1.Baines AT, Xu D, Der CJ. (2011). Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 3(14): 1787–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins MA, Pasca di Magliano M. (2014). Kras as a key oncogene and therapeutic target in pancreatic cancer. Front Physiol. 4:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vigil D, Cherfils J, Rossman KL, Der CJ (2010). Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer 10(12): 842–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos JL, Rehmann H, Wittinghofer A. (2007). GEFs and GAPs: critical elements in the control of small G proteins. Cell 129(5): 865–77. [DOI] [PubMed] [Google Scholar]
- 5.Xu S, Long BN, Boris GH, Chen A, Ni S, Kennedy MA (2017). Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras. Acta Crystallogr D Struct Biol 73: 970–984. [DOI] [PubMed] [Google Scholar]
- 6.Chen M, Peters A, Huang T, Nan X (2016). Ras Dimer Formation as a New Signaling Mechanism and Potential Cancer Therapeutic Target. Mini Rev Med Chem. 16(5):391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, Dyba MA, Stefanisko K, Tarasov SG, Gursoy A, Keskin O, Tarasova NI, Gaponenko V, Nussinov R. (2015). GTP-Dependent KRas Dimerization. Structure 23(7): 1325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock JF (2003). Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol 4(5): 373–384. [DOI] [PubMed] [Google Scholar]
- 9.Hancock JF, Parton RG (2005). Ras plasma membrane signalling platforms. Biochem. J 389(Pt 1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock JF, Magee AI, Childs JE, Marshall CJ (1989). All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57: 1167–1177. [DOI] [PubMed] [Google Scholar]
- 11.Hancock J, Paterson H, Marshall C (1990). A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63: 133–139. [DOI] [PubMed] [Google Scholar]
- 12.Ahearn I, Zhou M, Philips MR(2018). Posttranslational Modifications of RAS Proteins. Cold Spring Harb Perspect Med. 8(11). pii: a031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Wang Y, Vikis H, Johnson L, Liu G, Li J, Anderson W, Sills R, Hong H, Devereux T, Jacks T, Guan K, You M. (2001). Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet 29(1): 25–33. [DOI] [PubMed] [Google Scholar]
- 14.Inouye K, Mizutani S, Koide H, Kaziro Y (2000). Formation of the Ras Dimer Is Essential for Raf-1 Activation. J. Biol. Chem 275(6): 3737–3740. [DOI] [PubMed] [Google Scholar]
- 15.Güldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kötting C, Gerwert K (2012). N-Ras forms dimers at POPC membranes. Biophys J. 103(7):1585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin WC, Iversen L, Tu HL, Rhodes C, Christensen S, Iwig J, Hansen S, Huang W, Groves J (2014). H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. USA 111(8): 2996–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan X, Tamguney T, Collisson E, Lin L, Pitt C, Galeas J, Lewis S, Gray J, McCormick F, Chu S (2015). Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc Natl Acad Sci USA 112(26): 7996–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, Cobbert J, Lavoie H, Smith M, Rajakulendran T, Dowdell E, Okur MN, Dementieva I, Sicheri F, Therrien M, Hancock JF, Ikura M, Koide S, O’Bryan JP (2017). Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol. 13(1):62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer-Smith R, Li L, Prasad S, Koide A, Koide S, O’Bryan J. (2017). Targeting the α4-α5 interface of RAS results in multiple levels of inhibition. Small GTPases 10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrogio C, Köhler J, Zhou ZW, Wang H, Paranal R, Li J, Capelletti M, Caffarra C, Li S, Lv Q, Gondi S, Hunter JC, Lu J, Chiarle R, Santamaría D, Westover KD, Jänne PA (2018). KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS. Cell 172(4):857–868.e15. [DOI] [PubMed] [Google Scholar]
- 21.Prakash P, Sayyed-Ahmad A, Cho KJ, Dolino DM, Chen W, Li H, Grant BJ, Hancock JF, Gorfe AA (2017). Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers. Sci Rep. 7:40109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prior IA, Muncke C, Parton RG, Hancock JF (2003) Direct visualization of ras proteins in spatially distinct cell surface microdomains. J. Cell Biol 160(2): 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plowman SJ, Muncke C, Parton RG, Hancock JF (2005) H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 102(43): 15500–15505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian T, Harding A, Inder K, Plowman S, Parton R, Hancock J (2007). Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol 9(8): 905–914. [DOI] [PubMed] [Google Scholar]
- 25.Barcelo C, Paco N, Beckett A, Alvarez-Moya B, Garrido E, Gelabert M, Tebar F, Jaumot M, Prior I., Agell N (2013). Oncogenic K-Ras segregates at spatially distinct plasma membrane signaling platforms according to its phosphorylation status. J. Cell Sci 126: 4553–4559. [DOI] [PubMed] [Google Scholar]
- 26.Tian T, Plowman SJ, Parton RG, Kloog Y, Hancock JF (2010). Mathematical modeling of K-Ras nanocluster formation on the plasma membrane. Biophys. J 99(2): 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA (2007). Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1, 2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem 50(4): 674–684. [DOI] [PubMed] [Google Scholar]
- 28.Abankwa D, Gorfe AA, Inder K, Hancock JF (2010). Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. USA 107(3): 1130–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkar-Banerjee S, Sayyed-Ahmad A, Prakash P, Cho KJ, Waxham MN, Hancock JF, Gorfe AA (2017). Spatiotemporal Analysis of K-Ras Plasma Membrane Interactions Reveals Multiple High Order Homo-oligomeric Complexes. J Am Chem Soc 139(38):13466–13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Z, Tzivion G, Belshaw P, Vavvas D, Marshall M, Avruch J (1996). Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature 383(6596): 181–185. [DOI] [PubMed] [Google Scholar]
- 31.Nan X, Collisson EA, Lewis S, Huang J, Tamgüney TM, Liphardt JT, McCormick F, Gray JW, Chu S. (2013). Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. Proc Natl Acad Sci USA 110(46): 18519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, Ikura M (2015). Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA 112: 6625–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang Z, Marshall CB, Nishikawa T, Gossert AD, Jansen JM, Jahnke W, Ikura M (2018). Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site. Cell Chem Biol 25: 1327–1336.e4. [DOI] [PubMed] [Google Scholar]
- 34.Prakash P, Litwin D, Liang H, Sarkar-Banerjee S, Dolino D, Zhou Y, Hancock JF, Jayaraman V, Gorfe AA. (2019). Dynamics of Membrane-Bound G12V-KRAS from Simulations and Single-Molecule FRET in Native Nanodiscs. Biophys J. 116(2):179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillette WK, Esposito D, Abreu Blanco M, Alexander P, Bindu L, Bittner C, Chertov O, Frank PH, Grose C, Jones JE, Meng Z, Perkins S, Van Q, Ghirlando R, Fivash M, Nissley DV, McCormick F, Holderfield M, Stephen AG (2015). Farnesylated and methylated KRAS4b: high yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci Rep. 5: 15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agamasu C, Ghirlando R, Taylor T, Messing S, Tran TH, Bindu L, Tonelli M, Nissley DV, McCormick F, Stephen AG. (2019). KRAS Prenylation Is Required for Bivalent Binding with Calmodulin in a Nucleotide-Independent Manner. Biophys J. 116(6): 1049–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barklis E, McDermott J, Wilkens S, Schabtach E, Schmid MF, Fuller S, Karanjia S, Love Z, Jones R, Rui Y, Zhao X, Thompson D (1997). Structural analysis of membrane-bound retrovirus capsid proteins. EMBO J. 16(6): 1199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barklis E, McDermott J, Wilkens S, Fuller S, Thompson D (1998). Organization of HIV-1 capsid proteins on a lipid monolayer. J Biol Chem 273(13): 7177–80. [DOI] [PubMed] [Google Scholar]
- 39.McDermott J, Mayo K, Barklis E (2000). Three-dimensional organization of retroviral capsid proteins on a lipid monolayer. J Mol Biol. 302(1): 121–33. [DOI] [PubMed] [Google Scholar]
- 40.Mayo K, Vana ML, McDermott J, Huseby D, Leis J, Barklis E (2002). Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J Mol Biol. 316(3): 667–78. [DOI] [PubMed] [Google Scholar]
- 41.Mayo K, McDermott J, Barklis E (2002). Hexagonal organization of Moloney murine leukemia virus capsid proteins. Virology 298(1): 30–8. [DOI] [PubMed] [Google Scholar]
- 42.Mayo K, Huseby D, McDermott J, Arvidson B, Finlay L, Barklis E (2003). Retrovirus capsid protein assembly arrangements. J Mol Biol. 325(1): 225–37. [DOI] [PubMed] [Google Scholar]
- 43.Alfadhli A, Barklis RL, Barklis E (2009). HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate.Virology 387(2): 466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alfadhli A, Mack A, Ritchie C, Cylinder I, Harper L, Tedbury PR, Freed EO, Barklis E (2016). Trimer Enhancement Mutation Effects on HIV-1 Matrix Protein Binding Activities. J Virol. 90(12)5657–5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stark H, Chari A. (2016). Sample preparation of biological macromolecular assemblies for the determination of high-resolution structures by cryo-electron microscopy. Microscopy 65(1): 23–34. [DOI] [PubMed] [Google Scholar]
- 46.Drulygte I, Johnson R, Hesketh E, Hurdiss D, Scarff C, Porav S, Ranson N, Muench S, Thompson R (2018). Approaches to altering particle distributions in cryo-electron microscopy sample preparation. Acta Crystallogr D Struct Biol. 74(6): 560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gipson B, Zeng X, Zhang ZY, Stahlberg H (2007). 2dx--user-friendly image processing for 2D crystals. J Struct Biol. 157(1):64–72. [DOI] [PubMed] [Google Scholar]
- 48.Berman H, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, Shindyalov I, Bourne P (2000). RSCB: The Protein Data Bank. Nucleic Acids Res. 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurer T, Garrenton L, Oh A, Pitts K, Anderson D, Skelton N, Fauber B, Pan B, Malek S, Stokoe D, Ludlam M, Bowman K, Wu J, Giannetti A, Starovasnik M, Mellman I, Jackson P, Rudolph J, Wang W, Fang G (2012). Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA 109: 5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW (2012). Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation. Angew.Chem.Int.Ed.Engl 51: 6140–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503(7477): 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD (2015). Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res. 13: 1325–1335. [DOI] [PubMed] [Google Scholar]
- 53.Xiong Y, Lu J, Hunter J, Li L, Scott D, Choi HG, Lim SM, Manandhar A, Gondi S, Sim T, Westover KD, Gray NS (2017). Covalent Guanosine Mimetic Inhibitors of G12C KRAS. ACS Med Chem Lett 8: 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quevedo CE, Cruz-Migoni A, Bery N, Miller A, Tanaka T, Petch D, Bataille CJR, Lee LYW, Fallon PS, Tulmin H, Ehebauer MT, Fernandez-Fuentes N, Russell AJ, Carr SB, Phillips SEV, Rabbitts TH (2018). Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment. Nat Commun 9: 3169–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, Zhang Y, Sanchez-Martin M, Badgley MA, Huang CS, Tran TH, Akkiraju H, Brown LM, Nandakumar R, Cremers S, Yang WS, Tong L, Olive KP, Ferrando A, Stockwell BR (2017). Multivalent Small-Molecule Pan-RAS Inhibitors. Cell 168: 878–889.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Harrison RA, Li L, Zeng M, Gondi S, Scott D, Gray NS, Engen JR, Westover KD (2017). KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry. Structure 25: 1442–1448.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng M, Lu J, Li L, Feru F, Quan C, Gero TW, Ficarro SB, Xiong Y, Ambrogio C, Paranal RM, Catalano M, Shao J, Wong KK, Marto JA, Fischer ES, Janne PA, Scott DA, Westover KD, Gray NS (2017). Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C. Cell Chem Biol 24: 1005–1016.e3. [DOI] [PubMed] [Google Scholar]
- 58.Nnadi CI, Jenkins ML, Gentile DR, Bateman LA, Zaidman D, Balius TE, Nomura DK, Burke JE, Shokat KM, London N (2018). Novel K-Ras G12C Switch-II Covalent Binders Destabilize Ras and Accelerate Nucleotide Exchange. J Chem Inf Model 58: 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bery N, Cruz-Migoni A, Bataille CJ, Quevedo CE, Tulmin H, Miller A, Russell A, Phillips SE, Carr SB, Rabbitts TH (2018). BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions. eLife 7:37122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cruz-Migoni A, Canning P, Quevedo CE, Bataille CJR, Bery N, Miller A, Russell AJ, Phillips SEV, Carr SB, Rabbitts TH (2019). Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl. Acad. Sci. U.S.A 116: 2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fetics SK, Guterres H, Kearney BM, Buhrman G, Ma B, Nussinov R, Mattos C (2015). Allosteric Effects of the Oncogenic RasQ61L Mutant on Raf-RBD. Structure 23: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Unwin PN. (1974). Electron microscopy of the stacked disk aggregate of tobacco mosaic virus protein. II. The influence of electron irradiation of the stain distribution. J Mol Biol. 87(4):657–70. [DOI] [PubMed] [Google Scholar]
- 63.Serwer P (1977). Flattening and shrinkage of bacteriophage T7 after preparation for electron microscopy by negative staining. J Ultrastruct Res. 58(3): 235–43. [DOI] [PubMed] [Google Scholar]
- 64.Berriman J, Leonard KR (1986). Methods for specimen thickness determination in electron microscopy. II. Changes in thickness with dose. Ultramicroscopy 19(4): 349–66. [DOI] [PubMed] [Google Scholar]
- 65.Jésior JC, Wade RH (1987). Electron-irradiation-induced flattening of negatively stained 2D protein crystals. Ultramicroscopy 21(4): 313–9. [DOI] [PubMed] [Google Scholar]
- 66.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Kräusslich HG. (2000). Biochemical and structural analysis of isolated mature cores ofhuman immunodeficiency virus type 1. J Virol. 74(3): 1168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeager M, Dryden K, Ganser-Pornillos B (2013). Lipid monolayer and sparse matrix screening for growing two-dimensional crystals for electron crystallography: methods and examples. Methods Mol Biol 955: 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J ComputChem. 25(13):1605–12. [DOI] [PubMed] [Google Scholar]
- 69.Travers T, Lopez C, Van QN, Neale C, Tonelli M, Stephen AG, Gnanakaran S (2018). Molecular recognition of RAS/RAF complex at the membrane: role of RAF cysteine-rich domain. Scientific Reports 8: 8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Y, Prakash P, Liang H, Cho K-J, Gorfe A, Hancock J (2017). Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168: 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung J, Lee Y, Denson J, Gillette W, Stephen A, Groves J (2018). K-Ras4B remains monomeric on membranes over a wide range of surface densities and lipid compositions. Biophys J. 114: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lommerse P, Snaar-Jagalska B, Spaink H, Schmidt T (2005). Single-molecule diffusion measurements of H-Ras at the plasma membrane of live cells reveal microdomain localization upon activiation. J. Cell Sci 118: 1799–1809. [DOI] [PubMed] [Google Scholar]
- 73.Cho K, Kasai R, Park J, Chigurupati S, Heidorn S, van der Hoeven D, Plowman S, Kusumi A, Marais R, Hancock J (2012). Raf inhibitors target ras spatiotemporal dynamics. Curr. Biol 22: 945–955. [DOI] [PubMed] [Google Scholar]
- 74.Blazevits O, Mideksa Y, Solman M, Ligabue A, Ariotti N, Nakhaeizadeh H, Fansa E, Papageorfiou A, Wittinghofer A, Ahmadian M, Abankwa D (2016). Galectin-1 dimers scaffold Raf-effectors to increase H-Ras nanoclustering. Scientific Reports 6: 24165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin T, Lavoie H, Sahmi M, David M, Hilt C, Hammell A, Therrien M (2017). RAF inhibitors promote RAS-RAF interaction by allosterically disrupting RAF autoinhibition. Nature Comm. 8: 1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yee Y, Phelps C, Huang T, Mostofian B, Wu L, Zhang Y, Chang Y, Stork P, Gray J, Zuckerman D, Nan X (2019). High-throughput single-particle tracking reveals nested membrane nanodomain organization that dictates Ras diffusion and trafficking. bioRxiv: 10.1101/552075. [DOI] [PMC free article] [PubMed] [Google Scholar]